Abstract
A unique extracellular and thermostable cyclomaltodextrin glucanotransferase (CGTase) from the hyperthermophilic archaeon Thermococcus sp. strain B1001 produces predominantly (>85%) α-cyclomaltodextrin (α-CD) from starch (Y. Tachibana, et al., Appl. Environ. Microbiol. 65:1991–1997, 1999). Nucleotide sequencing of the CGTase gene (cgtA) and its flanking region was performed, and a cluster of five genes was found, including a gene homolog encoding a cyclomaltodextrinase (CDase) involved in the degradation of CDs (cgtB), the gene encoding CGTase (cgtA), a gene homolog for a CD-binding protein (CBP) (cgtC), and a putative CBP-dependent ABC transporter involved in uptake of CDs (cgtDE). The CDase was expressed in Escherichia coli and purified. The optimum pH and temperature for CD hydrolysis were 5.5 and 95°C, respectively. The molecular weight of the recombinant enzyme was estimated to be 79,000. The CDase hydrolyzed β-CD most efficiently among other CDs. Maltose and pullulan were not utilized as substrates. Linear maltodextrins with a small glucose unit were very slowly hydrolyzed, and starch was hydrolyzed more slowly. Analysis by thin-layer chromatography revealed that glucose and maltose were produced as end products. The purified recombinant CBP bound to maltose as well as to α-CD. However, the CBP exhibited higher thermostability in the presence of α-CD. These results suggested that strain B1001 possesses a unique metabolic pathway that includes extracellular synthesis, transmembrane uptake, and intracellular degradation of CDs in starch utilization. Potential advantages of this starch metabolic pathway via CDs are discussed.
Cyclomaltodextrins (CDs) are cyclic oligosaccharides consisting of α-1,4-linked 6-, 7-, or 8-glucopyranose units, usually referred to as α-, β-, or γ-CDs, respectively. CDs possess a unique torus shape and the polar hydroxyl groups are oriented toward the outside, keeping the interior cavity relatively hydrophobic. Therefore, CDs are soluble in water and the hydrophobic environment of the cavity enables them to form inclusion complexes with many organic and inorganic molecules, thereby changing the physical and chemical properties of the included compounds. This is the basis of broad applications in the food, cosmetic, and pharmaceutical industries (2, 28).
CDs are formed enzymatically from starch by the action of cyclomaltodextrin glucanotransferase (CGTase) [EC 2.4.1.19; 1,4-α-d-glucan 4-α-d-(1,4-glucano)-transferase (cyclizing)] (39). Cyclomaltodextrinase (CDase) [EC 3.2.1.54; cyclomaltodextrin dextrin-hydrolase (decyclizing)] is a unique enzyme which can hydrolyze CD and release the substance from CD inclusion complexes. After the first report of the CDase from Bacillus maceranse (8), CDases from various microorganisms such as Bacillus coagulans (23), Bacillus sphaericus E-244 (34, 35), Clostridium thermohydrosulfuricum strain 39E (recently reclassified as Thermoanaerobacter ethanolicus strain 39E) (38, 40), alkalophilic Bacillus sp. (50), Bacillus subtilis strain H-17 (24, 25), Flavobacterium sp. (3), B. sphaericus strain ATCC 7055 (15), Klebsiella oxytoca strain M5a1 (11, 12), and alkalophilic Bacillus sp. strain I-5 (22) were studied.
In K. oxytoca M5a1, the novel starch degradation pathway via CD, including the extracellular conversion of starch into CDs by CGTase and uptake of the CDs by a specific system followed by intracellular linearization by a CDase, was proposed. In addition, the genes responsible for starch metabolism were clustered on the chromosome. Until now, no other organisms possessing both CGTase and CDase for the synthesis and degradation of CDs have been found.
Recently, we have isolated a hyperthermophile, Thermococcus sp. strain B1001, which produces a unique CGTase that can catalyze predominantly the formation of α-CD (>85%), with small amounts of β- and γ-CDs, from starch (45). The cgtA gene encoding CGTase has been cloned and sequenced (48). In the present paper, we report the sequence analysis of the adjacent region of the CGTase gene containing the genes involved in transport and degradation of CDs. We also describe the purification and characterization of an extremely thermostable CDase and a CD-binding protein (CBP) from Thermococcus sp. strain B1001. Furthermore, a unique metabolic pathway of starch involving synthesis, transport, and degradation of CDs is proposed.
MATERIALS AND METHODS
Microorganisms, media, and growth conditions.
Thermococcus sp. strain B1001 was cultured as previously described (45). Escherichia coli strain JM109 (49) was used as a host for transformation and plasmid preparation and grown at 37°C in Luria-Bertani medium (30). E. coli BL21(DE3) and E. coli BL21-CodonPlus(DE3)-RIL (Novagen, Madison, Wis.) were used for heterologous expression of the target protein and were grown at 37°C in NZCYM medium (21, 30). Ampicillin (100 μg/ml) was added to the medium when necessary to select plasmid carrier.
Chemicals.
Soluble starch was purchased from Nacalai Tesque Co., Inc. (Kyoto, Japan). Maltose, maltotriose, maltotetraose, maltopentaose, and maltohexaose were obtained from Wako Pure Chemical Industries, Ltd. (Osaka, Japan), and maltoheptaose was obtained from Sigma (St. Louis, Mo.). CDs were obtained from Nihon Shokuhin Kako Co., Inc. (Tokyo, Japan), and pullulan was obtained from Hayashibara Co., Inc. (Okayama, Japan). Other biochemicals were standard commercial preparations.
DNA manipulation.
Restriction endonucleases, DNA polymerase, and T4 DNA ligase were purchased from Toyobo Co., Ltd. (Osaka, Japan). Southern blot analysis was performed according to established procedures (48). A Thermo Sequenase fluorescent-labeled primer cycle sequencing kit with 7-deaza dGTP was purchased from Amersham Pharmacia Biotech UK Ltd., (Little Chalfont, Buckinghamshire, England). Plasmids for DNA sequencing were extracted and purified by using a Wizard Plus Minipreps DNA Purification System (Promega, Madison, Wis.). Unless otherwise stated, DNA manipulations were performed essentially as described by Maniatis et al. (30).
Construction of plasmids pET-cgtB1 and pET-cgtB2.
The cgtB gene was amplified by PCR with three kinds of primers: primer 1, 5′-CATATGTATAAAATTTTCGGCTTTAAAGACAATGACTACC-3′; primer 2, 5′-CATATGCGTGAAAAGGGCGATCGTTGGTATATCAAGGTAGAGC-3′; and primer 3, 5′-CAGGAAACAGCTATGAC-3′ (M13 reverse primer). Primers 1 and 2 possess an additional NdeI site at the 5′-terminal regions, shown in italics in the sequences. In order to achieve efficient expression (20, 29), several rare codons which are not efficiently utilized in E. coli for N-terminal amino acids were replaced with codons which are frequently used in E. coli. GGA for Gly6 and Gly50, AGG for Arg47, AGA for Arg52, and ATA for Ile55 were replaced with GGC, CGT, CGT, and ATC, respectively. First, to construct pTY-331, plasmid pTY-33 (48) was digested with SacI (position 1) and BamHI (position 2521), and then the 2.5-kbp fragment harboring the entire coding region of CDase was inserted into the respective site of pUC19. As for pET-cgtB1 construction, PCR amplification was carried out with primers 1 and 3 using pTY-331 as a template. The amplified DNA was subcloned into vector pBluescript II SK(+) (Stratagene, La Jolla, Calif.) and checked by DNA sequencing. The insert DNA was digested with NdeI and BamHI and then inserted into the respective site of pET-25b(+) (Novagen). The resultant plasmid was designated pET-cgtB1. As for pET-cgtB2 construction, primer 2 was used instead of primer 1. Other procedures were carried out as for pET-cgtB1 construction.
Enzyme assay.
CDase activity was measured in the reaction mixture that contained 1% β-CD in 50 mM Britton-Robinson buffer (pH 5.5) and appropriately diluted enzyme. After incubation at 90°C for 5 min, the reducing sugar was measured by the dinitrosalicylic acid method (4). One unit of activity was defined as the amount of enzyme which released 1 μmol of reducing sugar equivalent to glucose per min. Protein concentration was measured by the BCA Protein Assay Kit (Pierce, Rockford, Ill.) with bovine serum albumin as the standard.
Purification of recombinant CDase.
CDase production by E. coli BL21(DE3) carrying pET-cgtB1 was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at mid-exponential growth phase and incubation continued for 4 h at 37°C. The cells (from 5 liters of culture) were centrifuged and the pellet was washed with buffer A (50 mM Tris-HCl buffer [pH 8.0]). Cells were disrupted by sonication and supernatant fraction was recovered by centrifugation at 48,384 × g for 30 min at 4°C. The supernatant was heat-treated at 85°C for 15 min and centrifuged again at 48,384 × g for 30 min at 4°C. The obtained supernatant was brought to 60% ammonium sulfate saturation and kept at 4°C overnight. The precipitate was collected by centrifugation at 48,384 × g for 30 min, dissolved in 40 ml of buffer A, and dialyzed overnight against the same buffer. The dialysate was applied to a weak anion-exchange column (DEAE Sepharose Fast Flow; Pharmacia, Uppsala, Sweden) previously equilibrated with buffer A, and CDase was eluted by a linear gradient of NaCl using a fast-performance liquid chromatography (FPLC) system (Pharmacia). The active fraction was loaded onto a high-resolution anion-exchange column (Mono Q HR 5/5; Pharmacia) equilibrated with buffer A and eluted with a linear gradient of NaCl.
TLC.
A mixture of oligosaccharides was analyzed by silica gel thin-layer chromatography (TLC). Aliquots (3 μl) of the sample were developed on a silica gel plate (Kieselgel 60; Merck Co., Berlin, Germany) with isopropyl alcohol-acetone-water (2:2:1 [vol/vol/vol]), and the oligosaccharides were detected by spraying the plate with aniline diphenylamine reagent (4 ml of aniline, 4 g of diphenylamine, 200 ml of acetone, and 30 ml of 85% phosphoric acid) and baking it.
Construction of plasmid pET-CBP.
In order to construct pTY-401, plasmid pTY-40 was digested with PstI (position 4158) and EcoRI (position 6425) and then the 2.3-kbp fragment was inserted into respective site of pUC18. As for pET-CBP construction, the cgtC gene was amplified by PCR with primer 3 and primer 4 (5′-CATATGAAGAAGGCACTGTTTGCTTTATTGTTG-3′) using pTY-401 as a template. The amplified DNA was subcloned into vector pBluescript II SK(+) and checked by DNA sequencing. The insert DNA was digested with EcoRI completely and then with NdeI partially. The 1.4-kbp fragment harboring the entire coding region of CBP was inserted into the respective site of pET-25b(+). The resultant plasmid was designated pET-CBP.
Purification of recombinant CBP.
CBP synthesis by E. coli BL21-CodonPlus(DE3)-RIL carrying pET-CBP was induced by 1 mM IPTG at mid-exponential phase and incubation continued for 6 h at 25°C. The cells (from 1 liter of culture) were centrifuged and the pellet was washed with buffer A. Cells were disrupted by sonication and the supernatant fraction was recovered by centrifugation at 48,384 × g for 30 min at 4°C. The supernatant was heat-treated at 80°C for 10 min and centrifuged again at 48,384 × g for 30 min at 4°C. The obtained supernatant was applied to a strong anion-exchange column (HiTrap Q; Pharmacia) previously equilibrated with buffer A, and CBP was eluted by the linear gradient of NaCl using an FPLC system.
Binding assay of CBP.
Purified CBP was applied to an α-CD-(epoxy)-Sepharose 6B affinity column (Pharmacia) previously equilibrated with buffer A. The column was washed with buffer A containing 1.0 M NaCl, and bound protein was eluted with buffer A supplemented with 1% α-CD. As another binding assay method, purified CBP was loaded onto an amylose resin (New England Biolabs, Inc., Beverly, Mass.) and then the column was washed with buffer A containing 1.0 M NaCl. Bound protein was eluted with buffer A containing 1% maltose. The washed and eluted fractions were applied to sodium dodecyl sulfate (SDS)-polyacrylamide gels and protein bands were detected by staining with Coomassie brilliant blue.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper appear in the GenBank database with the accession number AB034969.
RESULTS
Nucleotide sequence of the CDase gene and identification of the gene product.
The cgtA gene encoding CGTase of Thermococcus sp. strain B1001 was previously cloned and sequenced (48). Sequence analysis of the upstream region of cgtA revealed that another open reading frame (ORF) clustered with cgtA, as shown in Fig. 1. Computer inspection revealed that the ORF contained two putative ATG start codons at base positions 248 and 383 and a TAG termination codon at position 2228. The deduced amino acid sequence of the ORF contains four conserved regions identified with amylolytic enzymes as belonging to the α-amylase family (27, 46) (Fig. 2). A search with the BLAST program (43) revealed that the protein encoded by the ORF is homologous to neopullulanases (19, 26), CDases (22, 35), maltogenic amylase (6), and other amylolytic enzymes. It was suggested that the ORF encoded a kind of amylolytic enzyme. In order to specify the type of amylolytic characteristics and to identify an essential methionine as a translation start site, two types of plasmids, pET-cgtB1 and pET-cgtB2, for the ORF starting from each of two methionines, were constructed as explained above and the respective recombinant plasmids were used to transform E. coli BL21(DE3). The crude recombinant protein prepared from E. coli carrying the ORF translated from the first ATG hydrolyzed β-CD as a substrate, but the protein could not hydrolyze soluble starch and pullulan. By contrast, another recombinant protein translated from the second ATG did not exhibit hydrolytic activity. Furthermore, the first ATG is preceded by a putative ribosome-binding site complementary to the 3′ end of 16S rRNA from strain B1001 (45). Therefore, the larger ORF starting at position 248 (1,980 nucleotides coding for a protein of 660 amino acids, with a calculated molecular mass of 78,839 Da) was considered to be the gene encoding the amylolytic enzyme. The most putative type of the enzyme is CDase, because it hydrolyzed CDs efficiently but apparently did not hydrolyze soluble starch and pullulan in a 5-min reaction.
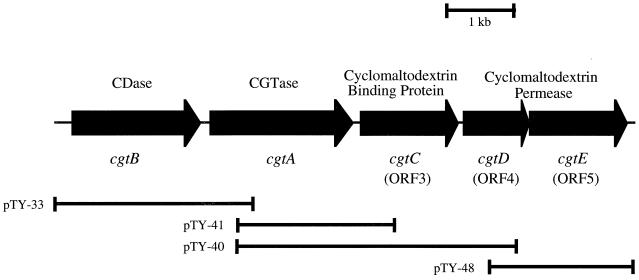
FIG. 1.
Structure of the gene cluster containing genes for synthesis, transport, and degradation of CDs in Thermococcus sp. strain B1001. Arrows show the localization of each gene and the orientation of the coding sequences. pTY-33, pTY-40, pTY-41, and pTY-48 indicate the regions cloned by the respective plasmids.
FIG. 2.
Comparison of the deduced amino acid sequences for the CDase proteins. The four regions conserved among the α-amylase family enzymes are boxed. Asterisks represent identical amino acid residues in all five polypeptides, and bullets indicate identical amino acid residues among organisms belonging to the domain Bacteria. Putative catalytic amino acid residues are indicated by circles. Polypeptides: B1001, Thermococcus sp. strain B1001 (AB034969); 39E, T. ethanolicus 39E (M88602); I-5, Bacillus sp. strain I-5 (U49646); E-244, B. sphaericus E-244 (X62576); and M5a1, K. oxytoca M5a1 (X86014).
Comparison of amino acid sequences.
The deduced amino acid sequence of the CDase from B1001 was compared with those of other CDases reported previously (Fig. 2), and a distinct region homologous with CDases from various bacteria was found (12, 22, 35, 38). In addition to the four highly conserved regions, Asp416, Glu442, and Asp507 residues as catalytic sites corresponding to Asp206, Glu230, and Asp297 of Taka-amylase A are very well conserved. Based on the results from site-directed mutagenesis, these three residues have been recognized as the catalytic sites in Taka-amylase A (33) and CDase (37). Therefore, it was suggested that the Asp416, Glu442, and Asp507 residues of the CDase from B1001 are directly involved in the catalytic site. Moreover, the CDase from B1001 possesses a unique N-terminal extension containing about 60 amino acid residues. Deletion of this N-terminal extended region caused a lack of CDase activity, suggesting that this extended region is important for the enzymatic activity of the CDase, although the mechanism is not clear.
Purification and characterization of the recombinant CDase.
CDase was purified from E. coli BL21(DE3) cells carrying plasmid pET-cgtB1 encoding the ORF from the first ATG codon. The recombinant CDase was purified by the following steps: preparation of cell lysate, heat treatment (85°C, 15 min), and ammonium sulfate fractionation (60% saturation), followed by two ion-exchange column chromatographies with DEAE Sepharose Fast Flow and Mono Q HR 5/5. The purity of the CDase was confirmed by migration of the protein as a single band with a molecular mass of approximately 79 kDa in an SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown). Hydrolysis of various substrates was analyzed with the purified enzyme (Table 1). The CDase showed a specific activity of 1,940 U/mg of protein.
TABLE 1.
Hydrolysis of various substrates by CDasea
| Substrate | Hydrolysis activity (U/mg of protein) | Relative activity (%) |
|---|---|---|
| α-CD | 1,080 | 55 |
| β-CD | 1,940 | 100 |
| γ-CD | 640 | 33 |
| Soluble starch | ND | |
| Pullulan | ND |
Substrates were dissolved into 50 mM Britton-Robinson buffer (pH 5.5) at a final concentration of 1%. Enzyme activity was measured as described in Materials and Methods. ND, not detectable (optical density at 535 nm of <0.05) after 5 min at 90°C.
The enzyme activities at various temperatures were compared by measuring the formation of the reducing sugar after 5 min of incubation of the mixture of the enzyme and β-CD in Britton-Robinson buffer (pH 5.5). The maximum enzyme activity was observed at 95°C, and even at 120°C almost 40% of the maximum activity was detected. The optimum pH for the reaction of the CDase on β-CD at 90°C was 5.5. In order to study the thermal stability of the CDase in the absence of substrate, the enzyme (in 50 mM Britton-Robinson buffer [pH 5.5]) was incubated at 90°C for various time periods, and the remaining activity was measured by the standard method. At 90°C, the half-life of the CDase was 2 h.
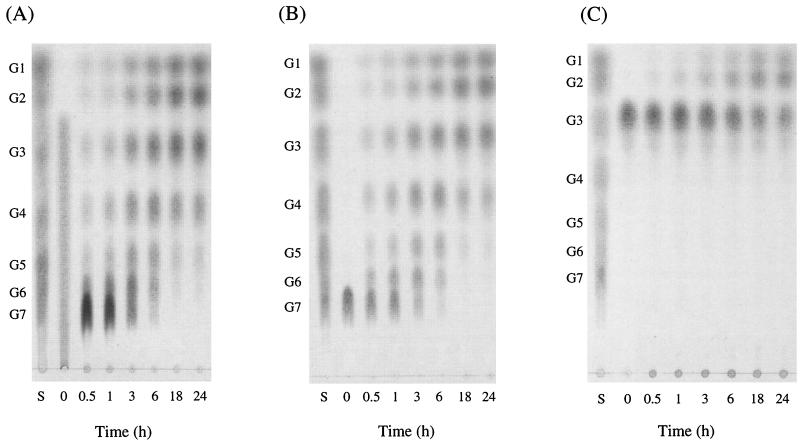
A number of linear maltodextrins and three types of CDs (α-, β-, and γ-CDs) were incubated with CDase for a prolonged time and samples were taken periodically. The spectrum of reaction products was assessed by TLC. Figure 3 shows the results when β-CD, maltoheptaose, and maltotriose were used as substrates. The analysis of hydrolyzed products suggested that these three CDs were initially hydrolyzed to the linear maltodextrins of each size and then they were subsequently degraded to the small maltodextrins. The relative reaction velocity decreased when maltodextrins with shorter chain lengths were used. Indeed, maltotetraose and maltotriose were very slowly hydrolyzed and maltose was not utilized as a substrate of the CDase. The end products of the CDase reaction were, therefore, glucose and maltose. Starch was hydrolyzed more slowly than maltotriose and an extremely small amount of degraded maltodextrins was detected after 18 h, whereas no increase in reducing ends was observed in the case of pullulan even after 24 h of incubation (results not shown).
FIG. 3.
Thin-layer chromatograms of hydrolysis products of β-CD (A), maltoheptaose (B), and maltotriose (C). Enzyme reactions were performed at the indicated periods and generated products were analyzed. Standards given are (from top): G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose; G6, maltohexaose; G7, maltoheptaose.
Nucleotide sequence of the gene for a putative CBP.
Gene walking towards the downstream region from the position of the cgtA was performed, a 4.2-kbp HindIII-HindIII (positions 2639 and 6840, respectively) fragment was cloned into pUC19, and the resultant plasmid was named pTY-40. Sequence analysis of the downstream region of cgtA revealed the existence of another ORF (ORF3) forming a gene cluster with cgtB-cgtA as shown in Fig. 1. ORF3, starting from ATG at nucleotide position 4779, was in the same direction as the cgtA. A possible ribosome-binding site (GAGG) was located 9 nucleotides upstream from the ATG. ORF3 ends with a TGA stop codon at position 6102. The nucleotide sequence of ORF3 consists of 1,323 nucleotides, encoding 441 amino acid residues with a calculated molecular mass of 48,513 Da. The deduced amino acid sequence of ORF3 exhibited 34, 32, and 34% identity with those of the periplasmic maltose-binding protein (MalE) of E. coli (9), the periplasmic CBP (CymE) of K. oxytoca M5a1 (12, 36), and the trehalose-maltose-binding protein of the hyperthermophilic archaeon Thermococcus litoralis (18), respectively. Archaea do not possess an outer membrane and periplasmic space. Therefore, these binding proteins were suggested to exist as soluble lipoproteins which allowed the anchorage of the binding proteins to the external surface of the cytoplasmic membrane. We found that the 25 amino acids in the N-terminal region of ORF3 show typical characteristics of signal peptides found in precursors of secretory proteins: (i) a hydrophobic core adjacent to the N terminus and (ii) the sequence VASGCIG, corresponding to consensus amino acid sequence of lipoprotein signal peptidase cleavage sites (LAAGCSS) (47). Thus, ORF3 is considered to encode a solute-specific binding protein. Following the putative signal cleavage site, a Gly-rich region (amino acids 25 to 28) and a remarkable Thr- and Ser-rich region (amino acids 29 to 63), which may function as a flexible linker, were found.
Identification of the binding protein.
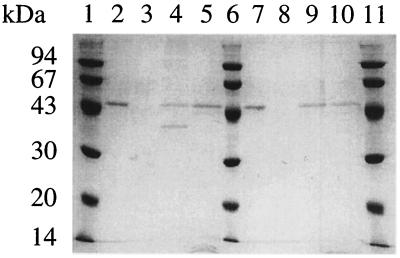
An expression vector for the putative binding protein (ORF3) was constructed and introduced into E. coli BL21-CodonPlus(DE3)-RIL. Heat treatment of the crude extract at 80°C for 10 min resulted in considerable purity of the target protein. The putative solute-binding protein was purified by anion-exchange column chromatography and found to migrate as a single protein band with a molecular mass of approximately 49 kDa in SDS-PAGE (Fig. 4, lane 2). Considering that ORF3 is located downstream of the cgtB-cgtA genes, the protein might be involved in the degradation process for CDs synthesized extracellularly. Thus, it was speculated that ORF3 encodes a CBP. The binding specificity of a protein derived from ORF3 was analyzed by measuring the binding to the α-CD affinity column. As shown in Fig. 4, the protein bound to the α-CD affinity column and was eluted by buffer containing α-CD. Furthermore, the protein bound to not only α-CD but also amylose resin, and it was eluted by maltose. When the bound protein fraction was eluted by α-CD and was heat-treated in an SDS-gel loading buffer at 100°C for 5 min, two bands were observed (lane 4). The smaller band disappeared when the eluted protein sample was treated under highly denaturing conditions (120°C, 5 min) (lane 5). In the fraction eluted by maltose (lane 9), a single band was observed even when treated in SDS-gel loading buffer at 100°C for 5 min, indicating that the structure of the putative CBP in the presence of α-CD is more thermostable than that bound to maltose. These results indicated that ORF3 might encode a CBP rather than a maltose-binding protein.
FIG. 4.
Binding assay of CBP. Proteins were electrophoresed on a 12% acrylamide gel and stained with Coomassie brilliant blue. Lanes: 1, 6, and 11, molecular mass standards (namely, rabbit muscle phosphorylase b [94 kDa], bovine serum albumin [67 kDa], egg white ovalbumin [43 kDa], bovine erythrocyte carbonic anhydrase [30.1 kDa], and soybean trypsin inhibitor [20.1 kDa]); 2 and 7, purified CBP used for binding experiment; 3, unbound fraction from α-CD affinity column; 4 and 5, bound fraction from α-CD affinity column (eluted fraction with α-CD); 8, unbound fraction from amylose resin; 9 and 10, bound fraction from amylose resin (eluted fraction with maltose). Bound protein to α-CD affinity column was eluted by α-CD and incubated at 100°C (lane 4) or 120°C (lane 5) for 5 min in an SDS-gel loading solution. Bound protein to amylose resin was eluted by the buffer containing maltose and incubated at 100°C (lane 9) or 120°C (lane 10) for 5 min in an SDS-gel loading buffer.
Nucleotide sequence of the genes for putative CBP-dependent permease proteins.
Starting from an ATG at position of 6151, there is another ORF (ORF4), but a putative stop codon could not be found in the cloned fragment. To obtain the DNA region encoding the entire ORF4, subsequent gene walking was performed, a 2.2-kbp EcoRI-SphI (positions 6425 and 8576, respectively) fragment was cloned into pUC19, and the resultant plasmid was named pTY-48 (Fig. 1). The nucleotide sequence of the entire ORF4 could be determined. ORF4 encodes a protein of 300 amino acids with molecular weight of 33,915. Immediately downstream of ORF4, ORF5, which encodes a protein of molecular weight 49,706 (447 amino acids), is located in the same direction. A search with the BLAST program revealed that the deduced amino acid sequences of ORF4 and ORF5 were homologous to the permease proteins of the maltose transport system encoded by malF and malG, respectively. MalF and MalG are inner membrane components of the maltose-binding protein-dependent maltose-maltodextrin ABC transporter (5). The deduced amino acid sequence of ORF4 shared 27, 31, and 36% identity with those of the maltose transport protein (MalF) of E. coli (13), the cyclomaltodextrin transport protein (CymF) of K. oxytoca M5a1 (12), and the trehalose-maltose transport protein (MalG) of T. litoralis (18), respectively. The deduced amino acid sequence of ORF5 shared 34, 36, and 34% identity with those of MalG of E. coli (7), CymG of K. oxytoca M5a1 (12), and MalG of T. litoralis (18), respectively. The predicted amino acid sequences of both ORF4 and ORF5 contain the consensus sequence EAAX2L/DGAX8IXLP that is found in membrane components of ABC transporters (41). In this conserved region, the amino acids are predominantly hydrophobic, with a consistent location at the C terminus. ORF4 and ORF5 may encode putative CBP-dependent membrane transporter proteins. Accordingly, the three genes (ORF3, ORF4, and ORF5) were designated cgtC, cgtD, and cgtE, respectively.
DISCUSSION
We have shown the presence of a cgtBACDE gene cluster on the Thermococcus sp. strain B1001 chromosome (Fig. 1). Respective genes encode functional proteins necessary for intracellular degradation (CgtB) of CDs, extracellular synthesis (CgtA), specific recognition (CgtC), and transmembrane transport (CgtD and CgtE). Purification and characterization of recombinant CDase (CgtB) and CBP (CgtC) were performed in the present study.
The CDase is very thermostable, with an optimum temperature for activity of 95°C. The CDases from other thermophilic sources, B. subtilis H-17 and T. ethanolicus 39E, have optimum temperatures of 65 to 68°C (24) and 65°C (38, 40), respectively. The strain B1001 CDase exhibited a half-life of 2 h at 90°C and the hydrolytic activity was maintained in the presence of substrates even after 18 h at 90°C (Fig. 3). The B1001 CDase is the most thermostable among the enzymes previously reported and characterized.
The cgtC gene was suggested to encode a kind of solute-binding protein by sequence similarity. Characterization of the purified recombinant CgtC revealed that the gene product is functional as a CBP. The CBP contains a long stretch of putative signal sequence in the N-terminal region, and the possible cleavage site might be modified by lipid, which may function as an anchor to the cytoplasmic membrane. The Thr- and Ser-rich region (SSPTQTTTTT repeat) was observed between the putative anchor domain and CD binding domain. It was reported for a pullulanase of T. thermosulfurigenes EM1 that the Thr-, Ser-, and Gly-rich region, due to the probable extended, flexible nature of its structure, would allow optimal orientation of the enzyme's catalytic site toward the substrate (31). Interestingly, several S-layer anchoring enzymes exhibit variations of Thr-rich regions, which are usually described as O-glycosylation regions. Indeed, in the case of an archaeal hyperthermostable type II pullulanase from Thermococcus hydrothermalis, the Thr-rich region has been suggested to be a target for intensive O glycosylation (10). It has also been reported that glucoamylase from Aspergillus awamori possesses this region for O glycosylation and the region is important for efficient degradation of insoluble starch granules (42). The Thr- and Ser-rich region forms a rather extended and flexible conformation susceptible to proteolytic digestion (32). Intense O glycosylation of this region would reduce the vulnerability of this structure. Based on these reports, the Thr- and Ser-rich region of strain B1001 may serve as a linker to maintain the flexibility of the CD binding domain on the surface of the cell membrane for efficient uptake of extracellular CDs. This is the first report of a solute-binding protein that includes this unique linker.
The cgtDE genes are located downstream of the cgtC gene, in the same direction. The deduced amino acid sequence of cgtD (CgtD) and cgtE (CgtE) showed similarity to those of MalF and MalG, parts of the maltose-binding protein-dependent ABC transport systems of E. coli. In addition, the organization of genes for CBP and the membrane transporting apparatus cluster (cgtCDE) is similar to that of E. coli and other bacterial binding protein-dependent ABC transport systems. In the case of the E. coli maltose-maltodextrin transport system, the maltose-binding protein (MalE) is located in the periplasmic space, two hydrophobic membrane proteins (MalF and MalG) form the translocation pore, and two additional subunits (MalK) are peripherally associated with the membrane proteins at the inner face of the membrane. Recently, an ABC transporter for maltose-trehalose was reported in the hyperthermophilic archaeon T. litoralis (17, 18). These recent reports and our experimental results strongly suggest that strain B1001 possesses the CBP-dependent ABC transport system for specific uptake and utilization of CD as a carbon source. In the organization of the T. litoralis ABC transporter for maltose-trehalose, the malEFG operon was very similar to that of E. coli but did not contain the E. coli malK homolog. The location of the malK gene is close to the malEFG gene in T. litoralis (17). The malK homolog was not found directly adjacent and distal to cgtE in strain B1001. The possibility that strain B1001 has the malK homolog encoding the ATPase subunit should be considered, though it is unclear that its gene is also a member of the cgtBACDE cluster.
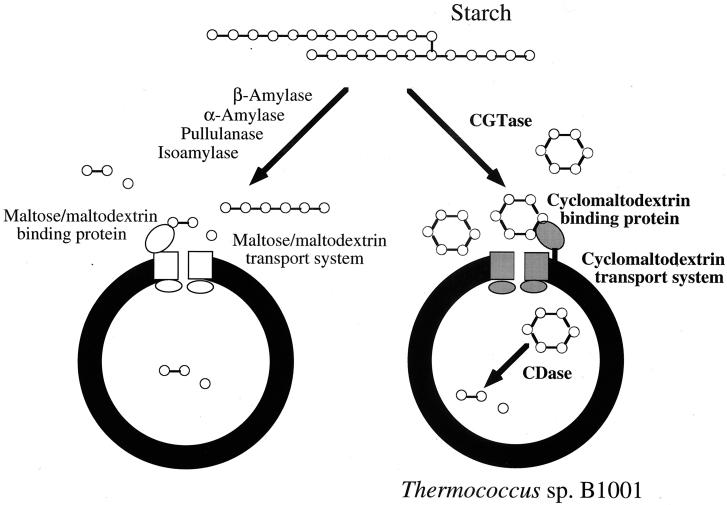
Typical signal peptide sequences (a positively charged N terminus followed by a stretch of hydrophobic residues) as secretory signals were found in the precursors of the CGTase (48) and CBP from strain B1001 and the α-amylase of Pyrococcus kodakaraensis strain KOD1 (44). It is generally assumed that signal peptides of Archaea are similar to those of Bacteria and Eucarya (1). However, comparison of the amino acid sequence of the CDase revealed that the gene does not encode a putative signal peptide, suggesting that the CDase of strain B1001 is an intracellular enzyme. Previously, we reported that strain B1001 produced a thermostable CGTase in the extracellular fraction (45). In the present study, the existence of a CBP-dependent ABC transport system for CD was implied. It has been reported that K. oxytoca M5a1 utilizes starch as a sole carbon and energy source via two metabolic pathways. The first pathway involves extracellular degradation of starch into linear maltodextrins by hydrolysis of the α-1,6-glycosidic bonds via the pullulanase and subsequent cleavage of the α-1,4-glycosidic linkages by disproportionation activity of the CGTase. Maltodextrins are transported and assimilated via a binding protein-dependent ABC transporter and intracellular hydrolytic enzymes. The second pathway, a novel starch degradation pathway, involves the extracellular conversion of starch into CDs by CGTase and uptake of the CDs by a specific binding and transporting system following intracellular linearization by a CDase (12). The genes involved in this starch utilization pathway are organized in two divergently oriented clusters in the chromosome of K. oxytoca M5a1. The existence of the CGTase, CBP, and CDase in strain B1001 indicates that a mechanism similar to that of the second pathway of K. oxytoca M5a1 exists as a starch assimilation system (Fig. 5). In addition, indirect evidence for this starch metabolism only through CDs is supported by the fact that no remarkable enzymatic activity to produce reducing sugar was observed in the culture of B1001. The unique starch metabolic pathway via CDs may be more advantageous for strain B1001 among hyperthermophiles growing in a high-temperature environment because synthesis of CDs allows effective and competitive exploitation of starch as a carbon source. CDs are not hydrolyzed by exo-type amylases such as glucoamylases and β-amylases because CDs have no nonreducing ends. The raw starch-binding domain of A. awamori exhibits binding affinity not only to raw starch but also to α-, β-, and γ-CDs, although the enzyme cannot hydrolyze CDs. Thus, the activity of the glucoamylase will be competitively inhibited by binding of CDs to the raw starch binding site of the enzyme (14, 16). CDs show various degrees of resistance and inhibition to hydrolytic enzymes produced by competitors, indicating that strain B1001 converts starch to CDs as inhibitors of amylolytic enzymes produced by competitors. The production of linear maltodextrins as a carbon source by competitors is inhibited by CDs. In addition, CBP binds not only to CDs but also to linear maltodextrins, although strain B1001 does not secrete a maltodextrin-producing enzyme. The fact that a CBP-dependent ABC transporter system also functions as the classical maltose-maltodextrin transporter indicates that strain B1001 might assimilate even maltodextrins produced by competitors as well as CDs. The cluster encoding extracellular CGTase, the CBP-dependent ABC transport system for CD, and intracellular CDase would provide benefits to the cell in the exploitation of carbon sources in a high-temperature environment where starch substrates are in alpha form (Fig. 5).
FIG. 5.
Proposed model for degradation of starch via the maltose-maltodextrin and the CD pathways.
ACKNOWLEDGMENTS
This work was supported in part by the Core Research for Evolutional Science and Technology (CREST) Program of the Japan Science and Technology Corporation.
REFERENCES
- 1.Albers S-V, Kornings W N, Driessen A J M. A short signal sequence in membrane-anchored proteins of Archaea. Mol Microbiol. 1999;31:1595–1597. doi: 10.1046/j.1365-2958.1999.01286.x. [DOI] [PubMed] [Google Scholar]
- 2.Bender H. Production, characterization, and application of cyclodextrins. Adv Biotechnol Proc. 1986;6:31–71. [Google Scholar]
- 3.Bender H. Purification and characterization of a cyclodextrin-degrading enzyme from Flavobacterium sp. Appl Microbiol Biotechnol. 1993;39:714–719. [Google Scholar]
- 4.Bernfeld H. Amylases α and β. Methods Enzymol. 1955;1:149–150. [Google Scholar]
- 5.Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–29. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha H J, Yoon H G, Kim Y W, Lee H S, Kim J W, Kweon K S, Oh B H, Park K H. Molecular and enzymatic characterization of a maltogenic amylase that hydrolyzes and transglycosylates acarbose. Eur J Biochem. 1998;253:251–262. doi: 10.1046/j.1432-1327.1998.2530251.x. [DOI] [PubMed] [Google Scholar]
- 7.Dassa E, Hofnung M. Sequence of gene malG in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePinto J A, Campbell L L. Purification and properties of the cyclodextrinase of Bacillus macerans. Biochemistry. 1968;7:121–125. doi: 10.1021/bi00841a016. [DOI] [PubMed] [Google Scholar]
- 9.Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- 10.Erra-Pujada M, Debeire P, Duchiron F, O'Donohue M J. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J Bacteriol. 1999;181:3284–3287. doi: 10.1128/jb.181.10.3284-3287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feederle R, Pajatsch M, Kremmer E, Böck A. Metabolism of cyclodextrins by Klebsiella oxytoca M5a1: purification and characterisation of a cytoplasmically located cyclodextrinase. Arch Microbiol. 1996;165:206–212. doi: 10.1007/BF01692863. [DOI] [PubMed] [Google Scholar]
- 12.Fiedler G, Pajatsch M, Böck A. Genetics of a novel starch utilisation pathway present in Klebsiella oxytoca. J Mol Biol. 1996;256:279–291. doi: 10.1006/jmbi.1996.0085. [DOI] [PubMed] [Google Scholar]
- 13.Frosshauer S, Beckwith J. The nucleotide sequence of the gene for malF protein, an inner membrane component of maltose transport system of Escherichia coli. J Biol Chem. 1984;259:10896–10903. [PubMed] [Google Scholar]
- 14.Fukuda K, Teramoto Y, Goto M, Sakamoto J, Mitsuiki S, Hayashida S. Specific inhibition by cyclodextrins of raw starch digestion by fungal glucoamylase. Biosci Biotechnol Biochem. 1992;56:556–559. doi: 10.1271/bbb.56.556. [DOI] [PubMed] [Google Scholar]
- 15.Galvin N M, Kelly C T, Fogarty W M. Purification and properties of the cyclomaltodextrinase of Bacillus sphaericus ATCC 7055. Appl Microbiol Biotechnol. 1994;42:46–50. [Google Scholar]
- 16.Goto M, Tanigawa K, Kanlayakrit W, Hayashida S. The mechanism of binding of glucoamylase I from Aspergillus awamori var. kawachi to cyclodextrins and raw starch. Biosci Biotechnol Biochem. 1994;58:49–54. doi: 10.1271/bbb.58.49. [DOI] [PubMed] [Google Scholar]
- 17.Greller G, Horlacher R, DiRuggiero J, Boos W. Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem. 1999;274:20259–20264. doi: 10.1074/jbc.274.29.20259. [DOI] [PubMed] [Google Scholar]
- 18.Horlacher R, Xavier K B, Santos H, DiRuggiero J, Kossmann M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi K, Ara K, Saeki K, Ozaki K, Kawai S, Ito S. Nucleotide sequence of the gene that encodes a neopullulanase from an alkalophilic Bacillus. Biosci Biotechnol Biochem. 1992;56:514–516. doi: 10.1271/bbb.56.514. [DOI] [PubMed] [Google Scholar]
- 20.Izumi M, Fujiwara S, Takagi M, Kanaya S, Imanaka T. Isolation and characterization of a second subunit of molecular chaperonin from Pyrococcus kodakaraensis KOD1: analysis of an ATPase-deficient mutant enzyme. Appl Environ Microbiol. 1999;65:1801–1805. doi: 10.1128/aem.65.4.1801-1805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeon S-J, Fujiwara S, Takagi M, Imanaka T. Pk-cdcA encodes a CDC48/VCP homolog in the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1: transcription and enzymatic characterization. Mol Gen Genet. 1999;262:559–567. doi: 10.1007/s004380051118. [DOI] [PubMed] [Google Scholar]
- 22.Kim T J, Shin J H, Oh J H, Kim M J, Lee S B, Ryu S, Kwon K, Kim J W, Choi E H, Robyt J F, Park K H. Analysis of the gene encoding cyclomaltodextrinase from alkalophilic Bacillus sp. I-5 and characterization of enzymatic properties. Arch Biochem Biophys. 1998;353:221–227. doi: 10.1006/abbi.1998.0639. [DOI] [PubMed] [Google Scholar]
- 23.Kitahata S, Taniguchi M, Beltran S D, Sugimoto T, Okada S. Purification and some properties of cyclodextrinase from Bacillus coagulans. Agric Biol Chem. 1983;47:1441–1447. [Google Scholar]
- 24.Krohn B M, Lindsay J A. Purification and characterization of a thermostable α-glucosidase from Bacillus subtilis high-temperature growth transformant. Curr Microbiol. 1991;22:273–278. doi: 10.1007/BF01577379. [DOI] [PubMed] [Google Scholar]
- 25.Krohn B M, Lindsay J A. Reclassification of a thermostable α glucosidase from Bacillus subtilis H-17 as a cyclomaltodextrinase. Enzyme Microb Technol. 1992;14:194–196. [Google Scholar]
- 26.Kuriki T, Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989;135:1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- 27.Kuriki T, Imanaka T. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87:557–565. doi: 10.1016/s1389-1723(99)80114-5. [DOI] [PubMed] [Google Scholar]
- 28.Loftsson T, Brewster M E. Pharmaceutical applications of cyclodextrins. 1. Drug solubilization and stabilization. J Pharm Sci. 1996;85:1017–1025. doi: 10.1021/js950534b. [DOI] [PubMed] [Google Scholar]
- 29.Makrides S C. Strategies for achieving high-level expression of genes in E. coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Matuschek M, Burchhardt G, Sahm K, Bahl H. Pullulanase of Thermoanaerobacterium thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene, composite structure of the enzyme, and a common model for its attachment to the cell surface. J Bacteriol. 1994;176:3295–3302. doi: 10.1128/jb.176.11.3295-3302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moens S, Vanderleyden J. Glycoproteins in prokaryotes. Arch Microbiol. 1997;168:169–175. doi: 10.1007/s002030050484. [DOI] [PubMed] [Google Scholar]
- 33.Nagashima T, Tada S, Kitamoto K, Gomi K, Kumagai C, Toda H. Site-directed mutagenesis of catalytic active-site residues of Taka-amylase A. Biosci Biotechnol Biochem. 1992;56:207–210. doi: 10.1271/bbb.56.207. [DOI] [PubMed] [Google Scholar]
- 34.Oguma T, Kikuchi M, Mizusawa K. Purification and some properties of cyclodextrin-hydrolyzing enzyme from Bacillus sphaericus. Biochim Biophys Acta. 1990;1036:1–5. doi: 10.1016/0304-4165(90)90205-b. [DOI] [PubMed] [Google Scholar]
- 35.Oguma T, Matsuyama A, Kikuchi M, Nakano E. Cloning and sequence analysis of the cyclomaltodextrinase gene from Bacillus sphaericus and expression in Escherichia coli cells. Appl Microbiol Biotechnol. 1993;39:197–203. doi: 10.1007/BF00228606. [DOI] [PubMed] [Google Scholar]
- 36.Pajatsch M, Gerhart M, Peist R, Horlacher R, Boos W, Böck A. The periplasmic cyclodextrin binding protein CymE from Klebsiella oxytoca and its role in maltodextrin and cyclodextrin transport. J Bacteriol. 1998;180:2630–2635. doi: 10.1128/jb.180.10.2630-2635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Podkovyrov S M, Burdette D, Zeikus J G. Analysis of the catalytic center of cyclomaltodextrinase from Thermoanaerobacter ethanolicus 39E. FEBS Lett. 1993;317:259–262. doi: 10.1016/0014-5793(93)81288-b. [DOI] [PubMed] [Google Scholar]
- 38.Podkovyrov S M, Zeikus J G. Structure of the gene encoding cyclomaltodextrinase from Clostridium thermohydrosulfuricum 39E and characterization of the enzyme purified from Escherichia coli. J Bacteriol. 1992;174:5400–5405. doi: 10.1128/jb.174.16.5400-5405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulley O A, French D. Studies on the schardinger dextrins. XI. The isolation of new schardinger dextrins. Biochem Biophys Res Commun. 1961;5:11–15. doi: 10.1016/0006-291x(61)90071-7. [DOI] [PubMed] [Google Scholar]
- 40.Saha B C, Zeikus J G. Characterization of thermostable cyclodextrinase from Clostridium thermohydrosulfuricum 39E. Appl Environ Microbiol. 1990;56:2941–2943. doi: 10.1128/aem.56.9.2941-2943.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saurin W, Köster W, Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 42.Semimaru T, Goto M, Furukawa K, Hayashida S. Functional analysis of the threonine and serine-rich Gp-I domain of glucoamylase I from Aspergillus awamori var. kawachi. Appl Environ Microbiol. 1995;61:2885–2890. doi: 10.1128/aem.61.8.2885-2890.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephen F A, Warren G, Webb M, Eugene W M, David J L. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Tachibana Y, Leclere M M, Fujiwara S, Takagi M, Imanaka T. Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J Ferment Bioeng. 1996;82:224–232. [Google Scholar]
- 45.Tachibana Y, Kuramura A, Shirasaka N, Suzuki Y, Yamamoto T, Fujiwara S, Takagi M, Imanaka T. Purification and characterization of an extremely thermostable cyclomaltodextrin glucanotransferase from a newly isolated hyperthermophilic archaeon, a Thermococus sp. Appl Environ Microbiol. 1999;65:1991–1997. doi: 10.1128/aem.65.5.1991-1997.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1→4)- and α-(1→6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- 47.Tam R, Saier M H. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto T, Fujiwara S, Tachibana Y, Takagi M, Fukui K, Imanaka T. Alteration of product specificity of cyclodextrin glucanotransferase from Termococcus sp. B1001 by site directed mutagenesis. J Biosci Bioeng. 2000;89:206–210. doi: 10.1016/s1389-1723(00)88740-x. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Yieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida A, Iwasaki Y, Akiba T, Horikoshi K. Purification and properties of cyclomaltodextrinase from alkalophilic Bacillus sp. J Ferment Bioeng. 1991;71:226–229. [Google Scholar]