Abstract
The hyperthermophilic archaeon Pyrococcus furiosus can utilize different β-glucosides, like cellobiose and laminarin. Cellobiose uptake occurs with high affinity (Km = 175 nM) and involves an inducible binding protein-dependent transport system. The cellobiose binding protein (CbtA) was purified from P. furiosus membranes to homogeneity as a 70-kDa glycoprotein. CbtA not only binds cellobiose but also cellotriose, cellotetraose, cellopentaose, laminaribiose, laminaritriose, and sophorose. The cbtA gene was cloned and functionally expressed in Escherichia coli. cbtA belongs to a gene cluster that encodes a transporter that belongs to the Opp family of ABC transporters.
Pyrococcus species are anaerobic hyperthermophilic members of the Archaea that are able to grow heterotrophically on a range of substrates. Pyrococcus furiosus (9) and Pyrococcus glycovorans (3) have been reported to grow on various sugars, including the β-glucoside cellobiose (20). On the other hand, Pyrococcus abyssi ST549 is unable to grow on cellobiose (12), despite the presence of a β-glucosidase (22). P. furiosus also utilizes the β-glucoside polymer laminarin (14), and metabolism of β-glucosides has been studied in some detail. Cellobiose is intracellularly hydrolyzed to two glucose molecules by the β-glucosidase CelB (20), while laminarin is first cleaved by the extracellular β-glucosidase, LamA, to yield smaller oligoglucosides. These are subsequently transported into the cell via an unknown mechanism and further hydrolyzed by CelB to glucose. The combined activity of CelB and LamA results in the complete hydrolysis of laminarin to glucose (14). Glucose is further metabolized by the modified Embden-Meyerhof pathway (26), which involves a glucokinase and phosphofructokinase, which are both ADP dependent (19).
To understand the energy yields during growth on β-glucosides, the mechanism of sugar uptake needs to be elucidated. In bacteria, cellobiose enters the cell either via a phosphoenolpyruvate-dependent phosphotransferase system (16, 18) or via a binding protein-dependent ATP-binding cassette (ABC) transporter (27). Analysis of the completed genome sequences of a variety of members of Archaea demonstrates that phosphoenolpyruvate-dependent phosphotransferase systems are absent in these organisms. In the thermoacidophile Sulfolobus solfataricus, sugars appear to be transported into the cell mainly via binding protein-dependent ABC transporter systems (1, 7). In the hyperthermophile Thermococcus litoralis, a trehalose-maltose ABC transport system has been described biochemically (31). The trehalose-maltose binding protein, TMBP, and the ATPase subunit, MalK, have been functionally expressed in Escherichia coli (13, 17). These binding protein-dependent transport systems exhibit an unusually high affinity for the sugar, with a Km in the submicromolar range. In the hyperthermophilic bacterium Thermotoga maritima, a high-affinity binding protein-dependent ABC transport system has been described for maltose, trehalose, and maltotriose (30). The abundance of such high-affinity transport systems in thermophilic organisms (both bacteria and archaea) suggests that they play a major role in sugar utilization in the nutrient-poor extreme environments in which these organisms thrive. In an effort to understand the metabolism of cellobiose in P. furiosus, we now report on a binding protein-dependent ABC transport system for oligo β-glucosides.
MATERIALS AND METHODS
Organisms and growth conditions.
P. furiosus Vc1 (DSM 3638) and P. abyssi GE5 (CNCM I-1302) cells were grown routinely at 80°C in modified Methanococcus medium (20) under anaerobic conditions in the presence of 5 mM carbohydrate or 0.2% (wt/vol) pyruvate. For P. abyssi, the medium was supplemented with 1% (wt/vol) elemental sulfur. Continuous monitoring of the growth of P. furiosus cells on different sugars was performed in a Cary 100 spectrophotometer (Varian, Mulgrave, Victoria, Australia) in microcuvettes under an N2-CO2 atmosphere at 90°C. Cells were grown in 750 μl of medium supplemented with 0.1% (wt/vol) sugar as indicated, and growth was monitored at 660 nm for 15 h. Cells grown on laminarin were monitored for 48 h. E. coli DH5α (9) and SF120 (2) cells were grown in Luria broth supplemented with the appropriate antibiotics at 37 or 25°C, respectively.
Chemicals.
Laminaribiose and laminaritriose were purchased from Dextra Laboratories (Reading, United Kingdom), sophorose was obtained from Sigma (Steinheim, Germany), and all other sugars were from Merck (Darmstadt, Germany). [3H]cellobiose was purchased from Amersham-Radiochemicals (Little Chalfont, Buckinghamshire, United Kingdom).
Transport and binding studies.
Cells grown overnight in 50 ml of medium were harvested under anaerobic conditions, washed once in growth medium without carbon source, and after resuspension, stored at room temperature until use. Transport assays were performed anaerobically at 80°C under a continuous flow of N2 gas by using 10 μg of cell protein/ml. [3H]cellobiose was added to a final concentration of 10 μM, and at different time points, samples were taken and washed twice with medium without carbon source by using BA85 nitrocellulose filters (Protran; Schleicher & Schuell, Dassel, Germany). The radioactivity retained on the filters was determined with FilterCount (Packard Bioscience B.V., Groningen, The Netherlands). The kinetic parameters of transport were estimated from triplicate measurements of the uptake for 10 s. For binding studies, 1 μM [3H]cellobiose was added to P. furiosus membranes or the purified protein (10 μg of protein per ml). Binding studies were performed aerobically at 60°C. After 3 min of incubation, reactions were terminated by the addition of 2 ml of ice-cold 0.1 M LiCl and samples were filtered and washed once with 2 ml of 0.1 M LiCl. The radioactivity retained by the filters was determined as described above.
Purification of binding proteins.
Cells were harvested and resuspended in 50 mM Tris-HCl (pH 7.5) and broken by a single pass through a French pressure cell at 600 lb/in2. Membranes were collected by ultracentrifugation for 45 min at 100,000 × g at 4°C. The pellet was resuspended in 50 mM Tris-HCl (pH 7.5) and washed once. Membranes were solubilized using 0.5% (vol/vol) Triton X-100 for 30 min at 37°C. Nonsolubilized material was removed by centrifugation (350,000 × g, 15 min, 4°C), and the supernatant was applied to a concanavalin A (ConA)-Sepharose (Pharmacia, Roosendaal, The Netherlands) column equilibrated with buffer A (25 mM Tris-HCl [pH 7.4], 500 mM NaCl, and 0.05% [vol/vol] Triton X-100). The column was washed thoroughly with buffer A, and bound glycoproteins were eluted using buffer A supplemented with 250 mM α-methyl-mannopyranoside. Fractions with cellobiose binding activity were pooled, dialyzed overnight against buffer B (25 mM Tris [pH 6.8] and 0.05% [vol/vol] Triton X-100), and applied on an HR5/5 MonoQ column (Pharmacia, Uppsala, Sweden) pre-equilibrated with buffer B. Proteins were eluted with a linear gradient of 0 to 500 mM NaCl in buffer B. The cellobiose binding protein (CbtA) eluted at 120 mM NaCl. Active fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, and stored at −80°C.
Cloning and expression of cbtA.
Oligonucleotide primers were designed based on the nucleotide sequence of the complete cbtA gene present in the P. furiosus database (http://wit.mcs.anl.gov/). The gene was amplified by PCR (forward primer, 5′-CCCCGATATCATGAAGAGACTCGTTGGTGTAC-3′; reverse primer, 5′-CCCCCGGATCCTTAAGATCTTCTCCTCCTT-3′), and the resulting 1.8-kb fragment was ligated in pBSKS (Stratagene, La Jolla, Calif.) to yield pSMK3, which was transformed to DH5α (15). pSMK3 was digested with BspHI and BamHI, and the insert was ligated into the expression vector pET302 (29) to yield pSMK4, which contained the cbtA gene with an N-terminal hexa-histidine tag. These expression plasmids were cotransformed with p1244 (21) into E. coli SF120 (2). The plasmid p1244 harbors tRNAs for the amino acids leucine, isoleucine, and arginine with rare codons. Cells were grown to an optical density at 660 nm of 0.8 and subsequently induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. Cells were harvested by centrifugation and broken by French press treatment at 800 lb/in2. Lysed cells were separated in a membrane and soluble fraction by ultracentrifugation (350,000 × g, 20 min, 4°C). The different fractions were analyzed by SDS-PAGE and Western blotting using His tag antibodies (Dianova GmbH, Hamburg, Germany) and assayed for [3H]cellobiose binding at 37 and 60°C.
Total RNA isolation and Northern analysis.
Total RNA was isolated from exponentially growing P. furiosus and P. abyssi cells by using the TRIZOL reagent (Gibco BRL Life Technologies, Breda, The Netherlands). P. abyssi RNA was treated with DNase I to remove coisolated DNA. For Northern blot analysis, 10 μg of total RNA was separated on formaldehyde–1.1% agarose gels and transferred to a Zeta-probe membrane (BIORAD, Veenendaal, The Netherlands) by capillary blotting. Primers were designed according to the gene sequences present in the P. furiosus (http://wit.mcs.anl.gov/) and P. abyssi (http://www.genoscope.cns.fr/Pab/) databases. Probes for cbtA (forward, 5′-CGCCCTCATGAAGAGACTCGTTGGTGT-3′; reverse, 5′-AACCTTAACCTCTTGGAGCC-3′), celB (forward, 5′-CTGGTTTCCAGTTTGAGATGGG-3′; reverse, 5′-TGGCTTTGGAAAAATTCTTGCCC-3′), RPF01470 (forward, 5′-ATGG GAGAATTGCCAATTGC-3′; reverse, 5′-TCAGCTCTTAATTGCGAGC-3′), PAB0627 (forward, 5′-ATGGAAAAACTAGTGGCAGCCATAGTTG-3′; reverse, 5′-TGAGACCCTCTTTGAGAACCACCC-3′) and PAB3089 (forward, 5′-ATGGGAGAGTTGCCAATTGC-3′; reverse, 5′-TCAGCTCTTAATAGCCAAC-3′) were digoxigenin labeled using PCR on genomic DNA. Detection was done with digoxigenin-alkaline phosphatase antibodies (Boehringer GmbH, Mannheim, Germany) and CDP-Star (Tropix Inc., Bedford, Mass.).
Other techniques.
For the determination of the N-terminal sequence of CbtA, the purified protein was electroeluted from SDS-PAGE gels and freeze-dried. Protein sequencing was performed by NAPS (Nucleic Acid/Protein Service Unit, Vancouver, Canada). DNA sequencing was performed by BioMedisch Technologisch Centrum (BMTC, University of Groningen, Groningen, The Netherlands). Glycoproteins in SDS-PAGE were stained using periodic acid-Schiff (Sigma) as previously described (24). Protein concentrations were determined using the DC Bio-Rad Kit (BIORAD).
RESULTS
Cellobiose uptake by P. furiosus
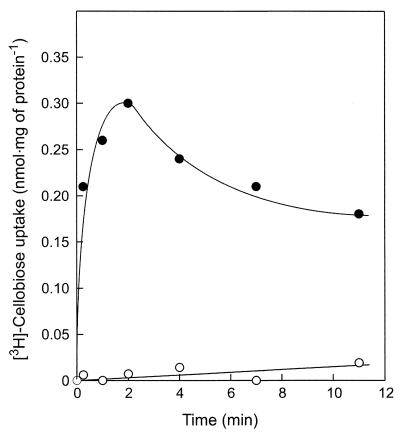
P. furiosus cells grown on cellobiose readily accumulate [3H]cellobiose when incubated at 80°C under anaerobic conditions (Fig. 1). Uptake was completely abolished by aerobic conditions (results not shown). Cellobiose uptake was strongly temperature dependent. Below 40°C, hardly any cellobiose was transported into the cell, while above 90°C, rapid metabolism resulted in a rapid decrease of the accumulated radioactivity. At 80°C, uptake of cellobiose occurred with a Km of 175 nM, indicating the presence of a high-affinity transport system. Cellobiose transport activity was found only in cells grown on cellobiose and was not observed in maltose-grown cells (Fig. 1). Uptake of [3H]cellobiose was completely inhibited by a 10-fold excess of nonlabeled cellobiose and cellotriose, but not by glucose, maltose, or lactose. These data suggest that P. furiosus contains a high-affinity transport system for cellobiose and cellotriose.
FIG. 1.
Cellobiose transport in P. furiosus cells. Cells were grown on cellobiose (●) or maltose (○) as the carbon and energy source, and the accumulation of 10 μM [3H]cellobiose was assayed under anaerobic conditions at 80°C.
Isolation and characterization of a cellobiose binding protein.
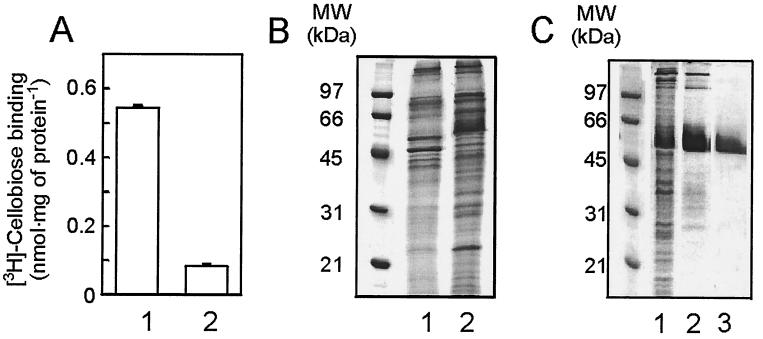
Membranes were isolated from cellobiose- and maltose-grown P. furiosus cells and were incubated with [3H]cellobiose. A high level of [3H]cellobiose binding was observed with membranes derived from cellobiose-grown cells, while membranes of maltose-grown cells showed only background binding (Fig. 2A). In contrast to cellobiose uptake, binding was not oxygen sensitive. Membranes derived from maltose- or cellobiose-grown cells were analyzed by SDS-PAGE and Coomassie brilliant blue staining. Comparison revealed the presence of a unique and abundant 55-kDa protein in the membranes of cellobiose-grown cells (Fig. 2B). The cellobiose binding protein was purified from Triton X-100-solubilized membranes by using [3H]cellobiose binding at 60°C to monitor the purification. The protein could be purified to homogeneity by ConA affinity chromatography followed by MonoQ anion-exchange chromatography. The binding activity corresponded to the 55-kDa protein which is now termed CbtA (Fig. 2C). Since CbtA binds to ConA while it is eluted with α-methyl-mannopyranoside, it appears to be glycosylated. The glycosylation was verified with periodic acid-Schiff staining of the protein after SDS-PAGE (not shown).
FIG. 2.
P. furiosus contains a cellobiose binding protein. (A) [3H]cellobiose binding to membranes derived from cells grown on cellobiose (lane 1) or maltose (lane 2). Binding studies were performed as described. (B) Coomassie brilliant blue-stained SDS-PAGE gel. Comparison between membranes derived from maltose-grown cells (lane 1) and membranes derived from cellobiose-grown cells (lane 2). (C) Coomassie brilliant blue-stained gel of purification of CbtA. Lane 1, membranes; 2, ConA fraction; 3, purified CbtA.
Purified CbtA binds cellobiose with a Kd of 45 nM and a Bmax of 0.7 nmol · mg of protein−1 at 60°C. The substrate specificity of CbtA was determined by means of competition for [3H]cellobiose binding. In 10-fold excess, nonlabeled cellobiose, cellotriose, cellotetraose, and cellopentaose appeared to be effective competitors for [3H]cellobiose binding to CbtA (Table 1). Competition was also observed with laminaribiose and laminaritriose, both building blocks of the polymer laminarin. The disaccharide sophorose was less effective as inhibitor, while the disaccharides β,β-trehalose and gentiobiose were ineffective. All of the sugars that effectively competed with [3H]cellobiose binding to CbtA also supported growth of P. furiosus (Table 1). These data suggest that CbtA is a broad-specificity β-glucoside binding protein.
TABLE 1.
Substrate specificity of the cellobiose binding protein CbtA and growth of P. furiosus on various sugars
| Competing substrate | Residual [3H]cellobiose bindinga (% [mean ± SEM] of control) | Growthb |
|---|---|---|
| Cellobiose | 11 ± 1 | + |
| Cellotriose | 12 ± 2 | + |
| Cellotetraose | 9 ± 1 | + |
| Cellopentaose | 10 ± 0.7 | + |
| Laminaribiose | 17 ± 3 | + |
| Laminaritriose | 12 ± 1 | ND |
| Sophorose | 33 ± 15 | + |
| Gentiobiose | 109 ± 26 | − |
| β,β-Trehalose | 89 ± 16 | − |
| Maltose | 100 ± 17 | + |
| Glucose | 85 ± 8 | − |
Binding of 200 nM [3H]cellobiose to purified CbtA was measured in the absence and presence of 2 μM concentrations of the indicated nonlabeled sugar substrates. Experiments were performed in triplicate with the indicated standard error of the mean.
Growth in the presence of 0.1% (wt/vol) of the indicated sugar as the sole carbon and energy source. +, growth; −, no growth; ND, not determined. Growth experiments were performed in duplicate.
Cloning and heterologous expression of CbtA.
N-terminal amino acid sequence analysis by Edman degradation of the purified CbtA yielded the amino acid sequence QEQELPR. Database searches of the P. furiosus genome (http://wit.mcs.anl.gov/) identified an open reading frame (ORF) (RPF00252) with an exact match. This ORF encoded an additional 20 amino acids at the N terminus, predicted to form a typical signal sequence. The calculated molecular mass of the mature protein is 70 kDa, which is substantially larger than the 55 kDa estimated for the purified CbtA by SDS-PAGE. This discrepancy is due to an incomplete denaturation of CbtA in SDS. After boiling for 30 min in 2% SDS, CbtA migrated as a 70-kDa protein on SDS-PAGE gels. Hydropathy analysis of CbtA indicates the presence of a hydrophobic domain at the carboxyl terminus that possibly functions as a membrane anchor. Strikingly, the hydrophobic domain is preceded by a serine/threonine-rich region that may function as a flexible linker to connect the catalytic domain to the membrane-anchoring region. Homology searches revealed that the protein belongs to the OppA family of binding proteins, with the highest homology to putative binding protein of various thermophilic members of Archaea and Bacteria. The cbtA gene is part of a gene cluster that includes four other genes (Fig. 3). The products of two of these ORFs, i.e., those of cbtB (RPF00251) and cbtC (RPF00250), are homologous to OppB and OppC, respectively. These proteins constitute the permease domain of the Opp system. The other two ORFs, cbtD (RPF00249) and cbtF (RPF00248), encode gene products that are homologous to OppD and OppF, the ATP-hydrolyzing subunits of the transport system. Upstream of cbtA and cbtB a putative TATA box is observed.
FIG. 3.
Genetic organization of the genes encoding the cellobiose transport system. The genes and their gene products are as follows: cbtA, the extracellular binding protein; cbtB and cbtC, permease domains; and cbtD and cbtF, the cytosolic ATP binding domains. The arrows indicate putative promoter regions. Boxes with identical shadings indicate homologous functions.
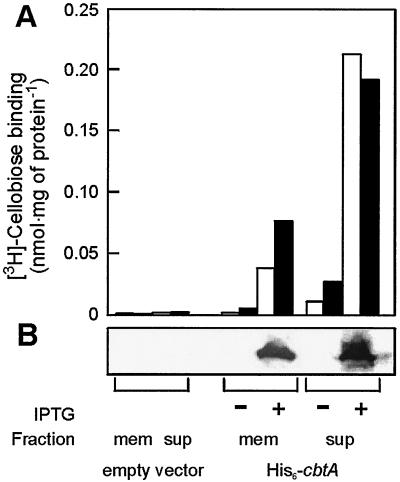
The cbtA gene was cloned with an N-terminal hexahistidine tag into an E. coli expression vector behind the trc promoter and transformed to E. coli strain SF120 together with p1244 (13). The latter plasmid bears tRNAs for the amino acids leucine, isoleucine, and arginine with codons that are rarely used by E. coli. Since E. coli uses a phosphoenolpyruvate transferase system for cellobiose uptake (10), functional expression of CbtA could conveniently be determined by [3H]cellobiose binding studies (Fig. 4A). While binding of cellobiose was absent in the soluble and membrane fraction of the lysed parental strain, significant binding levels were observed in the cells upon the induction of CbtA expression. The binding activity correlates with the presence of the protein in the various fractions, as evidenced by immunoblotting using a hexahistidine tag antibody (Fig. 4B). These results demonstrate that the P. furiosus cbtA gene encodes a cellobiose binding protein and that this protein can be functionally expressed in E. coli.
FIG. 4.
Expression of P. furiosus CbtA in E. coli SF120/1244. (A) Cellobiose binding activity at 37°C (white bars) and 60°C (black bars) using 500 nM [3H]cellobiose. (B) Western blot detection of His6-CbtA by using His antibodies in the membrane (mem) and supernatant (sup) of cell lysates after (+) and before (−) induction. The E. coli SF120/1244 cells that were used were cotransformed with p1244, which harbors the rare tRNA genes, and pET302 (empty vector) or pSMK4, which contains the cbtA gene with an N-terminal hexahistidine tag.
Cellobiose uptake by P. abyssi
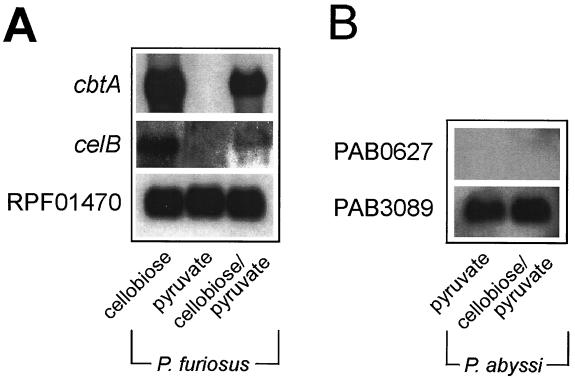
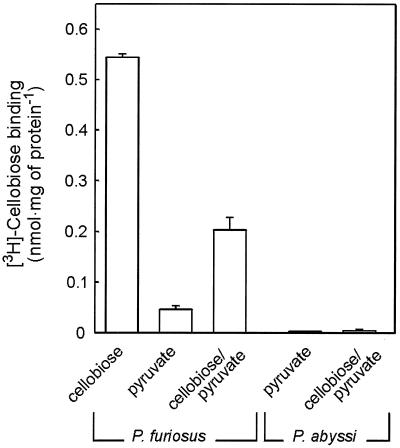
P. abyssi GE5 is unable to grow on cellobiose (8), but it does contain a gene cluster that is highly homologous to the cbt cluster of P. furiosus. ORFs PAB0627, PAB0628, PAB2363, PAB0630, and PAB0631 of the P. abyssi GE5 chromosome resemble cbtA, cbtB, cbtC, cbtD, and cbtF, respectively. The gene products are more than 79% identical. To determine if this putative cellobiose transport system is expressed in P. abyssi, cells were grown on pyruvate in the presence of cellobiose. Northern blotting shows that under these growth conditions, both cbtA and the structural gene of the β-glucosidase, celB, are expressed in P. furiosus, while no expression is seen when cells are grown on pyruvate only (Fig. 5A). Under similar growth conditions, it was not possible to detect the expression of the P. abyssi homolog of the P. fusiosus CbtA ORF, PAB0627 (Fig. 5B). Membranes derived from P. abyssi cells grown on pyruvate and a combination of pyruvate and cellobiose were inactive for [3H]cellobiose binding, while a high binding activity could be observed in membranes derived from P. furiosus cells grown on cellobiose and the combination of cellobiose and pyruvate. In cells grown solely on pyruvate, only minor binding activity could be detected (Fig. 6). These data suggest that P. abyssi is defective in the expression of the cellobiose transport system.
FIG. 5.
Northern blot analysis of total RNA extracted from P. furiosus (A) or P. abyssi (B) cells grown on cellobiose, pyruvate, or pyruvate-cellobiose. Histones (RPF01470, PAB3089) were used as internal controls for determining the total amount of RNA.
FIG. 6.
[3H]cellobiose binding to P. furiosus and P. abyssi membranes derived from cells grown on cellobiose, pyruvate, or pyruvate-cellobiose.
DISCUSSION
Here, we show that P. furiosus contains a high-affinity binding-protein-dependent ABC transport system for the uptake of cellobiose and most other β-glucosides. The cellobiose binding protein, CbtA, is a 70-kDa glycosylated protein. Strikingly, it is homologous to the di- and tripeptide binding proteins of the OppA family. So far, only the α-galactoside binding protein AgpA of Rhizobium meliloti was known to be a sugar-binding member of this family (11), which also includes nickel, opine, heme, and substituted sugar transporters (11, 28). The gene cluster encoding the cellobiose transporter includes genes that encode two distinct ATPases and two membrane domains. This architecture corresponds to what is generally observed for members of the Opp family of oligopeptide ABC transporters. ABC transporters for sugars usually contain only a single ATPase subunit that is thought to function as a homodimer.
Databank searches revealed the presence of many putative binding proteins of other thermophilic archaea and bacteria that are homologous to the cellobiose ABC transport system of P. furiosus. Nine out of 11 operons encoding ABC transporters present in the genome of the hyperthermophilic bacterium T. maritima encode members of the OppA family. It has been suggested that these transport systems are oligopeptide transporters (25), but based on our current finding and the locations of these operons in the vicinity of genes that are involved in sugar metabolism, it is more likely that some of these transporters are sugar transporters. The cellobiose transport system of the thermoacidophilic archaeon S. solfataricus also belongs to the Opp transporter family (7). Again, genes encoding sugar-metabolizing enzymes are located in the vicinity of the transport operon. In this respect, the gene upstream of the cbtA gene in P. furiosus encodes a β-mannosidase. Its specific physiological role is unclear (4).
Like oligopeptide binding proteins, CbtA binds a broad range of polymeric substrates. In contrast, sugar-binding proteins usually exhibit a narrow substrate specificity that is often limited to monosaccharides. Therefore, it may well be that the substrate binding pocket of CbtA more or less resembles that of the OppA family of binding proteins that can accommodate a range of short and long oligopeptides (6, 23).
The cbt gene cluster contains two putative TATA boxes, one upstream of cbtA and one upstream of the cbtB gene. The latter promoter most likely controls expression of the cbtBCDF genes. Northern analysis revealed larger amounts of cbtA transcript than of the cbtBCDF transcripts. The presence of two promoter regions presumably relates to the need for binding protein in excess to the transporter domains to allow efficient scavenging of the substrate at the external surface of the membrane.
P. abyssi GE5 harbors a gene cluster that shares a very high degree of homology with the cbtABCDF genes of P. furiosus. However, P. abyssi GE5 does not grow on cellobiose (12), which has been attributed to the lack of a gene encoding a β-glucosidase, CelB, needed to hydrolyze cellobiose to glucose (http://www.genoscope.cns.fr/Pab/). Another P. abyssi strain, ST549, does exhibit β-glucosidase activity (22) but is also unable to grow on cellobiose. Our data with P. abyssi GE5 indicate that the putative cellobiose transporter is not expressed when cells are grown on pyruvate in the presence of cellobiose. These conditions do, however, result in expression of the ctbABCDF genes of P. furiosus. It seems likely that P. abyssi GE5 is defective in a response regulator that triggers induction of the genes involved in cellobiose metabolism, including the transport system.
P. furiosus has not been reported to grow on cellulose, although the endoglucanase EglA exhibits hydrolytic activity against carboxymethyl cellulose (5). The organism, however, does grow on different cello-oligomers. EglA is an extracellular protein that exhibits a greater affinity for cellopentaose and cellohexaose compared to the shorter cello-oligomers (5). The long cello-oligomers will most likely first be hydrolyzed extracellularly to yield cellobiose, cellotriose, or cellotetraose. These compounds are then transported into the cell and hydrolyzed to glucose by CelB (20) and possibly other proteins. P. furiosus can also grow on the β-1,3-glucose polymer laminarin. This possibly involves the hydrolysis of laminarin into smaller laminari-oligomers by the extracellular enzyme LamA (14). The laminari-oligomers enter the cell via the cellobiose transport system and are then hydrolyzed to glucose by the intracellular β-glucosidase CelB (12). Our studies show that P. furiosus is able to grow on the β-glucoside sophorose. This compound also inhibits cellobiose binding to CbtA, and therefore it is likely that sophorose enters the cells via the cellobiose transporter. Sophorose metabolism requires an intracellular β-glucanase, but such an enzyme has not yet been reported for P. furiosus. The β-glucosides gentiobiose and β,β-trehalose are not recognized by CbtA, nor is P. furiosus able to grow on these substrates. CelB was shown to hydrolyze β-1,6-glycosidic bonds (T. Kaper, personal communication). Therefore, the inability of P. furiosus to grow on gentiobiose might be due to a lack of transport activity.
Summarizing, P. furiosus contains a binding protein-dependent ABC transport system for the uptake of cellobiose and a range of β-glucosides. This system is homologous to the OppA family of ABC transporters that are mainly used for oligopeptide transport. These transporters share the property that they are involved in the transport of oligomeric compounds, such as oligosaccharides and oligopeptides.
ACKNOWLEDGMENTS
This work was supported by the Earth and Life Sciences Foundation (ALW), which is subsidized by The Netherlands Organization for Scientific Research (NWO).
We thank Sonja V. Albers for helpful discussions; Neil Raven, PTL Microbial Products, CAMR, for the large-scale growth of Pyrococcus furiosus; John van der Oost and Ans Geerling for the initial help with the mRNA isolation; and Melchior Evers for advice on the RT-PCR.
REFERENCES
- 1.Albers S V, Elferink M G, Charlebois R L, Sensen C W, Driessen A J M, Konings W N. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J Bacteriol. 1999;181:4285–4291. doi: 10.1128/jb.181.14.4285-4291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baneyx F, Georgiou G. Construction and characterization of Escherichia coli strains deficient in multiple secreted proteases: protease III degrades high-molecular-weight substrates in vivo. J Bacteriol. 1991;173:2696–2703. doi: 10.1128/jb.173.8.2696-2703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbier G, Godfroy A, Meunier J R, Querellou J, Cambon M A, Lesongeur F, Grimont P A, Raguenes G. Pyrococcus glycovorans sp. nov., a hyperthermophilic archaeon isolated from the East Pacific Rise. Int J Syst Bacteriol. 1999;49(Pt. 4):1829–1837. doi: 10.1099/00207713-49-4-1829. [DOI] [PubMed] [Google Scholar]
- 4.Bauer M W, Bylina E J, Swanson R V, Kelly R M. Comparison of a beta-glucosidase and a beta-mannosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Purification, characterization, gene cloning, and sequence analysis. J Biol Chem. 1996;271:23749–23755. doi: 10.1074/jbc.271.39.23749. [DOI] [PubMed] [Google Scholar]
- 5.Bauer M W, Driskill L E, Callen W, Snead M A, Mathur E J, Kelly R M. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes β-1,4 bonds in mixed-linkage (1→3),(1→4)-β-d-glucans and cellulose. J Bacteriol. 1999;181:284–290. doi: 10.1128/jb.181.1.284-290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Detmers F J, Lanfermeijer F C, Abele R, Jack R W, Tampe R, Konings W N, Poolman B. Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc Natl Acad Sci USA. 2000;97:12487–12492. doi: 10.1073/pnas.220308797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elferink M G, Albers S V, Konings W N, Driessen A J M. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC-transporters. Mol Microbiol. 2001;39:1494–1503. doi: 10.1046/j.1365-2958.2001.02336.x. [DOI] [PubMed] [Google Scholar]
- 8.Erauso G, Reysenbach A-L, Godfroy A, Meunier J-R, Crump B, Partensky F, Baross J A, Marteinsson V, Barbier G, Pace N R, Prieur D. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch Microbiol. 1993;160:338–349. [Google Scholar]
- 9.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100oC. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 10.Fox C F, Wilson G. The role of a phosphoenolpyruvate-dependent kinase system in beta-glucoside catabolism in Escherichia coli. Proc Natl Acad Sci USA. 1968;59:988–995. doi: 10.1073/pnas.59.3.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gage D J, Long S R. α-Galactoside uptake in Rhizobium meliloti: isolation and characterization of agpA, a gene encoding a periplasmic binding protein required for melibiose and raffinose utilization. J Bacteriol. 1998;180:5739–5748. doi: 10.1128/jb.180.21.5739-5748.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godfroy A, Raven N D, Sharp R J. Physiology and continuous culture of the hyperthermophilic deep-sea vent archaeon Pyrococcus abyssi ST549. FEMS Microbiol Lett. 2000;186:127–132. doi: 10.1111/j.1574-6968.2000.tb09093.x. [DOI] [PubMed] [Google Scholar]
- 13.Greller G, Horlacher R, DiRuggiero J, Boos W. Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem. 1999;274:20259–20264. doi: 10.1074/jbc.274.29.20259. [DOI] [PubMed] [Google Scholar]
- 14.Gueguen Y, Voorhorst W G, van der Oost J, de Vos W M. Molecular and biochemical characterization of an endo-beta-1,3-glucanase of the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1997;272:31258–31264. doi: 10.1074/jbc.272.50.31258. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.Helaszek C T, White B A. Cellobiose uptake and metabolism by Ruminococcus flavefaciens. Appl Environ Microbiol. 1991;57:64–68. doi: 10.1128/aem.57.1.64-68.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horlacher R, Xavier K B, Santos H, DiRuggiero J, Kossmann M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajikawa H, Masaki S. Cellobiose transport by mixed ruminal bacteria from a cow. Appl Environ Microbiol. 1999;65:2565–2569. doi: 10.1128/aem.65.6.2565-2569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kengen S W M, de Bok F A, van Loo N D, Dijkema C, Stams A J, de Vos W M. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269:17537–17541. [PubMed] [Google Scholar]
- 20.Kengen S W M, Luesink E J, Stams A J, Zehnder A J. Purification and characterization of an extremely thermostable beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur J Biochem. 1993;213:305–312. doi: 10.1111/j.1432-1033.1993.tb17763.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim R, Sandler S J, Goldman S, Yokota H, Clark A J, Kim S H. Overexpression of archaeal proteins in Escherichia coli. Biotechnol Lett. 1998;20:207–210. [Google Scholar]
- 22.Ladrat C, Alayse-Danet A M, Barbier G, Dietrich J. A new thermostable glucose-activated β-glucosidase from the hyperthermophilic marine archaebacterium Pyrococcus abyssi: purification and characterization. J Mar Biotechnol. 1997;4:192–199. [Google Scholar]
- 23.Lanfermeijer F C, Detmers F J, Konings W N, Poolman B. On the binding mechanism of the peptide receptor of the oligopeptide transport system of Lactococcus lactis. EMBO J. 2000;19:3649–3656. doi: 10.1093/emboj/19.14.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuckin W F, McKenzie B F. An improved periodic acid fuchsin sulfite staining method for evaluation of glycoproteins. Clin Chem. 1958;4:476–483. [PubMed] [Google Scholar]
- 25.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 26.Schäfer T, Xavier K B, Santos H, Schönheit P. Glucose fermentation to acetate and alanine in resting cell suspensions of Pyrococcus furiosus: proposal of a novel glycolytic pathway based on 13C labelling data and enzyme activities. FEMS Microbiol Lett. 1994;121:107–114. [Google Scholar]
- 27.Schlosser A, Jantos J, Hackmann K, Schrempf H. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl Environ Microbiol. 1999;65:2636–2643. doi: 10.1128/aem.65.6.2636-2643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam R, Saier M H. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Does C, Manting E H, Kaufmann A, Lutz M, Driessen A J M. Interaction between SecA and SecYEG in micellar solution and formation of the membrane-inserted state. Biochemistry. 1998;37:201–210. doi: 10.1021/bi972105t. [DOI] [PubMed] [Google Scholar]
- 30.Wassenberg D, Liebl W, Jaenicke R. Maltose-binding protein from the hyperthermophilic bacterium Thermotoga maritima: stability and binding properties. J Mol Biol. 2000;295:279–288. doi: 10.1006/jmbi.1999.3367. [DOI] [PubMed] [Google Scholar]
- 31.Xavier K B, Martins L O, Peist R, Kossmann M, Boos W, Santos H. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1996;178:4773–4777. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]