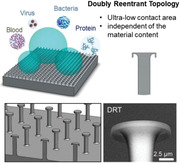
Abstract
The fomite transmission of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has drawn attention because of its highly contagious nature. Therefore, surfaces that can prevent coronavirus contamination are an urgent and unmet need during the coronavirus disease 2019 (COVID‐19) pandemic. Conventional surfaces are usually based on superhydrophobic or antiviral coatings. However, these coatings may be dysfunctional because of biofouling, which is the undesired adhesion of biomolecules. A superhydrophobic surface independent of the material content and coating agents may serve the purpose of antibiofouling and preventing viral transmission. Doubly reentrant topology (DRT) is a unique structure that can meet the need. This study demonstrates that the DRT surfaces possess a striking antibiofouling effect that can prevent viral contamination. This effect still exists even if the DRT surface is made of a hydrophilic material such as silicon oxide and copper. To the best of our knowledge, this work first demonstrates that fomite transmission of viruses may be prevented by minimizing the contact area between pathogens and surfaces even made of hydrophilic materials. Furthermore, the DRT geometry per se features excellent antibiofouling ability, which may shed light on the applications of pathogen elimination in alleviating the COVID‐19 pandemic.
Keywords: antibiofouling surfaces, COVID‐19 pandemic, doubly reentrant topology (DRT), superrepellency
The findings demonstrate that a unique fabricated doubly reentrant topology (DRT) structure carries remarkable superrepellent properties against biofouling of protein, blood, bacteria, and viruses. Moreover, this characteristic results from a highly minimized contact area and still exists even if the DRT surface is made of a hydrophilic material, such as silicon oxide.
1. Introduction
Since the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), developing strategies to prevent viral transmission has become an urgent and unmet need to alleviate the coronavirus disease 2019 (COVID‐19) pandemic. Although SARS‐CoV‐2 is mainly transmitted by exposure to infectious respiratory droplets and aerosol particles,[ 1 ] further studies have suggested prolonged survival of SARS‐CoV‐2 on inanimate objects.[ 2 ] This raises the issue of fomite transmission of SARS‐CoV‐2, which poses high biohazard risks to healthcare workers especially.[ 3 ] Therefore, special surfaces have been extensively developed to reduce viral transmission. Researchers have applied a variety of coating agents, including copper,[ 4 , 5 , 6 ] cationic polymers,[ 7 ] photodynamic polymers,[ 8 ] hydrogels,[ 9 ] nanoparticles,[ 4 , 5 , 10 ] graphene,[ 4 , 11 ] etc., on surfaces. Among them, copper has been reported to have an outstanding ability to kill micro‐organisms[ 12 ] and viruses such as SARS‐CoV‐2.[ 13 ] Thus, it has regained attention as an effective material for eliminating SARS‐CoV‐2.[ 14 ]
Besides, superhydrophobic surfaces are also considered an alternative strategy to reduce pathogen transmission. In the 1940s, Cassie and Baxter observed that water droplets roll off ducks’ feathers because of the feathers’ structure with densely packed particles.[ 15 ] Other natural examples include lotus leaves, Salvinia, mosquito eyes, and cicada wings.[ 16 ] Water droplets easily roll off these surfaces and can carry away dust or micro‐organisms, achieving a self‐cleaning effect and maintaining the function of surfaces. Subsequently, surfaces with randomly distributed or densely packed particles/pillars have been applied to various superhydrophobic systems.[ 17 ] The nonspecific adsorption or accumulation of undesired microbes and biomolecules on surfaces is also known as biofouling, which causes the degradation of materials and facilitates the transmission of pathogens.[ 18 ] This highly limits the applications of superhydrophobic surfaces. Current antibiofouling surfaces still mainly rely on specific coating agents with/without superhydrophobic properties. Degradation of these coatings will severely impact the surfaces’ antibiofouling performance. Thus, developing a novel antibiofouling surface independent of hydrophobic or antiviral coatings is essential to accomplish long‐term prevention of viral transmission. This kind of antibiofouling surface can be applied in different fields more broadly because it does not require specific coating agents.
Liu et al. designed a unique doubly reentrant topology (DRT) geometry with superrepellency. The DRT surface can repel liquids with extremely low surface energy (i.e., γ < 15 mJ m−2),[ 19 ] such as fluorinated solvents that can thoroughly wet almost all of the existing materials.[ 20 ] Without chemical modifications, the DRT surface, even made of hydrophilic materials like silicon dioxide (SiO2), could repel perfluorohexane (C6F14, also known as 3M Fluorinert FC‐72) with a surface energy of 10 mJ m−2.[ 20 ] These features make DRT a good candidate for antibiofouling surfaces independent of hydrophobic coatings. The DRT is a mushroom‐like structure, of which the second reentrant sidewall in nanoscale provides optimal upward force for liquid suspension. As long as the DRT geometry is intact, it can theoretically repel all kinds of liquids, even for extremely‐low‐energy liquids with an intrinsic contact angle (i.e., Young's angle) of almost zero. Unlike conventional methods of manufacturing superhydrophobic surfaces, a hydrophobic agent or coating is no longer required to fabricate DRT surfaces.[ 20 ] This novel design has been a breakthrough and provided new insights into the field of superhydrophobic materials. However, the antifouling potential of DRT surfaces against different biomolecules and their biomedical applications remain undetermined despite its reported superrepellency nature.
In this study, we validated the outstanding performance of the antibiofouling effect on DRT surfaces for the first time. The antibiofouling characteristics of DRT surfaces mainly result from the minimal contact area, whether hydrophobic or hydrophilic material. We also demonstrated that copper coating would not alter the DRT geometry or its superrepellent properties. As a result, the adherent bacteria or viruses on DRT surfaces were almost undetectable. Through the unique fabrication of the DRT geometry, surfaces can achieve ultimate blockage of biofouling and viral transmission, which is essential to alleviate the COVID‐19 pandemic.
2. Results and Discussion
2.1. Design and Characteristics of Different Surfaces
We used two other surfaces, i.e., plain surface (PS) and simple column surface (SCS), for comparison with the DRT surface (Figure 1 ). The PS represents characteristics of the pure material so that the intrinsic contact angle (i.e., Young's angle) and the biofouling ability of the material itself could be determined. The SCS, which is the most common design of a structured surface made of hydrophobic materials or coatings for enhancing liquid repellency, is a square array of circular posts. However, for many hydrophilic materials, including SiO2 and copper, used in this study, the SCS design would reduce the liquid repellency of the surface instead. For hydrophilic liquids, the downward surface tension on the SiO2‐SCS causes the structure to be immersed in the liquid, which further increases the contact area, as shown in Figure 1a; and Figure S1 (Supporting Information). The larger contact area leads to increased biofouling on the surface. In contrast, the upward force could be optimally achieved on the DRT surface to suspend liquids, enhancing the liquid repellency and decreasing the contact area, further reducing the fouling of biomolecules. The geometric parameters of DRT are shown in Figure S2 (Supporting Information). The fabrication process of the surfaces is described in the Experimental Section.
Figure 1.
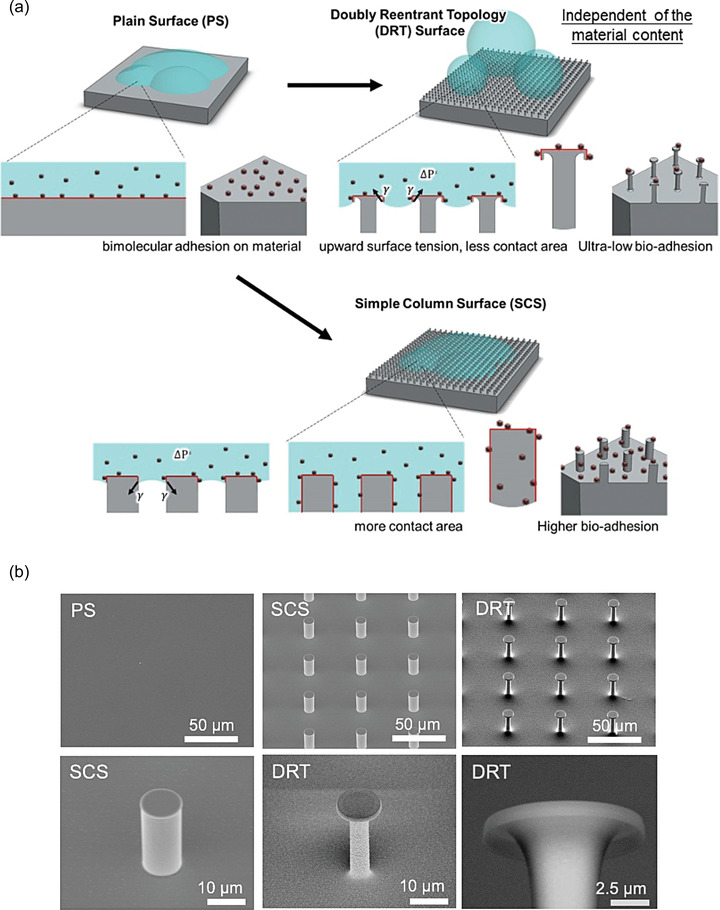
a) Illustration of liquids on the PS, SCS, and DRT surface. b) Scanning electron microscopy (SEM) imaging of different surfaces.
2.2. Contact Angle, Liquid Repellency, and Contact Area on Surfaces
The superhydrophobic property is characterized by the surface roughness of large contact angles (>150°) and low hysteresis angles.[ 21 ] The contact angle was measured through a syringe pump and a contact angle meter (Figure S3, Supporting Information). The test liquids included double‐distilled water (ddH2O), protein in phosphate‐buffered saline (PBS), blood, bacteria in PBS, and viruses in Dulbecco's modified eagle medium (DMEM). The photos of the static contact angle of all five solutions are shown in Figure 2a. For the SiO2‐PS, all five solutions’ intrinsic contact angles were 12.8° to 27.2°. For the SiO2‐SCS, the contact angles ranged from 7.1° to 40.3°. Finally, for the SiO2‐DRT surface, all the solutions were wholly suspended on surfaces with large contact angles ranging from 161.1° to 168.2°. As the contact angles increase, liquids are more likely to be suspended on a surface with less contact area. The contact angles should be at least 150° for liquids to be fully suspended on surfaces (i.e., superhydrophobic). Only the SiO2‐DRT surface can fulfill this requirement. We also tested the antibiofouling performance of the SiO2‐DRT surface at a tilted angle. As in Figure S4 (Supporting Information), the droplet of ddH2O and blood quickly rolled off the tilted SiO2‐DRT surface within 3.5 s.
Figure 2.
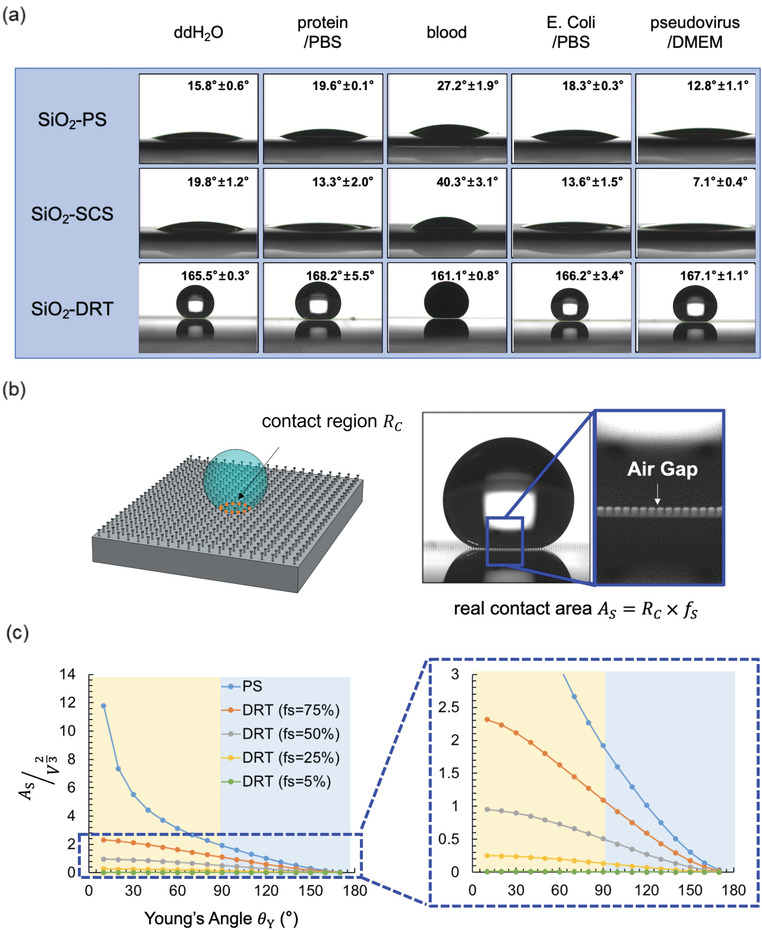
a) The contact angle of various biomolecules on different surfaces. b) The contact region of liquid on a surface. c) The ratio of the real contact area (i.e., liquid‐solid area) to the (liquid volume)2/3 versus intrinsic contact angle (i.e., Young's angle). Abbreviations: DRT, doubly reentrant topology; f s, the liquid–solid fraction; PS, plain surface.
The theoretical basis of liquid repellency on a smooth or structured surface is described below. Droplets can be categorized into the Wenzel‐droplets or the Cassie‐droplets, which completely wet surfaces or are suspended by surfaces.[ 22 ] A specially structured surface is required to reach the Cassie state for low‐energy fluids.[ 20 ] The contact angle of Cassie‐droplets can be evaluated by the Cassie–Baxter Equation (1)
| (1) |
| (2) |
where θ* is the contact angle on a surface, f s is the liquid–solid fraction, f g is the liquid–vapor fraction, θY is the intrinsic contact angle (also known as Young's angle), A s is the liquid–solid area, and A g is the liquid–vapor area. The SCS requires θY > 90° to suspend liquids. On the nanostructured DRT surface, the f s can be minimized to <6% so that the θ* can be successfully maximized to >150° even for θY ≈ 0°. The correlation between the contact angle and the contact region (in the macroscopic view, Figure 2b) can be described as
| (3) |
where R c is the contact region (in the macroscopic view), and V is the volume of the liquid. The detailed deduction is in the Supporting Information. The real contact area between the liquid and the surface (in the microscopic view), i.e., A s in Equation (2), equals R c × f s. By replacing R c with A s∕f s and θ* with θY according to Equation (1), we can get
| (4) |
The correlation between and the intrinsic contact angle (θY) can be illustrated in Figure 2c. Even if the intrinsic contact angle between a liquid and a surface is nearly zero, DRT surfaces can still exhibit repellency and increase their contact angle by decreasing f s. According to our deduction, DRT surfaces reduce the contact region (R c) and minimize the real contact area (A s). Especially for the condition of θY < 90°, the efficacy of reducing the As is even more prominent (Figure 2c).
2.3. Antibiofouling of Protein, Blood, Bacteria, and Viruses
The design of DRT could achieve superhydrophobicity by reaching remarkably decreased solid‐liquid fraction (f s) described in Equations (2). Thus, we tested the antibiofouling performance of the promising DRT surfaces since their applications have not been demonstrated.
We compared the adhesion capacity of protein (with fluorescence), blood (Figure 3a), bacteria, and viruses on the SiO2‐PS, SiO2‐SCS, and SiO2‐DRT surface. The surfaces’ fluorescence intensity could quantitatively measure the extent of fouling protein on the surfaces. Twenty microliters of the protein liquid at a 10 µg mL−1 concentration were placed on the tested surfaces for 5, 10, and 15 min. Then, we washed the tested surfaces with ddH2O and measured the fluorescence intensity on the surfaces using a fluorescence microscope. The fluorescence photography of surfaces is shown in Figure S5 (Supporting Information). For the SiO2‐PS, the fluorescence intensity increased over time and reached a plateau of 62.65 a.u. in ≈10 min. For the SiO2‐SCS, the fluorescence intensity was even higher than SiO2‐PS because the SCS structure was immersed in liquids (Figure 1a), resulting in increased contact area from the sidewalls of the columns. For the SiO2‐DRT surface, the fluorescence intensity was around 26.79 a.u. and did not significantly increase over time (Figure 3b). Overall, the fouling protein on the SiO2‐DRT surface was much less than that on the SiO2‐PS and SiO2‐SCS because of less fluorescence intensity.
Figure 3.
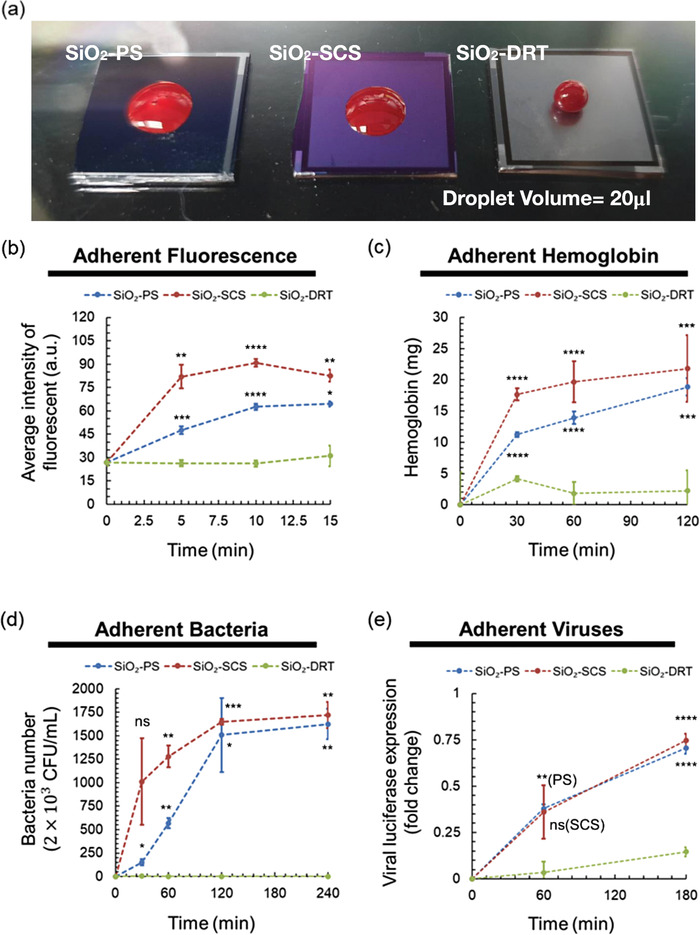
Antifouling effect of different surfaces. a) Blood was suspended on the DRT surface but not PS or SCS. Quantification of b) adherent protein (n = 3 at each point), c) blood (n = 5 at each point), d) bacteria (n = 3 at each point), and e) viruses (n = 3 at each point) on different surfaces. Data shown in are mean ± standard deviation (SD). Denotation: ns, p > 0.05; * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001 versus SiO2‐DRT alone.
We then compared fouling blood on different surfaces. Twenty microliters of blood were placed on the tested surfaces for 0.5, 1, and 2 h, respectively, and the adherent hemoglobin was quantified. Figure 3c shows the amount of hemoglobin from the adherent blood. The adherent hemoglobin was calculated by subtracting collected hemoglobin of washing PBS from the initial amount of loading hemoglobin, and the original raw data were in Figure S6 (Supporting Information). For the SiO2‐PS, the maximum adherent hemoglobin was about 18.86 mg. For the SiO2‐SCS, the curve was similar to that of the SiO2‐PS, and the maximum adherent hemoglobin was approximately 21.80 mg. For the SiO2‐DRT surface, the maximum adherent hemoglobin was only 2.26 mg. The fouling hemoglobin on the SiO2‐DRT surface was much less than that on the SiO2‐PS and SiO2‐SCS. We also tested if prolonged blood contact would alter the DRT geometry and its superrepellency. As in Figure S7 (Supporting Information), the SEM imaging demonstrated no alteration of the DRT geometry, and the contact angles were all >150° after 0‐, 12‐, 24‐, and 36‐h contact.
Next, we compared the antifouling ability of different surfaces against bacterial adhesion. Escherichia coli (E. coli) was utilized in the bacterial adhesion test. We assessed the bacteria fouling using the colony‐forming unit (CFU) method (Figure S8a, Supporting Information). Twenty microliters of E. coli in PBS solution were placed on the tested surfaces for 0.5, 1, 2, and 4 h (Figure 3d). The number of adherent bacteria on the SiO2‐DRT surface was minimal throughout the 4 h contact time. In contrast, the adherent bacteria were significantly more on the PS and SCS and increased over time. We also used the spectrophotometric method to measure the OD value of 600 nm wavelength to quantify adherent bacteria and obtained similar results (Figure S8b,c, Supporting Information). The fouling bacteria on the SiO2‐DRT surface was nearly undetectable and much less than those on the SiO2‐PS and SiO2‐SCS.
The virus adhesion test was conducted using SARS‐CoV‐2 pseudoviruses. SARS‐CoV‐2 pseudovirus that carries SARS‐CoV‐2 spike (S) protein is an artificial virus widely used to investigate SARS‐CoV‐2 virology and the viral entry into target cells.[ 23 ] The SARS‐CoV‐2 pseudovirus was obtained from the National RNAi Core of Academia Sinica in Taiwan and generated by the transfection of 293T cells using a lentiviral backbone plasmid encoding the fluorescent reporter protein and the S protein (Figure S9a, Supporting Information).[ 24 ] Twenty microliters of SARS‐CoV‐2 pseudovirus in DMEM solution (2 × 104 infectious units in 20 µL) were placed on the tested surfaces for 0.5, 1, 2, and 3 h, respectively. The viral fouling on the surfaces was examined by the expression of luciferase. Next, we subjected the adherent viruses to infecting ACE2‐overexpressing HEK293T cells that proliferated within the cells. The adherent viruses were estimated by subtraction of the collected viruses in washing PBS from the total loading amount of the viral solution. The viral RNA was extracted, and the viral amount was evaluated by measuring the luciferase expression using quantitative real‐time PCR (Figure S9b, Supporting Information). The adherent virus replicative activity indicated only slight fouling of SARS‐CoV‐2 pseudoviruses on the SiO2‐DRT surface. However, the viral fouling and replication increased over time on the SiO2‐PS and the SiO2‐SCS (Figure 3e).
2.4. SEM Images of Bacteria and Viruses on Different Surfaces
The SEM images of adherent bacteria on the SiO2‐PS, SiO2‐SCS, and SiO2‐DRT surface are shown in Figure 4 . E. coli were widely spread on the SiO2‐PS, and the putative morphology of E. coli could be observed. On the SiO2‐SCS, the adherent bacteria could be found on both the columns’ tops and sidewalls. As for the SiO2‐DRT surface, we could not find any adherent bacteria initially. Therefore, we concentrated the bacterial solution five hundred times and applied it to the SiO2‐DRT surface. The bacteria only adhered to the top of the SiO2‐DRT surface since the droplet only contacted the tops of the DRT structure. This mushroom‐like geometry could decrease bacterial fouling because of the minimal contact area.
Figure 4.
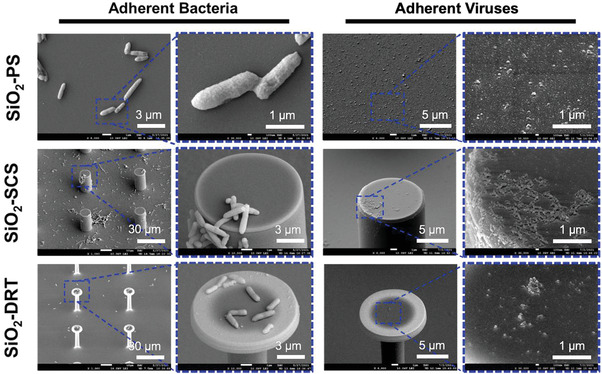
SEM images of bacteria and viruses on different surfaces. For the DRT surface, we concentrated the bacterial and viral solution of 500X and 100X, respectively, because the initial concentration could not yield visible pathogens on the DRT surface.
The SEM images of adherent viruses are also shown in Figure 4. To avoid confounding by the constituents and debris in the DMEM, we used ultracentrifuges to concentrate the viral particles. Next, we resuspended the pellet in PBS solution. The viral particles were spread over the surfaces after applying the SARS‐CoV‐2 pseudovirus/PBS solution. Fouling SARS‐CoV‐2 pseudoviruses could be observed on the SiO2‐PS and the tops and sidewalls of the SiO2‐SCS. However, we could not see the viral fouling on the SiO2‐DRT surface under the same condition. Thus, we applied a 100X concentrated SARS‐CoV‐2 pseudovirus/PBS solution onto this surface. Because of the minimal contact area, only a few amounts of viral particles could be observed on the tops of the SiO2‐DRT surface. Our findings indicated that the DRT geometry also effectively reduced the fouling of SARS‐CoV‐2 pseudoviruses.
2.5. Copper Coating for Reducing Transmission of Pathogens
Various coating agents with antibiofouling ability have been applied with superhydrophobic structures to improve performance.[ 22 , 25 ] Researchers have also developed several self‐healing materials.[ 18 ] The well‐defined sol–gel chemistry was utilized in the silica‐colloid‐based superhydrophobic coating, which could reduce the adsorption of common and highly pathogenic micro‐organisms, such as Staphylococcus aureus and Pseudomonas aeruginosa.[ 26 ] This makes silica‐colloid‐based coating a good application for biomedical devices. Antibiotics or other nature inhibitors could also be applied on the surfaces to kill micro‐organisms and viruses.[ 27 ] For example, Patir et al. impregnated crystal violet, a common disinfectant, on a slippery surface to prevent bacterial contamination.[ 28 ]
The use of copper and its alloys has been beneficial for reducing contact‐mediated infections. Although the detailed mechanisms of copper's antimicrobial/antiviral activity are not fully understood, copper ions released from the copper surface are believed to play a significant role in these actions.[ 29 ] The copper ions can cause depletion of sulfhydryls, such as cysteine and glutathione, leading to the oxidation of proteins and lipids as the following reactions
| (5) |
| (6) |
The resulting hydrogen peroxide subsequently participates in the Fenton reaction (7) and generates highly reactive hydroxyl radicals, leading to the biomolecular damage and inactivation of DNA or RNA[ 13 , 29 ]
| (7) |
Although the oxide formation on copper may occur after a long time, even in standard ambient conditions, Hans et al. showed that copper oxides, including CuO and Cu2O, still retained their outstanding antimicrobial properties.[ 30 ] Researchers also observed that the survival time of SARS‐CoV‐2 on copper is significantly reduced to less than 4 h compared to that on other materials such as steel (3–5 days), glass (4 days), wood (4‐5 days), and plastics (≥5 days).[ 31 ] In addition, compared with several existing materials, copper can be easily applied at a low cost. These advantages of copper have prompted us to coat it onto the DRT surface and examine if this combination (i.e., Cu‐DRT) would exhibit both the superrepellent ability and antibacterial/antiviral efficacy, leading to the scavenging and extinction of pathogens at the same time. We successfully coated copper on the DRT surface (i.e., Cu‐DRT, Figure 5a; and Figure S10a, Supporting Information) that did not alter the DRT geometry and its superrepellency. As expected, the Cu‐DRT showed large contact angles and low hysteresis angles (Figure S10b, Supporting Information).
Figure 5.
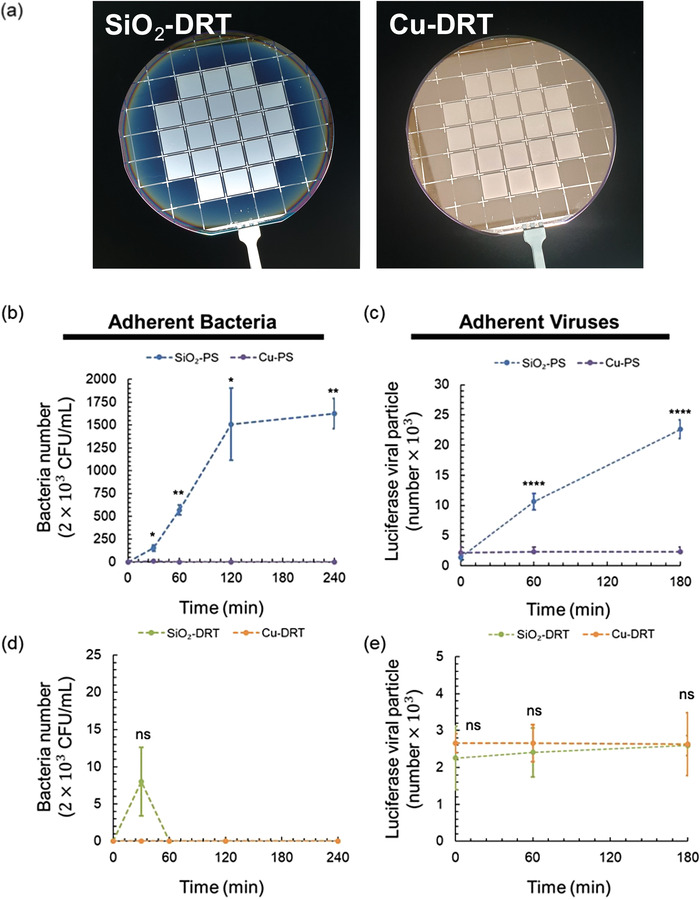
Comparison of adherent bacteria and viruses on SiO2 and Cu‐coated surfaces. a) The appearance of the SiO2‐DRT and Cu‐DRT surface. b) The adherent bacteria number on the SiO2‐PS and Cu‐PS (n = 3 at each point). c) The adherent viral particle number on the SiO2‐PS and Cu‐PS (n = 6 at each point). d) The adherent bacteria number on the SiO2‐DRT and Cu‐DRT surface (n = 3 at each point). e) The adherent viral particle number on the SiO2‐DRT and Cu‐DRT surface (n = 6 at each point). Data shown are mean ± SD. Denotation: ns, p > 0.05; * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001.
We compared the antifouling effect between the PS and the DRT surface with or without the copper coating. To examine the fouling of bacteria or viruses and the copper‐mediated scavenging potential against bacteria/viruses, the adherent bacteria or viral particles on the surfaces were detached by trypsinization. The bacterial fouling on surfaces with or without the copper coating was assessed using the CFU method. The viral fouling was evaluated by the luciferase reporter assay after their replication in host cells. For the Cu‐PS, no bacterial or viral activity was observed on the surface over time (Figure 5b,c). This result validated the outstanding antibacterial and antiviral performance of copper again. For the DRT surfaces, the adherent bacteria and viruses were consistently much less than those on PS (Figure 5d,e; and Figure S11, Supporting Information). The bacteria on the Cu‐DRT surface was undetectable over time, while some colonies were found on the SiO2‐DRT after 30 minutes of contact. The viral particle number was similar between the SiO2‐DRT and Cu‐DRT surface over time. By applying the unique DRT geometry, DRT surfaces can exhibit excellent antibiofouling ability against bacteria and viruses independent of materials and coatings. Cu coatings on DRT surfaces might help kill minimal residual bacteria and viruses to ensure the ultimate blockade of contamination.
Based on our results, the Cu‐DRT surface with the putative geometry could effectively reduce the fouling of bacteria and viruses. Even if there are sporadic residual bacteria and viruses on the Cu‐DRT surface, the copper coating will exhibit unusual activity that can ultimately extinguish the adherent bacteria and viruses. This kind of surface may have potential applications in biomedical devices or environments such as ceilings and walls of operation rooms to prevent bacterial and viral contamination.
3. Conclusion
We demonstrated that the DRT surfaces could significantly reduce biofouling and viral contamination. Furthermore, despite their constituents being made of hydrophilic materials such as SiO2 and copper, the DRT surfaces can still substantially minimize the contact area with liquid. Currently existing antibiofouling surfaces usually require hydrophobic or antiviral coatings such as copper,[ 4 , 5 , 6 ] cationic polymers,[ 7 ] photodynamic polymers,[ 8 ] hydrogels,[ 9 ] nanoparticles,[ 4 , 5 , 10 ] graphene,[ 4 , 11 ] etc. Unlike conventional surfaces, the antibiofouling and superrepellent abilities of DRT surfaces do not depend on their materials or coatings but on their unique geometry. This characteristic helps with the realization of various applications for DRT surfaces. The unique DRT geometry contributes to the antibiofouling performance of the DRT surfaces and results in a physical antibiofouling effect. Thus, coatings such as copper could be applied on DRT surfaces to potentially enhance antibiofouling ability. As described previously, copper is a promising candidate that can be coated on surfaces for contact‐killing pathogens, including SARS‐CoV‐2.[ 32 ] Although SARS‐CoV‐2 is transmitted predominantly by air‐borne routes, frequent environmental contamination of SARS‐CoV‐2 has been reported in patient care services such as emergency departments and intensive care units.[ 33 ] The possibility of SARS‐CoV‐2 fomite transmission cannot be entirely ruled out, and disinfection of frequently touched surfaces is still highly recommended.[ 34 ] The geometry‐based superrepellent properties exhibit unprecedented advantages for blocking the transmission of viruses or other micro‐organisms, which is crucial to alleviate the COVID‐19 pandemic.
4. Experimental Section
Test Liquid Preparation
To examine the antifouling potential of various fabricated surfaces, we prepared five tested liquids, including ddH2O, protein solution, a human blood sample, bacterial solution, and viral solution. The protein solution was prepared from the goat anti‐rabbit IgG (H + L) cross‐adsorbed secondary antibody, Alexa Fluor 647 (Thermo Fisher Scientific, MA), with a 2 mg mL−1 concentration. The blood sample was drawn from a healthy donor in an acid citrate dextrose (ACD) tube from BD Vacutainer. The peripheral blood from the healthy donor was collected following the Ethical and Institutional Review Board of Taipei Veterans General Hospital (ID No. 2020‐07‐036CC and 2020‐05‐004C). The informed written consent from the donor was obtained before the research. The bacterial solution was prepared by mixing Escherichia coli (E. coli) with PBS buffer. The E. coli was obtained from National RNAi Core from Academia Sinica in Taiwan. Once the bacteria stock solution (ECOS, Yeastern Biotech Co., Ltd., Taiwan) was thawed from a −80 °C freezer, we amplified it with TB buffer (Won‐Won Biotechnology Co., Ltd., Taiwan). We incubated it in the incubator shaker at 37 °C overnight. The bacteria mixture was collected after 18 h and centrifuged at 3000 rpm for 10 min. The pallet was resuspended in 1 mL PBS and was quantified by NanoPhotometer N60 (Implen Inc., CA) by regression analysis. The SARS‐CoV‐2 pseudovirus from National RNAi Core from Academia Sinica in Taiwan was used for the viral solution. The SARS‐CoV‐2 pseudovirus was packed using a lentiviral backbone, a plasmid‐encoding luciferase sequence, and a plasmid‐expressing spike (S) protein as the surface glycoprotein of the viral envelope. The minimal plasmid set of lentiviral protein (Tat, Gal‐Pol, and Rev) was used to assemble viral particles, and the CMV promoter was used to drive GFP expression.[ 24 ] The viral solution was prepared by mixing SARS‐CoV‐2 pseudoviruses with DMEM. We put 20 µL of solution (protein, hemoglobin, bacteria, and virus) on surfaces in each experiment.
Fabrication of the Plain Surface (PS), Simple Column Surface (SCS), Doubly Reentrant topology (DRT), and Copper‐Coated DRT (Cu‐DRT)
The fabrication process of the PS was only one step. First, one µm of silicon dioxide was deposited through thermal oxidation (Figure S12a, Supporting Information). The fabrication process of the SCS shown in Figure S12b (Supporting Information) comprises five main steps, starting from a silicon wafer. First, the 1‐µm silicon dioxide is deposited through thermal oxidation. Second, the photoresist layer was patterned through lithography. Third, the 1‐µm silicon oxide was anisotropically etched through the reactive‐ion etching (RIE). Fourth, the photoresist residue was removed through the piranha solution. Finally, the 0.3 µm silicon dioxide was grown through thermal oxidation.
The fabrication process of the DRT is shown in Figure S12c (Supporting Information), which started from a silicon wafer. Eight main steps were conducted as follows. First, the 1 µm silicon dioxide was deposited through thermal oxidation. Second, the photoresist layer was patterned through lithography. Third, the 1 µm silicon dioxide and the 1.5 µm silicon were anisotropically etched through RIE. Fourth, the photoresist residue was removed through the piranha solution. Fifth, the 0.3 µm silicon dioxide was deposited through thermal oxidation. Sixth, the 0.3 µm silicon dioxide was anisotropically etched through RIE. Seventh, the 25 µm silicon was anisotropically etched through RIE. And the 2 µm silicon was isotropically etched through RIE. Finally, 0.3 µm silicon dioxide was grown through thermal oxidation.
Evaluation of Protein Fouling on the Surfaces
The Goat anti‐Rabbit IgG (H+L) Cross‐Adsorbed Secondary Antibody, Alexa Fluor 647 (thermal fisher, 1:200), was used as the protein solution. The solution was spotted on the PS, SCS, and DRT surfaces at a volume of 20 µL. After different time intervals, PBS was used to wash the surface. The inverted microscope Olympus IX73 was used to capture the fluorescent pictures. The exposure time was 4 s. The adhesion of protein was quantified using the Image J software.
Evaluation of Hemoglobin Fouling on the Surfaces
The adhesion of blood was evaluated and calculated by the fixed amount of 20 µL of blood droplet minus recycled blood amount. The hemoglobin was detected using the BioVision Hemoglobin Colorimetric Assay (Catalog # K219, BioVision, Inc., CA). First, human blood (donated from a healthy donor) was spotted on surfaces at a volume of 20 µL. Then, after different time intervals, 180 µL PBS was added onto the surface to dilute and recycle the blood. Each collected blood sample was further added with an equal volume of ddH2O to adjust the concentration. The standard curve was constructed with the Hemoglobin Standard provided in the commercial kit, and 20 µL of each sample was added per well, followed by adding 180 µL Hemoglobin Detector to present the color complex. The absorbance at 575 nm was measured with SpectraMax M3 Multi‐Mode Microplate Reader (Molecular Devices, Sunnyvale, CA), and the concentration was calculated using the standard curve.
Evaluation of Bacterial Fouling on the Surfaces
The stocked DH5 alpha strain of E. coli was thawed at room temperature from −80 °C and cultured in LB broth (WonWon, Taiwan). After 18 h of shaking in 37 °C incubation, one mL of bacteria LB solution was taken and spun down (Kubota, Bunkyo‐ku, Tokyo) at 3000 rpm. Next, we discarded the supernatant and resolved the pellet in 500 µL PBS solution. The initial bacterial concentration was measured according to 600 nm wavelengths with the NanoPhotometer (IMPLEN, USA).
For the biofouling test, 20 µL of the prepared bacterial solution with fixed concentration was loaded onto various fabricated surfaces. At the end of incubation, 500 µL PBS washing solution was added to the remnant bacterial droplets. Then the bacteria‐containing washing buffer was collected and subjected to the following analyses and assays. The washing buffer was analyzed using NanoPhotometer to evaluate bacterial concentration at 600 nm wavelengths.
The 10cm Petri dishes (Falcon, USA) were covered with 10 mL of sterilized LB agar (Cyrusbioscience, Taiwan). The bacteria‐containing washing buffer from the fabricated surfaces was diluted using PBS buffer after gelation. Ten µL of the diluted washing buffer was resuspended in 100 µL LB buffer (the final dilution ratio = 1: 5 × 104) and spread onto the LB agar plates. After 18‐hour incubation under 37 °C, the adherent bacteria on the fabricated surfaces were detached using 200 µL trypsin and diluted in PBS buffer. Ten µL of the diluted buffer containing the detached bacteria was resuspended in 100 µL LB buffer (the final dilution ratio = 1: 2 × 104) and plated onto the LB agar plates.
Evaluation of SARS‐CoV‐2 Pseudovirus Fouling on the Surfaces
Considering ACE2 as the entry receptor for SARS‐CoV‐2 infection, we used ACE2‐overexpressing HEK293T cells for SARS‐CoV‐2 pseudovirus infection. The ACE2‐overexpressing HEK293T cells were seeded in 12‐well plates at a density of 2×105 cells per well and incubated for 24 h. PS, SCS, and DRT surfaces were cleaned according to the designated procedure, sterilized with 75% ethanol, and deposited in 12‐well plates. Subsequently, 20 µL SARS‐CoV‐2 pseudoviral solution (2 × 104 infectious unit µL−1) were loaded onto the surfaces and incubated. After incubation, 1 mL of fresh medium was used to resuspend and recycle the SARS‐CoV‐2 pseudovirus. The pseudovirus‐containing medium was supplemented with 8 µg mL−1 polybrene (Merck, Kenilworth, NJ), and then ACE2‐overexpressing HEK293T cells were shifted to this medium for SARS‐CoV‐2 pseudovirus infection. The 12‐well plate was centrifuged at 2000 rpm for 1 hour (Kubota, Bunkyo‐ku, Tokyo) to enhance infection and then replaced with a fresh medium for further incubation. After two days, cells were harvested, and the cell lysates were subjected to luciferase reporter assay (Catalog # E1500, Promega, Madison, WI) or quantitative real‐time PCR.
Quantitative Real‐Time PCR (qPCR)
The viral RNA was isolated from the SARS‐CoV‐2 pseudovirus using the Viral Nucleic Acid Extraction Kit II (Catalog No. VR100, Geneaid, Taipei, Taiwan) and stored in RNase‐free water. The cDNA was reverse‐transcribed with SuperScript III (Invitrogen, Waltham, MA) using GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Waltham, MA). The qPCR was performed on QuantStudio 3 real‐time PCR systems (Applied Biosystems, Life Technologies, Carlsbad USA) according to the reaction protocol: pre‐denaturation at 94 °C for 5 min, followed by 25–30 cycles of denaturation at 94 °C for 30 s, annealing at 58–62 °C for 30 s, and extension at 72 °C for 45 s. The mean Ct values were further taken to validate the virus RNA content on each chip. The primer sequences used for qPCR are qLuc_Forward: 5′‐TGA ACA TCA CGT ACG CGG AA‐3′; qLuc_Reverse 5'‐TCC GAT AAA TAA CGC GCC CA‐3'.
Luciferase Reporter Assay
Two days after infection by the viral samples from the surfaces, ACE2‐overexpressing HEK293T cells were harvested with the Passive Lysis Buffer for Promega luciferase assay (Catalog # E1500, Promega, Madison, WI, USA). The luciferase assay was conducted according to the manufacturer's instructions for use. The lysates were added with Luciferase Assay Reagent II (LAR II) to generate luminescence. For analysis, the luminescent photos were taken with UVP ChemStudio PLUS Imaging System (Analytik Jena AG, Thuringia, Germany). The luciferase signals were quantified using the Image J software.
Scanning Electron Microscopy (SEM) Photographing and Energy Dispersive Spectrometer (EDS)
After removing nonadherent bacteria or pseudoviruses by buffer washing, the remnant bacteria or pseudoviruses were fixed with paraformaldehyde for 30 min and dehydrated by increasing ethanol concentrations (70%, 80%, 90%, and 100%) at a 20 min interval. The surfaces with adherent micro‐organisms were air‐dried in a laminar flow hood overnight. Subsequently, the surface samples were coated with gold (JFC‐1200 Auto Fine Coater, Japan) and subjected to electron microscopy (JEOL JEM‐2000EXII, Japan). The elemental analysis of different surfaces in SEM was performed simultaneously using energy dispersive spectroscopy (EDS).
Statistical Analysis
The schematic diagrams were drawn using Solidworks and Microsoft PowerPoint. The line charts were plotted using Microsoft Excel. Data were expressed as mean ± standard deviation (SD). Statistical differences between 2 groups or among multiple groups were detected by an unpaired two‐tailed Welch's t‐test or an unpaired one‐way Brown–Forsythe and Welch's ANOVA with Dunnett's T3 multiple comparisons test (DRT surfaces as the control group), respectively, using Prism version 8 (Chicago, IL, USA). The criterion for significance was set as p < 0.05, and highly significant differences in the statistics were accepted, if p < 0.001. All data presented are representative of at least 3 independent experiments.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
M.‐S.L., Y.C., and P.‐C.T. contributed equally to this work. This study was funded by the Ministry of Science and Technology (MOST) (MOST 108‐2320‐B‐010 ‐019 ‐MY3; MOST 109‐2327‐B‐010‐007; MOST 109‐2327‐B‐016‐002; MOST111‐2321‐B‐A49‐007), Establishment of Regenerative Medicine and Cell Therapy Platform of Veterans General Hospital system (110VACS‐003) and Establishment of epidemic prevention and research platform in the veterans medical system for the control of emerging infectious diseases (110VACS‐007). Ministry of Health and Welfare (MOHW) (MOHW108‐TDU‐B‐211‐133001, MOHW109‐TDU‐B‐211‐114001), VGH, NTUH Joint Research Program (VN109‐16), VGH, TSGH, NDMC, AS Joint Research Program (VTA107‐V1‐5‐1, VTA108‐V1‐5‐3, VTA109‐V1‐4‐1), AS Clinical Research Center (IBMS‐CRC109‐P04), the “Cancer Progression Research Center, National Yang‐Ming University” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan, and the Ministry of Education through the SPROUT Project‐ Center For Intelligent Drug Systems and Smart Bio‐devices (IDS2B) of National Chiao Tung University Taiwan.
Lee M.‐S., Chien Y., Teng P.‐C., Huang X.‐Y., Lin Y.‐Y., Lin T.‐Y., Chou S.‐J., Chien C.‐S., Hsiao Y.‐J., Yang Y.‐P., Hsu W., Chiou S.‐H., Superrepellent Doubly Reentrant Geometry Promotes Antibiofouling and Prevention of Coronavirus Contamination. Adv. Mater. Technol. 2022, 2200387. 10.1002/admt.202200387
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Meyerowitz E. A., Richterman A., Gandhi R. T., Sax P. E., Ann. Intern. Med. 2021, 174, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goldman E., Lancet Infect. Dis. 2020, 20, 892.32628907 [Google Scholar]
- 3.a) Kanamori H., J. Infect. 2021, 82, e17; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kraay A. N. M., Hayashi M. A. L., Berendes D. M., Sobolik J. S., Leon J. S., Lopman B. A., Emergy Infect. Dis. 2021, 27, 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Das Jana I., Kumbhakar P., Banerjee S., Gowda C. C., Kedia N., Kuila S. K., Banerjee S., Das N. C., Das A. K., Manna I., Tiwary C. S., Mondal A., ACS Appl. Nano Mater. 2020, 4, 352. [Google Scholar]
- 5. Kumar S., Karmacharya M., Joshi S. R., Gulenko O., Park J., Kim G. H., Cho Y. K., Nano Lett. 2021, 21, 337. [DOI] [PubMed] [Google Scholar]
- 6. Khorsand Kheirabad A., Pan X., Long S., Kochovski Z., Zhou S., Lu Y., McInerney G., Yuan J., Nano Select. 2022, 3, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sinclair T. R., Robles D., Raza B., van den Hengel S., Rutjes S. A., de Roda Husman A. M., de Grooth J., de Vos W. M., Roesink H. D. W., Colloids Surf., A 2018, 551, 33. [Google Scholar]
- 8. Peddinti B. S. T., Scholle F., Ghiladi R. A., Spontak R. J., ACS Appl. Mater. Interfaces 2018, 10, 25955. [DOI] [PubMed] [Google Scholar]
- 9. Korogiannaki M., Jones L., Sheardown H., Langmuir 2019, 35, 950. [DOI] [PubMed] [Google Scholar]
- 10.a) de Dicastillo C. L., Correa M. G., Martínez F. B., Streitt C., Galotto M. J., Titanium Dioxide, IntechOpen, London: 2020; [Google Scholar]; b) Merkl P., Long S., McInerney G. M., Sotiriou G. A., Nanomaterials 2021, 11, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galante A. J., Yates K. A., Romanowski E. G., Shanks R. M. Q., Leu P. W., ACS Appl. Nano Mater. 2022, 5, 718. [Google Scholar]
- 12. Delgado K., Quijada R., Palma R., Palza H., Lett. Appl. Microbiol. 2011, 53, 50. [DOI] [PubMed] [Google Scholar]
- 13. Poggio C., Colombo M., Arciola C. R., Greggi T., Scribante A., Dagna A., Materials 2020, 13, 3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Hans M., Erbe A., Mathews S., Chen Y., Solioz M., Mücklich F., Langmuir 2013, 29, 16160; [DOI] [PubMed] [Google Scholar]; b) Culbertson E. M., Culotta V. C., Semin. Cell Dev. Biol. 2021, 115, 19. [DOI] [PubMed] [Google Scholar]
- 15. Cassie A. B. D., Baxter S., Nature 1945, 155, 21. [Google Scholar]
- 16. Liu M., Wang S., Jiang L., Nat. Rev. Mater. 2017, 2, 17036. [Google Scholar]
- 17. Feng L., Li S., Li Y., Li H., Zhang L., Zhai J., Song Y., Liu B., Jiang L., Zhu D., Adv. Mater. 2002, 14, 1857. [Google Scholar]
- 18. Liu S., Guo W., Adv. Funct. Mater. 2018, 28, 1800596. [Google Scholar]
- 19. Grigoryev A., Tokarev I., Kornev K. G., Luzinov I., Minko S., J. Am. Chem. Soc. 2012, 134, 12916. [DOI] [PubMed] [Google Scholar]
- 20. Liu T. L., Kim C. J., Science 2014, 346, 1096. [DOI] [PubMed] [Google Scholar]
- 21. Kreder M. J., Alvarenga J., Kim P., Aizenberg J., Nat. Rev. Mater. 2016, 1, 15003. [Google Scholar]
- 22. Yong J., Chen F., Yang Q., Huo J., Hou X., Chem. Soc. Rev. 2017, 46, 4168. [DOI] [PubMed] [Google Scholar]
- 23.a) Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F., Proc. Natl. Acad. Sci. USA 2020, 117, 11727; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Muik A., Wallisch A. K., Sanger B., Swanson K. A., Muhl J., Chen W., Cai H., Maurus D., Sarkar R., Tureci O., Dormitzer P. R., Sahin U., Science 2021, 371, 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Ahmad Mulyadi Lai H. I., Chou S. J., Chien Y., Tsai P. H., Chien C. S., Hsu C. C., Jheng Y. C., Wang M. L., Chiou S. H., Chou Y. B., Hwang D. K., Lin T. C., Chen S. J., Yang Y. P., Int. J. Mol. Sci. 2021, 22, 1320; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee C. Y., Huang C. H., Rastegari E., Rengganaten V., Liu P. C., Tsai P. H., Chin Y. F., Wu J. R., Chiou S. H., Teng Y. C., Lee C. W., Liang Y., Chen A. Y., Hsu S. C., Hung Y. J., Sun J. R., Chien C. S., Chien Y., Int. J. Mol. Sci. 2021, 22, 9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang P., Lin L., Zang D., Guo X., Liu M., Small 2017, 13, 1503334. [DOI] [PubMed] [Google Scholar]
- 26. Privett B. J., Youn J., Hong S. A., Lee J., Han J., Shin J. H., Schoenfisch M. H., Langmuir 2011, 27, 9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.a) Yu Q., Yuan Y., Wen J., Zhao X., Zhao S., Wang D., Li C., Wang X., Wang N., Adv. Sci. 2019, 6, 1900002; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun Z., Ostrikov K., Sustainable Mater. Technol. 2020, 25, e00203. [Google Scholar]
- 28. Patir A., Hwang G. B., Lourenco C., Nair S. P., Carmalt C. J., Parkin I. P., ACS Appl. Mater. Interfaces 2021, 13, 5478. [DOI] [PubMed] [Google Scholar]
- 29. Grass G., Rensing C., Solioz M., Appl. Environ. Microbiol. 2011, 77, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hans M., Erbe A., Mathews S., Chen Y., Solioz M., Mucklich F., Langmuir 2013, 29, 16160. [DOI] [PubMed] [Google Scholar]
- 31. van Doremalen N., Bushmaker T., Morris D. H., Holbrook M. G., Gamble A., Williamson B. N., Tamin A., Harcourt J. L., Thornburg N. J., Gerber S. I., Lloyd‐Smith J. O., de Wit E., Munster V. J., N. Engl. J. Med. 2020, 382, 1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.a) Carvalho A. P. A., Conte‐Junior C. A., Glob. Chall. 2021, 5, 2000115; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hosseini M., Chin A. W. H., Behzadinasab S., Poon L. L. M., Ducker W. A., ACS Appl. Mater. Interfaces 2021, 13, 5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye G., Lin H., Chen S., Wang S., Zeng Z., Wang W., Zhang S., Rebmann T., Li Y., Pan Z., Yang Z., Wang Y., Wang F., Qian Z., Wang X., J. Infect. 2020, 81, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewis D., Nature 2021, 590, 26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


