Abstract
NLRP6, a Nod-like receptor family member, has been shown to affect intestinal homeostasis and microbial colonization through organizing a huge protein complex called inflammasome. NLRP6 inflammasome promotes the cleavage and secretion of inflammatory cytokines or the cleavage of pore-forming Gasdermin D to initiate the inflammatory cell death called pyroptosis, which plays important roles in responding to pathogen invasion. However, questions about the ligand(s) that trigger NLRP6 inflammasome activation, or the mechanisms that how a ligand triggers NLRP6 inflammasome assembly, are emerging. In this mini-review, we summarize the current understandings of ligand recognition of NLRP6, the role of liquid-liquid phase separation in NLRP6 inflammasome assembly, and potential links with human health and diseases.
Keywords: NLRP6, inflammasome, phase separation, human disease
Introduction
Microbial infection induces complex interactions between the pathogen and the host. Pathogens express evolutionarily conserved structures, termed pathogen-associated molecular patterns (PAMPs), which are essential for their survival and pathogenicity. PAMPs recognition is the first step of defense against invading microbial pathogens.1 Recently more and more evidences support the crucial role of inflammasome activation in anti-microbial inflammation or sterile inflammation.2,3 NLRP6, one of the nucleotide-oligomerization domain-like receptor (NLR) family members that is expressed predominantly in intestine and also in liver, plays important roles in sensing and initiating the anti-bacterial and anti-viral immune response.4–8 In this review, the progress of pathogen recognition and mechanisms of NLRP6 inflammasome activation in innate immune defenses are elaborated. Moreover, the relationship between NLRP6 and human disease, as well as the possibilities of therapeutic applications with pathogen recognition and liquid-liquid phase separation (LLPS) are discussed as well.
NLRP6 as a sensor of bacterial components
Elinav et al. reported in 2011 the role of NLRP6 in maintaining the microbiota homeostasis, that deficiency of NLRP6 in mouse colonic epithelial cells results in reduced IL-18 levels and leads to dysbiosis through alterations in the composition of intestinal microbial community.4,6,9 In 2012, Anand et al. found that Nlrp6-deficient mice are resistant to infection with the bacterial pathogens Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli.5 Later in 2018, Hara et al. found that lipoteichoic acid (LTA) from Gram-positive bacteria binds and activates NLRP6 inflammasome, which exacerbates the Gram-positive bacterial infection. Biochemical experiments showed LTA directly binds NLRP6 at high affinity via the LRR domain and the glycerophosphate repeat (GPR) of LTA is critical for the induction of caspase-11 cleavage.8 In addition, a study also showed NLRP6 may recognize lipopolysaccharide (LPS), a component from Gram-negative bacteria. Leng et al. demonstrated that LPS directly binds to the LRR of the NLRP6 monomer and induces overall conformational changes and dimerization, then forms an inflammasome complex with ASC and caspase-1 to cleave pro-IL-1β and IL-18 into their biologically active forms.10 To date, 5 molecules (taurine, histamine, spermine, LTA, and LPS) have been reported to modulate NLRP6 inflammasome directly or indirectly in response to bacteria invasion. Levy et al. found that taurine is a microbiota-dependent positive inflammasome modulator while histamine and spermine were the two strongest suppressors of NLRP6 mediated IL-18 secretion.6 Later in 2021, Shen et al. found that only spermine exhibited weaker direct binding with NLRP6 (KD = 417 uM).11 More works are required to reveal the precise mechanisms of NLRP6 recognition of bacteria components.
Muller et al. found that microbiota depletion by antibiotics treatment led to NLRP6- and caspase 11–dependent loss of intrinsic enteric-associated neurons (iEANs), indicative of the involvement of NLRP6 inflammasome in microbiota depletion-associated death of iEANs.12 Also, another work from Daniel Mucida group showed Salmonella infection induced iEAN death is also dependent on NLRP6 and Casp11.13 Similarly, NLRP6 mediated pyroptosis triggered by bacterial component plays an important role in such iEANs death.
In the meantime, NLRP6 mediated signals are reported to be regulated by bacteria or bacteria products. Short chain fatty acids (SCFAs), one type of main metabolites produced by microbiota, ameliorate the fructose- and histamine-suppressed NLRP6 expression as well as colonic inflammasome activation, resulting in the protection against a high-fructose diet-induced hippocampal neuroinflammation and neuronal loss.14 Whether SCFAs or other bacterial metabolites can regulate NLRP6 inflammasome requires further studies.
NLRP6 as a sensor of viral components
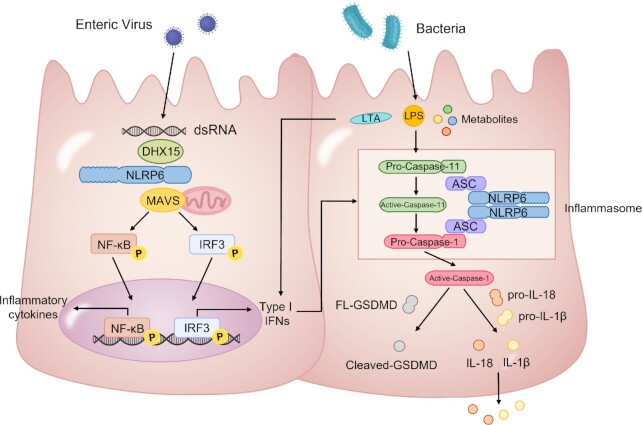
NLRP6 has also been reported to play a role in anti-RNA virus immune response. In 2015, Wang et al. found that Nlrp6−/− mice showed stronger susceptibility to EMCV infection, and higher viral loads in the intestine, upon intragastric infection of EMCV. Mechanistical study revealed that NLRP6 cooperates with DHX15 to recognize long double-stranded RNA (dsRNA) and thus activate interferon and interferon stimulated genes (ISGs) through mitochondrial antiviral signaling proteins (MAVS), to restrict enteric viruses infection.7 In 2017, Zhu et al. also showed that NLRP6 may directly bind to dsRNA by GST-pull down experiment.15 However, conclusive evidence still lacks to prove dsRNA as the direct ligand to activate NLRP6 inflammasome (Fig. 1).
Figure 1.
Previously reported NLRP6 mediated signaling pathways in response to microbes in the intestine. NLRP6 binds to viral dsRNA through DEAH-box helicase 15 (DHX15) and triggers the induction of type I interferons (IFNs) and inflammatory cytokines through mitochondrial antiviral signaling protein (MAVS). NLRP6 also recognizes bacterial components and induces processing of caspase-11, which promotes caspase-1 activation and IL-18/IL-1β maturation. Gasdermin D is cleaved by active caspase-1 and drives pyroptosis.
Role of phase separation in NLRP6 inflammasome activation
In a recent study published in Cell, Shen et al. clarified that NLRP6 binds to both dsRNA and LTA, but not dsDNA, ssRNA, LPS, or metabolites. Interestingly, upon dsRNA stimulation, dsRNA induces liquid-liquid phase separation (LLPS) of NLRP6 to promote inflammasome activation in intestine and liver (Fig. 1).11,16
First of all, they analyzed the binding dissociation constants (KD) of the potential ligands that have been reported, and found that NLRP6 directly interacts with dsRNA and LTA, with further increased affinity for longer dsRNA. The strong and selective binding ability of NLRP6 suggests that it may act as a specific PRR for various dsRNA species from pathogens. Since increased evidence has shown RNA and RNA binding proteins are involved in the LLPS formation,17 and of note, during infection, the RNA binding protein NLRP6 has been reported to form puncta, which is an important feature of the LLPS, they reasoned whether dsRNA and NLRP6 form LLPS to initiate the downstream signal.
Then they tried multiple in vitro and cell experiments and showed that both dsRNA and LTA can induce NLRP6 to form a dynamic, liquid-like condensed phase. Next Li R. identified a poly-lysine region in NLRP6 protein that is important for the multivalent interaction between NLRP6 by utilizing the Predictor of Natural Disordered Regions (PONDR) program and an optogenetic platform (optoDroplets). Mutation in this region compromises dsRNA-induced formation of NLRP6 puncta, GSDMD cleavage, and death of cells. Moreover, Li R. generated the poly-lysine mutant mice and found that LLPS of NLRP6 is important for defense against a positive-sense single-stranded RNA (+ssRNA) coronavirus in a model of murine hepatitis virus (MHV) infection, and also important for maintaining the microbiota-immune homeostasis in the gut.
Thus, it is proposed that LLPS of NLRP6 is a common response to ligand stimulation, which serves as a checkpoint to direct NLRP6 to distinct functional outcomes depending on the cellular context. The innovations of this paper are: (i) For the first time, it provided clear evidence that LLPS occurs during inflammasome activation; (ii) it showed clearly NLRP6, but not NLRP3/NLRC4, recognizes dsRNA to form LLPS as a platform, explaining the potential mechanism that how NLRP6 integrates multiple signaling (dsRNA, LTA, inflammasome, interferon); (iii) It demonstrated for the first time the physiological role of LLPS in mice by using the poly-lysine mutant mice in which the multivalent interaction between NLRP6 is affected.
NLRP6 in human diseases
Previous studies of NLRP6 were mostly focused on mouse models. However, based on the evidence that NLRP6 expression is highly conserved between human and mouse, a growing number of studies suggest that NLRP6 is closely associated with a variety of human diseases.
Consistent with the mouse model, the transcriptome and proteome analysis showed high expression of NLRP6 in the human gastrointestinal tract, indicating an important role of NLRP6 in maintaining gut homeostasis in humans.18 Tomuschat et al. found that the expression of NLRP6 in the colonic epithelium as measured by immunofluorescence decreases markedly in specimens from patients with Hirschsprung's disease (HSCR) compared with controls,19 indicating dysregulated microbial homeostasis in HSCR patients. Mukherjee et al. showed that the expression of NLRP6 deubiquitinating enzyme CYLD is downregulated in UC patients, which is negatively correlated with IL-18 levels in the colonic mucosa.20 It is possible that the regulatory mechanisms inhibiting excessive activation of NLRP6-mediated inflammation are defective in inflammatory bowel disease (IBD) patients.
Although most studies regarding NLRP6 are focused on intestine, the regulatory role of this molecule during infection and inflammation in other organs and cells has also been reported. Ghimire et al. found an increased expression of NLRP6 in neutrophils, macrophages, and epithelial cells in the lungs obtained from pneumonia patients. In Methicillin-resistant Staphylococcus aureus (MRSA) infection model, the up-regulated NLRP6 augments cell death and leads to less phagocytes to clear bacteria. This suggests that NLRP6 could have regulatory function in pulmonary host defense. In addition, higher expression of NLRP6 was observed in patients with pulpitis and apical periodontitis.21,22 Lu et al. found that NLRP6 suppresses the inflammatory response of human periodontal ligament cells by inhibiting NF-κB and ERK signal pathways, while Tian et al. preferred to explain this phenotype by the upregulation and activation of the NLRP6-CASPASE 4 pathway.22 A similar anti-inflammatory role of NLRP6 has been reported in rheumatoid arthritis patients. They found that silencing of NLRP6 can enhance pro-inflammatory cytokine production via activating NF-κB pathway.23
Role of NLRP6 in cancer has also been reported. Study showed lower expression of NLRP6 in gastric cancer surgical tissue compared with adjacent normal gastric tissues.24 Overexpression of NLRP6 in gastric cancer cells led to a significant decrease in cell proliferation, migration, and invasion, indicating that NLRP6 acts as a tumor suppressor.24 Another study also suggested that NLRP6 exerts inhibitory effects on gastric cancer cell growth by promoting the ubiquitination of GRP78.25 Liu et al. detected increased NLRP6 in the intestine of hepatocellular carcinoma (HCC) patients with C. albicans. They found the colonization of C. albicans promoted HCC growth in wild-type mice but didn't affect tumor growth in Nlrp6–/– mice,26 indicating a tumor-promoting role of NLRP6 in HCC patients. Furthermore, studies showed that NLRP6-dependent pyroptotic and inflammatory gene expression predicts HCC prognosis.27,28 The expression of these genes has also been used to predict the prognosis of cutaneous melanoma and the effect of anticancer therapies.29 Furthermore, NLRP6 inflammasome has also been suggested to enhance malignancy, immune evasion, and radioresistance of glioma cells.30
Thus, NLRP6 seems to act as an inflammation initiator in the intestinal epithelial cells through inducing inflammasome activation, pyroptosis, and IL-18 production, while act as a regulator of inflammation in the immune cells through inhibiting NF-κB pathway. In the neuron system, NLRP6 functioning as an inflammasome remains unclear in the context of sterile- and bacterial-induced inflammation. The role of NLRP6 in tumor cells is still controversial. More studies on mouse models and human disease patients are needed to validate whether NLRP6 is a promising target to treat inflammatory diseases or cancer.
Conclusions and perspectives
The role of NLRP6 in the maintenance of intestinal microbiota homeostasis and anti-RNA viral immune response has been reported recently. Viruses and bacteria activate NLRP6 with dsRNA or LTA respectively, and in response to that, LLPS mediated by the multivalent interaction between NLRP6, serves as a hub to integrate multiple signaling pathways and direct NLRP6 to distinct functional outcomes. NLRP6 is also found to be associated with human diseases, such as IBD and colorectal cancer.
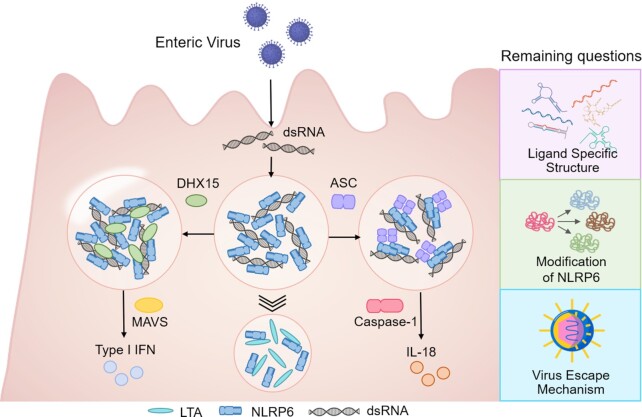
Although the underlying mechanisms of NLRP6 inflammasome activation, as well as the ligands recognition, are of great interests and biological significances, many of these aspects remain poorly characterized and questions unanswered. First, since dsRNA and LTA are completely different molecules, what is the molecular basis of the ligand recognition of NLRP6? Second, are there post-transcriptional modifications on NLRP6 protein that may be important for ligand recognition, LLPS formation, and inflammasome activation? Third, how is dsRNA/NLRP6 LLPS physiologically or pathologically regulated in cells, for example, interfered by other biological process or targeted by viruses for immune evasion (Fig. 2)?
Figure 2.
Recent understanding of the role of dsRNA/NLRP6 LLPS in signaling integration, and perspectives of the remaining questions. A brief model to hypothesize how NLRP6 LLPS may serve as a hub to integrate and regulate multiple signaling pathways, and the proposed future directions of the topic.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Key R&D Program of China (Grant No. 2018YFA0508000) (SZ), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29030101) (SZ), the National Natural Science Foundation of China (Grants No. 82061148013, 81822021, and 81821001) (SZ).
Contributor Information
Runzhi Li, Institute of Immunology, CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Science and Medicine, University of Science and Technology of China, Hefei 230027, China.
Yang Zan, Institute of Immunology, CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Science and Medicine, University of Science and Technology of China, Hefei 230027, China.
Kaiwen Sui, Institute of Immunology, CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Science and Medicine, University of Science and Technology of China, Hefei 230027, China.
Shu Zhu, Institute of Immunology, CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Science and Medicine, University of Science and Technology of China, Hefei 230027, China; School of Data Science, University of Science and Technology of China, Hefei 230026, China; Institute of Health and Medicine, Hefei Comprehensive National Science Center, Hefei 230051, China.
Conflict of interest
The authors declare no conflicts of interest. In addition, as an Editorial Board Member of Precision Clinical Medicine, the corresponding author Shu Zhu was blinded from reviewing ormaking decisions on this manuscript.
References
- 1. Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–73. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levy M, Shapiro H, Thaiss CA, et al. NLRP6: A multifaceted innate immune sensor. Trends Immunol. 2017;38:248–60. doi: 10.1016/j.it.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 3. Li RZ, Zhu S. NLRP6 inflammasome. Mol Aspects Med. 2020;76:100859. doi: 10.1016/j.mam.2020.100859. [DOI] [PubMed] [Google Scholar]
- 4. Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anand PK, Malireddi RKS, Lukens JR, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–93. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levy M, Thaiss CA, Zeevi D, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–43. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang P, Zhu S, Yang L, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–30. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hara H, Seregin SS, Yang D, et al. The NLRP6 Inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. Cell. 2018;175:1651–64..e14. doi: 10.1016/j.cell.2018.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wlodarska M, Thaiss CA, Nowarski R, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–59. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leng FW, Yin H, Qin S, et al. NLRP6 self-assembles into a linear molecular platform following LPS binding and ATP stimulation. Sci Rep. 2020;10:198. doi: 10.1038/s41598-019-57043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen C, Li R, Negro R, et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell. 2021;184:5759–74..e20. doi: 10.1016/j.cell.2021.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muller PA, Matheis F, Schneeberger M, et al. Microbiota-modulated CART(+) enteric neurons autonomously regulate blood glucose. Science. 2020;370:314–21. doi: 10.1126/science.abd6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matheis F, Muller PA, Graves CL, et al. Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell. 2020;180:64–78..e16. doi: 10.1016/j.cell.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li JM, Yu R, Zhang LP, et al. Dietary fructose-induced gut dysbiosis promotes mouse hippocampal neuroinflammation: a benefit of short-chain fatty acids. Microbiome. 2019;7:98, doi: 10.1186/s40168-019-0713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu S, Ding S, Wang P, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546:667–70. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen C, Li R, Negro R, et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell. 2021;184:5759–74..e20. doi: 10.1016/j.cell.2021.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boeynaems S, Alberti S, Fawzi NL, et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018;28:420–35. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gremel G, Wanders A, Cedernaes J, et al. The human gastrointestinal tract-specific transcriptome and proteome as defined by RNA sequencing and antibody-based profiling. J Gastroenterol. 2015;50:46–57. doi: 10.1007/s00535-014-0958-7. [DOI] [PubMed] [Google Scholar]
- 19. Tomuschat C, Virbel CR, O'Donnell AM, et al. Reduced expression of the NLRP6 inflammasome in the colon of patients with Hirschsprung's disease. J Pediatr Surg. 2019;54:1573–7. doi: 10.1016/j.jpedsurg.2018.08.059. [DOI] [PubMed] [Google Scholar]
- 20. Mukherjee S, Kumar R, Lenou ET, et al. Deubiquitination of NLRP6 inflammasome by Cyld critically regulates intestinal inflammation. Nat Immunol. 2020;21:626–35. doi: 10.1038/s41590-020-0681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu WL, Zhang L, Song DZ, et al. NLRP6 suppresses the inflammatory response of human periodontal ligament cells by inhibiting NF-kappaB and ERK signal pathways. Int Endod J. 2019;52:999–1009. doi: 10.1111/iej.13091. [DOI] [PubMed] [Google Scholar]
- 22. Tian XX, Li R, Liu C, et al. NLRP6-caspase 4 inflammasome activation in response to cariogenic bacterial lipoteichoic acid in human dental pulp inflammation. Int Endod J. 2021;54:916–25. doi: 10.1111/iej.13469. [DOI] [PubMed] [Google Scholar]
- 23. Lin Y, Luo ZQ. NLRP6 facilitates the interaction between TAB2/3 and TRIM38 in rheumatoid arthritis fibroblast-like synoviocytes. FEBS Lett. 2017;591:1141–9. doi: 10.1002/1873-3468.12622. [DOI] [PubMed] [Google Scholar]
- 24. Wang QQ, Wang CM, Chen JL. NLRP6, decreased in gastric cancer, suppresses tumorigenicity of gastric cancer cells. Cancer Management and Research. 2018;10:6431–44. doi: 10.2147/Cmar.S182980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang XY, Wu X, Wang Q, et al. NLRP6 suppresses gastric cancer growth via GRP78 ubiquitination. Exp Cell Res. 2020;395:112177. doi: 10.1016/j.yexcr.2020.112177. [DOI] [PubMed] [Google Scholar]
- 26. Liu ZR, Li Y, Li C, et al. Intestinal Candida albicans promotes hepatocarcinogenesis by up-regulating NLRP6. Frontiers in Microbiology. 2022;13:812771. doi: 10.3389/fmicb.2022.812771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L, Zhang X, Zhao W, et al. NLRP6-dependent pyroptosis-related lncRNAs predict the prognosis of hepatocellular carcinoma. Frontiers in Medicine. 2022;9:760722, doi: 10.3389/fmed.2022.760722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou TH, Wang T, Zeng K, et al. A nomogram based on a three pyroptosis gene model and clinical parameters for predicting prognosis of hepatocellular carcinoma. Gene. 2022;819:146243. doi: 10.1016/j.gene.2022.146243. [DOI] [PubMed] [Google Scholar]
- 29. Xu Y, Chen Y, Niu Z, et al. A novel pyroptotic and inflammatory gene signature predicts the prognosis of cutaneous melanoma and the effect of anticancer therapies. Frontiers in Medicine. 2022;9:841568, doi: 10.3389/fmed.2022.841568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu Y, Cao F, Xiong Y, et al. SP1 transcriptionally activates NLRP6 inflammasome and induces immune evasion and radioresistance in glioma cells. Int Immunopharmacol. 2021;98:107858, doi: 10.1016/j.intimp.2021.107858. [DOI] [PubMed] [Google Scholar]