Abstract
Proteolysis is involved in cell differentiation and the progression through the cell cycle in Caulobacter crescentus. We have constitutively expressed the transmembrane chemoreceptor McpA from a multicopy plasmid to demonstrate that McpA degradation is modulated during the cell cycle. The level of McpA protein starts to decrease only when the swarmer cells differentiate into stalked cells. The reduction in McpA protein levels is maintained until the stalked cells develop into predivisional cells, at which point the level returns to that observed in swarmer cells. The cell-cycle-regulated degradation of McpA does not require the last 12 C-terminal amino acids, but it does require three amino acids (AAL) located 15 residues away from the C terminus. The ClpXP protease is essential in C. crescentus for viability, and thus, we tested McpA degradation in xylose conditional mutants. The effect on McpA degradation occurred within two generations from the start of ClpX depletion. The conditional mutants' growth rate was only slightly affected, suggesting that ClpX is directly involved in McpA proteolysis.
Proteolysis is an important facet of programmed cellular processes in both eukaryotes and prokaryotes. In Caulobacter crescentus, proteolysis is involved in a number of important biological functions including chromosomal replication, cell division, the generation of asymmetry, and motility (3, 13, 14, 25, 26, 34). One of the first proteins in C. crescentus to be shown to undergo specific proteolysis was McpA, a receptor for the chemotaxis response. McpA, a cytoplasmic membrane protein, is a member of a large family of receptors (18). The cytoplasmic domain of McpA is highly conserved, whereas its transmembrane domains and periplasmic substrate-binding domain show little similarity. McpA is synthesized only in predivisional cells (1), where it is targeted to the cell pole that will form the flagellated pole of the swarmer cell (2). Since McpA is not synthesized in swarmer cells (1), its degradation guarantees that it does not reappear until the sessile stalked cells develop into predivisional cells. The C terminus of McpA is required for its proteolysis (3), but the highly conserved methylation and signaling domains are not (32). In Escherichia coli, the five amino acids at the C terminus in high-abundance chemoreceptors are required for binding the chemoreceptor methyltransferase (CheR) and the chemoreceptor methylesterase (CheB) (4, 6, 24). The McpA chemoreceptor has very similar amino acids at its C terminus, and since it is also methylated (1), it is conceivable that these residues are involved in binding CheR and CheB, but it is unclear whether they are involved in its proteolysis.
Five ATP-dependent proteases, Lon, ClpXP, ClpAP, ClpYQ, and FtsH, have been identified in the C. crescentus genome (23), but to date, only two have been extensively studied. The Lon protease is required for the degradation of the essential DNA methylase CcrM, but it is not needed for McpA proteolysis (34). The ClpXP protease is required for the degradation of the essential response regulator CtrA, a member of the OmpR family of DNA binding proteins (13). In C. crescentus, unlike in E. coli, the clpX and clpP genes are essential for cell viability (13). The Clp proteases are composed of two subunits, an ATP binding regulatory subunit (ClpA or ClpX) and a proteolytic subunit (ClpP). The regulatory subunit ClpX is responsible for substrate recognition, disassembly, and presentation to ClpP for degradation. Although there are a few exceptions (9, 22), most substrate recognition by ClpX occurs by binding to the disordered C terminus of its substrate (20). Since the extreme C terminus of McpA is required for proteolysis, ClpXP would be a good candidate for the McpA protease. In this study, we have tested the requirement of ClpXP in McpA degradation. Also, we have identified critical C-terminal amino acid residues that are important for the proteolysis of McpA, thus demonstrating that the CheR and CheB docking site is not necessary for degradation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are shown in Table 1. E. coli strains were cultured at 37°C in Terrific broth for liquid medium or Luria-Bertani medium for solid medium supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), or tetracycline (15 μg/ml) as required. C. crescentus strains were grown in PYE medium (0.2% [wt/vol] Bacto Peptone [Difco], 0.1% [wt/vol] yeast extract [Difco], 1 mM MgSO4, 0.5 mM CaCl2) and incubated at 28°C or at room temperature and supplemented with chloramphenicol (1 μg/ml), tetracycline (1 μg/ml), nalidixic acid (20 μg/ml), or kanamycin (5 μg/ml) as necessary and 20 mM xylose (PYEX) or 20 mM glucose (PYEG), where indicated. C. crescentus strains were also grown in minimal M2 medium supplemented with xylose (8). Plasmids were mobilized into C. crescentus from the E. coli strain S17-1 or from strain DH10B using MT607 as a conjugational helper strain.
TABLE 1.
Bacterial strains
| Strain | Genotype | Reference or source |
|---|---|---|
| E coli | ||
| DH10B | F−mcrA Δ(mrr-sdRMS-mcrBC) ΔlacX74 φ80dlacZ ΔM15 deoR recA1 endA1 araΔ139 rpsL | Gibco BRL |
| MT607 | MM294 recA56 pRK2013 aphAIΩTn9 | 28 |
| S17-1 | RP4 2-Tc::Mu Km::Tn7 | 31 |
| C. crescentus | ||
| UJ199 | NA1000 clpPΩaadA xylXΩpUJ174 | 13 |
| UJ200 | NA1000 clpXΩaadA xylXΩpUJ175 | 13 |
| MRKA208 | NA1000 Δbla6 | 32 |
| MRKA580 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 | 32 |
| MRKA800 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP3 | This work |
| MRKA852 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP305 | This work |
| MRKA843 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP307 | This work |
| MRKA801 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP311 | This work |
| MRKA803 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP313 | This work |
| MRKA849 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP382 | This work |
| MRKA841 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP384 | This work |
| MRKA930 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 xylXΩpXCP3ELD | This work |
| MRKA828 | NA1000 Δbla6 Δ(cagAI-cheYIII)che17 pXCP1 | This work |
Plasmid constructions.
Six primers were designed for PCR: BstXI (5′-ATC ATC GGG GTC ATC GAC GAA ATC GCC TTC-3′), CTD5 (5′-GGG GCC TGG GCG AGA ATT CAC GAA CCG CTG-3′), CTD7 (5′-TCC GAC CCG GAC GAA TTC AGG TGT TCA GAC-3′), CTD11 (5′-ATA TCG AAT TCC TTC TAG ACA TCC GAG GCG-3′), CTD13 (5′-CGG GGG CCT TCT AGA GGG CCG CCG AAC CGC-3′), and ID8 (5′-CGG ACC AGC TCC ACG TGC TCG CCC TTC A-3′). The PmlI restriction endonuclease site was included in the ID8 primer; the XbaI site was included in the CTD5, -7, -11, and -13 primers; and the BstXI site was included in the BstXI primer for subcloning the amplified products. These primers were used with plasmid pCHEΔ22 as a template in PCR to amplify products (ID8*, CTD7*, CTD5*, CTD13*, and CTD11*). These PCR products were subcloned into pXCP3 to construct plasmids pXCP38, pXCP307, pXCP305, pXCP313, and pXCP311 (Table 2), respectively. A further two primers, ID2 (5′-TGG CCC GCT TCC ACG TGG GCT CCG GTT CGT-3′) and ID4 (5′-CCC GTC CGG GTC ACG TGA GCG GTT CGG C-3′), were designed to create the internal deletion constructs pXCP382 and pXCP384, respectively. The PmlI site was included in the ID2 and ID4 primers for subcloning the amplified products. These primers were used with plasmid pCHEΔ22 DNA as a template to generate site-directed mutations (ID2* and ID4*) by following the manufacturer's instructions (Promega). These mutant plasmid DNAs (ID2* and ID4*) were then subcloned into pXCP3 to construct plasmids pXCP32 and pXCP34, respectively. The 0.4-kb BstXI-PmlI fragment of plasmid pXCP38 was cloned into plasmids pXCP32 and pXCP34 to generate plasmids pXCP382 and pXCP384 (Table 2), respectively. The latter two plasmids contained the mcpA gene with internal deletions. The following primers were used to mutagenize pXCP3 to generate pXCP3ELD: 5′-CGG AGC AGC GGT TCG GAG CTC GAC GCC CAG GC-3′ and 5′-GCC TGG GCG TCG AGC TCC GAA CCG CTG CTC-3′. All mutants were confirmed by DNA sequencing.
TABLE 2.
Plasmids
| Plasmid | Descriptiona | Reference or source |
|---|---|---|
| pCHE22 | 2.29-kb EcoRI fragment of mcpA was cloned into pBluescript II SK(+) | 3 |
| pCHE22ΔS | 1.7 kb of SacI deletion of pCHE22 | This work |
| pSNX228-1 | Narrow-host-range pUC-based plasmid with novel MCS and M13 ori (pLITMUS28 [NEB]) and 0.6-kb HindIII-XhoI xylX fragment inserted into SnaBI site of pNTP228 | This work |
| pAS21 | Broad-host-range medium-copy vector with xylXp and MCS | A. Stotz and U. Jenal |
| pXCP1 | 2-kb HindIII fragment of pMCP6 was cloned into pAS21 | This work |
| pXCP2 | 2-kb HindIII fragment of pMCP6 was cloned into pSNX228-1 | This work |
| pXCP3 | BglII-EcoRV deletion of pXCP2 | This work |
| pXCP311 | Deletion of mcpA C terminus of pXCP3 | This work |
| pXCP313 | Deletion of mcpA C terminus of pXCP3 | This work |
| pXCP305 | Deletion of mcpA C terminus of pXCP3 | This work |
| pXCP307 | Deletion of mcpA C terminus of pXCP3 | This work |
| pXCP382 | Internal deletion of mcpA of pXCP3 | This work |
| pXCP384 | Internal deletion of mcpA of pXCP3 | This work |
| pXCP3ELD | ELD mutant of pXCP3 | This work |
Abbreviations: MCS, multiple cloning site; NEB, New England Biolabs..
Cell synchronization.
C. crescentus strains were grown at 28°C in PYE medium containing antibiotics, xylose (20 mM), or glucose (20 mM) when required. The strains were synchronized by first concentrating them by centrifugation at 6,000 × g for 5 min at room temperature, the loose pellet was discarded, and the hard pellet was resuspended in 10 ml of 10 mM phosphate buffer, pH 7.0, to which 10 ml of Percoll (Pharmacia) was added. Cells were recentrifuged at 10,000 × g for 20 min at room temperature, and the lower swarmer band was removed. Swarmer cells were washed twice by resuspending them in 10 mM phosphate buffer, pH 7.0, and centrifuging them at 6,000 × g for 5 min at room temperature. Finally, the swarmer cells were resuspended in PYEX and allowed to progress through the cell cycle. Swarmer cells were resuspended in PYE supplemented with xylose or glucose and allowed to proceed through the cell cycle at 28°C.
ClpX and ClpP depletion experiments.
The strains UJ200 and UJ199 were grown first in permissive conditions, PYE containing 20 mM xylose (PYEX), to an A600 of 0.8 to 1.0. The cells were washed twice with PYE and then grown in PYE containing 20 mM glucose (PYEG) for 0, 3, 6, 9, and 12 h. At these times, swarmer cells were isolated and allowed to proceed through the cell cycle in PYEG.
Immunoblotting.
Immunoblotting was carried out as described previously (3). Polyclonal antiserum against McpA was used at a 1:5,000 dilution. Secondary antibody (horseradish peroxidase-conjugated anti-rabbit) (Roche) was diluted 1:3,000, and ECL (Amersham) was used as the detection system.
RESULTS
McpA proteolysis is cell cycle regulated.
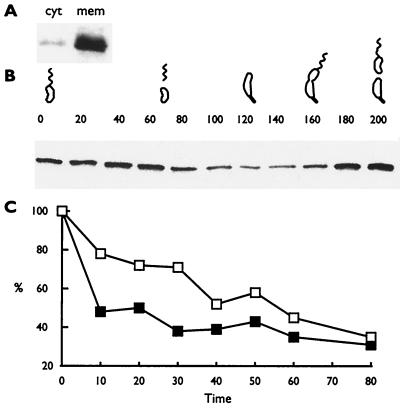
The wild-type mcpA gene is expressed only in predivisional cells, and thus, all the McpA is present prior to cell division (1). McpA is not synthesized in swarmer cells. Therefore, it is necessary to express McpA constitutively to show that the McpA protease activity is modulated during the cell cycle. We constructed the plasmid pXCP1, which has the mcpA gene under the control of the inducible xylose promoter on a multicopy plasmid. When the xylX promoter is induced in the presence of xylose, it is expressed constitutively throughout the cell cycle (21). The plasmid pXCP1 was introduced into strain MRKA580, which lacks the mcpA gene, resulting in the strain MRKA828. The level of McpA protein in MRKA828 is at least 10-fold higher than that in the wild-type strain MRKA208 (data not shown), and as in wild-type C. crescentus, the overexpressed McpA is membrane localized (Fig. 1A). Swarmer cells were isolated from strain MRKA828 and allowed to proceed through the cell cycle in minimal M2 medium containing 10 mM xylose. The level of the constitutively expressed McpA started to drop at 80 min into the cell cycle, when the swarmer cells were differentiating into stalked cells (Fig. 1B). At 160 min, when the cells had differentiated into predivisional cells, the level of McpA started to increase (Fig. 1B). The change in the steady-state levels of protein suggests that the activity of the protease involved in degrading McpA was modulated during the cell cycle. To test this hypothesis, we performed pulse-chase experiments at different times during the cell cycle. When cells were pulse-chased at 40 min into the cell cycle, McpA was degraded at a much higher rate than at the start of the cell cycle (Fig. 1C).
FIG. 1.
The activity of McpA proteolysis is modulated during the cell cycle. (A) Membrane localization of overexpressed McpA. cyt, cytoplasmic fraction; mem, membrane fraction. C. crescentus extracts were separated into cytoplasmic and membrane fractions as described by Shaw et al. (29). (B) Cell cycle immunoblots of the C. crescentus strain MRKA828, which overexpresses McpA constitutively throughout the cell cycle in M2X medium. The numbers above the cell cycle immunoblot represent the time in minutes at which samples were taken, and the C. crescentus drawings denote the progression through the cell cycle. Cell cycle progression was monitored by microscopic analysis. Equal amounts of protein were loaded in each lane. (C) A synchronized culture was labeled with 20 μCi of Tran35S-label (ICN)/ml for 4 min at 0 min (open boxes) or at 40 min (filled boxes) and then chased with 0.2% (wt/vol) Bacto Peptone, 0.1% (wt/vol) Bacto yeast extract, 0.5 mM methionine, and 0.05 mM cysteine. Samples were taken every 10 min except for the last time point. Equal counts were immunoprecipitated with McpA1 antisera. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were visualized by a Molecular Dynamics phosphorimager and quantified with IPLabScan software.
The extreme C terminus is not required for McpA proteolysis.
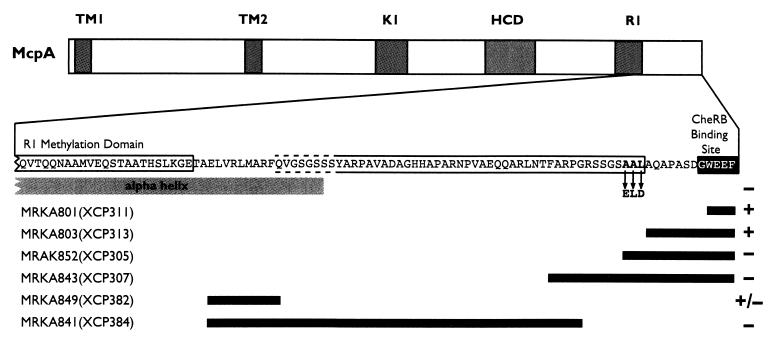
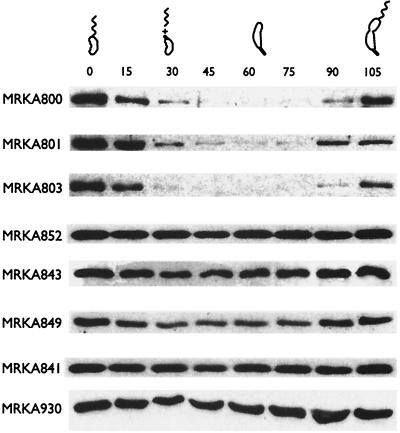
It has been previously shown using M2 epitope tags that the extreme C terminus of McpA is essential for its degradation (3). Because epitopes can perturb the localized structure of a protein, we decided to introduce ochre and amber mutations into mcpA in order to determine the C-terminal extent of the McpA degradation signal. Several C-terminal truncations of McpA were constructed (Fig. 2) and were fused to the xylXp inducible promoter and inserted into the chromosome of MRKA580 at the xylX locus. Deletion of the four C-terminal amino acids (WEEF) from McpA resulted in the strain MRKA801 (Fig. 2). The mutant McpA in MRKA801 was still degraded (Fig. 3), like the full-length McpA in the control strain MRKA800 (Fig. 3). The deletion of the last 12 C-terminal amino acids from McpA resulted in the strain MRKA803 (Fig. 2), which also degraded this McpA deletion during the swarmer-to-stalked-cell differentiation event (Fig. 3). Since it has been reported previously that proteins with nonpolar C termini are more vulnerable to proteolysis (12, 30), we decided to delete the next three-amino-acid sequence which contained three sequential hydrophobic amino acids (AAL), pXCP305 (Fig. 2). The plasmid pXCP305 was inserted into MRKA580 to make MRKA852, and the McpA deletion in this strain was not degraded (Fig. 3), suggesting that these nonpolar amino acids in the C-terminal region are required for McpA proteolysis. A further deletion of 25 C-terminal amino acids from McpA resulted in the strain MRKA843 (Fig. 2), and this McpA deletion was not degraded (Fig. 3).
FIG. 2.
C-terminal truncation and internal deletion constructs of the C terminus of mcpA. The figure shows a diagram of the McpA protein present in the various chromosomal deletions of the mcpA gene in the named C. crescentus strains (Table 1). All McpA protein derivatives are membrane localized (data not shown). TM1 and TM2 denote the transmembrane domains; K1 and R1 denote the methylation domains. HCD denotes the highly conserved domain. The black boxes denote the extent of the deletions in the mcpA gene present in the named C. crescentus strains (Table 1). The amino acids in boldface are the mutated residues in the pXCP3ELD mutant. The black-boxed amino acid residues are the CheRB pentapeptide docking site. The plus and minus signs indicate whether the McpA derivative is degraded. The gray box labeled alpha helix denotes the predicted α-helix derived from the Tsr structure (17). The open box around the amino acid sequence denotes the extent of the requirement for McpA degradation.
FIG. 3.
Cell cycle immunoblots of the mcpA mutants. Cell cycle immunoblotting using McpA antisera was performed as described in Materials and Methods. All the proteins expressed from the mutant mcpA genes were membrane localized like the wild-type protein (data not shown). The strain MRKA800 is the wild-type strain where the full-length mcpA gene is under the control of the xylXp promoter. The other strains (Table 1) have various mcpA mutations under the control of the xylXp promoter (Fig. 2). The numbers above the cell cycle immunoblots represent the times in minutes at which samples were taken, and the C. crescentus drawings denote the progression through the cell cycle in PYEX medium. Cell cycle progression was monitored by microscopic analysis. Equal amounts of protein were loaded in each lane.
The N-terminal extent of the requirement for McpA degradation.
We have already shown that the deletion of the methylation and signaling domains of McpA (Fig. 2) is not required for its degradation (32). Therefore, we constructed internal deletions from the methylation domain (R1) to the C terminus (Fig. 2) in order to determine the N-terminal extent of the requirement for degradation. An internal deletion of the amino acid residues 61 to 70 from the C-terminal end, located immediately downstream of the R1 methylation domain, resulted in the strain MRKA849 (Fig. 2). This McpA deletion was still degraded, as shown by the change in levels of McpA during the cell cycle (Fig. 3). However, the drop in McpA levels was not as obvious as that seen in the wild-type strain MRKA800 (Fig. 3), which suggests that it is degraded at a much lower rate. A further internal deletion including the amino acid residues 19 to 70 from the C terminus resulted in the strain MRKA841 (Fig. 2), and this McpA deletion was not degraded (Fig. 3). These data would suggest that the requirements for degradation are located after the conserved R1 methylation domain.
The C-terminal AAL amino acids are required for degradation.
The comparison of the McpA degradation data from strains MRKA852 and MRKA803 (Fig. 3) would suggest that the amino acids AAL are required for McpA degradation. Thus, these three amino acid residues in McpA were mutated to the amino acids ELD to create pXCP3ELD. The suicide plasmid pXCP3ELD was then conjugated into MRKA580, which created the strain MRKA930. Swarmer cells were isolated from the MRKA930 strain and were allowed to progress through the cell cycle. The immunoblot of cell cycle extracts from strain MRKA930 (Fig. 3) showed that the ELD derivative of McpA was not degraded. This suggests that the amino acid residues AAL at positions 643 to 645 in McpA are required for its degradation.
McpA proteolysis is dependent on ClpX.
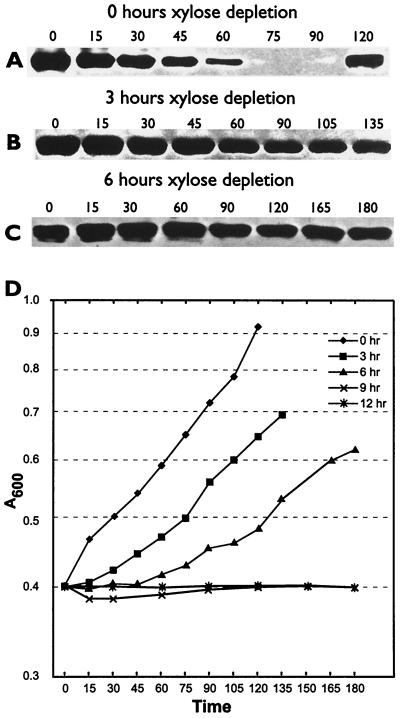
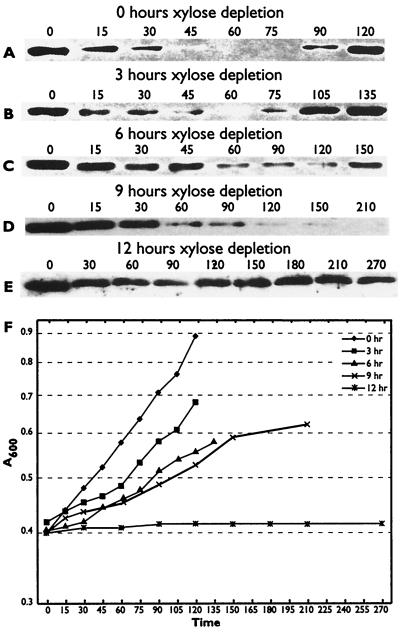
In the search for the McpA protease, we have shown elsewhere that the Lon (34) and ClpYQ (M. R. K. Alley, unpublished data) proteases are not required for McpA proteolysis. Since C-terminal hydrophobic amino acid residues are required for McpA proteolysis and hydrophobic amino acid residues at the extreme C terminus have been shown previously to be required for degradation of proteins via ClpXP (20), we decided to test the requirement for ClpXP in the degradation of McpA. The clpX gene is essential in C. crescentus (13); therefore, we had to use a clpX conditional strain. In the clpX conditional strain UJ200, the expression of ClpX is under the tight control of the xylose promoter xylXp, and so, once xylose is removed from the growth medium and replaced with glucose, the synthesis of ClpX is repressed. Thus, the levels of ClpX in the cell decrease with each subsequent cell division until, after four generations, most of the ClpX protein is removed (13). In order to examine the effects of ClpX depletion on McpA degradation, we set up a series of xylose depletion experiments by growing UJ200 for various times in the absence of xylose. At set times (0, 3, 6, 9, and 12 h), we isolated swarmer cells and performed cell cycle immunoblotting. At time zero, the degradation of McpA proceeded in a cell-cycle-dependent manner (Fig. 4A). The swarmer cells progressed through the cell cycle (Fig. 4D), with cell division taking place at 120 min. After 3 h of xylose depletion, or after approximately two generation times, McpA degradation had been virtually abolished (Fig. 4B). However, the swarmer cells still progressed through the cell cycle, with cell division occurring at 135 min, which is only slightly later than that for time zero (Fig. 4D). This would suggest that the effects on McpA degradation were due to ClpX depletion. After 6 h of xylose depletion, McpA was no longer degraded (Fig. 4C). Although the swarmer cells still progressed through the cell cycle, cell division occurred at 180 min, which was much later than that for 0 and 3 h of xylose depletion. Similarly, after 9 and 12 h of xylose depletion, no degradation of McpA was observed (data not shown), but the cells became filamentous and grew much more slowly (Fig. 4D).
FIG. 4.
ClpX is required for the cell-cycle-controlled proteolysis of McpA. (A to C) McpA cell cycle immunoblots of strain UJ200 after 0, 3, and 6 h of xylose depletion. The swarmer cells from UJ200 were isolated after 0, 3, and 6 h of xylose depletion and then allowed to proceed through the cell cycle at 28°C in PYEG. Cell cycle progression was monitored by microscopic analysis. Equal amounts of protein were loaded in each lane. The numbers above the figure represent the times in minutes at which samples were taken. (D) Growth curves of the UJ200 strain after xylose depletion for 0, 3, 6, 9, and 12 h. Cell division occurred at 120 min for 0 h of depletion, 135 min for 3 h of depletion, and 180 min for 6 h of depletion.
The requirement for ClpP in McpA degradation.
Since ClpX and ClpP form a protease complex, we tested whether ClpP is required for McpA degradation. We used the conditional clpP strain UJ199, because clpP is essential in C. crescentus (13). The expression of the essential clpP gene in UJ199 is under the tight control of the xylose-inducible promoter xylXp. As in the UJ200 strain, growth of UJ199 on glucose medium represses ClpP synthesis. After four generations, most of the ClpP protein in the cell has been removed (13). As described for UJ200, we set up a series of xylose depletion experiments by growing UJ199 in glucose-containing medium for various times to examine the effect on the cell cycle degradation of McpA. At zero time (Fig. 5A) and at 3 h (Fig. 5B) and 6 h (Fig. 5C) of xylose depletion, McpA was still degraded. As shown in Fig. 5F, the effect of xylose depletion on the growth of UJ199 went from cell division occurring at 120 min for time zero, to division occurring at 135 min for 3 h, to division occurring at 150 min for 6 h of xylose depletion. After 9 h of xylose depletion, which is equivalent to four generations, McpA was still degraded (Fig. 5D). After 12 h of xylose depletion, which is equivalent to five generations, McpA degradation was affected (Fig. 5E).
FIG. 5.
The effects of ClpP depletion on McpA degradation. (A to E) McpA cell cycle immunoblots of strain UJ199 after 0, 3, 6, 9, and 12 h of xylose depletion. The swarmer cells from UJ199 were isolated after 0, 3, 6, 9, and 12 h of xylose depletion and then allowed to proceed through the cell cycle at 28°C in PYEG. Cell cycle progression was monitored by microscopic analysis. Equal amounts of protein were loaded in each lane. The numbers above the figure represent the times in minutes at which samples were taken. (F) Growth curves of the UJ199 strain after xylose depletion for 0, 3, 6, 9, and 12 h. Cell division occurred at 120 min for 0 h of depletion, 135 min for 3 h of depletion, and 150 min for 6 h of depletion. In the 9-h xylose depletion experiment, no cell division had occurred by 210 min.
DISCUSSION
A large number of cell-cycle-regulated proteins are specifically degraded during the C. crescentus life cycle (11). These include not only the motility proteins FliF (14), McpA (3), CheYI (11), and CheD (11) but also proteins required for progression through the cell cycle, CtrA (7), FtsZ (16), and CcrM (34). Although a large number of protease substrates have been identified, cognate proteases are known for only two substrates, CcrM and CtrA. The Lon protease degrades CcrM (34), and ClpXP degrades CtrA (13). The CtrA response regulator is degraded at the same time as McpA and like McpA has a C-terminal requirement for its degradation (7). In this study, we have added McpA to the list of C. crescentus proteins that are degraded by ClpXP.
Cell-cycle-regulated ClpXP degradation.
Despite CtrA (7) and McpA degradation being modulated during the cell cycle, ClpX levels are constant (13), which would suggest that both McpA and CtrA are differentially recognized and targeted for degradation during the cell cycle by a yet unidentified regulator(s). It is at present unclear whether McpA and CtrA share a common ClpXP regulator. Both CtrA and McpA have critical C-terminal hydrophobic amino acids required for their degradation. However, CtrA requires its hydrophobic residues to be exposed at its extreme C terminus (7), while McpA is still degraded even when β-lactamase is fused to its C terminus (Alley, unpublished). In E. coli, ClpXP is involved in the degradation of a number of different substrates including, for example, the stationary-phase sigma factor RpoS (27), λO replication protein (33), and SsrA-tagged proteins (10). RpoS degradation requires the response regulator RssB (5), while the N terminus of the λO replication protein plays a critical role in its degradation (9). The ClpXP-mediated SsrA tagging system requires SspB, a ribosome-associated protein, which is regulated by nutrient stress (19). These examples demonstrate that ClpXP-mediated proteolysis can be regulated in various ways in E. coli, which would suggest that there are likely to be alternative ClpXP degradation pathways in C. crescentus. Therefore, showing that CtrA and McpA are degraded by the same protease does not explicitly guarantee that both proteins are regulated identically.
ClpP requirement.
Although the data for ClpP involvement are less strong than those for ClpX, the absence of any evidence for ClpX involvement with any other protease strongly suggests that the McpA protease is ClpXP. The best explanation for the weak effect observed with ClpP depletion is that in the absence of ClpP other proteases might be able to compensate for its loss. For example, in E. coli overproduction of ClpQY can compensate for the absence of Lon, ClpXP proteases, or low levels of FtsH protease in the degradation of the heat shock sigma factor ς32 (15).
The McpA degradation signal.
The crystal structure of the cytoplasmic domain of the E. coli chemoreceptor Tsr (17) suggests that the sequences C terminal to the R1 methylation domain in chemoreceptors form a surface-exposed flexible structure separate from the α-helices. The extreme C terminus contains the CheRB binding site (4, 35), which is exposed to allow CheR and CheB to bind the chemoreceptor. We have shown that the sequences located seven amino acids from this CheRB site (Fig. 2) are important for proteolysis, and therefore, we predict that this region, like the CheRB binding site, will be exposed. Our internal deletion data would suggest an upper limit on the size of the domain required for degradation of approximately 46 amino acids. However, not all these residues might be involved in McpA proteolysis, and most of the residues might be there just to form a linker to allow the critical residues to be exposed.
The identification of the McpA protease and the critical residues in McpA required for its degradation should enable us to dissect the processes involved in cell-cycle-regulated proteolysis.
ACKNOWLEDGMENTS
We thank Susan Jones, Naomi Ferguson, and Isabel Potocka for valuable discussions of the manuscript and Urs Jenal for strains UJ199 and UJ200 and plasmid pAS21.
This study was funded by a Wellcome Trust project grant (044761) to M.R.K.A. M.R.K.A. is a Royal Society University Research Fellow.
REFERENCES
- 1.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alley M R K, Maddock J R, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 3.Alley M R K, Maddock J R, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 4.Barnakov A N, Barnakova L A, Hazelbauer G L. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker G, Klauck E, Hengge-Aronis R. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc Natl Acad Sci USA. 1999;96:6439–6444. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djordjevic S, Stock A M. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- 7.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 8.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 9.Gonciarz-Swiatek M, Wawrzynow A, Um S J, Learn B A, McMacken R, Kelley W L, Georgopoulos C, Sliekers O, Zylicz M. Recognition, targeting, and hydrolysis of the λO replication protein by the ClpP/ClpX protease. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S, Roche E, Zhou Y, Sauer R T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grünenfelder B, Rummel G, Vohradsky J, Roder D, Langen H, Jenal U. Proteomic analysis of the bacterial cell cycle. Proc Natl Acad Sci USA. 2001;98:4681–4686. doi: 10.1073/pnas.071538098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman C, Thévenet D, Bouloc P, Walker G C, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenal U, Shapiro L. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 15.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of ς32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K K, Yokota H, Kim S H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 18.Le Moual H, Koshland D E., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 19.Levchenko I, Seidel M, Sauer R T, Baker T A. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 20.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 21.Meisenzahl A C, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςS subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 23.Nierman W C, Feldblyum T V, Laub M T, Paulsen I T, Nelson K E, Eisen J, Heidelberg J F, Alley M R K, Ohta N, Maddock J R, Potocka I, Nelson W C, Newton A, Stephens C, Phadke N D, Ely B, DeBoy R T, Dodson R J, Durkin A S, Gwinn M L, Haft D H, Kolonay J F, Smit J, Craven M B, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg S L, Venter J C, Shapiro L, Fraser C M. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okumura H, Nishiyama S, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quardokus E, Din N, Brun Y V. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweder T, Lee K H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor ςS by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma S B, Signer E R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990;4:344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- 29.Shaw P, Gomes S L, Sweeney K, Ely B, Shapiro L. Methylation involved in chemotaxis is regulated during Caulobacter differentiation. Proc Natl Acad Sci USA. 1983;80:5261–5265. [PMC free article] [PubMed] [Google Scholar]
- 30.Silber K R, Keiler K C, Sauer R T. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci USA. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 32.Tsai J W, Alley M R K. Proteolysis of the McpA chemoreceptor does not require the Caulobacter major chemotaxis operon. J Bacteriol. 2000;182:504–507. doi: 10.1128/jb.182.2.504-507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 34.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R K. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Li J, Li G, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]