Abstract
Abstract
Atrial fibrosis plays a key role in the initiation and progression of atrial fibrillation (AF). Atrial fibrosis is typically identified by a peak-to-peak amplitude of bipolar electrograms (b-EGMs) lower than 0.5 mV, which may be considered as ablation targets. Nevertheless, this approach disregards signal spatiotemporal information and b-EGM sensitivity to catheter orientation. To overcome these limitations, we propose the dominant-to-remaining eigenvalue dominance ratio (EIGDR) of unipolar electrograms (u-EGMs) within neighbor electrode cliques as a waveform dispersion measure, hypothesizing that it is correlated with the presence of fibrosis. A simulated 2D tissue with a fibrosis patch was used for validation. We computed EIGDR maps from both original and time-aligned u-EGMs, denoted as and , respectively, also mapping the gain in eigenvalue concentration obtained by the alignment, . The performance of each map in detecting fibrosis was evaluated in scenarios including noise and variable electrode-tissue distance. Best results were achieved by , reaching 94% detection accuracy, versus the 86% of b-EGMs voltage maps. The proposed strategy was also tested in real u-EGMs from fibrotic and non-fibrotic areas over 3D electroanatomical maps, supporting the ability of the EIGDRs as fibrosis markers, encouraging further studies to confirm their translation to clinical settings.
Graphical Abstract
Upper panels: map of from 3×3 cliques for Ψ= 0∘ and bipolar voltage map Vb-m, performed assuming a variable electrode-to-tissue distance and noisy u-EGMs (noise level σv = 46.4 μV ). Lower panels: detected fibrotic areas (brown), using the thresholds that maximize detection accuracy of each map 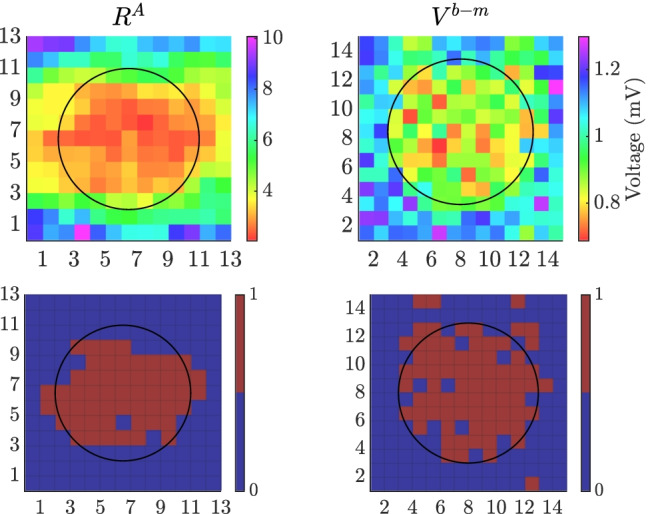
Keywords: Atrial fibrosis, Atrial fibrillation (AF), Bipolar electrograms (b-EGMs), Eigenvalue dominance ratio (EIGDR), Unipolar electrograms (u-EGMs)
Introduction
Atrial fibrosis represents a structural anomaly of the atrial myocardium [1]. It is characterized by an altered extracellular matrix activity caused by fibroblasts [2], which alters the electrical conduction and excitability of the tissue. Fibroblasts activation and proliferation, as well as their secretion of extracellular matrix proteins, such as collagen, characterize fibrotic tissue [3]. This structural remodeling mainly occurs during the reparative process to replace damaged myocardial parenchyma [4]. In addition to this replacement process, others, as reactive fibrosis to a trigger as inflammation, have been recognized as responsible for fibrosis. This makes detection of fibrotic tissue even more difficult and suggests the need for more specific imaging tools and markers to detect and quantify fibrosis [5].
Atrial fibrosis has been observed to be correlated to atrial fibrillation (AF) [6]. Despite the fact that AF represents the most common cardiac arrhythmia, its trigger mechanisms are not yet fully understood [7] and its causal relationship with atrial fibrosis is still challenging [1]. On the one hand, fibrosis-induced remodeling creates a substrate promoting AF [1, 6]; on the other hand, fibrosis can occur as a result of the electrical [1] as well as structural [4] atrial remodeling found in AF.
Atrial fibrosis is electrophysiologically characterized by low intracardiac electrograms (EGMs) amplitudes and conduction velocities [8], which may be measured with electroanatomical mapping (EAM) systems [9]. These allow displaying 3D voltage and activation time maps over a reconstruction of cardiac chambers anatomy and visualizing catheter position, so as to guide ablation procedures and treat arrhythmias with minimum radiation exposition [10]. Based on several studies, consensus exists on the choice of 0.5 mV as the threshold value of bipolar EGMs (b-EGMs) peak-to-peak amplitude to discriminate fibrotic areas in the atrium during sinus rhythm [11]. However, this procedure brings along several limitations. First of all, a peak-to-peak voltage measure disregards morphological and temporal information contained in the signal. Nevertheless, this reflects the possible presence of underlying abnormalities in the atria [12]. Secondly, bipolar voltage mapping may be influenced by other technical factors not related to the substrate, including the relative orientation between the recording electrode pair and wavefront propagation direction, electrode size, inter-electrode distance and b-EGMs filtering [12]. Bipolar voltage is also affected by the tissue-electrode contact, whose maintenance may be challenging in anatomically difficult sites (e.g., the pulmonary veins). Third, the definition of low-voltage areas has not been subjected to a standardization procedure and the voltage threshold has never received an histological validation [12].
In recent years, more attention has been paid to the role of fibrotic tissue on the initiation and perpetuation of AF than on its effects on the morphology of the EGMs [13]. In this sense, not many intracardiac signal processing methods based on EGM features have been proposed to detect fibrosis. Some studies have revealed the relationship of EGM morphology and tissue alterations, including ablation lesion formation [14]. Others have introduced a method to characterize the different fibrotic textures based on EGM fractionation due to the incidence of wavefront direction [15]. All these works have used in silico experiments for validation.
In this paper, we hypothesize that the waveform dispersion of neighbor unipolar EGMs (u-EGMs) is correlated with the presence of atrial fibrosis. Therefore, the aim of the work is to propose eigenvalue-based indices of waveform dispersion to identify fibrotic areas. They take into account the spatiotemporal relations of the u-EGM waveforms and overcome the limitations of the use of b-EGM voltage thresholding for the detection of fibrosis. Resulting maps, called eigenvalue dominance ratio (EIGDR) maps, were computed with a simulation setup, using two sizes of nearby electrode arrangements, referred as (22 and 33 electrodes), and three catheter orientations with respect to the tissue preferential direction (0, 30 and 45). The ability of each map was evaluated in detecting a fibrosis patch included in the simulated tissue. As a proof of concept, EIGDR values were also computed in clinical u-EGMs from patients, in four- and five-electrode cliques. They were correlated with the presence/absence of fibrosis based on late gadolinium enhancement-magnetic resonance imaging.
This paper is organized as follows: Section 2 presents the methodology and the datasets used for its validation. Section 3 contains the obtained results, whereas Sections 4, 5 and 6 include the discussion, limitations (with references to challenges and future perspectives) and conclusions of the work, respectively.
Methods
Atrial model
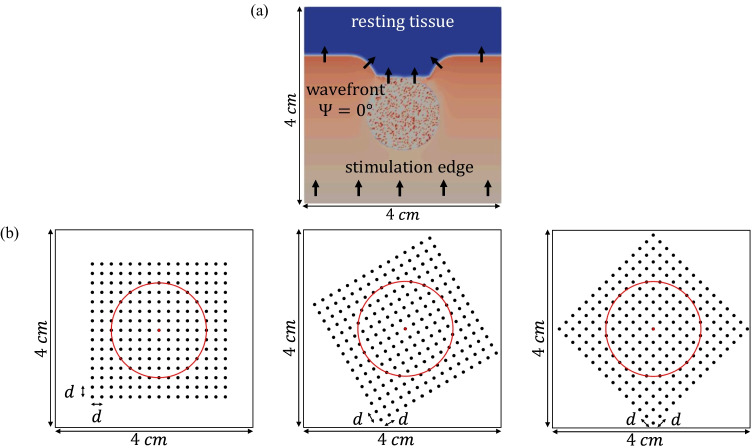
We simulated an atrial tissue slice of 44 cm by dividing it into adjacent square elements whose centers were separated 0.1 mm. Within the tissue slice, a circular patch with a diameter of 2 cm was defined, whose center was at the center of the 2D tissue, as shown in Fig. 1(a). Inside the patch, a fully transmural (from endo- to epicardial layer) pattern of diffuse fibrosis was randomly defined following a uniform distribution. The Maleckar model for myofibroblasts [16] was assigned to 20% of the nodes within the circular area. Although the percentage of atrial fibrosis is very patient-dependent, the density of 20% represents the threshold value between stage II and stage III according to the Utah classification [17] and therefore considered a realistic percentage for this study.
Fig. 1.
(a) Activation distribution at a particular time instant over the 2D tissue used in this work, including the fibrotic patch. Black arrows indicate propagation wavefront direction. (b) The three MEA orientations with respect to the tissue considered in this study: (leftmost), (middle) and (rightmost), where the red circle encompasses the fibrotic tissue area. It should be noted that representation in (a) refers to the relative orientation between tissue and propagation direction corresponding to
The cell model assigned to all the non-fibrotic nodes was a variant of the Courtemanche myocyte model [18] accounting for the atrial electrophysiological characteristics experimentally observed in the left atrium (LA) and the persistent AF (cAF) induced remodeling [19]. This electrical remodeling was introduced through the variation of the maximum conductances of the transient outward potassium current , the L-Type calcium current , the inward rectifier potassium current , the ultrarapid outward potassium current and the slow delayed rectifier potassium current (see Table 1), as in previous computational studies [19, 20]. Additionally, the diffusion tensor was adjusted to reproduce the conduction velocity in the LA, and was reduced by 30% in all elements of the patch with at least one fibroblast node [21], similarly to what was performed in [22].
Table 1.
Variation of the maximum conductances g for several ionic channels used to reproduce atrial electrical remodeling under persistent AF (cAF) conditions, accordingly to experimental studies reported in literature. As a comparison, g values have also been reported in control conditions
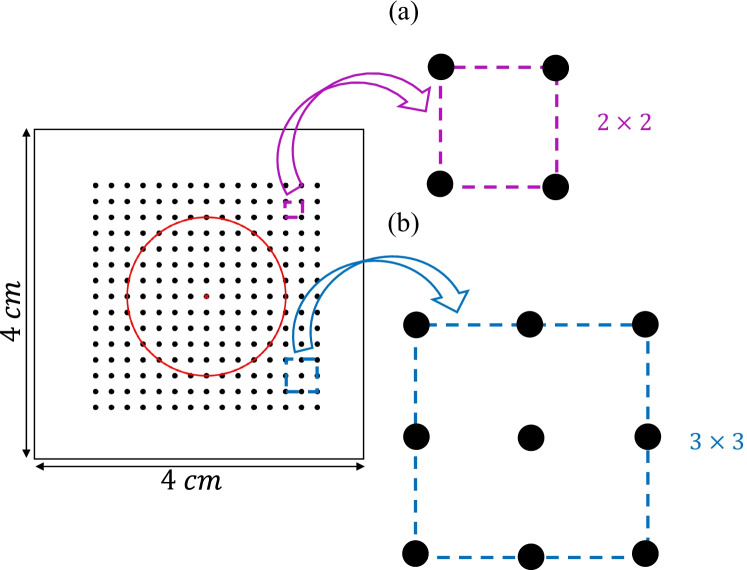
Simulations were run in ELVIRA software [23]. The monodomain formulation was solved using the operator splitting numerical scheme with a constant time step = 0.01 ms and a spatial resolution = 0.1 mm. Recording electrodes were distributed over the simulated anatomy mimicking a L L high-density multi-electrode array (MEA) (L = 15), where the inter-electrode distance was d = 2 mm. The simulated electrode grid was located so that its central electrode corresponds to the center of both the tissue slice and the fibrotic patch, and was rotated by an angle , , with respect to the tissue fiber direction. Fig. 1(b) shows the three MEA orientations over the simulated tissue geometry. Two generic cliques of different sizes were considered in this work as depicted in Fig. 2(a) and (b), forming four and nine-electrode square arrangements, respectively. Each clique location is referred to the lower left electrode, indexed as (i, j), within the MEA. The rest of electrodes in the clique are numbered from left to right and bottom to top, thus corresponding to locations , where , () for the , and , () for the arrangements, respectively.
Fig. 2.
Arrangements of four (a) and nine (b) electrodes (22 and 33 cliques, respectively) from the MEA
Synthetic signals
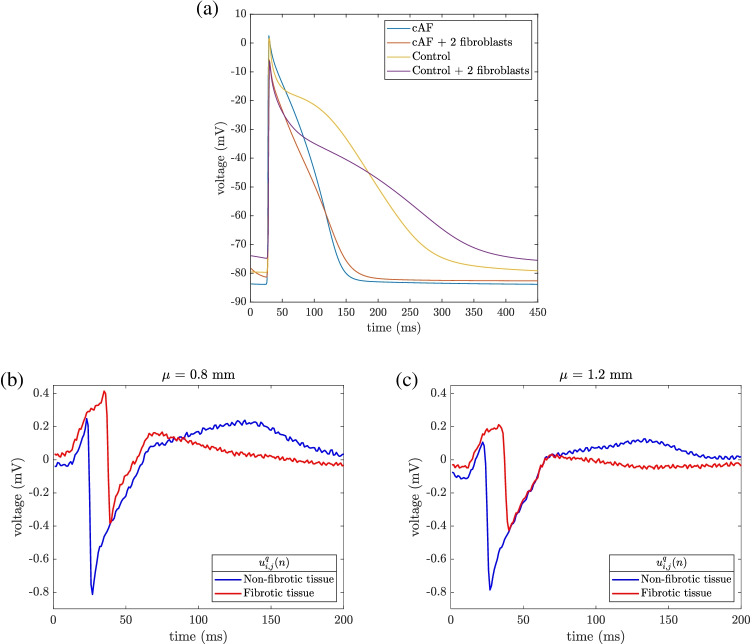
Unipolar EGMs, , were calculated as originated by the passage of the propagation wavefront by electrodes located at sites (i, j) of the MEA. We computed them in a volumetric tissue-blood model, assuming a temporal resolution of 1 ms, as done in [30]. First, extracellular potentials were obtained by an approximation of the bidomain formulation, considering the tissue immersed in a non-conductive bath. In order to reproduce fibrosis effects, inside the simulated fibrotic patch the single cardiomyocyte under cAF conditions was coupled with a randomly variable number of fibroblasts within the patch. Fig. 3(a) shows the action potentials registered in two different cardiomyocytes from the mesh, one outside the fibrotic patch and the other inside the fibrotic patch, where it is coupled with two fibroblasts. Electrical remodeling induced by cAF produces a 55% reduction in duration measured at 90% repolarization (248 vs. 111 ms), in concordance with experimental data [29]. Fibroblast coupling with cAF cardiomyocytes makes resting potential less negative (83 vs. 78 mV) and elongates the duration measured at 90% repolarization (111 vs. 120 ms). Then, u-EGMs were solved in the entire domain by the governing equations for a solid volume conductor and its boundary conditions at the tissue-blood interface.
Fig. 3.
Upper panel (a): action potentials (APs) in persistent atrial fibrillation (cAF), registered in two different nodes from the simulation mesh: in a cardiomyocyte outside the fibrotic patch (light blue line) and in a cardiomyocyte inside the fibrotic patch coupled with two fibroblasts (orange line). In order to show the effect of the applied electrical remodeling, APs were also shown in control conditions, from different simulations not including electrophysiological remodeling and not used in this work (yellow and purple lines, for uncoupled and coupled cardiomyocytes, respectively). Lower panel: Noisy unipolar EGMs () recorded in non-fibrotic, , and fibrotic tissue areas (blue and red line, respectively) when , at electrode-to-tissue distances of mm (b) and mm (c)
In order to take into account a realistic scenario, where the distance from the tissue may not be constant through the electrodes in the clique, we considered that the electrode-tissue distance, , varies following a normal distribution with mean = 1 mm and standard deviation = 0.1 mm. Two thousand different random configurations were simulated, where the distance of each electrode to the tissue was randomly and independently chosen following that distribution.
Synthetic u-EGMs were computed with sampling frequency of 1 kHz, duration of 0.5 s ( samples), and including one single activation (depolarization plus repolarization).
Simulated signals were corrupted with noise excerpts obtained from real u-EGMs, as previously done in [8]. Two thousand different noise segments were extracted from u-EGMs recorded with a multi-electrode PentaRay® catheter (Biosense-Webster, Inc., Diamond Bar, CA, USA) at intervals with no recorded EGMs. All noise segments were normalized to have standard deviations , corresponding to peak-to-peak amplitudes . They are concordant with observed average power in unipolar recordings [31] and guarantee a homogeneous power level. Different realizations, indexed by , of this recorded noise were randomly added to each one of the two thousand realizations of the simulated u-EGMs computed with a variable electrode-tissue distance within the MEA, generating as a result noisy unipolar signals , , . Noisy u-EGMs examples, in fibrotic and non-fibrotic tissue zones, are shown in Fig. 3(b) and (c), at two different electrode-tissue distances, respectively, with noise level corresponding to .
Clinical data
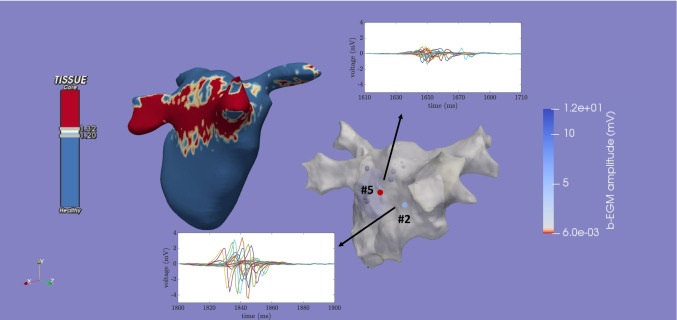
Intracavitary u-EGMs recorded during sinus rhythm with a PentaRay® catheter (Biosense-Webster, Inc., Diamond Bar, CA, USA) were used to evaluate performance of EIGDR approach with real signals. Clinical data were obtained from a patient with cAF, a slightly dilated LA (26 cm), a left ventricular ejection fraction of 58% and treated with anticoagulation and flecainide, registered at the Hospital Clínic, Barcelona, Spain. The data acquisition protocol was reviewed and approved by the Hospital Clinic Ethical Committee (Ethics approval number: HCB/2019/0881). The patient was informed and signed the consent form. A total of 758 mapping points (or catheter sites) were acquired at the anterior, posterior, lateral and septal wall, as well as the left atrial appendage and the pulmonary veins of the LA (left atrial regions) using the CARTO® 3 EAM system (Biosense-Webster, Inc., Diamond Bar, CA, USA), so as to reconstruct a real-time 3D anatomical map before the ablation procedure. Signals were acquired from the 20 poles distributed among the five branches of the catheter, Fig. 4, characterized by consecutive inter-electrode spacings d, at each branch, of 2, 6, and 2 mm, resulting in 20 u-EGMs associated at each mapping point. In order to evaluate performance of EIGDR-based markers, 38 catheter positions were selected by an operator-dependent visual approach. Specifically, we visually identified and manually annotated nineteen points clearly assignable to fibrotic and another nineteen points to non-fibrotic areas on the anatomic map. In order to guide this decision, the corresponding magnetic resonance image (MRI) was used as reference.
Fig. 4.
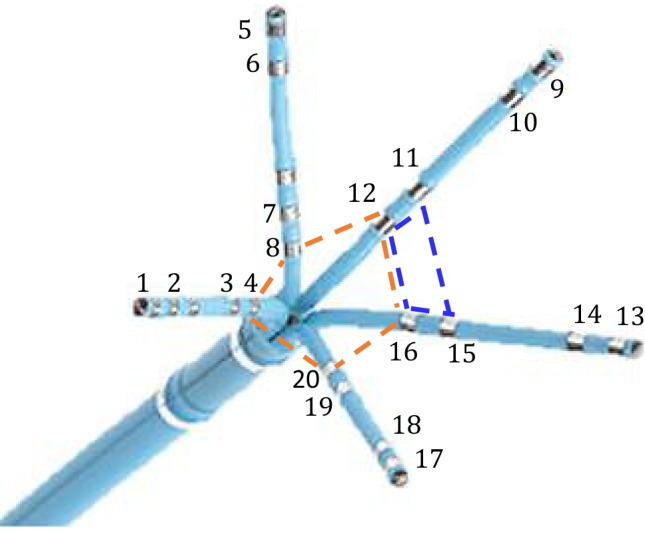
The PentaRay® catheter, where the 20 poles are highlighted. Two of the clique distributions considered around each catheter mapping point, with four (dashed blue line) and five (dashed orange line) electrodes, are also pointed out. This image was modified from the Biosense Webster catalog
Three of the catheter positions selected at fibrosis showed poor electrode-tissue contact for one or more splines of the catheter. In addition, mapping points located at borderline areas between fibrotic and healthy tissues over the MRI and/or the bipolar voltage map, which showed bipolar amplitudes unhealthy (< 0.1 mV) or poor healthy (< 0.5 mV), have been excluded from the analysis.
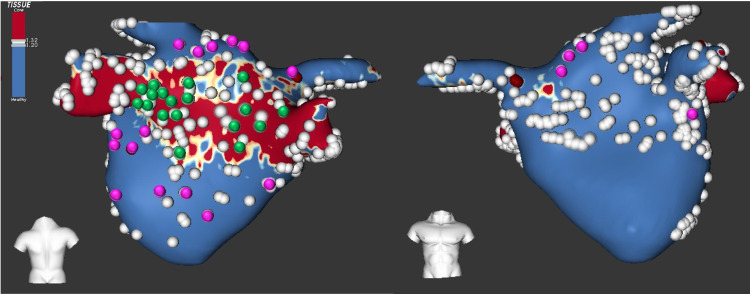
EAM data and MRI were co-registered with the ADAS 3D Medical imaging software (ADAS-3D, Barcelona, Spain), as shown in Fig. 5. That co-register was performed by manually selecting some landmarks (between six and ten) in specific areas (such as the pulmonary veins and the atrial appendage) of the meshes. In order to determine how pathological the tissue is, the methodology described in [32] was used. Following the image intensity ratio (IIR) based thresholding, a color-coded 3D mesh was automatically generated, showing in blue the healthy tissue (IIR<1.2) and in red the dense fibrosis (IIR>1.32) (Fig. 5).
Fig. 5.
Posteroanterior (left) and anteroposterior (right) views of color-coded 3D mesh of MRI (showing dense fibrosis in red and healthy tissue in blue) generated by ADAS 3D co-registered with all EAM mapping points provided by CARTO 3 (gray). The 38 mapping points selected over fibrotic and non-fibrotic areas to compute EIGDR and bipolar indices are highlighted in green and magenta, respectively
The u-EGMs were acquired with a sampling frequency of 1 kHz during 2.5 s (2500 samples) and include several activations, each containing the atrial depolarization, followed by the ventricular depolarization and repolarization. Unipolar signals were then high-pass filtered with 30 Hz cutoff frequency, using a third order Butterworth infinite impulse response filter, so as to reduce artefacts and emphasize more rapid components. An example of filtered u-EGMs at two different mapping points marked on the anatomical 3D mesh of the atrium is presented in Fig. 6, where the atrial activation was plotted. At each catheter position, u-EGMs were considered in the following five four-electrode cliques, according to the pole numbering in Fig. 4: (3,4,7,8), (7,8,11,12), (11,12,15,16), (15,16,19,20), (19,20,3,4) and the five-electrode clique (4,8,12,16,20), for their smaller inter-electrode spacing. As example, cliques (3,4,7,8) and (4,8,12,16,20) are depicted in Fig. 4.
Fig. 6.
3D reconstruction of the LA geometry (gray mesh) and corresponding co-registered MRI, showing the different regional distribution patterns of gadolinium (red areas: latest contrast enhancement, blue areas: absence of latest contrast enhancement). In the geometrical mesh, two of the mapping points acquired and considered in the analysis (point #5 at fibrosis, point #2 at non-fibrosis) are marked and color-coded according to their corresponding bipolar peak-to-peak amplitude. For each of them, the atrial activation windows extracted from the twenty filtered u-EGMs recorded with the PentaRay® catheter are also displayed. Note that not all displayed u-EGMs recorded at a particular catheter site belong to a clique, see Section 2.3, and therefore affect the EIGDR indices and bipolar amplitude computations
Eigenvalue analysis
Unipolar signals in the clique can be compactly represented by the following matrix, , as in [33]:
| 1 |
where the k-th column, , contains the samples of the unipolar signal , modeled later in Section 2.6:
| 2 |
We propose and assess EIGDR values from the clique u-EGMs in (1) to detect fibrosis. The N eigenvalues of the N N intra-signal correlation matrix were obtained from the following correlation matrix estimate within each clique:
| 3 |
The matrix is the intra-signal sample correlation matrix, whose eigenvalues reflect the degree of morphological variability among the signals in the clique.
For each clique, the ratio of the largest (i.e. dominant) eigenvalue and the remaining ones was estimated:
| 4 |
By comparing the first eigenvalue to the sum of the others, we are able to quantify the u-EGM energy percentage which can be explained by the shape of the first eigenvector. The ratio would be much higher than one if all u-EGMs were essentially identical to each other and all the waveforms can be explained with just the shape of the first eigenvector (i.e., low morphological variability). On the contrary, when waveform dispersion appears, the eigenvalues from to become higher, thus reducing the ratio .
Wave alignment
Eigenvalues of were computed from the original u-EGMs and after intra-clique time alignment, proposed to compensate the effect that different activation wavefront arrival times have on the EIGDR. In this case, unipolar signals were aligned according to Woody’s iterative procedure [34, 35]:
- at first l-th iteration, , the relative time delay between each unipolar signal and the u-EGM with the highest peak-to-peak amplitude within the clique (assumed as initial reference signal) was estimated by maximizing their cross-correlation:
5 - from the relative time delays, the average of the shifted signals within the clique was calculated:
6 - in each l-th iteration, , the cross-correlation between each and the average signal obtained in the previous iteration (assumed as updated reference signal) is maximized to find the updated relative time delays :
This process is repeated iteratively until the delay estimates no longer change.7
Eigenvalues of the intra-signal correlation matrix of the aligned u-EGMs were calculated in the same way as for their non-aligned version, thus leading to the formulation of the ratio (where the upper index denotes that the ratio comes from aligned u-EGMs within the clique) analogous to (4).
Unipolar signal modeling
The u-EGM signals within a clique located at (i, j) position within the MEA are indexed as , , . Assuming a plane wave propagation, the different electrodes in a clique are activated at different times, and therefore u-EGMs in the clique will be delayed versions among them, plus noise and non-homogeneous components. Therefore, they can be modeled, analogously to [36] for misaligned signal ensembles, as:
| 8 |
where:
s(n) is the u-EGM activation signal component assumed to be space invariant in the case of a plane wave propagation and homogeneous tissue free of fibrosis. Its energy is denoted as .
is the delay of the k-th u-EGM with respect to a time reference within the clique, introduced later. Delays are zero-mean and characterized by their variance in the normal tissue . In fibrotic areas, the reduction in conduction velocity with respect to healthy tissue increases the variance of the up to , where the factor > 1 in fibrosis and 1 in non-fibrotic tissue.
is a parameter accounting for u-EGM amplitude reduction between fibrotic, , and healthy, , tissues. It is modeled as a random variable with mean and variance (, in healthy tissue; , in fibrosis).
is a zero-mean fibrotic signal component across the clique with variance . In healthy tissue .
is the zero-mean noise component at the k-th u-EGM, with variance , Gaussian, white and uncorrelated with and .
Four different scenarios for each were considered in this study, as already proposed in [37], with/without fibrosis and with/without prior alignment. Their approximate theoretical eigenvalues and EIGDR were derived following parallel methodology to that used in [36] for repetitive signal ensemble alignment, as detailed below.
Prior alignment with no fibrosis
For perfectly aligned signals without fibrosis, i.e., , the intra-signal correlation matrix is given by:
| 9 |
where is the identity matrix and data vector , , is easily shown to be proportional to the first eigenvector of , whereas the remaining eigenvectors are chosen arbitrarily as long as they are orthogonal to the first. The eigenvalues are given by:
| 10 |
where is the signal energy. For real u-EGM, signal energy is much larger than noise energy, , and , resulting in an EIGDR of:
| 11 |
No prior alignment and no fibrosis
We now analyze the case of raw u-EGMs in the clique where misalignment of s(n) is assumed to be present in each k-th u-EGM, . Then, to estimate the eigenvalues we can approximate , for small , as [36]:
| 12 |
where and denote the first and second derivative of s(n), respectively. The intra-signal correlation matrix can be expressed as:
| 13 |
where and are the vector counterparts of and , respectively. It can be shown that the eigenvalues of are approximated by [36]:
| 14 |
where is the derivative signal energy. The resulting EIGDR is approximated by:
| 15 |
Note that when (i.e., perfect alignment) this equation becomes equal to (11).
Prior alignment and fibrosis
When u-EGMs are first aligned in fibrosis zones, each of them can be modeled as . The correlation matrix results in:
| 16 |
and their corresponding eigenvalues are:
| 17 |
which lead to:
| 18 |
where the parameters used have already been introduced at the beginning of this section and upper and lower indices in denote that the calculations are obtained from intra-clique aligned u-EGMs in fibrotic tissue, respectively.
No prior alignment with fibrosis
When raw u-EGMs come from fibrotic areas, each of them can be modeled as . In this case, the delay has larger standard deviation than in non-fibrotic areas, with the extra delay controlled by a multiplicative factor , thus resulting in the following correlation matrix:
| 19 |
which is similar to the case without fibrosis in (13) but with different proportionality factor and two different random components, corresponding to noise and fibrosis. The eigenvalues will similarly be approximated by:
| 20 |
The corresponding EIGDR is approximated by:
| 21 |
A summary of eigenvalues and the EIGDR are reported in Table 2 for the four scenarios.
Table 2.
Signal models for non-aligned (NA) and aligned (A) u-EGMs at non-fibrotic (NF) and fibrotic (F) areas, with their respective eigenvalues and eigenvalue dominance ratios EIGDR
| u-EGM | model | EIGDR | |
|---|---|---|---|
| NA, NF | |||
| A, NF | |||
| NA, F | |||
| A, F |
Note that if the inter-signal correlation matrix, , with , had been computed rather than the intra-signal correlation matrix , for the more general case with fibrosis and misalignment, and following a derivation parallel to the one presented in [36], the eigenvalues would have resulted in:
| 22 |
When computing the EIGDRs for this matrix the results are approximately the same as for .
In practice, the matrix is estimated as:
| 23 |
rather than with the theoretical expectations. From these estimates we observe that matrix is full rank while matrix (estimated as in (3)) is not (), circumstance that does not represent a limitation since no matrix inversions are required. In addition, as shown in [33], for the data-estimated autocorrelation matrices with , () and (), again showing equivalence of the EIGDR ratios for both matrices when estimated from the available data. Therefore just computational considerations can advise to use one or the other.
EIGDR-based fibrosis markers
According to the previous model, three main differential effects on signal shape, amplitude and arrival times are expected to occur in fibrotic as compared to non-fibrotic tissue:
higher morphology dispersion represented by the u-EGM signal component and quantified by ;
lower and less homogeneous signal amplitudes represented by and quantified by and (();
larger inter-signal misalignment within the clique as a result of slowed conduction and represented by delays with enlarged variance (, ) relative to healthy tissue ().
In order to determine which ratio to use for discriminating F and NF tissues, we first compare the EIGDR computed with no prior alignment of u-EGMs and no fibrosis, , with the case with fibrotic tissue, , see Table 2. We observe that , since the numerator in is smaller than in as a result of being larger than one, while the terms in denominator are larger, , and (, as a combination of the three effects introduced by fibrosis. This result suggests the use of the ratio , which becomes in fibrosis, with a thresholding strategy to discriminate if the clique is at fibrotic or healthy tissue.
The same analysis can be done when comparing the EIGDR with prior alignment of u-EGMs and fibrosis, , with respect to its counterpart with no fibrosis, . In this case, only the terms and ( are responsible of the difference, since has already been compensated for with alignment, resulting in . This suggests that may also be used as a thresholding strategy to discriminate fibrotic from healthy tissue.
In order to study which of the two options, or , is more sensitive to fibrotic tissue characteristics, we analyze how the difference between and evolves by varying . For that purpose, we compute the ratio , which under the proposed signal model can be approximated by:
| 24 |
Its partial derivative with respect to is:
| 25 |
Typically in fibrosis since conduction velocity reduction is less prominent ( for high fibrosis, [38, 39]) than voltage attenuation ( [12]) and consequently the product . This results in , meaning that the lower the misalignment the larger , implying higher EIGDR differences between fibrotic, , and healthy, , tissue. This justifies the advantage of pre-aligning u-EGMs in the cliques before EIGDR calculations, since the higher the misalignment , the lower and consequently the capacity of to discriminate between fibrosis and non-fibrosis, and suggests that is better suited fibrosis marker than .
Alternatively, we considered the ratio of EIGDR computed from perfectly aligned u-EGMs with respect to misaligned original u-EGM signals, representing the gain in eigenvalue concentration produced by alignment:
| 26 |
This expression has been estimated for the more general case including fibrosis (), so expressions and are used. Nevertheless, it can certainly be computed at cliques on any tissue, fibrotic or non-fibrotic, and its theoretical value when no fibrosis is present can be retrieved from (26) just by making , , , and . Sensitivity of to fibrosis has been analyzed by deriving (26) with respect to the fibrosis-induced parameters. Therefore, deriving with respect to to see how depends on the level of fibrosis, we obtain:
| 27 |
For small delays , , this expression results in , implying that gets reduced if the fibrotic component increases and suggesting the possibility of using as a fibrosis marker, like and . This behavior is also corroborated by computing the derivative of with respect to , taking into account u-EGM amplitude reduction in fibrosis:
| 28 |
This expression results for small delays , confirming that the larger the fibrosis (i.e., the smaller ), the smaller .
Nevertheless, under the same assumptions, the derivative of with respect to results in :
| 29 |
meaning that the larger the reduction of velocity due to fibrosis (i.e., the higher ), the greater , thus showing an opposite effect.
However, as already said before, we expect that fibrosis effects on u-EGM amplitude and morphology, expressed by and , respectively, are much more marked than those on conduction velocity given by [40]. Therefore, we expect the index to be reduced when fibrosis is more severe.
According to this analysis, three different EIGDR-based metrics revealed to be sensitive to the presence of fibrosis and therefore suitable to distinguish between fibrotic and non-fibrotic areas: , , which can be interpreted as measurements of the shape homogeneity of the u-EGMs before and after time alignment, respectively, and the ratio between both, . Maps of , and were computed by processing the complete MEA in the two clique sizes considered in the simulation study. Each map consists of color-coded pixels, representing EIGDR value at each clique. The configuration provides one EIGDR value for each square group of four electrodes with diagonal vertices at (i, j), and , giving a total of pixel maps for each proposed marker. The clique provides one EIGDR value at each squared group of nine electrodes with diagonal vertices at (i, j) and , , resulting in maps of pixels, for , and .
EIGDR with variable electrode-to-tissue distance
When we introduce variable electrode-to-tissue distance, we need to modify the model by replacing s(n) with , being a random variable with mean and variance indexing all the within the clique. Similar analysis as in previous subsections leads to obtain:
| 30 |
which just introduces a multiplying factor, , with respect to the ratios in (11) and (18), equal in both ratios, so preserving the fibrosis stratification value of biomarker. This occurs in contrast to b-EGM peak-to-peak marker, , which is largely modified by the variable electrode-to-tissue distance, but a random way at each electrode, reducing its value as a stratification marker. Analogously:
| 31 |
| 32 |
which approximately result in the same multiplying factor with respect to ratios (15) and (21). Note that for small , and then the approximation of a multiplying factor relating fixed with variable electrode-to-tissue distance holds. This again shows that preserves the fibrotic stratification value in variable electrode-to-tissue distance situations. The same analysis also applies to the ratio .
Also note that in presence of more complex fibrillatory propagation patterns, changes occurring in the u-EGM morphology from electrode to electrode can initially be thought as a planar wave propagating in different directions. This will also result in an extra k-dependent amplitude component into the signal in the model of (8), depending of the angle of the planar wave, and thus also evidence not to largely affect the EIGDR.
EIGDR in real data from PentaRay®
Values of , and were also computed within the four- and five-electrode cliques considered at each mapping point acquired by the PentaRay® catheter. In order to quantify the atrial activity related dispersion, an atrial depolarization window of 100 ms fixed length () was extracted from the last recorded activation at each recording site. Therefore, the proposed EIGDR-based markers were calculated using windowed signals in each clique, aligning them when required as explained in Section 2.5.
Voltage-based fibrosis markers
We also considered bipolar voltage maps based on the peak-to-peak amplitudes and of the b-EGMs in each of the two MEA directions, , , , and , , , as well as on their maximum , , . These peak-to-peak amplitude-based maps were considered and their performance for fibrosis detection were compared against EIGDR maps. Each color-coded pixel bipolar map presents the same resolution as cliques EIGDR maps, providing pixels when processing the whole MEA.
Regarding clinical data, for each mapping point we derived b-EGMs along the PentaRay® catheter branches from filtered u-EGMs. Peak-to-peak amplitudes were computed using atrial depolarization windows extracted from the last recorded activation of b-EGMs at each bipole.
Performance assessment for fibrosis detection
Both EIGDR and bipolar mapping strategies were estimated and tested for each noisy realization , , considered in this study. For each map type, results are reported by aggregating the three different MEA orientations. This aggregated version represents a scenario where the relative angle of the propagation direction with respect to the catheter was not known a priori, thus being more realistic.
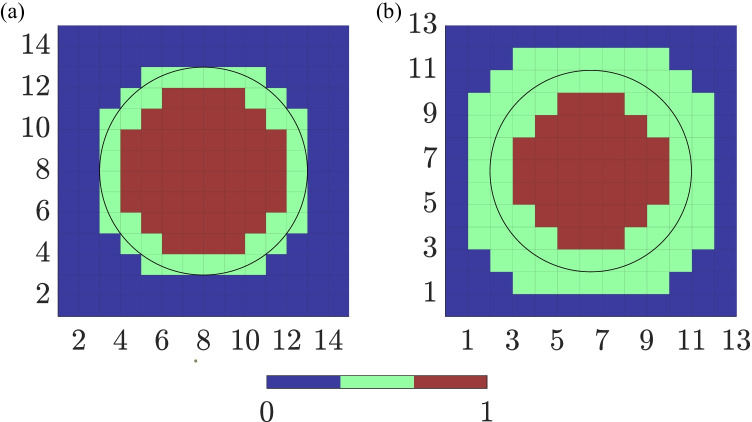
In order to quantitatively evaluate the ability of the different maps as markers for fibrosis detection, i.e. in discriminating pixels associated to the fibrotic patch from those related to non-fibrotic tissue, receiver operating characteristic (ROC) curves were used. Two ground-truth masks were created for that purpose, with the resolution of and maps, by labeling whether a clique (or an electrode pair in case of bipolar maps) lies within the fibrotic or the non-fibrotic area. In a first analysis, the ground-truth mask was created by assigning value 1 if the four electrodes within a clique lie in the fibrotic area, and value 0 if the four electrodes lie in the non-fibrotic area. In a similar way, the ground-truth mask was created by considering if the nine electrodes within a clique fully lie or not in fibrotic/non-fibrotic tissue. Cliques with some electrodes inside and some outside the fibrotic patch were not labeled and therefore discarded in the evaluation. The two ground-truth masks used in this study are shown in Fig. 7(a) and (b), for evaluating 1414 maps (both EIGDR and bipolar) and 1313 maps, respectively. Then, in a further analysis, two binary and ground-truth masks including those mixed cliques with electrodes inside and outside fibrosis region were considered for the evaluation. For that purpose, cliques whose distance between their central point and the center of the fibrotic patch was shorter than the radius of the patch were labeled as fibrotic. On the contrary, when this distance was longer than the radius, corresponding cliques were classified as non-fibrotic. For each EIGDR and bipolar mapping strategy, ROC curves were computed by varying the threshold for fibrosis identification, obtaining sensitivity and specificity in the detection of the fibrotic area [41]. In this context, true positive denotes the number of cliques correctly identified as fibrotic, false negative represents the number of missed cliques in the fibrotic area, true negative stands for the number of cliques correctly identified as non-fibrotic and false positive is the number of cliques incorrectly detected as fibrotic. The maximum detection accuracy (ACC), defined as the highest number of correctly identified cliques (fibrosis or non-fibrosis) divided by the total number of assessed cliques, was used as a measure of the overall fibrosis detection ability of each map. Values of ACC, as well as of the threshold corresponding to ACC, were computed for each map aggregated version considered in this work. Averaged results over the noisy realizations were then computed and evaluated for performance measurements.
Fig. 7.
(a) and (b) ground-truth masks for evaluating fibrosis detection ability of maps performed with 22 and 33 cliques, respectively. Green squares represent the pixels corresponding to cliques with some electrodes inside and some outside the fibrotic patch, i.e. those cliques lying in the border separating the fibrotic patch from non-fibrotic tissue, which were excluded from the evaluation
In the clinical data analysis, at each F and NF mapping point, median values of the three ratios , and were calculated over the six cliques considered. Analogously, the median and maximum values, and , respectively, among the five peak-to-peak bipolar amplitudes computed along the catheter branches and associated with the bipoles closest to the its center ((3,4), (7,8), (11,12), (15,16) and (19,20), according to Fig. 4), were computed at each mapping point in fibrosis and healthy tissue. In order to compare markers between F and NF tissues, median and interquartile range (IQR) over all the EIGDR-based indices and bipolar amplitudes were computed at both F and NF points, separately, as well as the p-values of the right-tailed Wilcoxon rank-sum test referring to the comparison of the metrics between the two areas. Finally, median and IQR of EIGDR indices and bipolar amplitudes were calculated over the six cliques and the five innermost bipoles, respectively, of all mapping points considered, at both F and NF areas, as well as their p-values referring to the global comparison of the metrics between those F and NF areas.
Results
Analysis of simulated data
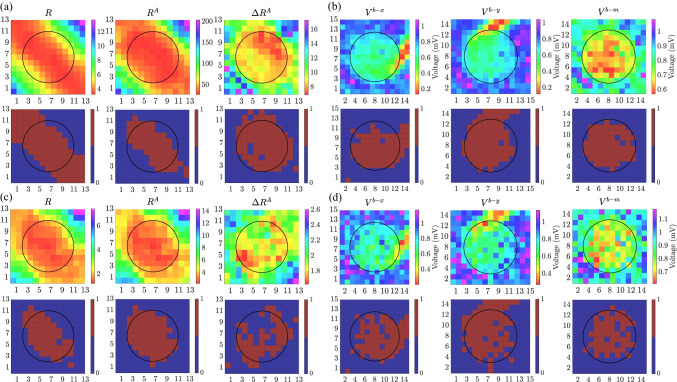
An example of the mapping strategies (computed with 33 cliques) for , variable electrode-to-tissue distance and without noise, is shown for EIGDR and bipolar maps at upper panels in Fig. 8(a) and (b), respectively. When noise is added at level of , results for one of the two thousand noisy realizations are presented at Fig. 8(c) and (d). In the lower panels, the fibrotic areas identified by using the thresholds that maximize the ACC are shown for each mapping strategy. Blue (brown) color inside the circle encompassing fibrotic patch denotes false negative (true positive), while outside denotes true negative (false positive) detection, respectively.
Fig. 8.
Upper panels: maps of , , from 33 cliques and bipolar voltage maps , , , for = , performed assuming a variable electrode-to-tissue distance and noise free ((a) and (b)) and noisy ((c) and (d), with noise level = 46.4 ) u-EGMs. Lower panels: detected fibrotic areas (brown), using the thresholds that maximize detection accuracy of each map
We reported values of , and computed from noisy () u-EGMs in the non-fibrotic clique : , and , as well as in the fibrotic clique : , and , which are consistent with derivations of the models presented in Section 2.7.
Results in this example illustrate that EIGDR maps performed from noise-free time-aligned u-EGMs, and , plotted at central and rightmost columns in Fig. 8(a), respectively, present fibrosis detection performance comparable to bipolar maps obtained as the maximum voltage of both MEA directions, , shown at rightmost column in Fig. 8(b). However, when u-EGMs are affected by noise, EIGDR maps (upper row at Fig. 8(c)) are more robust than bipolar maps (upper row at Fig. 8(d)), being the one showing the best fibrosis discrimination performance.
ACC values of all mapping strategies considered in this study are summarized in Table 3, assuming fixed or variable distance between MEA and tissue, and five different noise levels (reported as standard deviations and average peak-to-peak amplitudes ). For each map, thresholds having these maximum detection accuracy values were also reported in Table 4, where they were given in voltage units in case of b-EGM amplitude-based maps. Both ACC and threshold values were calculated and reported by aggregating the three catheter orientations with respect to the propagation direction. Despite this, bipolar voltage maps exhibit performance strongly dependent on the relative orientation between MEA and propagation direction (e.g., and for and , respectively, for fixed catheter-to-tissue distance and ). For , and are more affected by noise than EIGDR maps, both for fixed and variable electrode-to-tissue distance.
Table 3.
ACC of EIGDR and bipolar amplitude maps, reported jointly for the three MEA orientations and different scenarios, with fixed (FD) or variable (VD) electrode-to-tissue distance, corrupting u-EGMs with noise levels (). ACC values are presented as mean ± standard deviation except for fixed electrode-to-noise distance and () = 0.0
| Map | ACC (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fibrosis | Fixed electrode-to-tissue distance (FD) | Variable electrode-to-tissue distance (VD) | |||||||||
| marker | () | () | |||||||||
| 0.0 (0.0) | 5.8 (24.2) | 11.6 (48.4) | 23.2 (96.7) | 46.4 (193.5) | 0.0 (0.0) | 5.8 (24.2) | 11.6 (48.4) | 23.2 (96.7) | 46.4 (193.5) | ||
| u-EGM EIGDR clique | 74.0 | 74.7±0.7 | 77.0±0.9 | 81.1±1.2 | 83.3±1.6 | 73.2±1.0 | 74.1±1.0 | 76.4±1.1 | 80.9±1.2 | 83.3±1.6 | |
| 86.2 | 85.2±0.9 | 85.7±1.2 | 87.2±1.3 | 86.3±1.6 | 85.0±0.8 | 84.5±1.0 | 85.4±1.2 | 87.1±1.3 | 86.2±1.6 | ||
| 76.8 | 79.8±1.0 | 82.7±1.3 | 84.4±1.5 | 79.4±1.8 | 75.5±1.0 | 78.9±1.2 | 82.0±1.3 | 83.7±1.6 | 78.7±1.9 | ||
| u-EGM EIGDR clique | 78.4 | 78.4±0.2 | 78.5±0.4 | 82.7±1.4 | 87.9±2.1 | 78.1±0.4 | 78.1±0.4 | 78.4±0.6 | 82.6±1.4 | 87.9±2.1 | |
| 92.1 | 91.8±1.2 | 92.2±1.4 | 94.0±1.4 | 94.2±1.6 | 92.3±0.8 | 91.9±1.2 | 92.3±1.4 | 94.0±1.4 | 94.2±1.6 | ||
| 93.1 | 91.8±1.3 | 91.4±1.6 | 91.2±1.8 | 84.0±2.5 | 93.0±0.9 | 91.6±1.4 | 91.2±1.7 | 90.8±1.9 | 83.6±2.6 | ||
| b-EGM amplitude | 68.7 | 68.8±0.1 | 68.8±0.1 | 68.8±0.1 | 68.8±0.2 | 68.7±0.0 | 68.8±0.1 | 68.8±0.2 | 68.8±0.2 | 68.9±0.2 | |
| 90.8 | 90.9±0.3 | 90.9±0.4 | 88.7±0.7 | 82.5±1.0 | 86.8±1.0 | 86.9±1.0 | 87.3±1.0 | 86.5±1.0 | 81.6±1.1 | ||
| 96.2 | 96.2±0.3 | 96.1±0.4 | 93.4±0.7 | 86.9±1.1 | 92.5±1.1 | 92.6±1.1 | 92.8±1.0 | 91.6±1.0 | 86.1±1.2 | ||
Table 4.
Thresholds corresponding to the ACC values reported in Table 3, presented as mean ± standard deviation except for fixed electrode-to-noise distance and () = 0.0
| Map | Threshold | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fibrosis | Fix electrode-to-tissue distance (FD) | Variable electrode-to-tissue distance (VD) | |||||||||
| marker | () | () | |||||||||
| 0.0 (0.0) | 5.8 (24.2) | 11.6 (48.4) | 23.2 (96.7) | 46.4 (193.5) | 0.0 (0.0) | 5.8 (24.2) | 11.6 (48.4) | 23.2 (96.7) | 46.4 (193.5) | ||
| u-EGM EIGDR clique | 7.5 | 7.3±0.1 | 6.7±0.2 | 5.1±0.2 | 2.7±0.1 | 7.8±0.5 | 7.5±0.4 | 6.8±0.2 | 5.1±0.2 | 2.7±0.1 | |
| 86.8 | 69.1±3.3 | 39.3±2.1 | 14.5±0.7 | 4.3±0.2 | 87.3±2.9 | 67.7±3.4 | 38.7±2.1 | 14.4±0.7 | 4.3±0.2 | ||
| 9.1 | 7.5±0.3 | 4.9±0.2 | 2.5±0.1 | 1.5±0.0 | 9.1±0.7 | 7.4±0.3 | 4.9±0.2 | 2.5±0.1 | 1.5±0.0 | ||
| u-EGM EIGDR clique | 4.7 | 4.7±0.1 | 4.2±0.3 | 2.8±0.1 | 1.7±0.1 | 4.7±0.1 | 4.6±0.1 | 4.1±0.4 | 2.8±0.2 | 1.7±0.1 | |
| 36.9 | 32.1±1.0 | 23.1±0.8 | 10.7±0.4 | 3.5±0.1 | 36.1±0.6 | 31.8±1.0 | 22.9±0.8 | 10.7±0.4 | 3.5±0.1 | ||
| 9.8 | 8.6±0.2 | 6.4±0.1 | 3.5±0.1 | 1.9±0.0 | 9.7±0.2 | 8.6±0.2 | 6.4±0.1 | 3.5±0.1 | 1.9±0.0 | ||
| b-EGM amplitude (mV) | 0.01 | 0.02±0.00 | 0.03±0.01 | 0.05±0.02 | 0.11±0.05 | 0.01±0.00 | 0.02±0.01 | 0.04±0.01 | 0.07±0.02 | 0.12±0.05 | |
| 0.76 | 0.76±0.00 | 0.76±0.00 | 0.79±0.01 | 0.84±0.02 | 0.76±0.01 | 0.76±0.01 | 0.77±0.01 | 0.79±0.01 | 0.84±0.02 | ||
| 0.76 | 0.76±0.00 | 0.76±0.00 | 0.79±0.01 | 0.86±0.02 | 0.76±0.01 | 0.76±0.01 | 0.77±0.01 | 0.79±0.01 | 0.86±0.02 | ||
The selection of the thresholds for the EIGDR implies another challenge. This can be addressed by observing that values reached by at healthy tissue (11) can be obtained as the ratio between the estimated energy and N times the estimate of the noise variance , . The value of can be estimated from the data (e.g., by averaging the energy of the u-EGMs in the clique), while the noise variance can be estimated as the u-EGM signal variance in areas electrically silent. The threshold, , can be fixed to a value , where will control the trade-off between required sensitivity and specificity: small will provide high sensitivity and low specificity, and the reverse for large . Note that from results reported in this subsection for a non-fibrotic site such as with noise , we measured , while the optimum threshold reported in Table 4 for this noise level is 3.5, corresponding to a , which can be taken as a reference value. Similar analysis for the same noise level gives values estimates of 1.7 for and 0.39 for , as quantities to subtract to the estimates of and at non-fibrotic areas to derive usable threshold values in real clinical settings. These estimates will require additionally an estimate of the and , see Eqs. (15) and (26), which can be computed, e.g., from the standard deviation of estimated delays in a clique and from the derivative of the aligned and averaged u-EGMs in the clique, respectively.
Bipolar voltage map identifies the fibrotic area with an ACC of 96.2% when distance between MEA and tissue is fixed and u-EGMs are not affected by noise (). Nevertheless, this performance reduces to when the electrode-to-tissue distance is variable, and further when increasing noise level, reaching values 86.9±1.1% and 86.1±1.2% for the highest noise level (), in case of fixed and variable electrode-to-tissue distance, respectively. On the other hand, performed with 33 cliques is more robust to the effect of variable distance than , presenting and for fixed and variable catheter-to-tissue distance, respectively. In addition, it achieves when , for both fixed and variable distance scenarios, being consistent with example in Fig. 8. The same behavior has been observed when studying the three MEA orientations separately. For the highest noise level under test, and with 33 cliques, achieves greater ACC values than . In particular, reaches 95±2%, 95±3%, and 95±3% for , , and , respectively, both with fixed and variable electrode-to-tissue distances, while reaches 88±2%, 89±2%, and 90±2% with fixed distance, and 87±2%, 88±2% and 89 ±2%, with variable distance, for , , and , respectively. If the evaluation is performed without exclusion of cliques which have electrodes inside and outside the fibrotic patch, still provides higher ACC (83.0 ± 1.5%) than (81.2 ± 1.21%), in the largest noise contamination and with variable electrode-to-tissue distance. Moreover, when u-EGMs are not affected by noise, ACC goes from 80.2 % to 80.1 ± 0.6 % for while ACC reduces from 90.8 % to 87.2 ± 1.0 % for , from fixed to variable electrode-to-tissue distance.
Analysis of clinical data
Table 5 shows the median values and IQR of the different biomarkers computed at nineteen mapping points at fibrotic (F) and other nineteen at non-fibrotic (NF) areas. Two of them are depicted in Fig. 6. It can be observed that median values related to and indices evaluated at NF tissue are greater than their counterparts at F points. When u-EGMs are not previously time aligned, shows the following median [IQR] values: 2.45 [0.80] vs. 2.22 [1.47], at NF vs. F points, respectively. The same occurs when considering (7.35 [5.62] vs. 6.18 [4.08]), which revealed to be significantly lower at F than at NF areas (Wilcoxon rank-sum test, p-value<0.05).
Table 5.
Median values of EIGDR indices (,and) computed over the six cliques considered for the PentaRay® catheter, median () and maximum () bipolar amplitude computed over the five innermost electrode pairs along the splines of the catheter, at different mapping points, taken at fibrotic (F) and non-fibrotic (NF) areas, respectively. Median and interquartile range (IQR) were also performed among F and NF points, separately
| # Catheter site | (mV) | (mV) | ||||
|---|---|---|---|---|---|---|
| F | 1 | 3.42 | 4.79 | 1.04 | 0.04 | 0.60 |
| 2 | 6.18 | 6.18 | 1.00 | 0.90 | 4.46 | |
| 3 | 2.45 | 4.75 | 1.95 | 1.65 | 3.61 | |
| 4 | 3.25 | 8.03 | 2.18 | 0.10 | 2.31 | |
| 5 | 1.47 | 5.35 | 3.03 | 1.46 | 1.63 | |
| 6 | 2.33 | 6.46 | 4.22 | 1.35 | 1.42 | |
| 7 | 2.22 | 2.24 | 1.02 | 0.16 | 0.30 | |
| 8 | 2.42 | 9.52 | 4.30 | 0.39 | 1.53 | |
| 9 | 2.18 | 7.24 | 4.17 | 1.31 | 3.06 | |
| 10 | 2.19 | 8.05 | 4.31 | 1.30 | 1.43 | |
| 11 | 3.40 | 9.92 | 3.06 | 0.25 | 0.50 | |
| 12 | 1.77 | 16.2 | 9.83 | 1.91 | 3.52 | |
| 13 | 1.56 | 2.56 | 1.57 | 1.01 | 2.00 | |
| 14 | 2.60 | 13.4 | 3.94 | 0.44 | 0.51 | |
| 15 | 0.91 | 2.27 | 1.83 | 0.84 | 3.73 | |
| 16 | 1.05 | 1.29 | 1.38 | 0.45 | 1.20 | |
| 17 | 1.18 | 4.90 | 4.57 | 1.46 | 1.89 | |
| 18 | 6.07 | 6.31 | 1.00 | 0.06 | 0.09 | |
| 19 | 1.80 | 3.70 | 3.00 | 0.37 | 5.04 | |
| median/IQR | 2.22/1.47 | 6.18/4.08 | 3.00/2.78 | 0.84/1.05 | 1.63/2.66 | |
| NF | 1 | 4.54 | 8.08 | 2.44 | 1.42 | 2.06 |
| 2 | 1.47 | 15.4 | 12.4 | 3.45 | 7.93 | |
| 3 | 2.33 | 6.46 | 2.61 | 1.15 | 1.72 | |
| 4 | 3.63 | 7.33 | 1.81 | 0.22 | 0.80 | |
| 5 | 2.06 | 4.62 | 1.66 | 1.36 | 4.51 | |
| 6 | 2.29 | 7.51 | 3.29 | 0.99 | 2.51 | |
| 7 | 1.51 | 16.5 | 9.97 | 6.90 | 10.2 | |
| 8 | 1.92 | 9.69 | 5.03 | 0.24 | 1.43 | |
| 9 | 1.65 | 6.51 | 3.14 | 0.88 | 1.39 | |
| 10 | 2.69 | 14.1 | 4.90 | 0.46 | 0.84 | |
| 11 | 2.45 | 4.75 | 1.95 | 1.65 | 3.61 | |
| 12 | 2.70 | 5.10 | 1.86 | 0.86 | 2.03 | |
| 13 | 2.95 | 6.29 | 1.75 | 0.14 | 0.39 | |
| 14 | 4.31 | 16.5 | 2.96 | 0.21 | 1.54 | |
| 15 | 2.19 | 7.61 | 3.54 | 0.25 | 1.36 | |
| 16 | 3.17 | 6.72 | 1.76 | 1.10 | 1.78 | |
| 17 | 2.72 | 6.11 | 1.80 | 1.05 | 4.02 | |
| 18 | 2.23 | 12.5 | 4.03 | 0.67 | 2.20 | |
| 19 | 2.61 | 5.89 | 2.22 | 0.27 | 1.20 | |
| median/IQR | 2.45/0.80 | 7.33/5.62 | 2.61/2.09 | 0.88/1.05 | 1.72/2.09 | |
| p-value* | 0.17 | 0.03 | 0.26 | 0.35 | 0.32 |
* refers to the comparison of markers between F and NF areas
On the other hand, when considering each clique or bipolar measurement independently, as reported in Table 6, EIGDR markers based on the alignment of u-EGMs showed to be significantly greater at NF than their counterparts at F areas, assuming the following median [IQR] values: 7.42 [6.74] and 2.67 [3.25] vs. 5.85 [5.62] and 2.17 [3.12] for and at non-fibrotic and fibrotic tissue, respectively. These results are consistent with the theoretical model and overtake fibrosis discrimination performance of (Wilcoxon rank-sum test, p-value<0.05).
Table 6.
Median and interquartile range (IQR) of the EIGDR indices (, and ) computed individually on the six cliques of all catheter sites considered, and of bipolar amplitude values () computed individually on the five innermost electrode pairs of all catheter sites, at fibrotic (F) and non-fibrotic (NF) areas
| (mV) | |||||
|---|---|---|---|---|---|
| F | median/IQR | 2.14/2.13 | 5.85/5.62 | 2.17/3.12 | 0.55/1.26 |
| NF | median/IQR | 2.43/2.34 | 7.42/6.74 | 2.67/3.25 | 0.80/1.24 |
| p-value* | 0.08 | 0.0004 | 0.01 | 0.16 |
* refers to the comparison of markers between F and NF areas
Discussion
Clinical significance of the work
Detection of atrial fibrosis is capital for guiding catheter ablation strategies in AF. The typical intra-procedural assessment of atrial fibrosis by means bipolar voltage thresholding presents well-established limitations related to catheter-wavefront orientation, catheter-tissue contact, electrodes size and inter-electrode spacing, thus limiting its reliability as surrogate of fibrosis. Besides this, it is well-known that when using threshold-based approach, EGM morphology information and time relationship among adjacent electrodes are missing. Despite late gadolinium enhancement-magnetic resonance imaging represents the only non-invasive tool for atrial fibrosis diagnosis, its reproducibility remains under debate [42], as well as its utility in clinical settings [43].
In this work, we proposed u-EGMs eigenvalue dominance ratios (EIGDR) to quantify voltage waveform dispersion and investigated their performance as markers in discriminating fibrotic and non-fibrotic areas, by using a 2D simulated tissue including diffuse fibrosis. The hypothesis behind this approach is that underlying fibrosis in the atrium is reflected not only in the reduction of the waveform amplitude but also in the increased inter-signal dispersion in cliques of nearby electrodes, and that this dispersion will be insensitive to electrode-to-tissue distance as opposite to b-EGM amplitudes.
Performance evaluation of fibrosis markers with simulated data
We analyzed maps computed from noise-free u-EGMs, as well as from u-EGMs corrupted by homogeneous noise levels. As a first step, the distance between each electrode of a square MEA and tissue was assumed to be fixed at 1 mm. Then, in a further analysis, that distance was assumed to be variable following a normal distribution, so as to better approach the real situation where there is no guarantee of maintaining a perfect contact during the mapping.
Our results show that reducing misalignment among u-EGMs within the clique improves fibrosis detection ability of the proposed EIGDR-based index. This is in agreement with other studies where time alignment of b-EGMs has shown to be beneficial for electroanatomical mapping strategies robustness [8]. The index provides comparable fibrosis detection accuracy to the one of maximum bipolar voltage maps when u-EGMs are not affected by noise, and better when high noise levels are present (), for both fixed and variable electrode-to-tissue distances.
Results obtained by considering the three MEA orientations separately reinforce the consideration of as an index worth to be analyzed in extended studies with real recordings for discriminating between fibrotic and normal areas, also pointing out the larger impact of catheter orientation in bipolar amplitudes than in EIGDR metrics.
In addition, if the evaluation is performed without exclusion of cliques which have electrodes inside and outside the fibrotic patch, similar conclusions, with reduced difference ranges, can be drawn. The ratio still shows higher ACC in the largest noise contamination and with variable electrode-to-tissue distance. Moreover, EIGDR-based markers reveal to be more robust to the effect of variable distance than bipolar maps, especially when u-EGMs are not affected by noise.
Regarding threshold values corresponding to the maximum fibrosis detection accuracy, our findings reveal that the thresholds needed to maximize accuracy of bipolar maps and are greater than the one typically used in clinical settings (0.5 ), whereas presents lower voltage threshold. This is explained by the fact that there is no projection of wavefront propagation along the x-axis of the MEA when propagation orientation is .
Performance evaluation of fibrosis markers with clinical data
In the present study, we also tested the ability of the EIGDR-based markers to characterize the fibrotic substrate considering different mapping points acquired with a PentaRay® catheter within fibrotic and non-fibrotic areas over the LA anatomical map, using MRI as reference for that purpose.
The electrode clique organization, referred to a fixed structure catheter like the AdvisorTM HD Grid, was extended to a flexible structure catheter like the PentaRay®, where the inter-electrode spacing within the clique may vary at different acquisition points. Nevertheless, this does not represent a problem for the proposed method, as it is not dependent on electrode orientation.
Preliminary findings obtained from real u-EGMs in this paper reveal that the ratio based on time-aligned u-EGMs is the only EIGDR-based marker between F and NF mapping points, also showing better discrimination power than bipolar amplitudes and typically used in clinical settings. In addition, , together with , proved to be the only indices capable to globally discriminate fibrosis from non-fibrotic tissue, regardless of the mapping points and cliques/bipoles considered at each of them.
Limitations
Several limitations of this study need to be highlighted. First, we simulated a single scenario reproducing a simple propagation pattern in a 2D atrial model, which largely simplifies the real 3D anatomical and electrophysiological situations.
Although a single plane wavefront that propagates in a homogeneous tissue lends itself well to approximating the propagation during pacing, the previous considerations do not allow us to extend quantitatively the results to other propagation patterns, such as circular waves, wave collisions, reentrant wave fronts, among others, and model conditions, including conduction anisotropy or patchy fibrosis. Nevertheless, even if it is well-established that the underlying propagation pattern strongly influences EGMs morphology and their spatiotemporal information, we expect that it does not largely affect the local EIGDR computation. This is because we hypothesize that the correlation between the presence of fibrosis and the morphology dispersion of signals in electrode cliques is well modeled by a waveform assumed to be locally plane and homogeneous, irrespective of the global waveform distribution across the complete tissue. For the same reasons, we considered that the EIGDR approach would not be largely affected by the shape and size of the fibrotic patch. Note that the proposed intra-clique time alignment of the u-EGMs compensates the effect of different u-EGM arrival times on EIGDR. This leaves EIGDR to mostly represent spatial relationships differences among u-EGMs within each clique.
In this work, only the effect of broad-band noise affecting u-EGMs was considered, while specific periodic types of noise were not considered. It must be noted that far-field disturbances due to ventricular depolarization did not occur during atrial activation in sinus rhythm.
Lastly, results presented with real signals represent a proof of concept, but increased sample size need to be considered in order to elucidate whether the use of the EIGDR-based approach is advantageous for fibrosis detection in clinical settings.
Conclusions
In this paper we demonstrated that mapping strategies based on the EIGDR method are able to discriminate fibrotic from non-fibrotic tissue. In simulation, they attain comparable performance to map obtained by combining the b-EGMs amplitudes along the two directions of the MEA for low noise levels, when assuming both fixed and variable distance between the electrode grid and the tissue. Nevertheless, they outperform bipolar maps when higher noise levels are added. Moreover, performance of electrode cliques outperforms the cliques one and fibrosis detection benefits from the previous time alignment of u-EGMs. With clinical data, EIGDR approach showed promising results in discriminating fibrotic and non-fibrotic mapping points, especially when u-EGMs are previously aligned in time. Both scenarios studied lead to choose as EIGDR biomarker for fibrosis discrimination.
Acknowledgements
Computations were performed by ICTS NANBIOSIS (HPC Unit 27 at University of Zaragoza).
Biographies
Jennifer Riccio
received her Bachelor and Master’s Degrees in Biomedical Engineering at University of Naples “Federico II” in 2015 and 2018, respectively. She has been working since September 2018 towards the Ph.D degree in Biomedical Engineering at the Aragon Institute of Engineering Research (I3A) of the University of Zaragoza, Spain, within the Marie Skłodowska-Curie Innovative Training Networks (ITN) “MY-ATRIA” funded by the European Union’s Horizon 2020. Her research interests are focused on intracardiac signals processing for the diagnosis and treatment of atrial arrhythmias. More in detail, the project she is involved aims to characterize atrial propagation patterns and substrate for guiding atrial fibrillation (AF) ablation procedures and to predict ablation efficacy in terminating AF.
Alejandro Alcaine
received his B.Sc. and M.Sc. in Telecommunication Engineering in 2011 and his M.Sc. in Biomedical Engineering in 2012, both from University of Zaragoza (Zaragoza, Spain). In 2016, he got the Ph.D. in Biomedical Engineering under the supervision of Dr. Juan Pablo Martínez and Prof. Pablo Laguna at Aragón Institute of Engineering Research (I3A), University of Zaragoza (Zaragoza, Spain). After that, he was part of the BCN-MedTech Unit at Universitat Pompeu Fabra (Barcelona, Spain) as postdoctoral researcher and lecturer for two years. Spent half year as research engineer in the Arrhythmia Section of Hospital Clìnic (Barcelona, Spain) and one year as postdoctoral researcher at CIBER-BBN (Zaragoza, Spain). Nowadays, he is Associate Professor in Bioinformatics at Universidad San Jorge (Zaragoza, Spain). His research interests involve signal processing techniques applied to biological signals namely focused on its application to time-based detection and characterization of invasive intracardiac (EGM) signals for helping the guidance of ablation procedures for arrhythmia treatment as atrial fibrillation (AF) or ventricular tachycardia (VT) as well as machine learning-related applications to cardiology.
Sara Rocher
received her Bachelor’s Degree in Biomedical Engineering at Universitat Pompeu Fabra (2016) and her Master’s Degree in Biomedical Engineering (intensification in Advanced technologies in design of implants and tissue engineering) at Universitat Politècnica de València (2017). Currently, she is working on her Ph.D. in Technologies for Health and Well-being at the Ci2B (Centro de Investigación e Innovación en Bioingeniería) at Univeristat Politècnica de València. Her thesis is focused on the multiscale modeling of the human atria for the study of atrial fibrillation.
Laura Martinez-Mateu
is Assistant Professor at Universidad Rey Juan Carlos (Madrid, Spain), where she is teaching in the Bachelor’s degree in Biomedical Engineering (English) and in the Bachelor’s degree in Integral Design and Image Management. She is Senior Telecommunications Engineer, and received her M.Sc. in Biomedical Engineering and the Ph.D. in Technologies for Health and Well-being, both from the Polytechnic University of Valencia. Her research line focuses on the field of cardiac electrophysiology, specifically on computational modeling and simulation of arrhythmias and biomedical signal processing. Previously, she was an associate professor in the Department of Electronic Engineering at the Polytechnic University of Valencia, teaching subjects in the Bachelor’s and Master’s degrees in Telecommunication Engineering and in the Master’s degree in Industrial Engineering, as well as a postdoctoral researcher at the Polytechnic University of Valencia and the University of Michigan. She has participated as a researcher in 5 projects funded by regional, national and international public calls, published more than 30 scientific works and directed 8 final degrees’s and master’s projects.
Javier Saiz
became an Electronic Engineer (1986) and received his Ph D. Degree (1992) from the Universidad Politècnica de Valencia (UPV), Spain. He is currently Full Professor in the Electronic Department and in the Center for Research and Innovation in Bioengineering of the UPV. In 1995 he was a Postdoctoral Research Fellow at Department of Biomedical Engineering at Johns Hopkins University, Baltimore, where he was involved in developing mathematical models of action potential propagation in heart tissue. In 2010 he was Academic Visitor at Oxford University. His research interests include cardiac electrophysiology, computer modeling of cardiac electrical activity, biomedical signal processing and biomedical instrumentation. In these areas, he has published more than 370 scientific works and supervised 18 PhD students. Since 2017 is member of Royal Academy of Medicine of Valencia.
Eric Invers-Rubio
obtained his BSc in Biomedical Engineering in Universitat de Barcelona in 2017, and two years later, in 2019, he got his MSc in Biomedical Engineering focused in Radiology and Medical Imaging in Belgium, in Vrije Universiteit Brussel and Universiteit Gent. During his student years, he was able to participate in several research projects involving medical imaging data and radiology departments, such as during his MSc internship in Lima, Peru, and his MSc thesis in Brussels, Belgium. Nowadays, he is a PhD researcher part of the Personalize AF network in Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS) in Barcelona, Spain. His research interests are cardiac medical image and signal processing. As part of his current research, he is working with electrocardiographic imaging (ECGi) and MRI to non-invasively understand the electrophysiological and structural changing mechanisms of atrial fibrillation.
Maria S. Guillem
obtained her Master’s Degree in Biomedical Engineering (Northwestern University, NU, 2006) and Ph.D. in Electronics Engineering (Universitat Politècnica de València, UPV, 2009). She is the Deputy Director of ITACA Research Institute at UPV and head of the Cardiac Oriented Research Laboratory. She was a Fulbright fellow at NU and currently is an Associate Professor at the Electronics Engineering Department at UPV. Dr. Guillem is co-author of 40 international scientific research papers, more than 100 conference papers, 3 patents, and has participated in 40 national/international research projects, 7 of them as PI. Her research interests include the development of medical instrumentation and signal processing for the diagnosis and treatment of cardiac diseases. She participates in the Executive Board of the Consortium for Electrocardiographic imaging and the Board of Directors of Computing in Cardiology.
Juan Pablo Martínez
received the M.Sc. degree in Telecommunication Engineering in 1999, and the PhD degree in Biomedical Engineering in 2005, both at the University of Zaragoza. He is an Associate Professor in the Department of Electronic Engineering and Communications, and the Aragon Institute of Engineering Research (I3A) of the University of Zaragoza, where he is currently the coordinator of the Ph.D. Program in Biomedical Engineering. He is also a member of the Health Research Institute of Aragon (IIS-Aragon) and CIBER-BBN. His main interests are in the development of signal biomarkers for diagnosis, risk stratification and monitoring of cardiovascular diseases and other abnormal heart conditions, both using non-invasive and invasive bioelectrical signals.
Pablo Laguna
is Professor of Signal Processing and Communications in the Department of Electrical Engineering at the Engineering School, and a researcher at the Aragón Institute for Engineering Research (I3A), both at University of Zaragoza, Spain, where he is currently serving as director (2019 - ). He is also member of the Spanish Center for Biomedical Engineering, Biomaterial and Nano-medicine Research CIBER-BBN. His professional research interests are in Signal Processing, in particular applied to Biomedical applications.
Appendix
Table 7.
Table 7.
List of acronyms and symbols
| Acronyms | Symbols | ||
|---|---|---|---|
| A | aligned | transient outward potassium current | |
| ACC | maximum detection accuracy | L-Type calcium current | |
| AF | atrial fibrillation | inward rectifier potassium current | |
| b-EGM | bipolar electrogram | ultrarapid outward potassium current | |
| cAF | persistent atrial fibrillation | slow delayed rectifier potassium current | |
| EAM | electroanatomical mapping | constant time step in the monodomain formulation | |
| EGM | electrogram | spatial resolution in the monodomain formulation | |
| EIGDR | eigenvalue dominance ratio | L | number of electrodes in the MEA |
| F | fibrotic | d | inter-electrode distance in MEA and PentaRay® catheter |
| IIR | image intensity ratio | MEA-to-tissue rotating angle | |
| IQR | interquartile range | (i, j) | spatial coordinates for electrode location in the MEA |
| LA | left atrium | u-EGM model | |
| MEA | multi-electrode array | variable electrode-to-tissue distance in the MEA | |
| MRI | magnetic resonance image | mean of | |
| NA | non-aligned | SD of | |
| NF | non-fibrotic | N | number of u-EGM samples |
| ROC | receiver operating characteristic | SD of noise recording | |
| SD | standard deviation | peak-to-peak amplitude of real noise recording | |
| u-EGM | unipolar electrogram | q | index for noise recording realization |
| noisy u-EGM realization | |||
| K | number of u-EGM signals in a clique | ||
| k-th u-EGM signal in the clique | |||
| samples vector | |||
| u-EGMs samples matrix | |||
| intra-signal sample correlation matrix in the clique | |||
| intra-signal sample correlation matrix estimate in the clique | |||
| eigenvalue of | |||
| dominant-to-remaining eigenvalue ratio | |||
| highest peak-to-peak amplitude u-EGM in the clique | |||
| average u-EGM in the clique | |||
| s(n) | u-EGM activation signal component | ||
| energy of s(n) | |||
| delay of the k-th u-EGM with respect to a time reference within the clique | |||
| variance of ( 1 in non-fibrotic tissue; > 1 in fibrotic tissue) | |||
| u-EGM amplitude factor ( in non-fibrotic tissue; in fibrotic tissue) | |||
| mean of | |||
| variance of | |||
| zero-mean fibrotic signal component in | |||
| variance of | |||
| zero-mean Gaussian and white noise component at the k-th u-EGM | |||
| variance of | |||
| EIGDR with prior alignment and no fibrosis | |||
| first derivative of s(n) | |||
| second derivative of s(n) | |||
| vector counterpart of | |||
| vector counterpart of | |||
| EIGDR with no prior alignment and fibrosis | |||
| inter-signal correlation matrix in the clique | |||
| inter-signal correlation matrix estimate in the clique | |||
| eigenvalue of | |||
| gain in EIGDR by healthy tissue again fibrotic one, in a clique | |||
| gain in EIGDR produced by previous alignment in the clique | |||
| b-EGM along x direction of the MEA | |||
| b-EGM along y direction of the MEA | |||
| peak-to-peak amplitude of | |||
| peak-to-peak amplitude of | |||
| maximum between and | |||
| median value among all the bipolar amplitudes along the PentaRay® catheter splines | |||
| maximum value among all the bipolar amplitudes along the PentaRay® catheter splines | |||
| estimated noisy recording variance | |||
| threshold corresponding to ACC value | |||
| threshold offset from estimated EIGDR to control sensitivity to specificity performance | |||
| estimated from simulations |
Author contribution
JR, JPM and PL designed the study. SR, LM-M and JS performed the electrophysiological simulations. MG and EI-R provided the clinical data. PL and JPM formalized the methodology and supervised the project. JR developed the software and performed the required computations. AA provided the 3D EAM expertise. JR, JPM, PL and AA analyzed the results. All authors critically revised the manuscript and approved the submitted version.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by European Union’s Framework Programme for Research and Innovation Horizon 2020 (2014-2020) under the Marie Skłodowska-Curie Grant Agreements No. 766082 and No. 860974, by projects PID2019-105674RB-I00, PID2019-104881RB-I00 (MICINN) and Aragón Government (Reference Group Biomedical Signal Interpretation and Computational Simulation (BSICoS) T39-20R) cofunded by FEDER 2014-2020 “Building Europe from Aragon”, by fellowship ACIF/2018/174 and Grant PROMETEO/2020/043, both from Dirección General de Política Científica de la Generalitat Valenciana, and by DENIS Project (Volunteer Computer platform) supported through CoMBA 2021-2022 internal projects call from Universidad San Jorge.
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Declarations
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The clinical data were collected from a patient undergoing AF ablation, who also got a late gadolinium enhancement-magnetic resonance imaging acquisition prior to the ablation procedure. The data acquisition protocol was reviewed and approved by the Hospital Clinic Ethical Committee (Ethics approval number: HCB/2019/0881). The patient was informed and signed the consent form.
References
- 1.Platonov P. Atrial fibrosis: an obligatory component of arrhythmia mechanisms in atrial fibrillation? J Geriatr Cardiol. 2017;14(4):233–237. doi: 10.11909/j.issn.1671-5411.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xintarakou A, Tzeis S, Psarras S, Asvestas D, Vardas P (2020) Atrial fibrosis as a dominant factor for the development of atrial fibrillation: facts and gaps. Europace 22(3):342–351. 10.1093/europace/euaa009 [DOI] [PubMed]
- 3.Tzeis S, Asvestas D, Vardas P. Atrial fibrosis: translational considerations for the management of af patients. AER J. 2019;8(1):37–41. doi: 10.15420/aer.2018.79.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burstein B, Nattel S (2008) Atrial fibrosis: mechanisms and clinical relevance in atrial fibrillation. J Am Coll Cardiol 51(8):802–809. 10.1016/j.jacc.2007.09.064 [DOI] [PubMed]
- 5.de Boer RA, et al (2019) Towards better definition, quantification and treatment of fibrosis in heart failure. A scientific roadmap by the committee of translational research of the heart failure association (hfa) of the european society of cardiology. Eur J Heart Fail 21(3):272–285 [DOI] [PMC free article] [PubMed]
- 6.Everett TH, 4th, Olgin JE. Atrial fibrosis and the mechanisms of atrial fibrillation. Heart Rhythm. 2007;4(3 Suppl):S24–S27. doi: 10.1016/j.hrthm.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calkins H, et al (2017) 2017 hrs/ehra/ecas/aphrs/solaece expert consensus statement on catheter and surgical ablation of atrial fibrillation. Heart Rhythm 14(10):e275–e444. 10.1016/j.hrthm.2017.05.012 [DOI] [PMC free article] [PubMed]
- 8.Riccio J, Alcaine A, Rocher S, Martinez-Mateu L, Laranjo S, Saiz J, Laguna P, Martínez JP (2021) Characterization of atrial propagation patterns and fibrotic substrate with a modified omnipolar electrogram strategy in multi-electrode arrays. Front Physiol 12(674223). 10.3389/fphys.2021.674223 [DOI] [PMC free article] [PubMed]
- 9.Rodríguez-Mañero M, et al (2018) Validating left atrial low voltage areas during atrial fibrillation and atrial flutter using multielectrode automated electroanatomic mapping. JACC: Clin Electrophysiol 4(12):1541–1552. 10.1016/j.jacep.2018.08.015 [DOI] [PubMed]
- 10.Knackstedt C, Schauerte P, Kirchhof P (2008) Electro-anatomic mapping systems in arrhythmias. Europace 10(Suppl 3):iii28–iii34 [DOI] [PubMed]
- 11.Yamaguchi T, Fukui A, Node K (2019) Bipolar voltage mapping for the evaluation of atrial substrate: Can we overcome the challenge of directionality? J Atr Fibrillation 11(5):2116. 10.4022/jafib.2116 [DOI] [PMC free article] [PubMed]
- 12.Sim I, Bishop M, O’Neill M, Williams SE (2019) Left atrial voltage mapping: defining and targeting the atrial fibrillation substrate. J Interv Card Electrophysiol 56(3):213–227. 10.1007/s10840-019-00537-8 [DOI] [PMC free article] [PubMed]
- 13.Sánchez J, Luongo G, Nothstein M, Unger LA, Saiz J, Trenor B, Luik A, Dössel O, Loewe A (2021) Using machine learning to characterize atrial fibrotic substrate from intracardiac signals with a hybrid in silico and in vivo dataset. Front Physiol 12(699291). 10.3389/fphys.2021.699291 [DOI] [PMC free article] [PubMed]
- 14.Keller MW, Schuler S, Wilhelms M, Lenis G, Seemann G, Schmitt C, Dössel O, Luik A. Characterization of radiofrequency ablation lesion development based on simulated and measured intracardiac electrograms. IEEE Trans Biomed Eng. 2014;61(9):2467–2478. doi: 10.1109/TBME.2014.2322515. [DOI] [PubMed] [Google Scholar]
- 15.Campos FO, Wiener T, Prassl AJ, dos Santos RW, Sánchez-Quintana D, Ahammer H, Plank G, Hofer E. Electro-anatomical characterization of atrial microfibrosis in a histologically detailed computer model. IEEE Trans Biomed Eng. 2013;60(8):2339–2349. doi: 10.1109/TBME.2013.2256359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maleckar MM, Greenstein JL, Giles WR, Trayanova NA (2009) Electrotonic coupling between human atrial myocytes and fibroblasts alters myocyte excitability and repolarization. Biophys J 97(8):2179–2190. 10.1016/j.bpj.2009.07.054 [DOI] [PMC free article] [PubMed]
- 17.Chelu MG, King JB, Kholmovski EG, Ma J, Gal P, Marashly Q, AlJuaid MA, Kaur G, Silver MA, Johnson KA, Suksaranjit P, Wilson BD, Han FT, Elvan A, Marrouche NF (2018) Atrial fibrosis by late gadolinium enhancement magnetic resonance imaging and catheter ablation of atrial fibrillation: 5-year follow-up data. J Am Heart Assoc 7(23):e006313. 10.1161/JAHA.117.006313 [DOI] [PMC free article] [PubMed]
- 18.Courtemanche M, Ramirez RJ, Nattel S (1998) Ionic mechanisms underlying human atrial action potential properties: insights from a mathematical model. Am J Physiol 275(1):H301-H321. 10.1152/ajpheart.1998.275.1.H301 [DOI] [PubMed]
- 19.Martinez-Mateu L, et al (2018) Factors affecting basket catheter detection of real and phantom rotors in the atria: A computational study. PLoS Comput Biol 14(3):e1006017. 10.1371/journal.pcbi.1006017 [DOI] [PMC free article] [PubMed]
- 20.Tobón C, Villa CAR, Heidenreich E, Romero L, Hornero F, Saiz J (2013) A three-dimensional human atrial model with fiber orientation. Electrograms and arrhythmic activation patterns relationship. PLoS ONE 8(2):e50883. 10.1371/journal.pone.0050883 [DOI] [PMC free article] [PubMed]
- 21.Sánchez J, Gomez JF, Martínez-Mateu L, Romero L, Saiz J, Trenor B (2019) Heterogeneous effects of fibroblast-myocyte coupling in different regions of the human atria under conditions of atrial fibrillation. Front Physiol 10(847). 10.3389/fphys.2019.00847 [DOI] [PMC free article] [PubMed]
- 22.Almeida T, Nothstein M, Li X, Masè M, Ravelli F, Soriano D, Bezerra A, Schlindwein F, Yoneyama T, Dössel O, Ng G, Loewe A (2020) Phase singularities in a cardiac patch model with a non-conductive fibrotic area during atrial fibrillation. In: 2020 Computing in Cardiology (CinC). IEEE. 10.22489/CinC.2020.121
- 23.Heidenreich EA, Ferrero JM, Doblaré M, Rodríguez JF. Adaptive macro finite elements for the numerical solution of monodomain equations in cardiac electrophysiology. Ann Biomed Eng. 2010;38(7):2331–2345. doi: 10.1007/s10439-010-9997-2. [DOI] [PubMed] [Google Scholar]
- 24.Caballero R, de la Fuente MG, Gómez R, Barana A, Amorós I, Dolz-Gaitón P, Osuna L, Almendral J, Delpón FAE. In humans, chronic atrial fibrillation decreases the transient outward current and ultrarapid component of the delayed rectifier current differentially on each atria and increases the slow component of the delayed rectifier current in both. J Am Coll Cardiol. 2010;55(21):2346–54. doi: 10.1016/j.jacc.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Van Wagoner DR, Pond A, Lamorgese M, Rossie S, McCarthy P, Nerbonne J (1999) Atrial L-type Ca2+ currents and human atrial fibrillation. Circ Res 85(5):428–436. 10.1161/01.RES.85.5.428 [DOI] [PubMed]
- 26.Workman AJ, Kane K, Rankin A (2001) The contribution of ionic currents to changes in refractoriness of human atrial myocytes associated with chronic atrial fibrillation. Cardiovasc Res 52(2):226–235. 10.1016/S0008-6363(01)00380-7 [DOI] [PubMed]
- 27.Dobrev D, Graf E, Wettwer E, Himmel HM, Hála O, Doerfel C, Christ T, Schüler S, Ravens U. Molecular basis of downregulation of g-protein-coupled inward rectifying k(+) current (i(k, ach) in chronic human atrial fibrillation: decrease in girk4 mrna correlates with reduced i(k, ach) and muscarinic receptor-mediated shortening of action potentials. Circulation. 2001;104(21):2551–7. doi: 10.1161/hc4601.099466. [DOI] [PubMed] [Google Scholar]
- 28.Voigt N, Trausch A, Knaut M, Matschke K, Varró A, Wagoner DRV, Nattel S, Ravens U, Dobrev D. Left-to-right atrial inward rectifier potassium current gradients in patients with paroxysmal versus chronic atrial fibrillation. Circ Arrhythm Electrophysiol. 2010;3(5):472–80. doi: 10.1161/CIRCEP.110.954636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch RF, Zeng X, Grammer JB, Popovic K, Mewis C, Kühlkamp V (1999) Ionic mechanisms of electrical remodeling in human atrial fibrillation. Cardiovasc Res 44(1):121–131. 10.1016/S0008-6363(99)00178-9 [DOI] [PubMed]
- 30.Martinez-Mateu L, Romero L, Saiz J, Berenfeld O. Far-field contributions in multi-electrodes atrial recordings blur distinction between anatomical and functional reentries and may cause imaginary phase singularities - a computational study. Comput Biol Med. 2019;108:276–287. doi: 10.1016/j.compbiomed.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Unger LA, Oesterlein TG, Loewe A, Dössel O (2019) Noise quantification and noise reduction for unipolar and bipolar electrograms. In: 2019 Computing in Cardiology (CinC). IEEE.
- 32.Benito EM, et al. Left atrial fibrosis quantification by late gadolinium-enhanced magnetic resonance: a new method to standardize the thresholds for reproducibility. Europace. 2017;19(8):1272–1279. doi: 10.1093/europace/euw219. [DOI] [PubMed] [Google Scholar]
- 33.Castells F, Laguna P, Sörnmo L, Bollmann A, Roig JM. Principal Component Analysis in ECG signal processing. EURASIP J Adv Signal Process. 2007;2007(74580):1–21. doi: 10.1155/2007/74580. [DOI] [Google Scholar]
- 34.Woody C (1967) Characterization of an adaptive filter for the analysis of variable latency neuroelectric signals. Med Biol Engng 5:539–554. 10.1007/BF02474247
- 35.Sörnmo L, Laguna P (2005) Bioelectrical Signal Processing in Cardiac and Neurological Applications. Amsterdam: Elsevier (Academic Press)
- 36.Laguna P, et al. Eigenvalue-based time delay estimation of repetitive biomedical signals. Digit Signal Process. 2018;75:107–119. doi: 10.1016/j.dsp.2018.01.007. [DOI] [Google Scholar]
- 37.Riccio J, Rocher S, Martinez-Mateu L, Alcaine A, Saiz J, Martínez JP, Laguna, P (2020) Unipolar electrogram eigenvalue distribution analysis for the identification of atrial fibrosis. In: 2020 Computing in Cardiology (CinC). IEEE. 10.22489/CinC.2020.434 [DOI] [PMC free article] [PubMed]
- 38.Nezlobinsky T, Solovyova O, Panfilov AV (2020) Anisotropic conduction in the myocardium due to fibrosis: the effect of texture on wave propagation. Scientific Reports 10(764). 10.1038/s41598-020-57449-1 [DOI] [PMC free article] [PubMed]
- 39.Palacio LC, Ugarte JP, Saiz J, Tobón C (2021) The effects of fibrotic cell type and its density on atrial fibrillation dynamics: An in silico study. Cells 10(10). 10.3390/cells10102769 [DOI] [PMC free article] [PubMed]
- 40.Vigmond E, Pashaei A, Amraoui S, Cochet H, Hassaguerre M. Percolation as a mechanism to explain atrial fractionated electrograms and reentry in a fibrosis model based on imaging data. Heart Rhythm. 2016;13(7):1536–1543. doi: 10.1016/j.hrthm.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Metz CE (1978) Basic principles of roc analysis. Seminars in Nuclear Medicine 8(4):283–298. 10.1016/S0001-2998(78)80014-2 [DOI] [PubMed]
- 42.Laţcu DG, Bun SS, Arroyo RC, Wedn AM, Benaich FA, Hasni K, Enache B, Saoudi N (2019) Scar identification, quantification, and characterization in complex atrial tachycardia: a path to targeted ablation? Europace 21:i21–i26. 10.1093/europace/euy182 [DOI] [PubMed]
- 43.Caixal G, et al. Accuracy of left atrial fibrosis detection with cardiac magnetic resonance: correlation of late gadolinium enhancement with endocardial voltage and conduction velocity. Europace. 2021;23(3):380–388. doi: 10.1093/europace/euaa313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.