Abstract
Severe acute respiratory syndrome coronavirus (SARS‐CoV), Middle East respiratory syndrome coronavirus (MERS‐CoV), and the current severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) are the most impactful coronaviruses in human history, especially the latter, which brings revolutionary changes to human vaccinology. Due to its high infectivity, the virus spreads rapidly throughout the world and was declared a pandemic in March 2020. A vaccine would normally take more than 10 years to be developed. As such, there is no vaccine available for SARS‐CoV and MERS‐CoV. Currently, 10 vaccines have been approved for emergency use by World Health Organization (WHO) against SARS‐CoV‐2. Virus‐like particle (VLP)s are nanoparticles resembling the native virus but devoid of the viral genome. Due to their self‐adjuvanting properties, VLPs have been explored extensively for vaccine development. However, none of the approved vaccines against SARS‐CoV‐2 was based on VLP and only 4% of the vaccine candidates in clinical trials were based on VLPs. In the current review, we focused on discussing the major advances in the development of VLP‐based vaccine candidates against the SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2, including those in clinical and pre‐clinical studies, to give a comprehensive overview of the VLP‐based vaccines against the coronaviruses.
Keywords: Middle East respiratory syndrome coronavirus (MERS‐CoV), severe acute respiratory syndrome coronavirus (SARS‐CoV), severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), vaccines, virus‐like particle (VLP)
1. INTRODUCTION
Before the emergence of severe acute respiratory syndrome coronavirus (SARS‐CoV), human coronaviruses (HCoV) such as HCoV‐229E, HCoV‐NL63, HCoV‐OC43, and HCoV‐HKU1 were known to cause only mild illness in the upper respiratory tract which contributes to about 15%–30% of common cold. 1 SARS‐CoV was first identified in 2003 as the causative agent of the severe acute respiratory syndrome (SARS) epidemic, which could be traced back to the Guandong province, China in late 2002. 2 Early infection by SARS‐CoV generally results in flu‐like symptoms such as fever, chills, headaches, muscle aches, and diarrhea. SARS‐CoV infection was known to cause high fever (over 38 °C) and pneumonia, which could develop into severe acute respiratory syndrome, leading to respiratory failure and death without the aid of ventilator. SARS‐CoV was reported to infect over 8000 individuals across 29 countries, resulting in the death of 774 humans (9.6% fatality). 2 , 3 This has spurred the interest of scientists all over the world to gain an understanding of immunity to guide vaccine development against the coronaviruses. The epidemic, lasting for a period of 8 months, was declared to have ended by the World Health Organization (WHO) on the July 5, 2003. Control of SARS‐CoV infection was achieved mainly through public health measures and strict isolation of patients.
Middle East respiratory syndrome (MERS), also known as camel flu, is another deadly respiratory disease which emerged following the SARS‐CoV outbreak in 2003. The causative agent of MERS is yet another coronavirus known as MERS‐coronavirus (MERS‐CoV). The virus was first identified in Saudi Arabia in 2012. 4 As of February 2022, MERS‐CoV infections were reported in 27 countries, with 2585 laboratory confirmed cases and 890 associated deaths, which corresponded to ~34% fatality in identified cases. Out of the total reported cases, 2184 occurred in Saudi Arabia, with 812 associated deaths or ~37.2% fatality. 5
Dromedary camels have been recognized as the reservoir host for MERS‐CoV, where the virus was zoonotically transmitted to humans primarily in Saudi Arabia where human–camel interaction is frequent. 6 Most of the reported cases involving human‐to‐human transmissions were nosocomially acquired through close contacts between healthcare workers and the infected patients. 7 , 8 To date, no specific treatment or vaccine is available for MERS‐CoV infections. Despite the high death rates, MERS‐CoV did not receive much attention as it did not appear to be transmitted easily between humans. Although MERS‐CoV outbreaks were mostly endemic in the Arabian Peninsula, an outbreak in South Korea several years back started from a single businessman who had visited the Middle East and was infected by MERS‐CoV. Upon his return to South Korea, a series of human‐to‐human super‐spreading events were triggered, eventually leading to an outbreak involving 186 cases with 38 associated death (~20% fatality). 9 , 10 , 11 The virus transmission to second and third generation contacts had raised immediate concerns regarding multiple mutations in MERS‐CoV, which might have resulted in enhanced human‐to‐human transmissions. 12 , 13
Since late 2019, another novel pandemic‐causing coronavirus has emerged. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) was first isolated from Wuhan, Hubei Province in China. 14 Even though the mortality rate of SARS‐CoV‐2 infection was significantly lower (2% mortality) than that of SARS‐CoV (10% mortality) and MERS‐CoV (20–37% mortality), 5 , 15 , 16 the rate of transmission for SARS‐CoV‐2 was unexpectedly high, with its receptor‐binding domain (RBD) having 10–20‐folds higher binding affinity to human angiotensin converting enzyme 2 (hACE2) receptor compare to the RBD of SARS‐CoV. 17 As of December 28, 2021, SARS‐CoV‐2 has infected over 282 million people worldwide, resulted in deaths of over 5 million. 16 By April 1, 2022, SARS‐CoV‐2 has infected over 488 million people, causing over 6 million deaths. 18 The mortality rate of SARS‐CoV‐2 has reduced significantly, from 1.8% (2019–2021) to 0.5% (2022–present) through the use of vaccines.
A vaccine usually takes 10–15 years to be developed until it is approved for human use. However, due to the severity of this pandemic, development of COVID‐19 vaccine was occurring at warp speed, particularly the clinical phases which have speeded up greatly. To date, 10 vaccines have been approved for use by the World Health Organization (WHO), which are BNT162b2 (Pfizer‐BioNTech), mRNA‐1273 (Moderna), AZD1222 (AstraZeneca), Ad26.COV2.S (Janssen), Covishield (Serum Institute of India), BBIBP‐CorV (Sinopharm), Covaxin (Bharat Biotech), Coronavac (Sinovac), COVOVAX (Serum Institute of India), and Nuvaxovid (Novavax). 19 Moreover, there are currently 153 vaccine candidates under clinical trials and 196 candidates are in pre‐clinical trials. 5 Although the SARS‐CoV‐2 pandemic brought very devastating social and economic impacts on the world, it had also brought revolutionary changes to vaccine development, including the rapid issuance of clinical trial guidelines, expedited regulatory reviews and approval processes. In addition, the combined clinical trials had accelerated the process of bringing novel COVID‐19 vaccines into clinical applications. 20 It is also worth mentioning that this pandemic has allowed the use of the first mRNA‐based vaccine (Comirnaty by Pfizer‐BioNTech) in human history of vaccination.
Of all the approved COVID‐19 vaccines, none are virus‐like particle (VLP)‐based. In the current review, we focus on discussing the major advances in development of VLP‐based vaccine candidates against the SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2, including those in clinical and preclinical studies. Scopus and Google search engine were used to achieve full coverage of journal articles related to the VLP‐based vaccines against these coronaviruses. Additionally, latest advancements of the candidate vaccines for SARS‐CoV‐2 cross‐checked with WHO COVID‐19 vaccine tracker and landscape were discussed. This review aims to give a comprehensive overview of the VLP‐based vaccines against the coronaviruses.
2. MOLECULAR CHARACTERISTICS OF THE β‐CORONAVIRUSES
SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2 are β‐coronaviruses which infect humans. These viruses contain a single‐stranded, positive‐sense RNA genome of 29.7, 30.1, and 29.9 kb for SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2, respectively. All these coronaviruses contain a cap structure at the 5′‐UTR which comprises a leader transcription‐regulating sequence (TRS) and a 3′ poly(A) tail. 21 The whole genome functions directly as mRNA which encodes for the production of open reading frames (ORFs) of ORF1a and ORF1b, which will then be processed into their respective non‐structural protein (NSP)s 1–16 by the viral NSP3 (PL proteinase) and NSP5 (3CL protease). The structural proteins on the other hand, is encoded by subgenomic RNAs (sgRNAs) which are produced during the synthesis of negative‐sense RNA strands, where the viral RNA‐dependent RNA polymerase (RdRp) can pause on each of the TRS present at the 5′ end of the structural proteins, followed by the detachment and relocation of the RdRp to the leader TRS located at the 5′‐UTR, thereby creating a major deletion on the NSP and part of the structural proteins. 22 , 23 This creates a set of mRNAs which will be transcribed into structural proteins, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, as well as the accessory proteins specific to each of the coronavirus. 24 , 25
During virus assembly, the N proteins bind and coat the viral genome, forming the ribonucleoprotein (RNP) complex which then buds into the endoplasmic reticulum‐Golgi intermediate compartment (ERGIC) containing the S, E, and M proteins on the membrane, forming a complete virion which will be released from the host cell through exocytosis. 22 , 23 S protein is the most significant component responsible for viral attachment, fusion, and entry into its host. 8 Therefore, S protein is the main target used in the development of vaccines against coronaviruses. Native S protein is homotrimeric in nature, where each of the subunit is consisted of two domains, known as S1 and S2. S1 contains a N terminal domain (NTD) and a RBD which is responsible for the viral attachment to its host receptors. While the RBD of SARS‐CoV and SARS‐CoV‐2 recognizes human angiotensin‐converting enzyme 2 (ACE2) which is present at high density in the epithelia of the lung and small intestine, MERS‐CoV attaches to its host through dipeptidyl peptidase 4 (DPP4) which is more abundant in the lower respiratory tract. 26 , 27 , 28 S2 on the other hand, mediates membrane fusion to allow virus entry into its hosts. 29
Most of the vaccines including the candidate vaccines in clinical trials use the whole spike (S) protein or at least the RBD of the spike 1 (S1) subunit. RBD, a 223 amino acid (a.a.) protein subunit, plays a direct role in SARS‐CoV‐2 infection as it binds to the human ACE2 receptor which is present in most of the human cell types including the lung and gastrointestinal epithelial cells, which then facilitates the viral entry into the hosts. 27 Therefore, antibodies which target the RBD region of the S1 protein were shown to be potent neutralizing antibodies. 30 , 31
3. CHARACTERISTICS OF VIRUS‐LIKE PARTICLES
Virus‐like particles (VLPs) are non‐infectious nanoparticles formed from one or few of the viral structural proteins which resemble the morphology of the native viruses but are devoid of viral genomic materials. VLPs were first identified in 1968 in the sera of patients with Down syndrome, leukemia, and hepatitis. The first two VLPs were derived from the hepatitis B virus (HBV) and expressed in Escherichia coli and Saccharomyces cerevisiae during the mid‐1980s. The self‐assembly drivers of a VLP are viral envelope or capsid proteins. 32 The VLP is primarily held together by electrostatic and hydrophobic forces, further enhanced by other molecular interactions such as disulfide bonds, forming strong but slightly flexible structures.
In general, there are two types of VLPs, one contains only the viral capsid or nucleocapsid proteins, whereas the other contains viral glycoproteins embedded in the lipid membrane which is usually derived from its hosts. While the non‐enveloped VLPs often contain rigid capsid structures, which are icosahedral or rod shape, the enveloped VLPs are roughly spherical which do not have a fixed shape and are of wide size range. VLPs can be further divided into different groups based on their structural complexity. Capsid proteins can be arranged up to three layers. Single‐protein VLPs have a relatively simple structure, while multi‐protein VLPs contain unique structural components such as the presence of several distinct capsid layers. 33 Other VLPs such as those derived from human immunodeficiency virus (HIV)‐1 and influenza virus have a lipid envelope that comprises viral surface antigens encircling the capsid structure.
As there are often hundreds of copies of the capsids or glycoproteins forming each VLP, desired epitopes can be fused to the VLPs for high density display which could help to induce stronger immune responses. For example, hepatitis B virus core antigen (HBcAg) VLP is formed from either 180 (T = 3) or 240 (T = 4) of the HBcAg capsid protein subunits and is capable of displaying up to 240 copies of foreign epitopes per VLP. 34 , 35 , 36
3.1. VLP as platform for vaccine development
To date, many platforms have been established in attempt to obtain vaccines with high efficiency and safety profile. Inactivated vaccines are produced via chemical or heat stress treatment. 37 The safety level of inactivated vaccines is higher compared to attenuated vaccines. However, their immunogenicity is generally reduced due to the inactivation treatment. Protein subunit vaccines on other hand are relatively safe and capable of inducing immune memory responses. 38 However, adjuvant is almost compulsory. While viral vector vaccines deliver antigen‐coding genes to target cells, 39 repeated use of the same viral component will result in reduced efficiency due to rapid clearance of the viral vector by host immune system developed during the previous administration. Additionally, safety concern toward viral vector vaccine especially the replicating one is debatable. Meanwhile, mRNA vaccines and DNA vaccines deliver genetic materials to the target cells to generate recombinant viral antigens which induce specific immune responses. 40 Although mRNA vaccines are safe, and the production is relatively fast and inexpensive, cold chain systems is required due to the instability of RNA. Although DNA vaccines are stable, it is generally less effective. 41 , 42 As VLP is robust and self‐adjuvanting, VLP‐based vaccines could easily become one of the most effective vaccine against coronaviruses.
VLPs have been used as vaccines against hepatitis B virus (Engerix® and Recombivax HB®) and human papillomavirus (Cervarix™ and Gardasil®). 43 In addition, VLP‐based vaccines such as hepatitis E virus (HEV) Hecolin and HBV ABX203 (HeberNasvac®) have also been licensed for use in humans for prevention and treatment of hepatitis E and chronic hepatitis B, respectively. 44 The VLP resembles a pathogen and contains conserved pathogen‐associated molecular patterns (PAMPs) that can be readily recognized by the host immune system. Genetic engineering is used to produce VLPs by cloning the viral structural genes and they are expressed in prokaryotic or eukaryotic expression systems. 33 The VLPs are produced to display the antigenic determinants of target pathogens on their surface. The goal of VLP‐based vaccines is to raise immune responses against the displayed antigens which can either be peptides or whole antigens. VLPs may induce strong innate and adaptive immune responses against the conjugated antigens. 33 VLPs can also be loaded with immune modulators, such as innate immune system stimuli, to provoke more effective immune responses. Hence, they are ideal carriers for antigen delivery and safer than whole pathogen‐based vaccines such as the live‐attenuated viral vaccines.
The construction of a recombinant VLP‐based vaccine takes advantage of the knowledge of the nucleotide sequences of viral structural proteins with self‐assembling ability to display the antigenic determinants of pathogens. 45 , 46 , 47 , 48 , 49 , 50 The expression systems for individual heterologous proteins have been developed based on the prokaryotic cells, yeast, insect cells, plants, and mammalian cells, which allow the production of VLPs carrying the target antigens both on laboratory and industrial scales. 38 , 51 , 52 , 53 , 54 , 55 High yield production of VLPs with upscaling potential have been established and can support the use of these technologies for pandemic settings. 51 , 52 , 53 , 56
Different expression systems have their own advantages and disadvantages. For instance, bacterial expression system such as E. coli is particularly useful in producing non‐enveloped VLPs at high yield within the shortest period. However, they are not suitable for producing enveloped VLPs, prone to misfolding, and are incapable of performing post‐translational modification (PTM), which is important for many eukaryotic‐based antigens such as the S protein of SARS‐CoV‐2. Mammalian cell expression systems on other hand can perform all PTMs, with the downside of being costly and time‐consuming. Therefore, strategies which combine the two expression systems have been explored, where the SARS‐CoV‐2 antigens were produced in mammalian cells, which were then conjugated to heterologous VLPs produced in E. coli. 57 , 58 However, not all eukaryotic‐based immunogenic epitopes require PTMs. For example, the SARS‐CoV HRC1 B‐cell epitope and SARS‐CoV‐2 receptor binding motif (RBM) displayed on VLPs were produced in E. coli and still induced virus neutralizing antibodies when used to immunize mice and rabbits. 53 , 59 , 60
As in mammalian cell, yeast, insect, and plant expression systems can also be used to produce both the enveloped and non‐enveloped VLPs, with the capability to perform PTMs. However, these expression systems often glycosylate proteins differently compared to human and mammalian cells. For example, yeasts tend to perform high‐mannose glycosylation, whereas insect cell glycosylation contains α1,3‐linked fucose residues while lacks terminal sialic acid. 61 , 62 Regardless, VLP‐based vaccine candidates produced in yeasts, plant and insect cells have made their way to clinical trials, 63 , 64 , 65 , 66 suggesting that the types of glycosylation may not be detrimental to the immunogenicity of SARS‐CoV‐2 epitopes.
Thus far, 110 viral proteins from 35 viral families were capable of assembly into VLPs. 38 , 67 Several VLP‐based vaccines are currently undergoing different phases of production and approval. Among the advantages of VLP vaccines are their high specificity, efficiency, and good pharmacokinetic characteristics. A previous study has shown that VLPs could reach the lymphatic nodes in less than 10 min when compared to other particle mixtures bearing the antigens. 68 Undoubtedly, VLP‐based vaccines offer new possibilities in the development of immunoprophylaxis strategies, including injection‐free formulations. The injection‐free vaccine can be administered via intranasal or oral routes. This is particularly important for the large‐scale animal breeding industry, as it is extremely laborious to perform injections of large numbers of animals. 33 , 69
3.2. VLP as immunogen
Viral capsids are made up of repetitive protein structures that can trigger innate immunity and lead to the production of neutralizing antibodies by B cells. 38 Dendritic cells (DCs) are an important component of antigen‐presenting cells (APCs) which link the innate and adaptive immunity. 70 DCs can take up particles sized from 100–500 nm such as VLPs through macropinocytosis and phagocytosis. DCs interact with VLPs through the same pattern recognition receptors (PRRs) such as toll‐like receptors (TLRs) and C‐type lectin receptors (CLRs). 70 Upon administration, VLP‐based vaccines are recognized by APCs such as DCs and transported to secondary lymphoid tissues such as the spleen. 71 The recognition and uptake of VLPs by DCs promote the maturation of DCs (Figure 1). Such events lead to the production of pro‐inflammatory factors such as TNF‐α and IL‐1β to recruit more APCs and increase lysosomal proteolytic activity in the DCs. 72 Subsequently, VLP‐based vaccines are processed into small peptides and presented as MHC–peptide complex on the surface of DCs. Simultaneously, B and T cells are activated by lymphocyte costimulatory molecules such as CD40, CD80, and CD86 present on the DC surface. The MHC class II‐peptide and costimulatory proteins activate CD4+ T‐helper cells to promote the proliferation and differentiation of both B and T cells. 73
FIGURE 1.
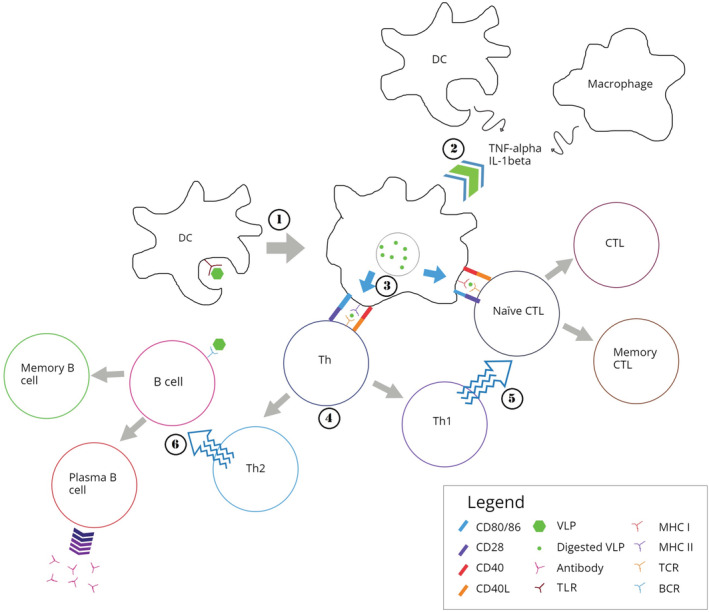
Immune responses induced by virus‐like particles (VLPs). 1: VLP interacts with pattern recognition receptors (PRRs) presence on the dendritic cell (DC), such as the toll‐like receptor (TLR). DC then engulf the VLP through phagocytosis or micropinocytosis. 2: This leads to the maturation of DC, which then secretes pro‐inflammatory cytokines such as TNF‐α and IL‐1β to recruit more antigen presenting cells (APCs), including DCs and macrophages. 3: VLP taken up by the DC are then enzymatically digested into short peptides, which binds to major histocompatibility complex class I (MHC I) and class II (MHC II) and are transported onto surface of the DC. 4: Short peptide displayed on MHC II, together with CD40 and CD80/86 then interact with T cell receptor (TCR), CD40L, and CD28 presence on naïve helper T cell (Th), respectively. This promotes the proliferation and differentiation of Th cells into type 1 (Th1) and type 2 (Th2) Th cells. 5: Similarly, peptides displayed on MHC I of the DC, along with CD40 and CD80/86 interact with TCR, CD40L, and CD28 presence on the naïve cytotoxic T lymphocyte (CTL). Aided by Th1, naïve CTL proliferates and differentiates into effector and memory CTLs, providing immediate and long‐lasting cellular immunity, respectively. 7: Naïve B cell on other hand interacts with intact VLP carried over by blood stream or DC via B cell receptor (BCR). Aided by Th2, the B cell differentiates into plasma B cells which actively secrete antibodies, and memory B cells which provides long‐lasting humoral immunity against the antigen presence on the VLP
VLP‐based vaccines are excellent candidates that are responsible for the stimulation of both cellular and humoral immunity. For instance, two doses of HBcAg‐zDIII (Zika virus envelope protein domain III) VLP vaccine could trigger both humoral and cellular immunity. 50 Another study also revealed that porcine parvovirus (PPV) VLP‐based vaccine was able to activate MHC I and II pathways which stimulate humoral and cellular immunity. 74 In addition, there are several reports of VLP‐based vaccines that could stimulate antibody production and cytotoxic T cell activation independent of T‐helper cells. 75 VLP‐based vaccines were reported to induce humoral and cellular immunity even without adjuvant, although adjuvants could significantly increase their immunogenicity. 73
4. DEVELOPMENT OF VLP VACCINES AGAINST CORONAVIRUSES
Scientists from all around the world have been working relentlessly to develop vaccines against SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2, due to the severity of these infectious diseases. Various VLP‐based vaccine candidates have been reported (Table 1). Apart from SARS‐CoV‐2, most of the reported studies are at the preliminary stage of development.
TABLE 1.
Preliminary studies on VLP vaccine candidates targeting the SARS‐CoV. MERS‐CoV, and SARS‐CoV‐2
| Features | Target viruses | Expression systems | References |
|---|---|---|---|
| Peptide‐based nanoparticle displaying SARS‐CoV HRC1 B‐cell epitope induced neutralizing antibodies which inhibited virus infection in vitro | SARS‐CoV | E. coli | Pimentel et al. 59 |
| Micellular nanoparticles based on SARS‐CoV S protein adjuvanted with matrix M1 induced neutralizing antibodies against SARS‐CoV. | SARS‐CoV | BEVS | Coleman et al. 76 |
| VLP formed from chimeric SARS‐CoV S protein carrying IAV HA, coexpressed with IAV M1 protein, protected mice from lethal SARS‐CoV challenge. | SARS‐CoV | BEVS | Liu et al. 77 |
| Chimeric VLP formed from mouse hepatitis virus (MHV) E, M, N proteins and SARS‐CoV S protein protected mice from SARS‐CoV challenge but resulted in pulmonary immunopathology. | SARS‐CoV | Mammalian cell | Lokugamage et al. 78 ; Tseng et al. 79 |
| VLP based on SARS‐CoV N protein induced high level of cytotoxic T cell responses when coadministered with plasmids encoding SARS‐CoV N protein and XIAP. | SARS‐CoV | Mammalian cell | Azizi et al. 80 |
| Micellular nanoparticles based on MERS‐CoV S protein adjuvanted with matrix M1 induced neutralizing antibodies against MERS‐CoV in a dose‐dependent manner. | MERS‐CoV | BEVS | Coleman et al. 76 ; Coleman et al. 81 |
| VLP formed from chimeric MERS‐CoV S protein carrying IAV HA, coexpressed with IAV M1 protein, induced antibodies capable of neutralizing pseudovirus of MERS‐CoV when adjuvanted with alum/CpG. | MERS‐CoV | BEVS | Lan et al. 82 |
| VLP formed from MERS‐CoV S, E, and M proteins adjuvanted with alum induced virus‐neutralizing antibodies and Th1‐mediated immune responses in rhesus macaques. | MERS‐CoV | BEVS | Wang et al. 83 |
| MERS‐CoV RBD displayed on parvovirus VP2 VLP induced pseudovirus neutralizing antibodies. When adjuvanted with poly(I:C), the VLP induced both Th1 and Th2 cell‐mediated immune responses. | MERS‐CoV | BEVS | Wang et al. 84 |
| MERS‐CoV RBD fused to ferritin‐based nanoparticle induced antibodies which inhibit the interaction between MERS RBD and hDPP4 receptor in a competitive ELISA. | MERS‐CoV | E. coli | Kim et al. 85 |
| Transmembrane region‐truncated MERS‐CoV S protein (SΔTM) produced in silkworm larvae assembled into nanoparticle and was able to bind to hDPP4. MERS‐CoV VLP were prepare by surfactant treatment and mechanical extrusion from Bm5 cell coexpressing MERS‐CoV S, E, and M proteins. | MERS‐CoV | Silkworm larvae, silk moth cell line | Kato et al. 86 |
| Mice primed with recombinant adenovirus serotype 5 encoding MERS‐CoV S protein, followed by boosters with MERS‐CoV S protein‐based VLP induced neutralizing antibodies, Th1, and Th2 immune responses, protected mice against virus challenge. | MERS‐CoV | Mammalian cell, BEVS | Jung et al. 87 |
| Transchromosomic bovine immunized with inactivated virus or MERS‐CoV S‐based micellular VLP produced fully human polyclonal IgG capable of reducing viral load in mouse model to near or below limit of detection when administarted before or after virus infection. | MERS‐CoV | Mammalian cells, transchromosomic bovine | Luke et al. 88 |
| MERS‐CoV RBD chemically cross‐linked to PLGA nanoparticle encapsulating the cyclic diguanylate monophosphate protected mice against lethal MERS‐CoV challenge. | MERS‐CoV | BEVS | Lin et al. 89 |
| Single dose of VSV replicon vaccine carrying SARS‐CoV‐2 RBD fused to glycoprotein of RABV protected mice from SARS‐CoV‐2 challenge. | SARS‐CoV‐2 | Mammalian cells | Hennrich et al. 90 |
| SARS‐CoV‐2 RBD (mammalian expressed) conjugated to SpyCatcher003‐mi3 VLP (E. coli expressed) via SpyTag/SpyCatcher technology induced neutralizing antibodies in mice and pigs. | SARS‐CoV‐2 | Mammalian cell, E. coli | Tan et al. 57 |
|
SARS‐CoV‐2 RBM displayed on bacteriophage AP205 VLP induced SARS‐CoV‐2 neutralizing antibodies. |
SARS‐CoV‐2 | E. coli | Liu et al. 60 |
| SARS‐CoV‐2 RBM fused to immunologically optimized cucumber mosaic virus VLP (CuMVTT) induced neutralizing antibodies in rabbits and mice. | SARS‐CoV‐2 | E. coli | Mohsen et al. 53 |
| SARS‐CoV‐2 RBD produced in mammalian cell chemically cross‐linked to CuMVTT produced in E. coli induced neutralizing antibodies in mice. | SARS‐CoV‐2 | Mammalian cell, E. coli | Zha et al. 58 |
| Prefusion‐stabilized SARS‐CoV‐2 S protein ectodomain (S2P) displayed on Newcastle disease VLP induced higher neutralizing antibodies than soluble S2P in mice. | SARS‐CoV‐2 | Mammalian cell | Yang et al. 91 |
| VLPs formed from co‐expressing influenza M1 protein with SARS‐CoV‐2 S or S1 induced neutralizing antibodies which partially inhibited binding of SARS‐CoV‐2 RBD to hACE2. | SARS‐CoV‐2 | BEVS | Chu et al. 92 |
|
SARS‐CoV‐2 S, E, and M co‐expressed in HEK‐293 cells assembled into VLP mimicking the actual virus. |
SARS‐CoV‐2 | Mammalian cell | Swann et al. 93 |
| SARS‐CoV‐2 S, E, M, and N co‐expressed in HEK‐293 T and Vero E6 cells assembled into VLPs mimicking the actual virus. | SARS‐CoV‐2 | Mammalian cells | Xu et al. 94 |
| SARS‐CoV‐2 S, E, and M co‐expressed in Saccharomyces cerevisiae platform (D‐Crypt™) self‐assembled into VLP mimicking the actual virus. | SARS‐CoV‐2 | Yeast | Arora et al. 95 |
Abbreviations: E, envelope; HA, haemagglutinin; hACE2, human angiotensin converting enzyme 2; IAV, influenza A virus; M, membrane; M1, matrix 1 protein; N, nucleocapsid; PLGA, poly(lactic‐co‐glycolic acid); RBD, receptor‐binding domain; RBM, receptor‐binding motif; S, spike; VLP, virus‐like particle.
4.1. VLP vaccines against SARS‐CoV
Although there was no upbreak being reported since July 2003, there are published data regarding VLP‐based candidate vaccines for SARS‐CoV. A VLP that displayed a recombinant SARS‐CoV B‐cell epitope from the C‐terminal heptad repeat (HRC1) of the S protein was developed Pimentel et al. 59 When 10 μg of the peptide nanoparticle without adjuvant was injected intraperitoneally into the mice, positive humoral response was observed. 59 The baculovirus–insect cell expression system or baculovirus expression vector system (BEVS) was widely used to produce the SARS‐CoV VLP vaccine containing the S protein. 76 The vaccine (1 μg of SARS‐CoV S) containing matrix M1 adjuvant was able to induce neutralizing antibody responses in BALB/c mice. In another study, a recombinant SARS‐CoV VLP chimeric vaccine containing the SARS‐CoV S protein and the influenza M1 protein was produced using BEVS. 77 The SARS‐CoV VLP vaccine (0.8 μg of S protein) in the absence of an adjuvant when administered intramuscularly or intranasally was able to protect mice from lethal challenge of SARS‐CoV. A dosage of 4 μg of S protein was observed to reduce virus titers in lungs to below detectable level, protected mice from weight loss, and elicited high levels of neutralizing antibodies against SARS‐CoV. 77
Besides, mammalian cells have also been chosen for the production of VLPs. Lokugamage and colleagues developed VLPs propagated in mammalian cells as SARS‐CoV vaccine candidates. 78 The chimeric VLPs produced from mammalian cells in the study were more immunogenic than SARS‐CoV VLPs produced with BEVS. The chimeric VLPs were produced by the co‐expression of SARS‐CoV S, E, M, and N proteins of the murine coronavirus (mouse hepatitis virus) in 293 T cells. When female BALB/c mice were immunized with the chimeric VLPs at 1 μg, SARS‐CoV replications within the lungs were suppressed. 78 The vaccination of another chimeric SARS‐CoV VLP (composed of S, E, M, and N) in BALB/c and C57BL/6 mice induced serum neutralizing antibody in a dosage‐dependent manner, which were further boosted with the use of alum adjuvant. 79 The notable eosinophils observed in the histology of the SARS‐CoV infected lung treated with VLPs indicated activation of both the innate and adaptive immune responses. 96
The SARS‐NC gene encodes the N protein (46 kDa) that is responsible for virus replication and cell cycle disruption in the host. Such a protein is highly immunogenic and has been targeted for the design of an effective vaccine. 80 Transmission electron microscopy revealed that the N proteins expressed in CHO cells self‐assembled into VLPs. A high level of specific SARS‐CD8+ T‐cell response and antibody titers were demonstrated in mice immunized with the N protein, which was further enhanced through co‐administration of recombinant plasmids encoding the N protein, recombinant plasmids encoding XIAP, and montanide/CpG. 80
4.2. VLP vaccines against MERS‐CoV
Numerous MERS‐CoV VLP‐based vaccines have been developed and evaluated in animals. BEVS was used to produce MERS‐CoV VLP vaccine containing the S protein. 76 The vaccine formulated with Matrix‐M™ adjuvant (saponin‐based) was able to induce neutralizing antibody responses in BALB/c mice. Subsequent study using the same vaccine revealed that 10 μg of MERS‐CoV S nanoparticles were able to produce significantly higher titers of anti‐MERS‐CoV S antibodies in vaccinated mice when compared to lower dosages (1 and 3 μg). 81 Similarly, a chimeric VLP containing modified MERS‐CoV S protein (fused to HA531–568 of H5N1) and the avian influenza matrix 1 protein (H5N1) was developed using BEVS. 82 Significant S‐specific IgGs and neutralizing antibodies were detected in BALB/c mice which were intramuscularly injected with 1 μg of chimeric VLPs of MERS‐S adjuvanted with 100 μl of alum and 10 μg of CpG. 82 A MERS‐CoV VLP vaccine made up of S, E, and M proteins was shown to stimulate IgG production and virus‐neutralizing antibodies with titers up to 1:40 in rhesus macaques. 83 The same research group evaluated a new chimeric VLP vaccine consisted of RBD of MERS‐CoV S protein and the canine parvovirus VP2 structural protein, where the chimeric VLP induced RBD‐specific humoral and cellular immune responses in mice. 84
Apart from BEVS, Kim and colleagues presented a novel bacterial VLP of MERS‐CoV antigen using ferritin as a molecular scaffold for self‐assembly. 85 The MERS‐CoV RBD fused with the RNA‐interaction domain (of human lysyl‐tRNA synthetase) and bacterioferritin was expressed in Escherichia coli. The sera of mice immunized with the nanoparticle was able to inhibit the interaction between MERS RBD and hDPP4 receptor in a competitive ELISA. More recently, Kato et al. 86 constructed VLPs which contained MERS‐CoV S, E, and M proteins produced in silkworm larvae and Bm5 cell line via surfactant treatment and mechanical extrusion. Compared to cultured cells, silkworm larvae were useful for recombinant protein production as they could be raised using simple artificial diet, with the capacity to mass‐produce recombinant proteins, including the VLPs. 97 , 98 , 99
Various alternative approaches have been adopted to construct VLPs for MERS vaccines. Jung and colleagues developed a heterologous prime‐boost immunization strategy combining recombinant adenovirus serotype 5 delivering the MERS‐CoV S gene (Ad5/MERS) and the MERS S protein nanoparticles with alum adjuvant. The vaccine had been demonstrated to be effective in mice by inducing humoral and cellular immune responses (production of neutralizing antibodies and Th1 cell activations against MERS‐CoV) which protected mice against MERS‐CoV challenge. 87 Another study used transchromosomic bovine immunized with MERS‐CoV clade B S protein VLP vaccine (Al‐Hasa strain) to produce human polyclonal IgGs, which was shown to reduce the viral load when administered 24 and 48 h after virus infection, demonstrating the potential application of the antibodies for treatment of MERS. 88
More recently, Lin et al. 89 developed a VLP‐based vaccine candidate using poly(lactic‐co‐glycolic acid) (PLGA) nanoparticles displaying the MERS‐CoV RBD via chemical conjugation while encapsulating the cyclic diguanylate monophosphate, an established stimulator of interferon genes (STING) agonist. The PLGA‐based VLP vaccine was shown to protect transgenic mice against lethal MERS‐CoV challenge. To date, there is no approved vaccine for MERS‐CoV, with only a handful of candidate vaccines in the early clinical phases (Phases I and II), including BVRS‐GamVac‐Combi, VTP‐500 (ChAdOx1), INO‐4700 MERS‐CoV, and MVA MERS. 100
4.3. VLP vaccines against SARS‐CoV‐2
4.3.1. SARS‐CoV‐2 VLP vaccine candidates in preliminary stage
The current COVID‐19 pandemic has challenged the international scientific community to find an effective vaccine against SARS‐CoV‐2 infections in the shortest possible time frame. A safe two‐in‐one replicon and VLP minispike vaccine for SARS‐CoV‐2 was developed by Hennrich et al. 90 In the study, the chimeric minispike [comprising RBD linked to the transmembrane stem‐anchor sequence derived from rabies virus (RABV) glycoprotein] was incorporated into the vesicular stomatitis virus (VSV) construct which lacked its native glycoprotein (VSVΔG), namely VSVΔG‐minispike‐eGFP. 90 When rescued with VSV G, the minispike displayed SARS‐CoV‐2 RBD as trimeric protein on the host cell surface membrane and also on the envelope of secreted non‐infectious VLPs. The VSVΔG containing the minispike is non‐propagating and could induce strong immune response. A single dose (1 × 106 infectious particles) of the VSV replicon vaccine (VSVΔG‐minispike‐eGFP), when complemented with VSV G, stimulated high titers of SARS‐CoV‐2 neutralizing antibodies in transgenic K18‐hACE2 mice which protected the mice against SARS‐CoV‐2 challenge. 90
Tan et al. 57 constructed a COVID‐19 nanoparticle vaccine candidate based on the display of RBD on a synthetic VLP platform, SpyCatcher003‐mi3, using SpyTag/SpyCatcher technology. In a prime‐boost regimen, low dose of RBD‐SpyVLP was able to induce high levels of neutralizing antibodies against SARS‐CoV‐2 in mice and pigs. Such RBD‐SpyVLP was also reactive to monoclonal antibodies from convalescent patients which recognized crucial epitopes on the RBD of the RBD‐Spy VLPs. Furthermore, it is thermostable and can be distributed globally in lyophilized form. 57 Liu et al. 60 constructed a vaccine candidate based on the bacteriophage AP205 VLP displaying the spike RBM of SARS‐CoV‐2. The RBM was fused to the C‐terminus of AP205 VLPs consisting of dimerized capsid proteins and was expressed in E. coli. The vaccine was able to induce the production of high levels of IgG antibodies that could recognize eukaryotically expressed RBD and S protein of SARS‐CoV‐2. In addition, the antibodies were capable of neutralizing SARS‐CoV‐2 and could be produced at large scale for immunization. 60
Mohsen et al. 53 constructed a COVID‐19 vaccine candidate by genetically fusing the RBM to cucumber mosaic virus (CuMVTT), where the mosaic vaccine, namely CuMVTT‐RBM was produced in E. coli. The CuMVTT‐RBM displaying about 70–90 RBM antigens per VLP was highly immunogenic in rabbits and mice, where it induced neutralizing antibodies which cross‐react with mutant RBDs of other variants of concern (wildtype, K417N, E484K, N501Y, K417N/E484K/N501Y, and L452R/E484Q), with avidity surpassing those of the convalescent human sera. In addition, Mohsen et al. 53 demonstrated a proof of concept where 2‐L fermentation volume yielded 0.5 g of CuMVTT‐RBM, equivalent to 2.5 million doses in 1000‐L fermentation. Moreover, the CuMVTT‐RBM VLP‐based vaccine was stable for at least 14 months when stored at 4°C. 53 The study by Mohsen et al. 53 was in fact based on an earlier study by Zha et al., 58 where RBD instead of RBM was produced in eukaryotic cells and chemically cross‐linked to CuMVTT produced in E. coli using succinimidyl 6‐(beta‐maleimidopropionamido) hexanoate (SMPH). The highly repetitive RBD displayed on the CuMVTT VLPs induced high levels of RBD‐specific neutralizing antibodies in mice which blocked the binding of SARS‐CoV‐2 to the hACE2 receptor. 58 The Newcastle disease virus‐like particles (NDVLPs) displaying the prefusion‐stabilized SARS‐CoV‐2 S ectodomain (S2P), namely the S2P‐NDVLP were investigated as a vaccine candidate. 91 A primary injection of mice with S2P‐NDVLP showed significantly higher neutralizing titer (geometric mean ID50 of 386) compared to mice immunized with the soluble S2P protein (geometric mean ID50 of 17). Two weeks after a boost immunization with S2P‐NDVLP, the neutralizing titers increased to 2125–4552 when dosages of between 2 and 250 μg were used.
While other studies made use of S RBD protein, Chu et al. 92 constructed an influenza matrix 1 protein‐based VLP produced in insect cells to display full S, S1, or S2 of the SARS‐CoV‐2. The VLP expressing the full S and S1 successfully elicited virus‐neutralizing antibodies capable of partially inhibiting the binding of RBD to hACE2 in a surrogate virus neutralization test.
In addition to the display of SARS‐CoV‐2 S protein on heterologous VLPs, few other studies reported the production of SARS‐CoV‐2 VLPs that co‐expressed the S, E, and M proteins of the virus. Swann et al. 93 produced the SARS‐CoV‐2 VLPs co‐expressing the viral proteins S, M, and E in mammalian HEK‐293 T cells which mimic the actual virus. The VLPs were demonstrated to maintain the structural integrity (confirmed using atomic force microscopy) upon drying in ambient condition, which could favor the transport and storage of the VLPs when applied as vaccine. Another study reported by Xu et al. 94 revealed that the expressions of M and E proteins were crucial for the efficient formation and release of SARS‐CoV‐2 VLPs. Xu et al. 94 expressed the S, M, E, and N proteins in both HEK‐293 T and Vero E6 cells, where the morphology of the VLPs produced in Vero E6 cells were reported to be more stable and unified compared to those produced in HEK‐293 T cells, as TEM analysis showed uniform‐sized VLPs with distinctive corona‐like structure, indicating optimal spike trimers formation on SARS‐CoV‐2 envelope. 94 An engineered Saccharomyces cerevisiae platform (D‐Crypt™) to co‐express the three proteins (S, E, and M), which subsequently self‐assembled into SARS‐CoV‐2 VLPs was also reported. 95 However, the immunogenicities of these SARS‐CoV‐2 VLPs were not reported.
4.3.2. SARS‐CoV‐2 VLP vaccine candidates in clinical phase
A total of 153 SARS‐CoV‐2 vaccine candidate is undergoing clinical development and 196 candidates are in pre‐clinical development as of April 2022. 5 Six out of the vaccines currently undergoing clinical evaluations are VLP‐based (Table 2). When compared with SARS‐CoV and MERS, the SARS‐CoV‐2 VLP‐based vaccines have been developed at a more rapid pace due. The process of vaccine research and commercial development has an average time span of 10 to 15 years with a 22% chance of completing the clinical phases (Clinical trials Phase I, II, and III) which requires an investment of at least US$ 0.8 billion. 102 The recent outbreak of SARS‐CoV‐2 has shortened the timeline to 12–18 months with the establishment of new public/private funding and coordinated programs to counteract the SARS‐CoV‐2 pandemic globally. These programs included WHO's Solidarity Vaccine Trial, Operation Warp Speed (OWS), and Accelerating COVID‐19 Therapeutic Interventions and Vaccines (ACTIV) partnerships as well as Rapid Acceleration of Diagnostics (RADx) by the National Institutes of Health (NIH). Candidate vaccines in human clinical trials were subjected to evaluations of safety, adverse side effects, tolerability, immunogenicity, and effective dosage in Phase I. 103 In Phase II, the immunogenicity, safety and efficacy would be further investigated in larger number of participants. Following that, the vaccine would be evaluated in thousands of individuals for its efficacy and adverse side effects in Phase III trials. To date, 9 COVID‐19 vaccines have passed Phase III trials and have been approved for emergency use globally by the WHO. 19 The final stage, Phase IV trials, would be conducted after receiving approvals from national regulatory agencies. Pharmacovigilance would involve further monitoring of the vaccine in a wider population over a longer duration.
TABLE 2.
VLP vaccines targeting SARS‐CoV‐2 in clinical trials
| Vaccine type | Adjuvant | Vaccine name | Groups | Dosage and route of administration | Phases | Location | Registration no. |
|---|---|---|---|---|---|---|---|
| Alum adsorbed VLP vaccine expressing HexaPro‐S, M, N, E proteins of the virus. Vaccine‐Wuhan; Vaccine‐Alpha variant; Vaccine‐Wuhan+Alpha variant | Alum + K‐3CpG ODN | SARS‐CoV‐2 VLP Vaccine | The Scientific and Technological Research Council of Turkey + Nobel Pharmaceuticals + MonitorCRO | 2 doses; SC | I, II | Turkey | NCT04818281; NCT04962893 |
| VLP vaccine expressing S2 subunit | MF59 | ABNCoV2 Vaccine | Bavarian Nordic + Radbound University | 2 doses; IM | I, II | Netherlands | NCT04839146; NCT05077267 |
| RBD antigen is conjugated to the hepatitis B surface antigen | Alum + CpG 1018 | COVIVAXX; RBD SARS‐CoV‐2 HBsAg VLP | Serum Institute of India + Accelagen Pty + SpyBiotech | 2 doses; IM | I/II | Australia | ACTRN12620000817943; ACTRN12620001308987 |
| Plant recombinant coronavirus‐like particle | CpG 1018/ AS03 | CoVLP SARS‐CoV‐2 | Medicago Inc. | 2 doses; IM | I, I/II, II, II/III | Canada | NCT04450004; NCT05065619; NCT04662697; NCT04636697 |
| Enveloped VLP (eVLP) of SARS‐CoV‐2 S glycoprotein | Alum | VBI‐2902a | VBI Vaccines Inc. | 2 doses; IM | I/II | Canada | NCT04773665 |
| RBD from SARS‐CoV‐2 and VLP vector | Alum | LYB001 | Yantai Patronus Biotech Co., Ltd. | 3 doses; IM | I, II/III | China | NCT05125926, NCT05137444 |
Source: Adapted from WHO. 101
Abbreviations: E, envelope; IM, intramuscular; M, membrane; N, nucleocapsid; RBD, receptor‐binding domain; S, spike; SC, subcutaneous; VLP, virus‐like particle.
COVIVAXX is a conjugated SpyCatcher::VLP vaccine that is developed by using SpyBiotech's patented SpyCatcher/SpyTag superglue protein technology to present the RBD of SARS‐CoV‐2 on the surface of the hepatitis B surface antigen (HBsAg) VLPs. 104 The vaccine targeted the RBD of the S protein of SARS‐CoV‐2 which was fused to the SpyTag peptide. SpyCatcher/SpyTag technology has been broadly used in vaccine development which allows high density display of antigens on VLPs through irreversible covalent bonds at specific orientation/epitope presentations. 105 Since HBsAg VLP is a licensed vaccine that has demonstrated a high level of immunogenicity and safety in humans, it can serve as a very attractive universal plug and play carrier for any antigen of interest, such as in malaria. 65 The HBsAg‐SpyCatcher component in the vaccine was produced in Hansenula polymorpha, and the RBD‐SpyTag component was produced in Pichia pastoris. The vaccine can be produced at an industrial scale. In animals, a 100% seroconversion was reported and high levels of antibody titers against the RBD of the S protein were detected. 106 Currently, a Phase I/II trial (ACTRN12620000817943; ACTRN12620001308987) is ongoing in Australia. Nonclinical studies demonstrated that pre‐existing immunity against the HBsAg did not affect the immunogenicity of the VLPs. 104 The COVIVAXX vaccine only required normal refrigeration temperatures of 2 to 8 °C. Clinical Phase I study was randomized and placebo‐controlled, involving healthy adults aged 18–45 years. The safety and immunogenicity outcomes following administration of 5 and 25 μg of the vaccine/ placebo were evaluated. This was followed by a Phase II study involving a separate group of healthy participants, aged 18–79 years and the safety and immunogenicity outcomes following doses of 5 or 25 μg of vaccine administered 28 days apart were compared with the placebo. A total of 280 participants were enrolled in the clinical study. The Phase III trial is expected to be conducted in India and Europe.
Another potential SARS‐CoV‐2 VLP‐based vaccine is Medicago's CoVLP produced in the Australian weed, Nicotiana benthamiana. 63 The transient production of vaccines by plants is a rapid, scalable, and effective technology in response to pandemics caused by influenza viruses or SARS‐CoV‐2. The CoVLP vaccine contains self‐assembled VLPs which are composed of trimers of stabilized pre‐fusion S protein on the VLP surface, formulated in ASO3 and CpG adjuvants. 64 The safety and immunogenicity data of the CoVLP SARS‐CoV‐2 was obtained from the phase II clinical trial. 107 In Phase II clinical study, the vaccine was administered as two intramuscular doses 21 days apart at three different dosages (3.75, 7,5, or 15 μg), alone or adjuvanted with either AS03 or CpG1018. Twenty‐one days after the administration of the second dose of adjuvanted CoVLP SARS‐CoV‐2, cellular (IFNγ and IL‐4) and humoral (anti‐S IgG and neutralizing antibodies) responses were detected. The study also revealed that the AS03‐adjuvanted vaccine (3.75 μg) induced 10‐fold higher neutralizing antibody titers when compared to sera from subjects recovering from COVID‐19 infection. No severe adverse events were reported, and reactogenicity was generally mild to moderate and short in duration. A simple dose of CoVLP induced good IFNγ response in both adults (aged 18–64) and older adults (aged 65 and above), although a stronger IFNγ and IL‐4 responses were observed in the younger adults. 107 CoVLP SARS‐CoV‐2 is currently in a Phase III clinical trial (NCT04636697) which enrolled 30,918 participants in North America, Latin America, and Europe. The Phase III trial is an event‐driven, randomized, observer‐blinded, crossover placebo‐controlled design that will evaluate the efficacy and safety of the CoVLP formulation, compared to placebo. Another plant‐based vaccine candidate known as KBP‐201 produced by the British American Tobacco (BAT) is currently in Phase I/II clinical trial (NCT04473690). Although KBP‐201 consists of chimeric VLP (cVLP) formed from the chemical conjugation of RBD and modified tobacco mosaic virus (TMV), the vaccine candidate is classified under protein subunit instead of VLP‐based vaccine. 5 , 108
VBI Vaccines Inc. has joined the National Research Council in a SARS‐CoV‐2 vaccine development program. 109 , 110 Murine leukemia virus (MLV)‐based enveloped virus‐like particles (eVLPs) were used to produce SARS‐CoV‐2 vaccine candidates, VBI‐2900 which consists of 2 vaccines: VBI‐2901 and VBI‐2902. VBI‐2901 is a trivalent pan‐coronavirus vaccine expressing the SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV S proteins, while VBI‐2902 is a monovalent vaccine that expresses the prefusion‐stabilized form of the SARS‐CoV‐2 S protein. Preclinical data demonstrated high potency of antigen expression by eVLPs in mice. 110 Modified S protein containing the ectodomain of SARS‐CoV‐2 S fused with the transmembrane cytoplasmic terminal domain of VSV‐G enabled high yields and density of S expression on MLV‐Gag eVLPs. A single dose of eVLPs adjuvanted with Alum phosphate (VBI‐2902a) induced high level of neutralizing antibody responses compared to SARS‐CoV‐2 patients. Data also revealed that VBI‐2902a was safe and highly efficacious in a hamster challenge model. VBI‐2902a was subjected to ongoing clinical evaluation as a single‐dose vaccine against SARS‐CoV‐2, along with VBI‐2905a. The Phase I/II study (NCT04773665) of VBI‐2902a initiated in March 2021 was randomized, observer‐blind, and placebo‐controlled. The study evaluated the safety, tolerability, and immunogenicity of VBI‐2902a at 1 and 2 doses (28 days interval) regimens at (5 μg of S protein per dose) compared to placebo. The initial Phase I stage had enrolled 61 healthy, unvaccinated adults aged 18–54 years. The data obtained showed that 5 μg VBI‐2902a was well‐tolerated and induced robust immune responses in all the subjects at levels greater than those observed in convalescent patient sera. Besides, there is no safety concerns that had been linked to the vaccine.
ABNCoV2 is another clinically tested SARS‐CoV‐2 VLP‐based vaccine developed under Bavarian Nordic. 66 , 111 ABNCoV2 was developed by combinary of both unique Drosophila S2 insect cell protein production (ExpreS2ion) by ExpreS2ion Biotechnologies and proprietary capsid VLP (cVLP) of AdaptVac produced in E. coli. The RBD antigens produced in Drosophila melanogaster cells were displayed on the Acinetobacter phage AP205 capsid‐like particles produced in E. coli through a split‐protein Tag/Catcher to ensure unidirectional and high‐density display of the RBD antigens. Preclinical data revealed that ABNCoV2 was highly immunogenic in mouse model upon immunization, where a single injection of the ABNCoV2 vaccine in mice elicited virus neutralization antibody titers similar to those found in SARS‐CoV‐2 convalescent patients. When further enhanced with a booster dose, the virus neutralization titers could rise above 1:10,000. 66 Phase I clinical trial (NCT04839146) assessed the safety and tolerability of two doses (dose ranges from 6 to 70 μg) of ABNCoV2, formulated with and without the MF59 adjuvant in healthy adult volunteers in a single center, open labeled trial. Initial data from the human trial had confirmed its ability to induce strong and broad‐spectrum antibody levels, superior to those of the currently approved vaccines, while also providing a favorable safety profile. It is worth highlighting that the data confirmed the ability of ABNCoV2 vaccine to trigger the production of neutralizing antibodies against several variants of SARS‐CoV2, including Wuhan, Alpha, Beta, and Delta variants. 112 Recently, Phase II clinical trial (NCT05077267) has started to recruit participants to evaluate the safety, tolerability, and immunogenicity of the ABNCoV2 vaccine (at dosage of 100 μg) in both seronegative and seropositive participants.
While most of the VLP‐based vaccines in clinical trial are based on the RBD or the S proteins of SARS‐CoV‐2, a VLP vaccine harboring the M, N, E, and HexaPro S antigens of the virus adjuvanted with K‐3CpG ODN was developed by The Scientific and Technological Research Council of Turkey. 113 The route of administration of the vaccine was distinct from other clinically tested VLP vaccines, where this SARS‐CoV‐2 vaccine is injected subcutaneously. 5 , 114 Phase I trial (NCT04818281) of the SARS‐CoV‐2 vaccine was designed as a double‐blinded, randomized, placebo‐controlled, and involved two different doses (10 and 40 μg). On the other hand, the Phase II trial (NCT04962893) recruited ~330 adults between 18 and 59 years who were healthy or had medically stable chronic diseases. The subjects were divided in a 1:1:1 ratio to receive two doses of VLP vaccine for Wuhan (40 μg) or VLP vaccine for Wuhan+Alpha variant (40 μg) or VLP vaccine for Alpha (British) variant (40 μg) with 21 days interval between immunizations.
The most recent addition to the vaccine candidates in clinical trials being LYB001 developed by Yantai Patrous Biotech, which consists of RBD displayed on VLP vector formulated in aluminum hydroxide. 5 LYB001 is designed as a three‐dose vaccine, where randomized, double‐blind, placebo‐controlled Phase I (NCT05125926) and Phase II/III (NCT05137444) clinical trials are to be carried out. Phase I clinical trial is estimated to involve 100 adult participants of 18–59 years old to evaluate the safety, reactogenicity, and immunogenicity profile of LYB001 in high dose (50 μg) upon favorable 7‐day safety and reactogenicity profile in low dose (25 μg). Phase II/III trial is estimated to enroll 1900 adult participants for immunogenicity and safety tests, where Phase III trial will be open‐labeled for extended safety evaluation, which will be completed upon 360‐day safety observation of all participants following third dose of vaccination.
4.3.3. SARS‐CoV‐2 VLP vaccine candidates in preclinical phase
There are 18 VLP‐based COVID‐19 vaccine candidates currently in preclinical stage (Table 3). ContiVir, a spin‐off from Max Planck Institute, has produced a corona VLP vaccine candidate. 115 The corona VLP is produced using ContiVirs innovative technologies which enable efficient production of the VLP using a fully continuous tubular bioreactor, and a size‐based capture chromatography step known as Steric Exclusion Chromatography (SXC) for purification of the corona VLP. 115 , 116 The IrsiCaixa AIDS Research Institute, the Barcelona Supercomputing Center (BSC), and the Animal Health Research Center (IRTA‐CReSA) have launched a project focusing on the development of the SARS‐CoV‐2 vaccine, with the display of SARS‐CoV‐2 S protein on the VLP of HIV. 117 Another preclinical project by Navarrabiomed used human and BEVS which secrete VLPs through the constitutive expressions of SARS‐CoV‐2 S, M, and E proteins. Additionally, they also use nonreplicative lentivirus pseudotyped with S protein and expressing the rest of the structural proteins. BIRB796 was also incorporated into the vaccine formulation as an adjuvant to enhance the T‐cell responses. 118 Similarly, a recombinant S protein‐presenting baculovirus vector has been used by Tampere University. 119 Another potential vaccine developed by Saiba GmbH used the patented CuMVTT technology for the display of RBD, where the highly repetitive RBD attached to CuMVTT significantly elevated antibodies capable of neutralizing SARS‐CoV‐2. 53 , 58 , 120 , 121 Despite being one of the earliest SARS‐CoV‐2 VLP‐based vaccine candidate, the vaccine candidate was not able to proceed to clinical trial due to the lack of funding, 121 which led them to switch focus in developing a “second generation” vaccine which is better in terms of safety, efficiency, stability, cost effectiveness, and transportability. 122
TABLE 3.
Preclinical VLP vaccines targeting SARS‐CoV‐2
| Type of candidate vaccine | Coronavirus target | Same platform for non‐Coronavirus candidates | Developers |
|---|---|---|---|
| VLP | SARS‐CoV‐2 | Max Planck Institute for Dynamics of Complex Technical Systems | |
| Virus‐like particle‐based dendritic cell (DC)‐targeting vaccine | SARS‐CoV‐2 | University of Manitoba | |
| VLP | SARS‐CoV‐2 | Bezmialem Vakif University | |
| Enveloped VLP (eVLP) | SARS‐CoV‐2, SARS‐CoV, & MERS‐CoV | CMV, GBM, Zika | VBI Vaccines Inc. |
| S protein integrated in HIV VLPs | SARS‐CoV‐2 | IrsiCaixa AIDS Research/IRTA‐CReSA/Barcelona Supercomputing Centre/Grifols | |
| VLP + Adjuvant | SARS‐CoV‐2 | Mahidol University/ The Government Pharmaceutical Organization (GPO)/Siriraj Hospital | |
| VLPs, lentivirus and baculovirus vehicles | SARS‐CoV‐2 | Navarrabiomed, Oncoimmunology group | |
| RBD displayed on VLP | SARS‐CoV‐2 | Saiba GmbH | |
| ADDomerTM multiepitope display | SARS‐CoV‐2 | Imophoron Ltd and Bristol University's Max Planck Centre | |
| Unknown | SARS‐CoV‐2 | Doherty Institute | |
| VLP | SARS‐CoV, SARS‐CoV‐2 | OSIVAX | |
| eVLP | SARS‐CoV‐2 | Malaria | ARTES Biotechnology |
| VLPs peptides/whole virus | SARS‐CoV‐2 | Univ. of Sao Paulo | |
| VLPs produced in baculovirus expression system | SARS‐CoV‐2 | Tampere University | |
| Plant‐derived VLP | SARS‐CoV‐2 | Shiraz University | |
| Myxoma virus co‐expressing S, M, N and E proteins | SARS‐CoV‐2 | Arizona State University | |
| Plasmid‐driven production of VLPs containing S, M, N and E proteins of SARS‐CoV‐2 | SARS‐CoV‐2 | Arizona State University | |
| VLP with RCB | SARS‐CoV‐2 | Berna Biotech Pharma | |
| A vaccine booster for COVID‐19 (CoVEG) | SARS‐CoV‐2 | ExcepGen |
Source: Adapted from WHO. 101
Abbreviations: CMV, cytomegalovirus; E, envelope; GBM, glioblastoma; HIV, human immunodeficiency virus; M, membrane; N, nucleocapsid; RBD, receptor‐binding domain; RCB, Reference Cell Bank; S, spike; VLP, virus‐like particle.
Meanwhile, Berna Biotech Pharma and Swiss Biotech Center are working together to develop a COVID‐19 vaccine based on RBD VLPs expressed from the human cell line platform which had demonstrated impressive efficacy in in vivo models. 123 , 124 Researchers from Arizona State University had also developed at least two VLP vaccine platforms: M20‐234 L where myxoma virus was employed to express parts of all the four major structural proteins: S, M, E, and N; and plasmid‐driven production of VLPs containing these proteins. 5 , 125 , 126 Another potential VLP‐based vaccine candidate produced by Imophoron in collaboration with the University of Bristol was based the on ADDomer™ multiepitope display system, where penton base proteins from adenovirus capable of forming dodecahedron VLPs were used to display the S protein of SARS‐CoV‐2, 127 , 128 with heat‐stable characteristics where cold chain storage was not required. 129
Almost all of the vaccines use the coronavirus strain‐specific S surface antigen. OSIVAX had teamed up with 3P Biopharmaceuticals to produce VLP‐based vaccine using oligoDOM® technology of OSIVAX (OVX313), based on a novel proprietary self‐assembling protein sequence with positively charged tail to display the highly conserved N protein of SARS‐CoV‐2 which could trigger potent B‐ and T‐cell immune responses. 130 OSIVAX aims to develop a broad‐spectrum coronavirus vaccine, OVX‐CoV, which could target SARS‐CoV and SARS‐CoV‐2. 5 , 130 Meanwhile, another SARS‐CoV‐2 S protein‐based VLP vaccine targeting the DCs are being developed in the University of Manitoba, where pseudotyped VLP (PVLP) of Delta variant had induced higher levels of TNF‐α, IL‐1β, and IL‐6 in human macrophages and DCs. 131 , 132 ARTES Biotechnology has begun the development of SARS‐CoV‐2 vaccine candidates based on its technology platforms: SplitCore and METAVAX®. 133 SplitCore used splitted HBcAg VLPs (cVLPs) as antigen presentation vehicles to display S and N proteins of SARS‐CoV‐2, 133 , 134 while METAVAX used eVLPs based on the duck hepatitis B small surface antigen produced in H. polymorpha to display S and N proteins on its N‐ and C‐terminals. 133 , 135 While VBI‐2902a and VBI‐2905a are in Phase I/II clinical trials, another eVLP vaccine candidate, VBI‐2901 from VBI Vaccines Inc. which express the S proteins of SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV is still in the preclinical stage. 110 Ghorbani et al. 136 from Shiraz University used in silico method to identify epitopes for construction of an ideal vaccine candidate against SARS‐CoV‐2 in plant‐derived VLPs. 136 , 137 However, the actual biological study has not been reported or disclosed. 5 Other than those mentioned above, there are three more VLP‐based vaccine candidates currently in the preclinical stage according to the COVID‐19 vaccine tracker and landscape published by WHO, which are from Mahidol University, Doherty Institute, University of Sao Paulo, and ExcepGen. 5 However, details of these projects have not been released to the public.
5. CONCLUSION
Despite the advantage of VLPs as vaccine platform, it did not receive much attention compared to protein subunit, viral‐vector, DNA, RNA, and inactivated virus vaccines. VLP‐based vaccines have been proven effective against HBV (Engerix®, Recombivax HB®, and HeberNasvac®), HEV (Hecolin), and human papillomavirus (Cervarix™ and Gardasil®). Out of the six VLP‐based vaccines in clinical phase, only CoVLP Medicago has reached Phase III trial and is estimated to be completed by April 2022. As VLP is highly immunogenic, self‐adjuvanting, and versatile, VLP‐based vaccines could easily become one of the most effective vaccines against coronaviruses, should it be developed with greater efforts.
AUTHOR CONTRIBUTIONS
Chean Yeah Yong: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); supervision (equal); validation (equal); writing – original draft (equal); writing – review and editing (equal). Winnie Pui Pui Liew: Data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); writing – original draft (equal). Hui Kian Ong: Conceptualization (equal); methodology (equal); validation (equal); visualization (equal); writing – original draft (equal). Chit Laa Poh: Conceptualization (equal); formal analysis (equal); supervision (equal); validation (equal); visualization (equal); writing – review and editing (equal).
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/btpr.3292.
ACKNOWLEDGMENTS
This work was supported by the International Research Network Grant Scheme, Sunway University [STR‐IRNGS‐SMLS‐CVVR‐01‐2021].
Yong CY, Liew WPP, Ong HK, Poh CL. Development of virus‐like particles‐based vaccines against coronaviruses. Biotechnol. Prog. 2022;e3292. doi: 10.1002/btpr.3292
Funding information International Research Network Grant Scheme, Sunway University, Grant/Award Number: STR‐IRNGS‐SMLS‐CVVR‐01‐2021
Contributor Information
Chean Yeah Yong, Email: yongcheanyeah@hotmail.com.
Chit Laa Poh, Email: pohcl@sunway.edu.my.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Mesel‐Lemoine M, Millet J, Vidalain PO, et al. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J Virol. 2012;86:7577‐7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Enserink M. SARS: a pandemic prevented. Science. 2003;302:2045. [DOI] [PubMed] [Google Scholar]
- 3. Chan‐Yeung M, Xu R‐H. SARS: epidemiology. Respirology. 2003;8:S9‐S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814‐1820. [DOI] [PubMed] [Google Scholar]
- 5. WHO . MERS situation update, February 2022. Accessed April 1, 2022. http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html
- 6. Gossner C, Danielson N, Gervelmeyer A, et al. Human‐dromedary camel interactions and the risk of acquiring zoonotic Middle East respiratory syndrome coronavirus infection. Zoonoses Public Health. 2016;63:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chowell G, Abdirizak F, Lee S, et al. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015;13:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cauchemez S, Nouvellet P, Cori A, et al. Unraveling the drivers of MERS‐CoV transmission. Proc Natl Acad Sci U S A. 2016;113:9081‐9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. WHO . Middle East respiratory syndrome coronavirus (MERS‐CoV)—Saudi Arabia. 2021. Accessed September 2021. https://www.who.int/emergencies/disease-outbreak-news/item/2021-DON333
- 10. Ki HK, Han SK, Son JS, Park SO. Risk of transmission via medical employees and importance of routine infection‐prevention policy in a nosocomial outbreak of Middle East respiratory syndrome (MERS): a descriptive analysis from a tertiary care hospital in South Korea. BMC Pulm Med. 2019;19:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ki M. 2015 MERS outbreak in Korea: hospital‐to‐hospital transmission. Epidemiology and Health. 2015;37:e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Liu D, Shi W, et al. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: Phylogenetics and coalescence analysis. MBio. 2015;6:e01280‐e01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oh M‐D, Park WB, Park S‐W, et al. Middle East respiratory syndrome: what we learned from the 2015 outbreak in the Republic of Korea. Korean J Intern Med. 2018;33:233‐246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England). 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. WHO . Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Accessed August 18, 2021. https://www.who.int/publications/m/item/summary-of-probable-sars-cases-with-onset-of-illness-from-1-november-2002-to-31-july-2003
- 16. Worldometer . COVID‐19 coronavirus pandemic. 2021. Accessed December 2021. https://www.worldometers.info/coronavirus/.
- 17. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367:1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Worldometer . COVID‐19 coronavirus pandemic. 2022. Accessed April 2022. https://www.worldometers.info/coronavirus/
- 19. WHO . 10 vaccines granted emergency use listing (EUL) by WHO. 2022. Accessed April 2022. https://covid19.trackvaccines.org/agency/who/
- 20. Kovac M. How COVID‐19 is changing the approach for the vaccine development process. 2021. Accessed September 2021. https://www.ppd.com/blog/how-covid-19-is-changing-the-approach-for-the-vaccine-development-process/.
- 21. Fehr AR, Perlman S. Coronaviruses: An overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, eds. Coronaviruses: Methods and Protocols. Springer New York; 2015:1‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and Functions of Coronavirus Replication–Transcription Complexes and their Relevance for SARS‐CoV‐2 Drug Design. Molecular Cell Biology; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295:12910‐12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui J, Li F, Shi Z‐L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruan Y, Wei CL, Ling AE, et al. Comparative full‐length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet (London, England). 2003;361:1779‐1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyerholz DK, Lambertz AM, McCray PB Jr. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol. 2016;186:78‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581:221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Min L, Sun Q. Antibodies and vaccines target RBD of SARS‐CoV‐2. Front Mol Biosci. 2021;8:671633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Piccoli L, Park YJ, Tortorici MA, et al. Mapping neutralizing and Immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell. 2020;183:1024‐1042.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rezaee F, Linfield DT, Harford TJ, Piedimonte G. Ongoing developments in RSV prophylaxis: a clinician's analysis. Curr Opin Virol. 2017;24:70‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Syomin BV, Ilyin YV. Virus‐like particles as an instrument of vaccine production. Mol Biol. 2019;53:323‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoon KY, Tan WS, Tey BT, Lee KW, Ho KL. Native agarose gel electrophoresis and electroelution: a fast and cost‐effective method to separate the small and large hepatitis B capsids. Electrophoresis. 2013;34:244‐253. [DOI] [PubMed] [Google Scholar]
- 35. Zahmanova G, Mazalovska M, Takova K, et al. Efficient production of chimeric hepatitis B virus‐like particles bearing an epitope of hepatitis E virus capsid by transient expression in Nicotiana benthamiana . Life. 2021;11:64. https://www.mdpi.com/2075-1729/11/1/64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karpenko LI, Ivanisenko VA, Pika IA, et al. Insertion of foreign epitopes in HBcAg: how to make the chimeric particle assemble. Amino Acids. 2000;18:329‐337. [DOI] [PubMed] [Google Scholar]
- 37. Gaglani M, Vasudevan A, Raiyani C, et al. Effectiveness of trivalent and quadrivalent inactivated vaccines against influenza B in the United States, 2011–2012 to 2016–2017. Clin Infect Dis. 2021;72:1147‐1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nooraei S, Bahrulolum H, Hoseini ZS, et al. Virus‐like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J Nanobiotechnology. 2021;19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y, Wang L, Cao H, Liu C. SARS‐CoV‐2 S1 is superior to the RBD as a COVID‐19 subunit vaccine antigen. J Med Virol. 2021;93:892‐898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu MA. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines (Basel). 2019;7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiang ZQ, Ertl HCJ. Next generation of rabies vaccines. In: Jackson AC, ed. Rabies (Third Edition). Academic Press; 2013:527‐541. [Google Scholar]
- 42. Mallapaty S. India's DNA COVID vaccine is a world first ‐ more are coming. 2021. Accessed July 2022. https://www.nature.com/articles/d41586-021-02385-x [DOI] [PubMed]
- 43. Zepeda‐Cervantes J, Ramirez‐Jarquin JO, Vaca L. Interaction between virus‐like particles (VLPs) and pattern recognition receptors (PRRs) from dendritic cells (DCs): toward better engineering of VLPs. Front Immunol. 2020;11:1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang X, Wang X, Zhang J, Xia N, Zhao Q. Escherichia coli‐derived virus‐like particles in vaccine development. NPJ Vaccines. 2017;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balke I, Zeltins A. Use of plant viruses and virus‐like particles for the creation of novel vaccines. Adv Drug Deliv Rev. 2019;145:119‐129. [DOI] [PubMed] [Google Scholar]
- 46. Yong CY, Yeap SK, Goh ZH, et al. Induction of humoral and cell‐mediated immune responses by hepatitis B virus epitope displayed on the virus‐like particles of prawn nodavirus. Appl Environ Microbiol. 2015;81:882‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yong CY, Yeap SK, Ho KL, Omar AR, Tan WS. Potential recombinant vaccine against influenza a virus based on M2e displayed on nodaviral capsid nanoparticles. Int J Nanomedicine. 2015;10:2751‐2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ninyio NN, Ho KL, Ong HK, et al. Immunological analysis of the hepatitis B virus “a” determinant displayed on chimeric virus‐like particles of Macrobrachium rosenbergii nodavirus capsid protein produced in Sf9 cells. Vaccine. 2020;8:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ong HK, Yong CY, Tan WS, et al. An influenza a vaccine based on the extracellular domain of matrix 2 protein protects BALB/c mice against H1N1 and H3N2. Vaccine. 2019;7:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang M, Lai H, Sun H, Chen Q. Virus‐like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci Rep. 2017;7:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vicente T, Roldão A, Peixoto C, Carrondo MJT, Alves PM. Large‐scale production and purification of VLP‐based vaccines. J Invertebr Pathol. 2011;107:S42‐S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Citiulo F, Crosatti C, Cattivelli L, Biselli C. Frontiers in the standardization of the plant platform for high scale production of vaccines. Plants (Basel). 2021;10:1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mohsen MO, Balke I, Zinkhan S, et al. A scalable and highly immunogenic virus‐like particle‐based vaccine against SARS‐CoV‐2. Allergy. 2022;77:243‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bahar MW, Porta C, Fox H, Macadam AJ, Fry EE, Stuart DI. Mammalian expression of virus‐like particles as a proof of principle for next generation polio vaccines. NPJ Vaccines. 2021;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Byrne B. Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol. 2015;32:9‐17. [DOI] [PubMed] [Google Scholar]
- 56. Kis Z, Shattock R, Shah N, Kontoravdi C. Emerging technologies for low‐cost, rapid vaccine manufacture. Biotechnol J. 2019;14:1800376. [DOI] [PubMed] [Google Scholar]
- 57. Tan TK, Rijal P, Rahikainen R, et al. A COVID‐19 vaccine candidate using SpyCatcher multimerization of the SARS‐CoV‐2 spike protein receptor‐binding domain induces potent neutralising antibody responses. Nat Commun. 2021;12:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zha L, Chang X, Zhao H, et al. Development of a vaccine against SARS‐CoV‐2 based on the receptor‐binding domain displayed on virus‐like particles. Vaccine. 2021;9:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pimentel TA, Yan Z, Jeffers SA, Holmes KV, Hodges RS, Burkhard P. Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem Biol Drug des. 2009;73:53‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu X, Chang X, Rothen D, et al. AP205 VLPs based on dimerized capsid proteins accommodate RBM domain of SARS‐CoV‐2 and serve as an attractive vaccine candidate. Vaccine. 2021;9:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wildt S, Gerngross TU. The humanization of N‐glycosylation pathways in yeast. Nat Rev Microbiol. 2005;3:119‐128. [DOI] [PubMed] [Google Scholar]
- 62. Geisler C, Jarvis D. Insect cell glycosylation patterns in the context of biopharmaceuticals. Post‐Translational Modification of Protein Biopharmaceuticals; Wiley, 2009:165‐191. [Google Scholar]
- 63. Ward BJ, Gobeil P, Séguin A, et al. Phase 1 randomized trial of a plant‐derived virus‐like particle vaccine for COVID‐19. Nat Med.Wiley, 2021;27:1071‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ward BJ, Gobeil P, Séguin A, et al. Phase 1 trial of a candidate recombinant virus‐like particle vaccine for COVID‐19 disease produced in plants. medRxiv. 2020;2020.11.04.20226282. [Google Scholar]
- 65. Kulkarni P, Raut S, Patki P, et al. Immunogenicity of a new, low‐cost recombinant hepatitis B vaccine derived from Hansenula polymorpha in adults. Vaccine. 2006;24:3457‐3460. [DOI] [PubMed] [Google Scholar]
- 66. Fougeroux C, Goksøyr L, Idorn M, et al. Capsid‐like particles decorated with the SARS‐CoV‐2 receptor‐binding domain elicit strong virus neutralization activity. Nat Commun. 2021;12:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chroboczek J, Szurgot I, Szolajska E. Virus‐like particles as vaccine. Acta Biochim pol. 2014;61:531‐539. [PubMed] [Google Scholar]
- 68. Mohsen MO, Gomes AC, Cabral‐Miranda G, et al. Delivering adjuvants and antigens in separate nanoparticles eliminates the need of physical linkage for effective vaccination. J Control Release. 2017;251:92‐100. [DOI] [PubMed] [Google Scholar]
- 69. Fournier JM, Gaudry D, Moreau Y, Balençon FR. Dry aerosol vaccination against Newcastle disease. I. Safety and activity controls on chickens. Dev Biol Stand. 1976;33:269‐272. [PubMed] [Google Scholar]
- 70. Sharma PK, Dmitriev IP, Kashentseva EA, et al. Development of an adenovirus vector vaccine platform for targeting dendritic cells. Cancer Gene Ther. 2018;25:27‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hodgins B, Pillet S, Landry N, Ward BJ. Prime‐pull vaccination with a plant‐derived virus‐like particle influenza vaccine elicits a broad immune response and protects aged mice from death and frailty after challenge. Immunity & Ageing. 2019;16:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fiebiger E, Meraner P, Weber E, et al. Cytokines regulate proteolysis in major histocompatibility complex class II‐dependent antigen presentation by dendritic cells. J Exp Med. 2001;193:881‐892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Karakus U, Thamamongood T, Ciminski K, et al. MHC class II proteins mediate cross‐species entry of bat influenza viruses. Nature. 2019;567:109‐112. [DOI] [PubMed] [Google Scholar]
- 74. Ji P, Liu Y, Chen Y, et al. Porcine parvovirus capsid protein expressed in Escherichia coli self‐assembles into virus‐like particles with high immunogenicity in mice and Guinea pigs. Antiviral Res. 2017;139:146‐152. [DOI] [PubMed] [Google Scholar]
- 75. Gomes AC, Mohsen MO, Mueller JE, Leoratti F, Cabral‐Miranda G, Bachmann MF. Early transcriptional signature in dendritic cells and the induction of protective T cell responses upon immunization with VLPs containing TLR ligands—a role for CCL2. Front Immunol. 2019;10:1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Coleman CM, Liu YV, Mu H, et al. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32:3169‐3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu YV, Massare MJ, Barnard DL, et al. Chimeric severe acute respiratory syndrome coronavirus (SARS‐CoV) S glycoprotein and influenza matrix 1 efficiently form virus‐like particles (VLPs) that protect mice against challenge with SARS‐CoV. Vaccine. 2011;29:6606‐6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lokugamage KG, Yoshikawa‐Iwata N, Ito N, et al. Tseng C‐TK, Peters C, Makino S, chimeric coronavirus‐like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine. 2008;26:797‐808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tseng CT, Sbrana E, Iwata‐Yoshikawa N, et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PloS One. 2012;7:e35421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Azizi A, Aucoin S, Tadesse H, et al. A combined nucleocapsid vaccine induces vigorous SARS‐CD8+ T‐cell immune responses. Genetic Vaccines and Therapy. 2005;3:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Coleman CM, Venkataraman T, Liu YV, et al. MERS‐CoV spike nanoparticles protect mice from MERS‐CoV infection. Vaccine. 2017;35:1586‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lan J, Deng Y, Song J, Huang B, Wang W, Tan W. Significant spike‐specific IgG and neutralizing antibodies in mice induced by a novel chimeric virus‐like particle vaccine candidate for Middle East respiratory syndrome coronavirus. Virol Sin. 2018;33:453‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang C, Zheng X, Gai W, et al. MERS‐CoV virus‐like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017;8:12686‐12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang C, Zheng X, Gai W, et al. Novel chimeric virus‐like particles vaccine displaying MERS‐CoV receptor‐binding domain induce specific humoral and cellular immune response in mice. Antiviral Res. 2017;140:55‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kim YS, Son A, Kim J, et al. Chaperna‐mediated assembly of ferritin‐based Middle East respiratory syndrome‐coronavirus nanoparticles. Front Immunol. 2018;9:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kato T, Takami Y, Deo VK, Park EY. Preparation of virus‐like particle mimetic nanovesicles displaying the S protein of Middle East respiratory syndrome coronavirus using insect cells. J Biotechnol. 2019;306:177‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jung S‐Y, Kang KW, Lee E‐Y, et al. Heterologous prime–boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine. 2018;36:3468‐3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Luke T, Wu H, Zhao J, et al. Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS‐CoV in vivo. Sci Transl Med. 2016;8:326ra21‐326ra21. [DOI] [PubMed] [Google Scholar]
- 89. Lin LCW, Huang CY, Yao BY, et al. Viromimetic STING agonist‐loaded hollow polymeric nanoparticles for safe and effective vaccination against Middle East respiratory syndrome coronavirus. Adv Funct Mater. 2019;29:1807616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hennrich AA, Sawatsky B, Santos‐Mandujano R, et al. Safe and effective two‐in‐one replicon‐and‐VLP minispike vaccine for COVID‐19: protection of mice after a single immunization. PLoS Pathog. 2021;17:e1009064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang Y, Shi W, Abiona OM, et al. Newcastle disease virus‐like particles displaying prefusion‐stabilized SARS‐CoV‐2 spikes elicit potent neutralizing responses. Vaccine. 2021;9:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chu K‐B, Kang H‐J, Yoon K‐W, et al. Influenza virus‐like particle (VLP) vaccines expressing the SARS‐CoV‐2 S glycoprotein, S1, or S2 domains. Vaccine. 2021;9:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Swann H, Sharma A, Preece B, et al. Minimal system for assembly of SARS‐CoV‐2 virus like particles. Sci Rep. 2020;10:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xu R, Shi M, Li J, Song P, Li N. Construction of SARS‐CoV‐2 virus‐like particles by mammalian expression system. Front Bioeng Biotechnol. 2020;8:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Arora K, Rastogi R, Arora NM, et al. Multi‐antigenic virus‐like particle of SARS CoV‐2 produced in Saccharomyces cerevisiae as a vaccine candidate. bioRxiv. 2020. [Google Scholar]
- 96. Stambas J, Lu C, Tripp RA. Innate and adaptive immune responses in respiratory virus infection: implications for the clinic. Expert Rev Respir Med. 2020;14:1141‐1147. [DOI] [PubMed] [Google Scholar]
- 97. Minkner R, Park EY. Purification of virus‐like particles (VLPs) expressed in the silkworm Bombyx mori. Biotechnol Lett. 2018;40:659‐666. [DOI] [PubMed] [Google Scholar]
- 98. Kato T, Kajikawa M, Maenaka K, Park EY. Silkworm expression system as a platform technology in life science. Appl Microbiol Biotechnol. 2010;85:459‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mitsudome T, Xu J, Nagata Y, et al. Expression, purification, and characterization of endo‐β‐N‐acetylglucosaminidase H using baculovirus‐mediated silkworm protein expression system. Appl Biochem Biotechnol. 2014;172:3978‐3988. [DOI] [PubMed] [Google Scholar]
- 100. Carlson R. MERS vaccines 2022. Accessed July 2022. https://www.coronavirustoday.com/mers-vaccines
- 101. WHO . COVID‐19 vaccine tracker and landscape. 2021. Accessed December 2021. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 102. Pronker E, Weenen T, Commandeur H, Osterhaus A, Claassen H. The gold industry standard for risk and cost of drug and vaccine development revisited. Vaccine. 2011;29:5846‐5849. [DOI] [PubMed] [Google Scholar]
- 103. Zimmer C, Corum J, Wee S‐L, Kristoffersen M. Coronavirus vaccine tracker. 2022. Accessed August 2022. Available at: https://www.nyttimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html
- 104. Kaur H, Kaur M, Bhattacharyya A, et al. Indian contribution toward biomedical research and development in COVID‐19: a systematic review. Indian Journal of Pharmacology. 2021;53:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Marini A, Zhou Y, Li Y, et al. A universal plug‐and‐display vaccine carrier based on HBsAg VLP to maximize effective antibody response. Front Immunol. 2019;10:2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. covidX. COVIVAXX. 2021. Accessed December 2021. https://www.covidx.eu/covivaxx
- 107. Gobeil P, Pillet S, Séguin A, et al. Interim report of a phase 2 randomized trial of a plant‐produced virus‐like particle vaccine for COVID‐19 in healthy adults aged 18–64 and older adults aged 65 and older. medRxiv. 2021. [Google Scholar]
- 108. Maharjan PM, Choe S. Plant‐based COVID‐19 vaccines: current status, design, and development strategies of candidate vaccines. Vaccine. 2021;9:992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. VBI . VBI vaccines announces collaboration with the National Research Council of Canada to develop pan‐coronavirus vaccine candidate targeting COVID‐19, SARS, and MERS. 2020. https://www.vbivaccines.com/press-releases/nrc-pan-coronavirus-vaccine-collaboration/
- 110. Fluckiger A‐C, Ontsouka B, Bozic J, et al. An enveloped virus‐like particle vaccine expressing a stabilized prefusion form of the SARS‐CoV‐2 spike protein elicits highly potent immunity. Vaccine. 2021;39:4988‐5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bavarian Nordic . Bavarian Nordic reports positive topline phase 2 results for its covid‐19 vaccine candidate. 2021. Accessed February 2022. https://www.bavarian-nordic.com/investor/news/news.aspx?news=6440
- 112. Expres2ion . Positive topline results reported for the phase II COVID‐19 clinical trial with the ABNCoV2 vaccine. 2021. https://news.cision.com/expres2ion‐biotechnologies/r/positive‐topline‐results‐reported‐for‐the‐phase‐ii‐covid‐19‐clinical‐trial‐with‐the‐abncov2‐vaccine,c3466288
- 113. Motamedi H, Ari MM, Dashtbin S, et al. An update review of globally reported SARS‐CoV‐2 vaccines in preclinical and clinical stages. Int Immunopharmacol. 2021;96:107763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Abdulla ZA, Al‐Bashir SM, Al‐Salih NS, Aldamen AA, Abdulazeez MZ. A summary of the SARS‐CoV‐2 vaccines and technologies available or under development. Pathogens. 2021;10:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. MaxPlanck . Virus‐like particles as possible COVID‐19 vaccine. 2020. Accessed December 2021. https://www.mpg.de/15393185/virus-like-particles-as-possible-covid-19-vaccine
- 116. Marichal‐Gallardo P, Pieler MM, Wolff MW, Reichl U. Steric exclusion chromatography for purification of cell culture‐derived influenza a virus using regenerated cellulose membranes and polyethylene glycol. J Chromatogr A. 2017;1483:110‐119. [DOI] [PubMed] [Google Scholar]
- 117. IrsiCaixa . Project for the Development of Antibodies, Drugs and a Vaccine against Coronavirus. 2020. Accessed September 9, 2021. https://www.irsicaixa.es/en/news/project-development-antibodies-drugs-and-vaccine-against-coronavirus
- 118. Navarrabiomed . SARS‐CoV‐2 Biosecurity Vaccine Development Platforms. 2020. Accessed September 9, 2021. http://eu‐isciii.es/covidfundinspain/sars‐cov‐2‐biosecurity‐vaccine‐development‐platforms/
- 119. Tampere University . Special COVID‐19 funding for five projects at Tampere University. 2020. Accessed September 9, 2021. https://www.tuni.fi/en/news/special-covid-19-funding-five-projects-tampere-university
- 120. Saiba . Saiba AG selects AGC biologics to develop and produce COVID‐19 vaccine. 2020. Accessed December 2021. https://www.saiba‐biotech.com/2020/06/16/saiba‐ag‐selects‐agc‐biologics‐to‐develop‐and‐produce‐covid‐19‐vaccine/
- 121. Saiba . Saiba AG shows encouraging preclinical data for its COVID‐19 VLP‐based vaccine. 2020. Accessed December 2021. https://www.saiba-biotech.com/2020/03/25/saiba-ag-shows-encouraging-preclinical-data-for-its-covid-19-vlp-based-vaccine/
- 122. Saiba . Saiba AG focuses on a future COVID‐19 vaccine with advantages over first to market vaccine candidates. 2020. Accessed December 2021. https://www.saiba-biotech.com/2020/09/16/saiba-ag-focuses-on-a-future-covid-19-vaccine-with-advantages-over-first-to-market-vaccine-candidates/
- 123. BernaBiotechPharmaGmbH . Corona vaccine COVID‐19.2021. Accessed September 2021. https://www.berna-swiss.com/covid-19-1
- 124. Nessi E. Berna Biotech Pharma and Swiss Biotech Center to develop second‐generation COVID‐19 vaccine. 2021. Accessed December 2021. https://www.s-ge.com/en/article/news/20212-lifesciences-swissbiotechcenter
- 125. Innovations S . ASU is actively engaged in the fight against COVID‐19. 2021. Accessed December 2021. https://www.skysonginnovations.com/stories/asu-covid-technologies/
- 126. Kandikattu HK, Yadavalli CS, Venkateshaiah SU, Mishra A. Vaccine efficacy in mutant SARS‐CoV‐2 variants. Int J Cell Biol Physiol. 2021;4:1‐12. [PMC free article] [PubMed] [Google Scholar]
- 127. University‐of‐Bristol . New vaccine platform used to develop COVID‐19 candidates. 2020. Accessed September 9, 2021. http://www.bristol.ac.uk/news/2020/april/covid-19-vaccine-platform.html
- 128. Sari‐Ak D, Bufton J, Gupta K, et al. VLP‐factory™ and ADDomer(©): self‐assembling virus‐like particle (VLP) technologies for multiple protein and peptide epitope display. Curr Protoc. 2021;1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lansdowne LE. Developing temperature‐stable ADDomer vaccines for untreatable diseases. 2021. Accessed December, 2021. https://www.technologynetworks.com/biopharma/blog/developing-temperature-stable-addomer-vaccines-for-untreatable-diseases-356788
- 130. 3PBiopharmaceuticals . Osivax and 3P Biopharmaceuticals team up to develop a new universal vaccine against current and future variants of SARS‐COV‐2. 2021. December, 2021. https://3pbio.com/wp-content/uploads/2021/11/PressRelease-Osivax-Covid19-2021r.pdf
- 131. Silva DD. University of Manitoba a treatment for COVID‐19 will be found, reassures UM virologist. 2020. Accessed September 9, 2021. https://news.umanitoba.ca/a‐treatment‐for‐covid‐19‐will‐be‐found‐reassures‐um‐virologist/
- 132. Ao Z, Ouyang MJ, Olukitibi TA, Yao X. SARS‐CoV‐2 Delta spike protein enhances the viral fusogenicity and inflammatory cytokine production. bioRxiv. 2022;25:104759. 10.1016/j.isci.2022.104759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. ARTES . ARTES joins global combat against corona. 2020. Accessed December 2021. https://artes-biotechnology.biz/artes-joins-global-combat-against-corona/
- 134. ARTES . SplitCore—the latest milestone in ARTES´ vaccine development offer. 2020. Accessed December 2021 http://artes‐biotechnology.biz/splitcore‐the‐latest‐milestone‐in‐artes‐vaccine‐development‐offer/
- 135. ARTES . METAVAX®. 2013. Accessed December 2021. https://artes-biotechnology.biz/metavax-3/
- 136. Ghorbani A, Zare F, Sazegari S, Afsharifar A, Eskandari M, Pormohammad A. Development of a novel platform of virus‐like particle (VLP)‐based vaccine against COVID‐19 by exposing epitopes: an immunoinformatics approach. New Microbes and New Infections. 2020;38:100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hemmati F, Hemmati‐Dinarvand M, Karimzade M, et al. A worthy platform to produce vaccine against SARS‐CoV‐2. Biotechnol Lett. 2022;44:45–57. 10.1007/s10529-021-03211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


