Abstract
To study the involvement of DNA replication in UV-induced illegitimate recombination, we examined the effect of temperature-sensitive dnaB mutations on illegitimate recombination and found that the frequency of illegitimate recombination was reduced by an elongation-deficient mutation, dnaB14, but not by an initiation-deficient mutation, dnaB252. This result indicates that DNA replication is required for UV-induced illegitimate recombination. In addition, the dnaB14 mutation also affected spontaneous or UV-induced illegitimate recombination enhanced by the recQ mutation. Nucleotide sequence analyses of the recombination junctions showed that DnaB-mediated illegitimate recombination is short homology dependent. Previously, Michel et al. (B. Michel, S. Ehrlich, and M. Uzest, EMBO J. 16:430–438, 1997) showed that thermal treatment of the temperature-sensitive dnaB8 mutant induces double-stranded breaks, implying that induction of illegitimate recombination occurs. To explain the discrepancy between the observations, we propose a model for DnaB function, in which the dnaB mutations may exhibit two types of responses, early and late responses, for double-stranded break formation. In the early response, replication forks stall at damaged DNA, resulting in the formation of double-stranded breaks, and the dnaB14 mutation reduces the double-stranded breaks shortly after temperature shift-up. On the other hand, in the late response, the arrested replication forks mediated by the dnaB8 mutation may induce double-stranded breaks after prolonged incubation.
Illegitimate recombination (IR) takes place between DNA sequences that have little or no homology and is a major cause of chromosomal aberrations, such as deletions or translocations. IR can be classified into two types, short-homology-independent IR (SHIIR) and short-homology-dependent IR (SHDIR). SHIIR occurs between sequences having virtually no homology and is mediated by DNA topoisomerases (2, 3, 24). SHDIR is induced by UV irradiation or other DNA-damaging agents and requires short regions of homology between recombination sites (24, 25, 28).
SHDIR usually takes place at a low frequency, but it is greatly enhanced by genotoxic agents (11, 18). It is known that this type of recombination is enhanced by RecE and RecJ and is suppressed by DNA polymerase I, SbcB, RecQ, and UvrAB (1, 7, 9, 10, 25, 27). Previous studies have suggested that DNA ends are processed by 5′-to-3′ exonucleases, such as RecE or RecJ, and as a result DNAs with a 3′ overhang are formed. These molecules anneal with other molecules carrying a complementary overhang, and then end joining takes place. It has also been found that DNA-binding proteins affect this type of IR. Fis and IHF (integration host factor) enhance SHDIR, and H-NS suppresses it (21, 22).
It is known that a double-stranded break (DSB) can result from arrest of a replication fork, which may be caused by DNA secondary structure, DNA lesions, or DNA-bound proteins (5, 6, 8). The involvement of Rep and DnaB helicases in DSB formation indicates that there is a link between DNA replication stoppage and the formation of DSBs (13). In addition, RuvABC, which catalyze branch migration and cleavage of Holliday junctions, are responsible for DSB formation at stalled replication forks (20).
In this study, to determine the role of DNA replication in UV-induced IR, we examined the effects of dnaB(Ts) mutations on IR. One of the mutants used, a dnaB14 mutant, had a defect in the elongation process during DNA replication (12), and another mutant, a dnaB252 mutant, had a defect in initiation of DNA replication (19). We found that the frequency of UV-induced IR was reduced by the elongation-deficient mutation in the dnaB14 mutant but not by the initiation-deficient mutation in the dnaB252 mutant. Thus, we concluded that processivity of DnaB helicase is a prerequisite for UV-induced IR. Below we discuss a model for the possible function of DnaB in UV-induced IR.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli strains used in this study are described in Table 1. Most of the strains used in this study are derivatives of E. coli K-12 which contain one unit of the λ cI857 prophage; the exceptions are Ymel and the P2 lysogen. Ymel was used for titration of the total number of λ phage, and the P2 lysogen was used for titration of the number of λ Spi− phage.
TABLE 1.
E. coli strains used in this study
| Strain | Parent | Relevant genotype or phenotype | Source or constructionc |
|---|---|---|---|
| Ymel | supE supF | Our collection | |
| WL95 | P2 | Amersham | |
| FA77a | dnaB14 | H. Ogawa | |
| HI1747a | FA77 | mal::Tn9 | FA77 × P1 · 594 |
| HI1748a | FA77 | dnaB14 mal::Tn9 | FA77 × P1 · 594 |
| HI1756a | FA77 | mal::Tn9 (λ cI857)b | HI1747 |
| HI2151a | FA77 | dnaB14 mal::Tn9 (λ cI857)b | HI1748 |
| HI2158a | FA77 | mal::Tn9 (λ cI857 xis1)b | HI1747 |
| HI2159a | FA77 | dnaB14 mal::Tn9 (λ cI857 xis1)b | HI1748 |
| HI2152 | 594 | mal::Tn9 | 594 × P1 · AQ207 |
| HI2153 | 594 | dnaB252 mal::Tn9 | 594 × P1 · AQ207 |
| HI2154 | 594 | mal::Tn9 (λ cI857)b | HI2152 |
| HI2155 | 594 | dnaB252 mal::Tn9 (λ cI857)b | HI2153 |
| HI2700 | FA77 | recQ1802 mal::Tn9 (λ cI857)b | HI1756 × P1 · HI2034 |
| HI2701 | FA77 | recQ1802 dnaB14 mal::Tn9 (λ cI857)b | HI2151 × P1 · HI2034 |
Strain derived from FA77 (12).
All of the λ prophages contained a single copy and carried the c1857 mutation.
P1 · 594, P1 · AQ207, and P1 · HI2034 were P1 phages prepared with E. coli strains 594, AQ207, and HI2034, respectively.
Measurement of the frequency of λ Spi− phage induced spontaneously or by UV irradiation.
E. coli λ cI857 or a derivative of this strain was grown at 30°C in λ YP broth. If necessary, 2 ml of the culture was irradiated with a UV lamp (15 W) with a wavelength of 253.6 nm. Thermal induction of λ prophage was carried out by incubation at 42°C for 15 min. The culture was then incubated at 37°C for 2 to 4 h. The titer of λ Spi− phage was measured on a lawn of E. coli WL95 (P2). The number of all λ phage was determined on a lawn of E. coli Ymel. The frequency of IR was calculated by dividing the number of λ Spi− phage by the total number of all λ phage (11).
Measurement of the frequency of λ Spi− phage in a lysogen carrying λ xis1 prophage.
E. coli λ cI857 xis1 or a derivative of this strain was thermally induced, and phage lysate was prepared as described above. The frequency of IR was calculated by dividing the number of λ Spi− phage by the number of cells before prophage induction (21).
Independent isolation of λbio transducing phages induced by UV irradiation.
E. coli λ lysogen was irradiated with UV as described above. The culture was then divided into 50 small tubes. Each tube containing 0.5 ml of the culture was then incubated at 42°C for 15 min. The cultures were then incubated at 37°C for 2 h. Phage lysates were plated on a lawn of E. coli WL95. A plaque derived from each tube was picked, suspended in M9 buffer, and plated on a lawn of Ymel to isolate a single clone.
Determination of the locations of recombination junctions in λ Spi− phages.
The locations of recombination junctions were determined by PCR by using multiple combinations of primers. λbio transducing phages were identified by using a mixture of several sets of primers. The method used in this study has been described previously by Ukita and Ikeda (25). The recombination junctions were then sequenced and analyzed with an ABI PRISM 310 genetic analyzer (Perkin-Elmer).
Sequence analysis of the mutation site in the dnaB14 mutant.
The mutant dnaB gene was amplified by PCR performed with oligomers B1FH (5′-CAA TTC GTG TTG CCA TGT G-3′) and B4RB (5′-TCA CAA CAG TTG CCG CTT G-3′) and template DNA extracted from a dnaB mutant. The amplified 1.6-kb fragment contained a region from 100 bp upstream of the ATG codon to 50 bp downstream of the stop codon of the dnaB gene. The fragment was directly sequenced by using the oligomers mentioned above, as well as B3F (5′-TGG AGA TGC CAT CAG AAC AC-3′) and B2R (5′-TTC CAG CGT ACG GTA ATC CGA AAG C-3′).
RESULTS
Characterization of the dnaB mutations.
To study the role of DNA replication in IR, we examined the effects of two temperature-sensitive mutations on IR. One mutation, the dnaB14 mutation, has a defect in elongation of DNA replication (12), and the other mutation, the dnaB252 mutation, has a defect in initiation of DNA replication (19). Because the dnaB14 mutation site is not known yet, we determined the nucleotide sequence of the mutant gene and found that it contains a point mutation consisting of the following base substitution: the 46th G from the first ATG is changed to an A. This finding indicates that the mutated DnaB protein has a D16K amino acid substitution. On the other hand, the dnaB252 mutation has been demonstrated previously to be a base substitution consisting of a change of the 869th G to an A. The mutant protein therefore has a G299E amino acid substitution (19).
Effects of the dnaB mutations on IR.
We next tested the effects of the dnaB mutations on IR during prophage induction. To do this, the wild-type strain and the strain with a heat-sensitive mutation in the dnaB gene were lysogenized by infection of λ cI857 and subjected to the λ Spi− assay. This assay utilizes the Spi− phenotype as a marker for λbio transducing phages (11). Specialized transducing phages generated from the λ prophage by IR usually contain the E. coli genes gal and bio, which are adjacent to the phage genome. Thus, most λbio transducing phages are defective in the red-gam region of λ phage DNA and can form plaques on an E. coli P2 lysogen lawn (Spi− phenotype), whereas normal λ phage cannot. Thus, it is possible to select λ Spi− phage from the phage pool. The number of λ Spi− phage is assumed to be the same as the number of λbio transducing phages since previous experiments have shown that most λ Spi− phages are λbio phages (11, 28).
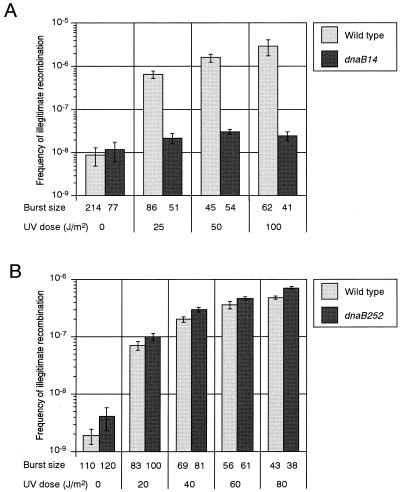
It is known that UV irradiation greatly enhances the frequency of IR. Using the system described above, we examined the effects of dnaB mutations on UV-induced IR. After UV irradiation of the λ lysogen, cultures were incubated at 42°C for 15 min for thermal induction of λ prophage. The cultures were then incubated at 37°C for 2 to 4 h. The yield of all phages was not significantly affected by the dnaB14 mutation. On the other hand, the frequency of IR in the dnaB14 mutant was approximately 100-fold lower than that in the wild type (Fig. 1A). Next, we examined the effect of the dnaB252 mutation on IR and found that the frequency of IR in the dnaB252 mutant was comparable to that in the wild type (Fig. 1B). We have also tried to examine the effects of other elongation-defective mutations, including dnaB8, dnaB42, dnaB266, and dnaB432 (19), but we could not recover all λ phages from the mutants under our assay conditions (data not shown). We also examined the effects of a dnaA(Ts) mutation on IR. DnaA is known to play a role in the initiation of DNA replication. As expected, the dnaA(Ts) mutation did not affect the frequency of IR (data not shown). These results indicate that DnaB helicase participates in IR, implying that progression of replication forks through damaged DNA is a prerequisite for DSB formation.
FIG. 1.
Effects of dnaB(Ts) mutations on UV-induced IR. An E. coli λ cI857 lysogen (HI1756 or HI2154) or a dnaB(Ts) mutant (HI2151 or HI2155) was treated with several different doses of UV irradiation. A λ lysogen was then thermally induced at 42°C and incubated at 37°C to prepare phage lysate. The frequency of IR was measured as described in Materials and Methods. The values are averages based on four independent measurements. The error bars indicate standard errors. (A) dnaB14; (B) dnaB252.
Effect of the dnaB14 mutation on formation of λ Spi− phages in an E. coli λ xis1 lysogen.
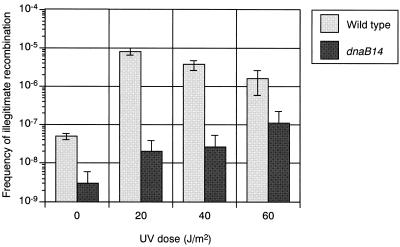
The results described above imply that DnaB helicase directly participates in IR. However, it is also possible that the transient defect in DnaB function during prophage induction may temporarily stop DNA replication. As a result, Int-mediated excision of λ prophage may take place during this stop in replication, resulting in the loss of λ prophage and a reduction in the formation of λbio transducing phage. To rule out this possibility, we examined the effect of the dnaB14 mutation on formation of λ Spi− phage in an E. coli strain carrying an excision-deficient λ prophage. Since Int-mediated excision is not observed in the λ xis1 mutant, λ xis1 prophage should not be lost during thermal induction even in the dnaB(Ts) mutant (21). The results obtained indicated that the frequency of IR in the dnaB14 mutant carrying λ xis1 prophage was lower than that in the wild type, confirming that DnaB helicase is directly involved in IR (Fig. 2).
FIG. 2.
Effect of the dnaB14 mutation on UV-induced IR during induction of λ xis1 lysogen. E. coli λ cI857 xis1 lysogen (HI2158) or a dnaB(Ts) mutant (HI2159) was irradiated with UV, and phage lysate was prepared after thermal induction. The frequency of IR was determined as described in Materials and Methods.
Distribution and nucleotide sequences of junctions formed by UV-induced IR in the dnaB14 mutant.
We determined the distribution of recombination junctions of λ Spi− phages derived from the wild type and the dnaB14 mutant. As shown in a previous study (28), approximately one-half of the recombinants are formed by recombination at hotspot I sites. These sites are located in the bioC gene of E. coli and the gam gene of λ DNA. In addition, the relative rates of recombination at hotspots II and III were 13 and 8%, respectively (Table 2). On the other hand, in the recombinants derived from the dnaB14 mutant, the relative rates of recombination at hotspots I and II were 17 and 66%, respectively (Table 2). Recombination at hotspot III was not detected in the phages derived from this mutant. These results indicate that the dnaB14 mutation preferentially suppressed IR at hotspots I and III.
TABLE 2.
Distribution of recombination sites of λbio transducing phages formed during UV irradiationa
| Strain | Relevant mutation | No. of phages tested | % of recombination at:
|
|||
|---|---|---|---|---|---|---|
| Hotspot I | Hotspot II | Hotspot III | Nonhotspot sites | |||
| HI1756 | Wild type | 53 | 55 | 13 | 8 | 24 |
| HI2151 | dnaB14 | 30 | 17 | 66 | 0 | 17 |
The distributions of recombination sites of recombinant phages derived from the wild type and the dnaB14 mutant were determined by PCR. The junctions were classified into four classes: hotspot I, hotspot II, hotspot III, and nonhotspot sites. The dose of UV irradiation was 100 J/m2.
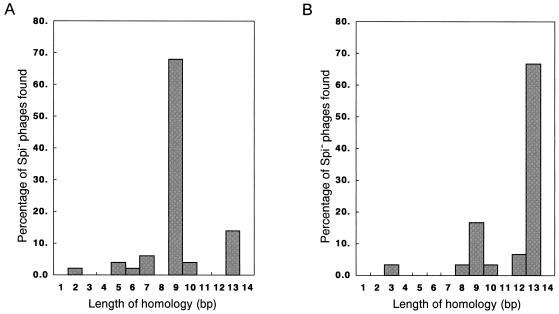
Next, we determined the nucleotide sequences of the junctions of λbio transducing phages. PCR fragments containing the junctions were analyzed by direct sequencing. The results showed that λbio transducing phages derived from both the wild type and the dnaB14 mutant required short regions of homology between the E. coli and λ DNAs (Fig. 3). The average lengths of the homologous regions for the parental recombination sites were 9.1 bp in the wild type and 11.7 bp in the dnaB14 mutant. These results indicate that DnaB-mediated IR is short homology dependent.
FIG. 3.
Distribution of lengths of overlapping nucleotides at recombination junctions. The y axis indicates the number of λbio transducing phages, and the x axis indicates the number of overlapping nucleotides at the recombination junction (i.e., the number of overlapping nucleotides in the λ and E. coli genomes). (A) Distribution of the lengths of overlapping nucleotides at the junctions derived from wild-type strain HI1756. (B) Distribution of the lengths of overlapping nucleotides at the junctions derived from dnaB14 mutant HI2151.
Effect of the dnaB14 recQ double mutation on IR.
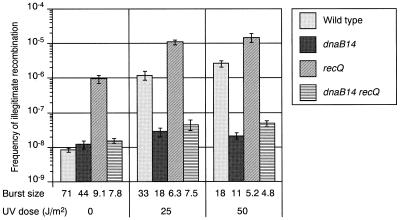
In a previous study, we showed that the frequency of IR is increased by the recQ mutation with or without UV irradiation (10). This result indicates that RecQ is a suppressor of IR. To analyze the functional relationship between RecQ helicase and DnaB helicase in IR, we determined the frequency of IR in the recQ dnaB14 double mutant. The frequency of IR in the double mutant was lower than that in the recQ single mutant, with or without UV irradiation (Fig. 4). These results indicate that the increased IR in the recQ mutant is suppressed by the dnaB14 mutation irrespective of UV irradiation.
FIG. 4.
Effect of the recQ dnaB14 double mutation on IR. The frequency of λ Spi− phages was measured in a recQ single mutant, HI2700, and in a recQ dnaB14 double mutant, HI2701, with or without UV irradiation as described in the legend to Fig. 1.
DISCUSSION
The dnaB14 mutation is located at amino acid 16 (Glu is changed to Lys), which occurs in the amino-terminal fragment (fragment III) that is about 12 kDa long (amino acid 15 to amino acid 126 or 128). This domain is known to be essential for DnaB helicase activity and for DnaB interaction with certain primases, such as DnaC, and other primosomal proteins (14, 15). It is also known that the dnaB14 mutation causes a defect in elongation during DNA replication (12) and that the dnaB252 mutation, which contains a G299E amino acid substitution, has a defect in initiation of DNA replication (19). Upon thermal treatment of bacteria for a short time, the dnaB14 mutation did not significantly affect the yield of all λ phages but greatly affected the frequency of UV-induced IR. On the other hand, the dnaB252 mutation did not affect UV-induced IR. These results indicated that DnaB is involved in UV-induced IR, implying that progression of the replication fork and collision of the replication fork with a damaged region of DNA are essential for IR.
DnaB interacts with many replication proteins, such as DnaC, DnaG, DNA polymerase III, and λ P protein, resulting in the formation of a replisome. When a DNA lesion is present on a chromosome, DNA polymerase III stalls if the lesion is located on the leading strand during DNA synthesis. On the lagging strand, however, DNA polymerase III arrested at the lesion relocates to the next RNA primer site, resulting in continued DNA synthesis (23). In the replisome, the role of DnaB is to unwind DNA at replication forks. An important property of this process is that the helicase activity of DnaB is not suppressed by the presence of DNA lesions (16). It is therefore possible for a long single-stranded DNA region to be formed at the stalled replication fork. Such a single-stranded DNA region could be cleaved by an endonuclease. The reduced frequency of UV-induced IR in the dnaB14 mutant suggests that the mutant DnaB protein has a low processivity and therefore a low probability of collision with damaged DNA, which would result in lower levels of DSBs.
Michel et al. (13) showed that thermal treatment of the temperature-sensitive dnaB8 mutant induces DSBs in DNA. This observation apparently contradicts the results of the present work. However, we suggest that thermal treatment of the dnaB8 mutant may have resulted in irreversible arrest of the replication forks for long period at a nonpermissive temperature, consequently causing DSB formation at the stalled replication sites. On the other hand, a reduced frequency of UV-induced IR in the dnaB14 mutant was detected immediately after temperature shift-up. Therefore, we propose a model in which the dnaB mutants may exhibit two types of responses, early and late responses, for DSB formation. In the early response, replication forks stall at damaged DNA, resulting in DSB formation, and the dnaB14 mutation affects the DSBs shortly after temperature shift-up. On the other hand, in the late response, arrested replication forks mediated by the dnaB8 mutation may induce DSBs after prolonged incubation.
Two major models, the break and join model and the slipped mispairing model, have been proposed for IR. Both models have been used to explain SHDIR in general. In the latter model, a replication fork slips down a region of the DNA strand during DNA replication, resulting in a deletion or other mutation. The former model consists of three steps: DNA DSB, DNA end processing, and end joining. Recently, Onda et al. (17) showed that reduction or augmentation of ligase activity affects the frequency of UV-induced SHDIR, as well as the lengths of homologous sequences at the recombination junctions in SHDIR. The results of these authors strongly support the idea that UV-induced IR is mediated by the break and join mechanism. It is therefore likely that the role of DnaB in promotion of UV-induced IR is formation of DSBs in DNA.
Since it is known that linear chromosomal DNA accumulates spontaneously in a recB or recC mutant (13), enhanced IR might be expected in such mutants. However, this is not the case. The frequency of IR in a recB recC double mutant with or without UV irradiation was comparable to that in the wild type (Y. Ogata and H. Ikeda, unpublished results). It is therefore possible that linear chromosomal DNA is not an appropriate substrate for λbio phage formation. Since it is also known that low-molecular-weight DNAs are abundant in UV-induced cells and the number of DSBs per genome is between 20 and 30 at a UV dose of 100 J/m2 (4), there is another possibility, that the low-molecular-weight DNAs are substrates for IR. Wang and Smith (26) proposed a model in which low-molecular-weight DNAs may be produced as a result of breaks in the parental DNA opposite unrepaired DNA daughter strand gaps. The role of DnaB in formation of DSBs or low-molecular-weight DNAs is being investigated in our laboratory.
There are two types of IR that lead to formation of λbio transducing phages, SHDIR and SHIIR. Nucleotide sequence analyses of the recombination junctions in the wild type showed that one-half of the junctions were formed at the hotspot I site and the other half were formed at nonhotspot sites, as described previously (28). Most junctions contain short regions of homology in the recombination sites, indicating that DnaB plays a role in promoting SHDIR. We also found that the relative rate of UV-induced recombination in the dnaB14 mutant is reduced at hotspots I and III and increased at hotspot II. This result suggests that DSBs that occur at hotspot I and III sites may be relatively dependent on DnaB. The reason for the preference for DSB sites remains to be determined.
The experiment with the recQ dnaB14 double mutant showed that the effect of the dnaB14 mutation is dominant over the effect of the recQ mutation in the IR pathway. We have previously shown that the recQ mutation enhances IR. Based on the properties of RecQ helicase, we previously proposed that this protein may disrupt recombination intermediates produced by annealing of complementary single-stranded ends (10). These results imply that DnaB helicase plays a role in the introduction of DSBs into DNA at an early step of the IR pathway, while RecQ helicase may act as a suppressor of IR in a later step of this pathway. Moreover, we showed that overproduction of DnaB increased the frequency of IR and that there is a synergy between overproduction of DnaB and the recQ mutation for enhancement of recombination (29). This result is consistent with the proposed functions of DnaB and RecQ in our model of IR (10).
ACKNOWLEDGMENTS
We thank T. Kogoma, A. Kaidoh, and H. Ogawa for sending bacterial strains and for helpful advice, J. Inselberg for critical reading of the manuscript, and S. Omura for encouragement throughout this work.
This work was supported by Grants-in-Aid for Scientific Research (B) and Scientific Research on Priority Areas (B) to H.I. from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Allgood N, Silhavy T. Escherichia coli xonA (sbcB) mutant enhances illegitimate recombination. Genetics. 1991;127:671–680. doi: 10.1093/genetics/127.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashizawa Y, Yokochi T, Ogata Y, Shobuike Y, Kato J, Ikeda H. Mechanism of DNA gyrase-mediated illegitimate recombination: characterization of Escherichia coli gyrA mutations that confer hyper-recombination phenotype. J Mol Biol. 1999;289:447–458. doi: 10.1006/jmbi.1999.2758. [DOI] [PubMed] [Google Scholar]
- 3.Bierne H, Ehrlich S D, Michel B. Deletions at stalled replication forks occur by two different pathways. EMBO J. 1997;16:3332–3340. doi: 10.1093/emboj/16.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonura T, Smith K C. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K12. J Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y, Kogoma T. The mechanism of recA polA lethality: suppression by RecA-independent recombination repair activated by the lexA(def) mutation in Escherichia coli. Genetics. 1995;139:1483–1494. doi: 10.1093/genetics/139.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connelly J, Leach D. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 7.Coukell M, Yanofsky C. Increased frequency of deletions in DNA polymerase mutants of Escherichia coli. Nature. 1970;228:633–635. doi: 10.1038/228633a0. [DOI] [PubMed] [Google Scholar]
- 8.Cox M, Goodman M, Kreuzer K, Sherratt D, Sandler S, Marians K. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 9.Hanada K, Iwasaki M, Ihashi S, Ikeda H. UvrA and UvrB suppress illegitimate recombination: synergistic action with RecQ helicase. Proc Natl Acad Sci USA. 2000;97:5989–5994. doi: 10.1073/pnas.100101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda H, Shimizu H, Ukita T, Kumagai M. A novel assay for illegitimate recombination in Escherichia coli: stimulation of λbio transducing phage formation by ultraviolet light and its independence from recA function. Adv Biophys. 1995;31:197–208. doi: 10.1016/0065-227x(95)99392-3. [DOI] [PubMed] [Google Scholar]
- 12.McMilin K D, Russo V E A. Maturation and recombination of bacteriophage lambda DNA molecules in the absence of DNA replication. J Mol Biol. 1972;68:49–55. doi: 10.1016/0022-2836(72)90261-6. [DOI] [PubMed] [Google Scholar]
- 13.Michel B, Ehrlich S, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama N, Arai N, Bond M, Kaziro Y, Arai K. Nucleotide sequence of dnaB and the primary structure of the DnaB protein from Escherichia coli. J Biol Chem. 1984;259:97–101. [PubMed] [Google Scholar]
- 16.Oh E, Grossman L. Helicase properties of the E. coli UvrAB protein complex. Proc Natl Acad Sci USA. 1987;84:3638–3642. doi: 10.1073/pnas.84.11.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onda M, Yamaguchi J, Hanada K, Asami Y, Ikeda H. Role of DNA ligase in the illegitimate recombination that generates λbio transducing phages. Genetics. 2001;158:29–39. doi: 10.1093/genetics/158.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raha M, Hutchinson F. Deletions induced by gamma rays in the genome of Escherichia coli. J Mol Biol. 1991;220:193–198. doi: 10.1016/0022-2836(91)90001-m. [DOI] [PubMed] [Google Scholar]
- 19.Saluja D, Godson G. Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43, and dnaB454. J Bacteriol. 1995;177:1104–1111. doi: 10.1128/jb.177.4.1104-1111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seigneur M, Bidnenko V, Ehrlich S, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 21.Shanado Y, Kato J, Ikeda H. Fis is required for illegitimate recombination during formation of λbio transducing phage. J Bacteriol. 1997;179:4239–4245. doi: 10.1128/jb.179.13.4239-4245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanado Y, Hanada K, Ikeda H. Suppression of gamma ray-induced illegitimate recombination in Escherichia coli by DNA binding protein H-NS. Mol Gen Genet. 2001;265:242–248. doi: 10.1007/s004380000399. [DOI] [PubMed] [Google Scholar]
- 23.Shavitt O, Litvneh Z. The β subunit modulates bypass and termination at UV lesions during in vitro replication with DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989;264:11275–11281. [PubMed] [Google Scholar]
- 24.Shimizu H, Yamaguchi H, Ashizawa Y, Kohno Y, Asami M, Kato J, Ikeda H. Short-homology-independent illegitimate recombination in Escherichia coli: distinct mechanism from short-homology-dependent illegitimate recombination. J Mol Biol. 1997;266:297–305. doi: 10.1006/jmbi.1996.0794. [DOI] [PubMed] [Google Scholar]
- 25.Ukita T, Ikeda H. Role of the recJ gene product in UV-induced illegitimate recombination at the hotspot. J Bacteriol. 1996;178:2362–2367. doi: 10.1128/jb.178.8.2362-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T V, Smith K C. Postreplicational formation and repair of DNA double-strand breaks in UV-irradiated Escherichia coli uvrB cells. Mutat Res. 1986;165:39–44. doi: 10.1016/0167-8817(86)90007-6. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi H, Hanada K, Asami Y, Kato J, Ikeda H. Control of genetic stability in Escherichia coli: the SbcB 3′-5′ exonuclease suppresses illegitimate recombination promoted by the RecE 5′-3′ exonuclease. Genes Cells. 2000;5:101–109. doi: 10.1046/j.1365-2443.2000.00309.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi H, Yamashita T, Shimizu H, Ikeda H. A hotspot of spontaneous and UV-induced illegitimate recombination during formation of λbio transducing phages. Mol Gen Genet. 1995;248:637–643. doi: 10.1007/BF02191702. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita T, Hanada K, Iwasaki M, Yamaguchi H, Ikeda H. Illegitimate recombination induced by overproduction of DnaB helicase in Escherichia coli. J Bacteriol. 1999;181:4549–4553. doi: 10.1128/jb.181.15.4549-4553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]