Abstract
The genomic DNA of bacteria is contained in one or a few compact bodies known as nucleoids. We describe a simple procedure that retains the general shape and compaction of nucleoids from Escherichia coli upon cell lysis and nucleoid release from the cell envelope. The procedure is a modification of that used for the preparation of spermidine nucleoids (nucleoids released in the presence of spermidine) (T. Kornberg, A. Lockwood, and A. Worcel, Proc. Natl. Acad. Sci. USA 71:3189–3193, 1974). Polylysine is added to prevent the normal decompaction of nucleoids which occurs upon cell lysis. Nucleoids retained their characteristic shapes in lysates of exponential-phase cells or in lysates of cells treated with chloramphenicol or nalidixate to alter nucleoid morphology. The notably unstable nucleoids of rifampin-treated cells were obtained in compact, stable form in such lysates. Nucleoids released in the presence of polylysine were easily processed and provided well-defined DNA fluorescence and phase-contrast images. Uniform populations of nucleoids retaining characteristic shapes could be isolated after formaldehyde fixation and heating with sodium dodecyl sulfate.
The genomic DNA of Escherichia coli is localized in one or two compact bodies, known as nucleoids, per cell (3, 15, 31, 36, 38, 40). There have been many attempts to isolate representative nucleoids outside of cells (24, 25). However, the various DNA-containing structures that have been isolated generally have failed to meet this goal in two important ways. First, the nucleoid DNA underwent the partial decompaction that is associated with cell lysis (21). Second, large amounts of residual envelope remained with the nucleoid DNA, so that many such preparations are more accurately described as lysed or broken cell preparations than as isolated nucleoids (see footnote 6 of reference 20).
Evaluation of the released nucleoids is made difficult by a lack of information on the structure of the original nucleoids within cells. Here we use retention of the general shape of nucleoids in initial cells as a criterion for evaluation of a new procedure for nucleoid isolation. There have been few reports on the shapes of isolated nucleoids; the doublet-shaped high-salt nucleoids (nucleoids released in the presence of high salt concentrations) described by Hecht et al. (10) and Van Ness and Pettijohn (33) and the large DNA double structure found in high-salt nucleoids of a DNA gyrase mutant by Steck and Drlica (28) are the only instances of which we are aware.
We have used the distinctively shaped nucleoids formed in cells that have been exposed to antibiotics (4, 5, 11, 12) as test objects. The three antibiotics used here, chloramphenicol, nalidixate, and rifampin, all promote the coalescence of nucleoids in cells and can cause changes in nucleoid morphology and stability:
The nucleoid coalescence caused by chloramphenicol (17, 32) has been attributed to a loss of cotranslational insertion linkages between the DNA and the cell envelope when protein synthesis is inhibited; the loss of these expanding forces then presumably allows coalescence and fuller compaction caused by the opposing cellular forces (37, 40). The stable, round shape of nucleoids in chloramphenicol-treated cells (11, 17, 27, 32, 35) provides structures that can be readily identified by light microscopy. In addition, nucleoids from chloramphenicol-treated cells are unusually stable (20, 21), further adding to their utility as test objects.
Nalidixate treatment can cause the nucleoids within cells to form elongated, crystalline bodies (14). Nalidixate is an inhibitor of DNA gyrase (8, 29); presumably for this reason, nalidixate-treated cells often contain nucleoids which have failed to segregate properly (28). The resulting DNA-containing structures provide distinctive test objects.
Rifampin causes two opposing effects. Cellular nucleoids are transformed into compact axial rods (4). In contrast to this form within cells, nucleoids removed from rifampin-treated cells have such a strong tendency to unfold (6, 23) that isolation of compact nucleoids from rifampin-treated cells is a test in itself for the nucleoid isolation procedure.
In the following study, we describe conditions which released compact nucleoids with characteristic cellular shapes. The conditions were derived from the effects of polylysine on nucleoid unfolding that were noted briefly in an earlier study (21). It is anticipated that preparations of compact nucleoids will have applications in studies of nucleoid structure and segregation and in gene localization.
MATERIALS AND METHODS
Materials.
Low-gelling temperature agarose (type VII), chloramphenicol, 4′,6-diamidino-2-phenylindole (DAPI), crystallized chicken egg white lysozyme (EC 3.2.1.17), E. coli DNA, sodium deoxycholate, spermidine · 3HCl, Brij 58 (polyoxyethylene 20 cetyl ether), rifampin, sodium nalidixate, sucrose, and poly-l-lysine (molecular weight,∼9,000) were purchased from Sigma Chemical Co.; diethylmalonic acid (98%) was purchased from Aldrich Chemical Co.
Cell growth and antibiotic treatments.
Exponential-phase E. coli K-12 C600 was grown at 37°C in 180 ml of Luria-Bertani medium (26) in a 500-ml Erlenmeyer flask in a rotary shaker (New Brunswick G-25D; 275 rpm). The DNA of the cells was fluorescently labeled (1) by addition of 0.5 μg of DAPI/ml at an A600 of 0.17. At an A600 of ∼0.5, the culture was chilled by swirling the flask in ice-water for 5 min, and the absorbance of the chilled culture was measured. Aliquots of chilled culture equal to 27.6 ml × (0.5/A600) were centrifuged for 10 min at 10,000 × g and 3°C, and each pellet was resuspended at 0°C in 0.5 ml of 20% sucrose–0.1 M NaCl–10 mM Tris-HCl buffer (pH 8.1). The resuspended cells were immediately lysed to minimize autolytic changes (9). Cells for treatment with antibiotics were grown and prelabeled with DAPI as described above. Chloramphenicol (final concentration, 30 μg/ml), sodium nalidixate (40 μg/ml), or rifampin (40 μg/ml) was added at an A600 of 0.25, and shaking at 37°C was continued for 1 h before the cells were harvested and resuspended as described above.
Polylysine-spermidine lysis procedure.
Resuspended cells (1 volume) were incubated for 1 min at 0°C with 1/5 volume of a solution containing 120 mM Tris · HCl buffer (pH 8.1), 50 mM sodium EDTA (pH 7), and 400 μg of lysozyme/ml; a lower lysozyme concentration in the same medium was used in some experiments. Polylysine (1/5 volume of an aqueous solution of 5 mg of poly-l-lysine · HBr/ml; molecular weight, ∼9,000) was then added. After incubation for 1 min at 0°C, 1/5 volume of 1% Brij 58–0.4% sodium deoxycholate–10 mM sodium EDTA (pH 7)–10 mM spermidine · 3HCl was added. The turbid mixture was incubated for 5 min at 37°C and rechilled. The partially clarified lysate was stored at 5°C. Lysates prepared with or without polylysine had a pH of about 6.2 to 6.7.
Large cellular components in the lysate, i.e., released nucleoids and lysed and unlysed cells, were separated from the excess reagents used in the lysis procedure and from released cytoplasmic materials by centrifugation through a layer of buffered sucrose. Lysate (1.5 ml) was layered over 8 ml of 15% sucrose in solution A (20 mM sodium diethylmalonate buffer [pH 7.1], 5 mM MgCl2, 1 mM β-mercaptoethanol) and centrifuged for 20 min at 10,000 × g (8,000 rpm) in a swinging-bucket rotor (HB-4 rotor, Sorvall) at 3°C. The pellet was resuspended in 500 μl of 15% sucrose in solution A by trituration and gentle agitation over a period of several minutes, yielding a nonviscous, opalescent mixture.
Sucrose gradient centrifugation of SDS-treated, HCHO-fixed samples.
Relatively homogeneous suspensions of nucleoids were isolated from formaldehyde (HCHO)-fixed preparations that had been treated with sodium dodecyl sulfate (SDS). The resuspended pellet described above was fixed in 2% HCHO for 1 h at 0°C and kept for ≥1 day at 5°C. SDS (3%) was added, and the samples were heated for 30 min at 50°C, chilled, and stored at 5°C. Precipitated SDS which formed at lower temperatures was redissolved by brief warming to room temperature, and samples of 0.95 ml were layered over sucrose gradients (gradient volume, 10.0 ml; 15 to 30% sucrose in solution A). After centrifugation for 15 min at 5,000 rpm in a swinging-bucket rotor (SW40 rotor; Beckman) at 5°C, fractions were collected by piercing the bottoms of the tubes and allowing the contents to drip into tared tubes; fraction volumes were calculated from fraction weights and solution densities of control gradients. Fractions were analyzed for DNA by fluorometry; aliquots (0.5 ml; containing ≤2 μg of DNA diluted in 10 mM sodium phosphate buffer [pH 7.4]–0.01% SDS) were mixed with 1.0 ml of 10 mM sodium phosphate buffer (pH 7.4)–0.15 μg of DAPI/ml, and their fluorescence was measured with a Hoefer DynaQuant 200 fluorometer with a cuvette (Amersham Pharmacia Biotech) using E. coli DNA as the standard.
Light microscopy.
Samples of 3 to 5 μl were added to glass slides and briefly mixed by use of a pipette tip with an equal volume of 2% low-gelling-temperature agarose−20 mM sodium diethylmalonate buffer (pH 7.1) that had been liquefied and equilibrated to 37°C. Coverslips were immediately added and fixed in place with Vaspar (22) applied along the edges. The slides were monitored by phase-contrast microscopy; DNA (DAPI) fluorescence or combined phase-contrast and DNA fluorescence exposures were made at intervals.
A Zeiss Axioskop microscope (model 20 with a 100-W mercury arc) with phase-contrast and epifluorescence optics and Zeiss filter set 02 (for DAPI images) was used with a Snappy video capture device (Play Inc.) for images from a Panasonic color digital camera (model GP-KR222). A Sony color video monitor (model PVM 14N2U) was used for image selection.
Electron microscopy.
Samples for electron microscopy were fixed with an equal volume of buffered 4% HCHO–1% glutaraldehyde (fixative solution) for 1 h at 0°C and centrifuged for 10 min at 12,000 × g and 3°C, and the pellets were stored in fresh aliquots of fixative solution. Subsequent steps were carried out at Paragon Bioservices, Inc., Baltimore, Md. Pellets were postfixed in buffered OsO4 for 1 h; after embedding was done with epoxy resin, sections of ∼70 nm were stained first with 2% uranyl acetate and then with 0.2% lead citrate and examined in a Zeiss electron microscope (model 10C).
RESULTS AND DISCUSSION
Polylysine-spermidine lysis procedure for nucleoid release.
The polylysine-spermidine lysis procedure is based upon the ability of polylysine to prevent nucleoid decompaction upon cell lysis (21). The procedure incorporates the detergent lysis protocol described by Kornberg et al. (13) with two important modifications. First, polylysine was added before lysis in order to retain nucleoid shape. Second, the temperature of the detergent lysis step was increased from 10°C to 37°C to promote increased release of nucleoids from the residual cell envelope. The increased release at the higher temperature may result from more effective detergent action on membranes (16) and/or increased autolytic effects.
The lysis procedure is rapid. The cells, generally prelabeled with DAPI, were resuspended at 0°C in a medium causing limited plasmolysis. After treatment for 1 min at 0°C with egg white lysozyme, polylysine was added, and the suspension was kept for 1 min at 0°C. A detergent-spermidine-EDTA mixture was added, and the suspension was kept for 5 min at 37°C, during which time its turbidity decreased substantially as lysis proceeded.
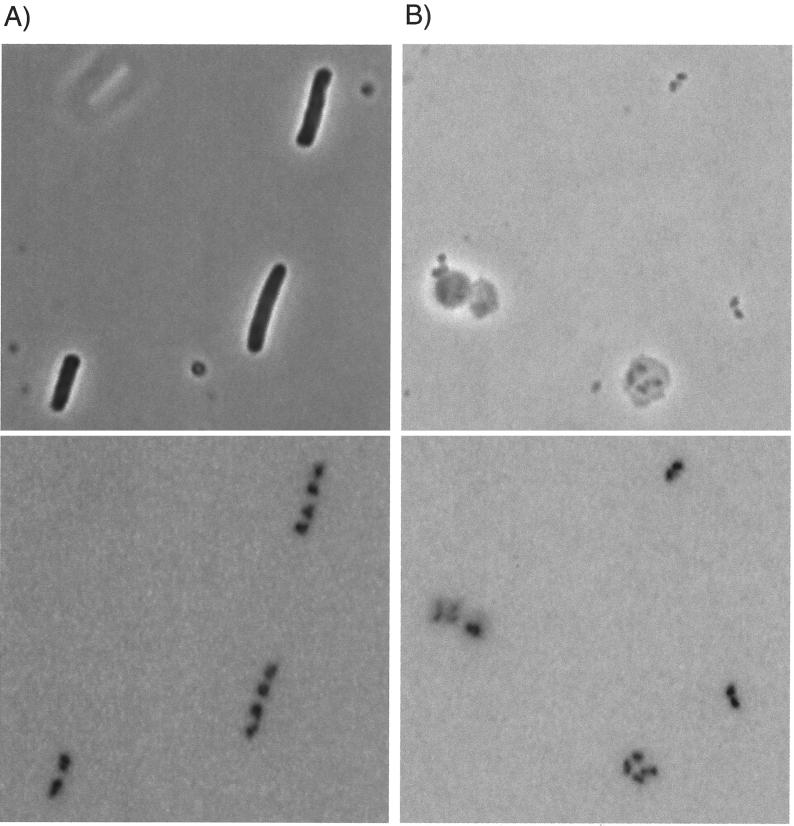
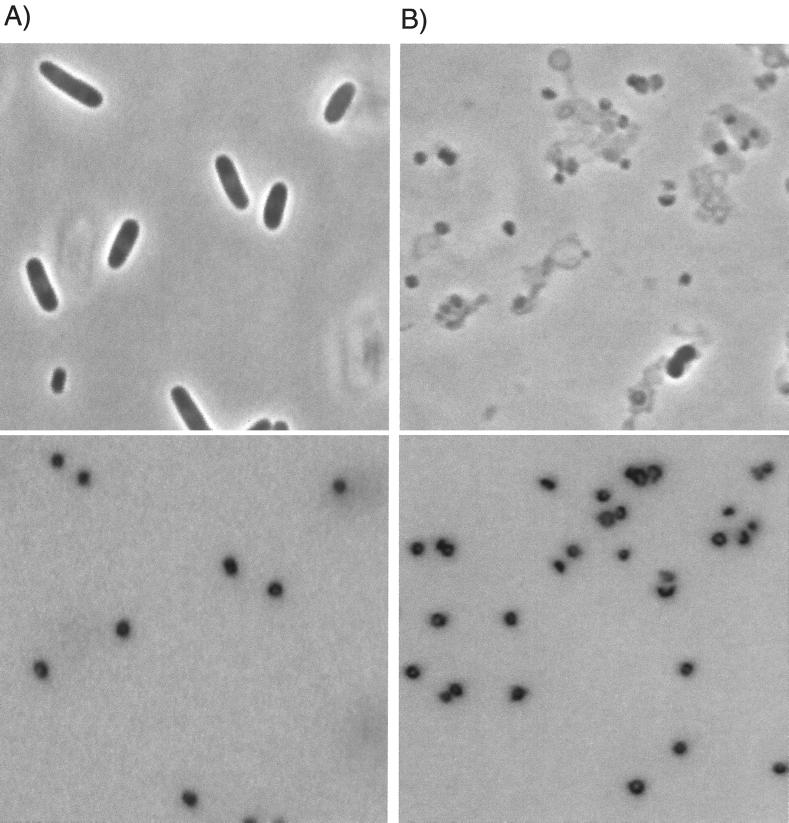
Applications of the polylysine-spermidine lysis procedure to exponential-phase cells and to chloramphenicol-treated cells of E. coli C600 are shown in Fig. 1 and 2, respectively. The top pictures in both figures are phase-contrast images used to visualize cells, cell ghosts, nucleoids, and larger debris; the bottom picture of each pair is a DAPI fluorescence image of the same microscope field used to show DNA localization. The characteristic dumbbell shape of the nucleoids in exponential-phase cells (15, 31) is illustrated in Fig. 1A, and the rounded nucleoids of chloramphenicol-treated cells (11, 17, 27, 32, 35) are shown in Fig. 2A.
FIG. 1.
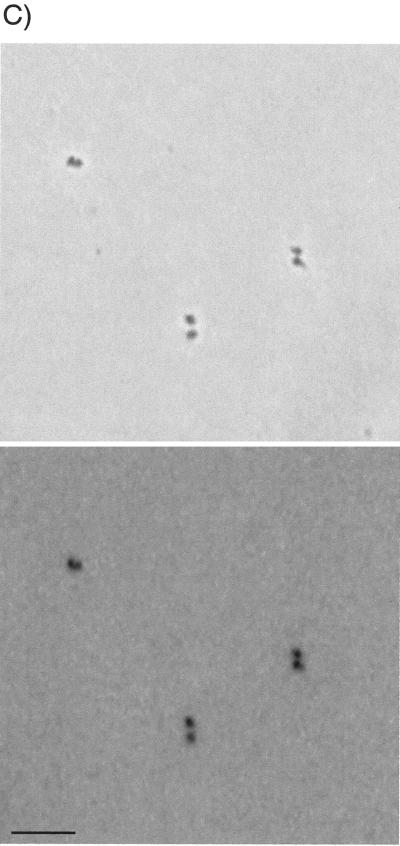
Comparison by light microscopy of exponential-phase E. coli cells with a polylysine-spermidine lysate of those cells and with nucleoids isolated from the lysate. (Upper panels) Phase-contrast images. (Lower panels) DNA fluorescence images of the same fields. The color of fluorescence images is inverted. (A) Exponential-phase cells of E. coli. (B) Polylysine-spermidine lysate of exponential-phase cells. (C) SDS-treated, HCHO-fixed nucleoids isolated by sucrose gradient centrifugation from the lysate used in panel B. See Fig. 5A for fractions pooled. Bar, 5 μm.
FIG. 2.
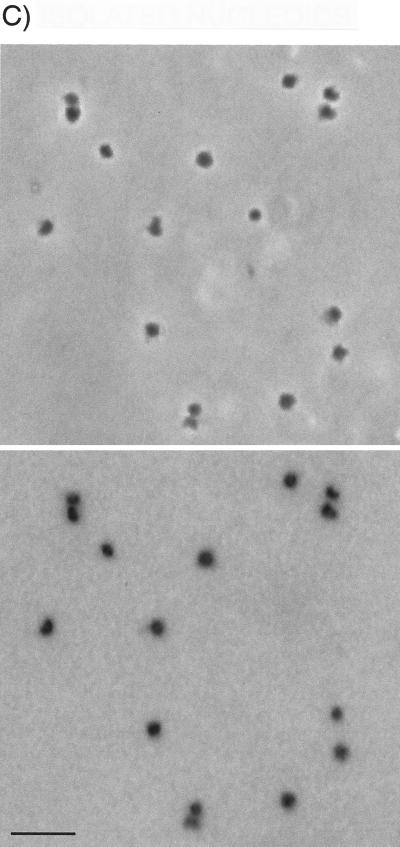
Comparison by light microscopy of chloramphenicol-treated E. coli cells with a polylysine-spermidine lysate of those cells and with nucleoids isolated from the lysate. (Upper panels) Phase-contrast images. (Lower panels) DNA fluorescence images of the same fields. The color of fluorescence images is inverted. (A) Chloramphenicol-treated cells of E. coli. (B) Polylysine-spermidine lysate of chloramphenicol-treated cells. (C) SDS-treated, HCHO-fixed nucleoids isolated by sucrose gradient centrifugation from the lysate used in panel B. See Fig. 5B for fractions pooled. Bar, 5 μm.
Lysates of exponential-phase cells and chloramphenicol-treated cells are shown in Fig. 1B and 2B, respectively. About 20 to 80% of the cell nucleoids were released from cell envelope material in such preparations; the remaining nucleoids were within cell ghosts or in the small fraction of unlysed cells. Characteristic shapes of the nucleoids within the cells were preserved upon lysis, both for the nucleoids released from cell envelopes and for those retained within cell ghosts.
The nucleoids in lysates of exponential-phase cells appeared to be more compact than the nucleoids within the corresponding cells (Fig. 1A versus 1B). In contrast, the sizes of the nucleoids in lysates of chloramphenicol-treated cells were similar to those of the cellular nucleoids from which they came (Fig. 2A versus 2B), possibly because of their already higher degree of compaction due to antibiotic exposure.
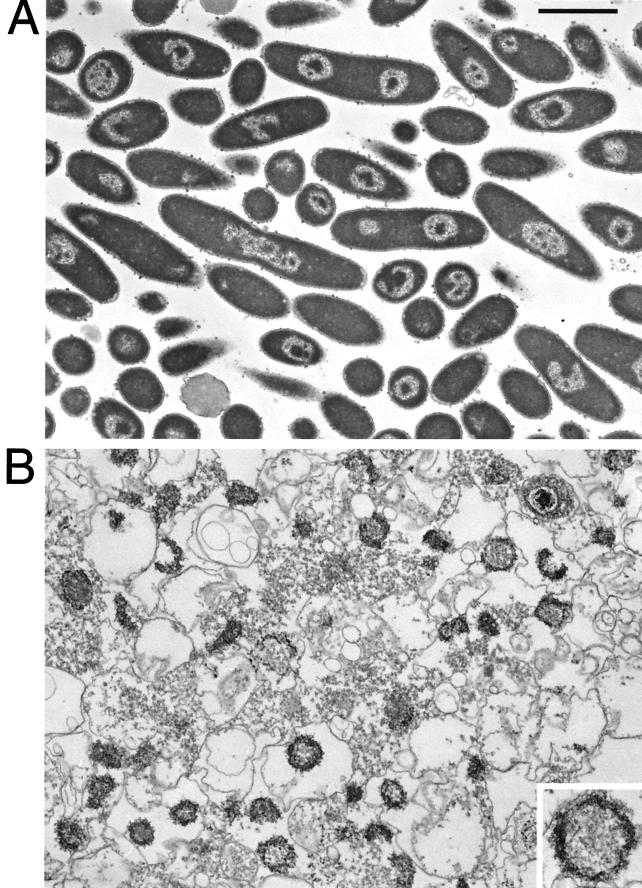
Retention of nucleoid size and shape after lysis of chloramphenicol-treated cells was also seen by electron microscopy. Thin sections of pellets of chloramphenicol-treated cells and from a lysate of those cells are shown in Fig. 3A and B, respectively; the shapes and dimensions of the nucleoids released from chloramphenicol-treated cells were similar to those of the nucleoid voids in the sections of the cells. Images of the rounded nucleoids formed in response to chloramphenicol in both Fig. 2 and 3 suggested a puckered toroidal model for the chloramphenicol-treated nucleoid, as if the round shape of the nucleoid had been distorted to fit within the cell. Such a model has not been proposed previously, to our knowledge, and is being further evaluated.
FIG. 3.
Electron microscopy of chloramphenicol-treated E. coli cells and a polylysine-spermidine lysate of those cells. Shown are thin sections of pellets obtained from chloramphenicol-treated cells (A) or from a polylysine-spermidine lysate of those cells (B). Bar, 2 μm. (Inset) Threefold-higher magnification.
Nucleoids in lysates of exponential-phase cells were stable for at least several days at 5°C and could be transferred by pipette, mixed by swirling, and so forth without noticeable changes in their appearance. Some preparations slowly accumulated aggregates, suggesting that some unfolding was occurring (18). The stability and ease of manipulation of these preparations are in contrast to the extreme lability of high-salt nucleoid preparations (39) or, to a lesser extent, of spermidine nucleoid preparations (18, 19).
Nucleoids that had been sedimented through a sucrose cushion were further tested for stability and sensitivity to several agents. Nucleoids from chloramphenicol-treated cells were very stable, appearing unchanged after 7 weeks at 5°C or after heating for 5 min at 65°C; characteristic round shapes were present after 5 min at 100°C. Nucleoids from both exponential-phase cells and chloramphenicol-treated cells were degraded by pancreatic DNase and extensively unfolded in the presence of trypsin or at high NaCl concentrations. Pancreatic RNase had no obvious effect on nucleoid morphology (S. B. Zimmerman and L. D. Murphy, unpublished data).
Polycation effects.
Three polycations, polylysine, lysozyme, and spermidine, are added in relatively large amounts in the polylysine-spermidine lysis procedure.
(i) Lysozyme.
Lysozyme was required for cell lysis. Final concentrations in the lysates of between 5 and 50 μg of lysozyme/ml produced efficient lysis with all of the cell preparations used here. Other strains of E. coli, other species, or cells grown under other conditions may require different lysozyme exposures. The current procedure does not lyse stationary-phase cells; adaptation of the efficient technique of Witholt et al. (34) for spheroplast preparation from stationary-phase cells may be useful for such samples but has not been tested.
(ii) Spermidine.
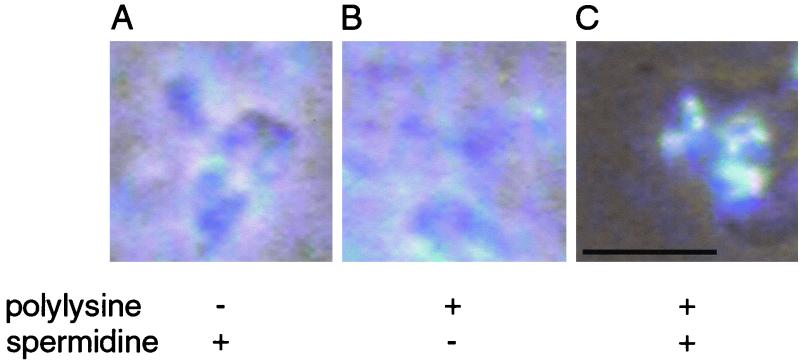
The diffuse DNA released in the absence of spermidine (Fig. 4B) may be contrasted with the compact DNA in its presence (Fig. 4C). A stabilizing effect of spermidine on isolated high-salt nucleoids was shown by Flink and Pettijohn (7).
FIG. 4.
Effects of polylysine and spermidine on lysis of exponential-phase E. coli. Lysates were prepared from exponential-phase E. coli using 0.34 mg of polylysine/ml as described in Materials and Methods except polylysine omitted (A), spermidine omitted (B), or no omissions (C). Bar, 5 μm. Simultaneous phase-contrast and DNA fluorescence images are shown without color inversion to emphasize the diffuse DNA fluorescence in panels A and B.
(iii) Polylysine.
The concentration of polylysine present at the time of cell lysis was critical in determining the efficiency of lysis and the recovery of compact nucleoids. At low concentrations of polylysine (a final concentration of <0.2 mg/ml in the lysates), lysis released diffuse clouds of DNA (Fig. 4A); similar diffuse images were obtained for lysates of antibiotic-treated cells that were made in the absence of polylysine (data not shown). At ∼0.3 to 0.8 mg of polylysine/ml, lysis released compact nucleoids (Fig. 4C). The efficiency of cell lysis decreased as the polylysine concentration increased. At polylysine concentrations of over 1.0 mg/ml, cells clumped and were not lysed (data not shown).
The mechanisms of the cation effects are certain to be complex. All three polycations presumably contribute to the compaction and stabilization of nucleoids by charge neutralization and cross-linking of DNA. The addition of polylysine (final concentration, 0.7 mg/ml) inhibited the activity of endogenous DNase in the lysates or added pancreatic DNase I (Zimmerman and Murphy, unpublished). Polycation effects on cell autolysins and/or membrane stability are also likely, given the strong stabilizing effects of both polylysine and spermidine on spheroplasts and protoplasts (30).
Isolation of HCHO-fixed nucleoids from polylysine-spermidine lysates.
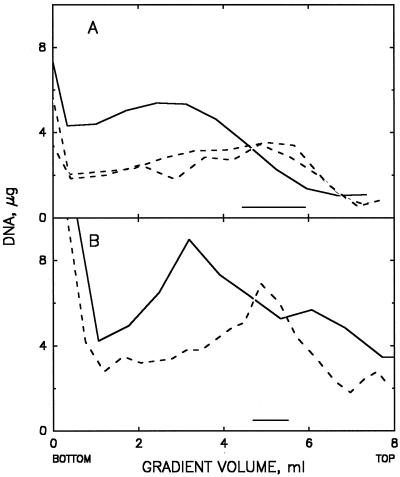
The larger objects in the lysates could be quickly separated from the bulk of released cytoplasmic materials and unbound reagents by sedimentation through a layer of buffered 15% sucrose. The free nucleoids, ghosts, and unlysed cells formed a pellet which was readily resuspended without obvious changes in the microscopic appearance of these materials. Attempts to separate the released nucleoids from ghosts or unlysed cells by sucrose gradient centrifugation of the redispersed pellets from the 15% sucrose step gradient were partially successful. Preparations from either exponential-phase or chloramphenicol-treated cell lysates gave broad sedimentation zones (Fig. 5A or B, respectively) in which the proportion of free nucleoids relative to that of nucleoids in ghosts or unlysed cells was increased severalfold (data not shown).
FIG. 5.
Sucrose gradient centrifugation of polylysine-spermidine lysates of exponential-phase or chloramphenicol-treated E. coli. Samples applied to the gradients were prepared by centrifugation of polylysine-spermidine lysates through buffered 15% sucrose; pellets were redispersed and fixed with HCHO as described in Materials and Methods (solid lines). Broken lines represent samples that were further incubated in 3% SDS for 30 min at 50°C before application to the gradients; the two broken lines in panel A are duplicate samples. (A) Samples from a lysate of exponential-phase cells (prepared with a final concentration of 5 μg of lysozyme/ml in the lysate); 360 μg of DNA was added to the gradient. (B) Samples from a lysate of chloramphenicol-treated cells; 530 μg of DNA was added to gradient. Horizontal bars indicate the fractions pooled (from the samples shown by the broken lines) that were used for Fig. 1C and 2C, respectively.
Relatively homogeneous preparations of isolated nucleoids that were microscopically free of envelope materials could be easily obtained from either exponential-phase cells or chloramphenicol-treated cells by use of HCHO-fixed polylysine-spermidine lysates. HCHO-fixed nucleoids were remarkably stable to exposure to SDS, retaining their shape and compaction after heating for 30 min at 50°C in 3% SDS; this treatment solubilized virtually all of the visible envelope remnants in HCHO-fixed lysates. In contrast, SDS exposure of unfixed nucleoids led to their immediate unfolding, accompanied by an enormous increase in viscosity.
Sucrose gradient centrifugation profiles of heated, SDS-treated, HCHO-fixed material from exponential-phase cells and chloramphenicol-treated cells are shown in Fig. 5A and B, respectively. About 10 to 30% of the nucleoids applied to the gradients could be recovered in fractions which retained the unaggregated, characteristic shapes shown in Fig. 1C and 2C, respectively. Many of the remaining nucleoids in the gradients appeared to be aggregates of the same characteristic units.
Lysates of cells exposed to nalidixate or rifampin.
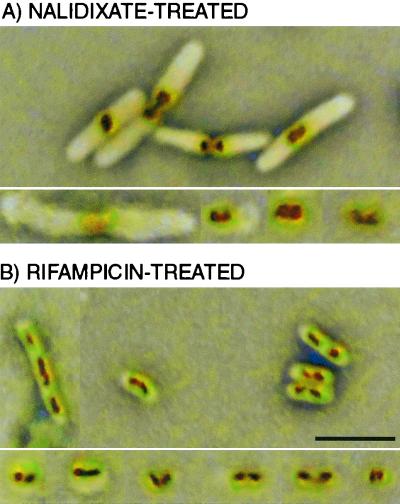
Nalidixate treatment caused most of the dividing cells to retain their DNA in a distinctively shaped structure that was positioned on either side of the demarcation between the dividing cells (Fig. 6A, upper panel). Such apparently incompletely resolved structures were found in the polylysine-spermidine lysates of these cells, both within cell envelopes and as free bodies (Fig. 6A, lower panel).
FIG. 6.
Comparison by light microscopy of rifampin- or nalidixate-treated E. coli cells with components of polylysine-spermidine lysates of those cells. (A) Nalidixate treatment. (B) Rifampin treatment. (Upper panels) Cells after antibiotic exposures. (Lower panels) Selected images from lysates of those cells. Images are simultaneous phase-contrast and DNA fluorescence exposures shown with inverted color. Bar, 5 μm.
Rifampin-treated cells were tested because of the instability of their nucleoids upon cell lysis under previous protocols (6, 23). The compact nucleoids of rifampin-treated cells (Fig. 6B, upper panel) were, indeed, recovered as compact bodies in polylysine-spermidine lysates of these cells, again occurring in part within cell envelopes and in part as free nucleoids (Fig. 6B, lower panel).
Applications of the polylysine-spermidine lysis procedure.
The long history of nucleoid isolation from E. coli (24, 25) suggests the importance ascribed to successful isolation procedures. The question becomes not only how to isolate a nucleoid that retains significant attributes of its cellular structure but how to determine if that has actually been accomplished. We have used morphology as a criterion. Nucleoids prepared by the polylysine-spermidine procedure retained the general nucleoid shapes that were present in the cells from which they came and had levels of DNA compaction similar to or greater than those in the cells. Such properties support the use of polylysine-spermidine nucleoids for a number of purposes.
(i) Structural studies.
Both the released nucleoids and the ghost-enclosed nucleoids are stable, and their components are accessible to many structural probes. Their nucleic acids can be inventoried and probed with nucleases to test for repetitive structures. The presence or the absence of “histone-like” and other proteins will be of interest. Comparisons between nucleoids from exponential-phase cells and nucleoids from cells exposed to various antibiotics or nucleoids from mutants with mutations in various constituents of the nucleoid may provide structural insights. The well-defined images of the compact released nucleoids offer the possibility of viewing nucleoids during decompaction caused by a variety of modalities. Several treatments of nucleoids from chloramphenicol-treated cells appeared to cause an isometric expansion in a preliminary survey.
(ii) Gene localization.
Nucleoids in polylysine-spermidine lysates may be useful in gene localization studies, such as those using fluorescence in situ hybridization or green fluorescent protein derivatives.
(iii) Characterization of unstable nucleoid configurations.
The effects of polycations may be useful for the isolation of otherwise unstable nucleoids, as illustrated here with nucleoids of rifampin-treated cells.
(iv) Size and shape markers.
The regular geometrical shapes of the released nucleoids, particularly those from chloramphenicol-treated cells, suggest their use as standard particles. The uniform particles present after SDS treatment of HCHO-fixed nucleoids may be useful in calibrations or tests of theories of sedimentation and in studies of the effects of morphological changes on transport or optical properties. Additional forms may be generated by variations in antibiotic exposure or by cross-linking.
(v) Nucleoid visualization.
The nucleoids in polylysine-spermidine lysates, both released and retained within ghosts, are sharply defined and can be visualized in dilute media by phase-contrast microscopy. Even nucleoids within spheroplasts, such as those prepared by the method of Birdsell and Cota-Robles (2), but in the presence of polylysine are very clearly defined by phase-contrast microscopy (Zimmerman and Murphy, unpublished).
ACKNOWLEDGMENTS
The comments of Gary Felsenfeld, Martin Gellert, and J. L. Rosner are very much appreciated.
REFERENCES
- 1.Åkerlund T, Bernander R, Nordström K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 2.Birdsell D C, Cota-Robles E H. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J Bacteriol. 1967;93:427–437. doi: 10.1128/jb.93.1.427-437.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drlica K, Bendich A J. Chromosome, bacterial. In: Lederberg J, editor. Encyclopedia of microbiology. 2nd ed. Vol. 1. San Diego, Calif: Academic Press, Inc.; 2000. pp. 808–821. [Google Scholar]
- 4.Dworsky P, Schaechter M. Effect of rifampin on the structure and membrane attachment of the nucleoid of Escherichia coli. J Bacteriol. 1973;116:1364–1374. doi: 10.1128/jb.116.3.1364-1374.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dworsky P. Einfluss von Inhibitoren der Protein- und Nucleinsäuresynthese auf die Gestalt des Nucleoids von Escherichia coli. Z Allg Mikrobiol. 1974;14:3–28. [PubMed] [Google Scholar]
- 6.Dworsky P. Unfolding of the chromosome of Escherichia coli after treatment with rifampicin. Z Allg Mikrobiol. 1975;15:243–247. doi: 10.1002/jobm.3630150404. [DOI] [PubMed] [Google Scholar]
- 7.Flink I, Pettijohn D E. Polyamines stabilise DNA folds. Nature. 1975;253:62–63. doi: 10.1038/253062a0. [DOI] [PubMed] [Google Scholar]
- 8.Gellert M, Mizuuchi K, O'Dea M H, Itoh T, Tomizawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godson G N. A technique of rapid lysis for the preparation of Escherichia coli polyribosomes. Methods Enzymol. 1967;12A:503–516. [Google Scholar]
- 10.Hecht R M, Taggart R T, Pettijohn D E. Size and DNA content of purified E. coli nucleoids observed by fluorescence microscopy. Nature. 1975;253:60–62. doi: 10.1038/253060a0. [DOI] [PubMed] [Google Scholar]
- 11.Kellenberger E, Ryter A, Séchaud J. Electron microscope study of DNA-containing plasms. II. Vegetative and mature phage DNA as compared with normal bacterial nucleoids in different physiological states. J Biophys Biochem Cytol. 1958;4:671–678. doi: 10.1083/jcb.4.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellenberger E. The physical state of the bacterial nucleus. Symp Soc Gen Microbiol. 1960;10:39–66. [Google Scholar]
- 13.Kornberg T, Lockwood A, Worcel A. Replication of the Escherichia coli chromosome with a soluble enzyme system. Proc Natl Acad Sci USA. 1974;71:3189–3193. doi: 10.1073/pnas.71.8.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin-Zaidman S, Frenkiel-Krispin D, Shimoni E, Sabanay I, Wolf S G, Minsky A. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc Natl Acad Sci USA. 2000;97:6791–6796. doi: 10.1073/pnas.090532397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason D J, Powelson D M. Nuclear division as observed in live bacteria by a new technique. J Bacteriol. 1956;71:474–479. doi: 10.1128/jb.71.4.474-479.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer M, De Jong M A, Woldringh C L, Nanninga N. Factors affecting the release of folded chromosomes from Escherichia coli. Eur J Biochem. 1976;63:469–475. doi: 10.1111/j.1432-1033.1976.tb10249.x. [DOI] [PubMed] [Google Scholar]
- 17.Morgan C, Rosenkranz H S, Carr H S, Rose H M. Electron microscopy of chloramphenicol-treated Escherichia coli. J Bacteriol. 1967;93:1987–2002. doi: 10.1128/jb.93.6.1987-2002.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy L D, Zimmerman S B. Isolation and characterization of spermidine nucleoids from Escherichia coli. J Struct Biol. 1997;119:321–335. doi: 10.1006/jsbi.1997.3883. [DOI] [PubMed] [Google Scholar]
- 19.Murphy L D, Zimmerman S B. Stabilization of compact spermidine nucleoids from Escherichia coli under crowded conditions: implications for in vivo nucleoid structure. J Struct Biol. 1997;119:336–346. doi: 10.1006/jsbi.1997.3884. [DOI] [PubMed] [Google Scholar]
- 20.Murphy L D, Zimmerman S B. Multiple restraints to the unfolding of spermidine nucleoids from Escherichia coli. J Struct Biol. 2000;132:46–62. doi: 10.1006/jsbi.2000.4306. [DOI] [PubMed] [Google Scholar]
- 21.Murphy L D, Zimmerman S B. A limited loss of DNA compaction accompanying the release of cytoplasm from cells of Escherichia coli. J Struct Biol. 2001;133:75–86. doi: 10.1006/jsbi.2001.4331. [DOI] [PubMed] [Google Scholar]
- 22.Murray R G E, Doetsch R N, Robinow C F. Determinative and cytological light microscopy. In: Gerhardt P, et al., editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. p. 26. [Google Scholar]
- 23.Pettijohn D E, Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harbor Symp Quant Biol. 1973;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Pettijohn D E. Prokaryotic DNA in nucleoid structure. Crit Rev Biochem. 1976;4:175–202. doi: 10.3109/10409237609105458. [DOI] [PubMed] [Google Scholar]
- 25.Pettijohn D E. The nucleoid. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 158–166. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. p. A.1. [Google Scholar]
- 27.Schaechter M, Laing V O. Direct observation of fusion of bacterial nuclei. J Bacteriol. 1961;81:667–668. doi: 10.1128/jb.81.4.667-668.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steck T R, Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984;36:1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- 29.Sugino A, Peebles C L, Kreuzer K N, Cozzarelli N R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor C W. Stabilization of protoplasts and spheroplasts by spermine and other polyamines. J Bacteriol. 1962;83:1101–1111. doi: 10.1128/jb.83.5.1101-1111.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valkenburg J A C, Woldringh C L, Brakenhoff G J, van der Voort H T M, Nanninga N. Confocal scanning light microscopy of the Escherichia coli nucleoid: comparison with phase-contrast and electron microscope images. J Bacteriol. 1985;161:478–483. doi: 10.1128/jb.161.2.478-483.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Helvoort J M L M, Kool J, Woldringh C L. Chloramphenicol causes fusion of separated nucleoids in Escherichia coli K-12 cells and filaments. J Bacteriol. 1996;178:4289–4293. doi: 10.1128/jb.178.14.4289-4293.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Ness J, Pettijohn D E. A simple autoradiographic method for investigating long range chromosome substructure: size and number of DNA molecules in isolated nucleoids of Escherichia coli. J Mol Biol. 1979;129:501–508. doi: 10.1016/0022-2836(79)90509-6. [DOI] [PubMed] [Google Scholar]
- 34.Witholt B, Boekhout M, Brock M, Kingma J, van Heerikhuizen H, de Leij L. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976;74:160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]
- 35.Woldringh C L, Nanninga N. Organization of the nucleoplasm in Escherichia coli visualized by phase-contrast light microscopy, freeze fracturing, and thin sectioning. J Bacteriol. 1976;127:1455–1464. doi: 10.1128/jb.127.3.1455-1464.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woldringh C L, Nanninga N. Structure of nucleoid and cytoplasm in the intact cell. In: Nanninga N, editor. Molecular cytology of Escherichia coli. London, England: Academic Press Ltd.; 1985. pp. 161–197. [Google Scholar]
- 37.Woldringh C L, Jensen P R, Westerhoff H V. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol Lett. 1995;131:235–242. doi: 10.1111/j.1574-6968.1995.tb07782.x. [DOI] [PubMed] [Google Scholar]
- 38.Woldringh C L, Odijk T. Structure of DNA within the bacterial cell: physics and physiology. In: Charlebois R L, editor. Organization of the prokaryotic genome. Washington, D.C.: American Society for Microbiology; 1999. pp. 171–187. [Google Scholar]
- 39.Worcel A, Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerman S B, Murphy L D. Macromolecular crowding and the mandatory condensation of DNA in bacteria. FEBS Lett. 1996;390:245–248. doi: 10.1016/0014-5793(96)00725-9. [DOI] [PubMed] [Google Scholar]