Abstract
Halohydrin dehalogenases, also known as haloalcohol dehalogenases or halohydrin hydrogen-halide lyases, catalyze the nucleophilic displacement of a halogen by a vicinal hydroxyl function in halohydrins to yield epoxides. Three novel bacterial genes encoding halohydrin dehalogenases were cloned and expressed in Escherichia coli, and the enzymes were shown to display remarkable differences in substrate specificity. The halohydrin dehalogenase of Agrobacterium radiobacter strain AD1, designated HheC, was purified to homogeneity. The kcat and Km values of this 28-kDa protein with 1,3-dichloro-2-propanol were 37 s−1 and 0.010 mM, respectively. A sequence homology search as well as secondary and tertiary structure predictions indicated that the halohydrin dehalogenases are structurally similar to proteins belonging to the family of short-chain dehydrogenases/reductases (SDRs). Moreover, catalytically important serine and tyrosine residues that are highly conserved in the SDR family are also present in HheC and other halohydrin dehalogenases. The third essential catalytic residue in the SDR family, a lysine, is replaced by an arginine in halohydrin dehalogenases. A site-directed mutagenesis study, with HheC as a model enzyme, supports a mechanism for halohydrin dehalogenases in which the conserved Tyr145 acts as a catalytic base and Ser132 is involved in substrate binding. The primary role of Arg149 may be lowering of the pKa of Tyr145, which abstracts a proton from the substrate hydroxyl group to increase its nucleophilicity for displacement of the neighboring halide. The proposed mechanism is fundamentally different from that of the well-studied hydrolytic dehalogenases, since it does not involve a covalent enzyme-substrate intermediate.
Halogenated aliphatics constitute an important class of environmental pollutants. Various microorganisms have evolved that are able to degrade some of these compounds and use them as sole sources of carbon and energy. Such organisms are of importance for bioremediation of polluted soil, groundwater, and wastewater. In most cases, specialized enzymes, designated dehalogenases, catalyze the cleavage of the carbon-halogen bonds, which is a key detoxification reaction. Hydrolytic dehalogenases have been studied extensively, which has resulted in detailed insight into the structure and mechanism of several enzymes of this class (8, 33). For other dehalogenases, structural and mechanistic data are hardly available.
Halohydrin dehalogenases, also referred to as haloalcohol dehalogenases or halohydrin hydrogen-halide lyases, occur in the degradation pathways of halopropanols and 1,2-dibromoethane, where they catalyze the nucleophilic displacement of a halogen by a vicinal hydroxyl group in halohydrins, yielding an epoxide, a proton, and a halide ion (7, 22, 30, 31). These enzymes also efficiently catalyze the reverse reaction, the halogenation of epoxides, and the dehalogenation of vicinal chlorocarbonyls to hydroxycarbonyls (2, 14, 31). The interest in halohydrin dehalogenases increased when it was found that the dehalogenation of halohydrins may proceed with high enantioselectivity, making these enzymes useful catalysts for the production of optically pure epoxides and halohydrins (1, 14–16).
In this study, we report the cloning of three bacterial halohydrin dehalogenase genes. Sequence analysis suggested that these proteins are similar to proteins of the short-chain dehydrogenase/reductase (SDR) family. The amino acids Ser132, Tyr145, and Arg149 were identified as the catalytic residues and are proposed to play a role highly similar to that of the conserved residues involved in the redox reaction catalyzed by the SDR family proteins.
MATERIALS AND METHODS
Materials.
All chemicals were purchased from Acros Chimica, Merck, Aldrich, or Sigma. Molecular biology enzymes were purchased from Boehringer. Oligonucleotide primers were supplied by Eurosequence BV, Groningen, The Netherlands.
Strains and growth conditions.
Agrobacterium radiobacter strain AD1 and Arthrobacter sp. strain AD2 were maintained on nutrient broth at 30°C. Mycobacterium sp. strain GP1 was maintained on selective plates with 1-propanol or 1,2-dibromoethane as a carbon source as described before (22). Escherichia coli strains HB101 (6), JM101 (35), and BL21(DE3) (28) were grown at 37°C in Luria-Bertani medium. For selection of recombinants carrying plasmids, the appropriate antibiotic was added at the following concentrations: 50 μg ml−1 for kanamycin, 50 μg ml−1 for ampicillin, and 12.5 μg ml−1 for tetracycline. E. coli BL21(DE3) grown at 17°C was used for high-level expression of recombinant halohydrin dehalogenase driven by the T7 promoter in plasmid pGEF+ (25).
Construction and screening of genomic libraries of halohydrin dehalogenase-producing bacteria.
Procedures for the isolation and manipulation of DNA were performed essentially as described by Sambrook et al. (24). The construction of a genomic library of Mycobacterium sp. strain GP1 was described before (22). The same procedure was used to construct a genomic library of A. radiobacter strain AD1 in the cosmid vector pLAFR3 (27). Restriction analysis of plasmids isolated from 16 transduced E. coli HB101 clones showed that all plasmids contained inserts. About 1,000 transductants were screened for halohydrin dehalogenase activity by monitoring halide production with 1,3-dichloro-2-propanol as a substrate as described before (22).
PCR and construction of expression vectors for halohydrin dehalogenase genes.
For overexpression, the halohydrin dehalogenase genes were amplified by PCR under conditions described before (32). The hheC gene was amplified from a plasmid preparation of pAD1–9B2 with the forward primer PFHheC (5′-ATCTGACCATGGCAACCGCAATTG-3′) and the reverse primer PRHheC (5′-CCCAACGGATCCACGAACCACGGC-3′) (NcoI and BamHI sites underlined, start codon shown in boldface, and substituted nucleotides shown in italics). The hheBGP1 gene starting with the second possible start codon was amplified from recombinant cosmid pGP1–4B5 (22) with the forward primer PF1HheBGP1 (5′-AAAACCATGGCTAACGGAAGACTGGCAGGC-3′) and reverse primer PRHheBGP1 (5′-GGGCTGTGGATCCTCTCAGGTGGCCCAGCCGCC-3′). The hheAAD2 gene was directly amplified from cells of Arthrobacter sp. strain AD2 with the forward primer PFHheAAD2 (5′-GAACCATGGTGATCGCCCTCGTGAC-3′) and the reverse primer PRHheAAD2 (5′-TGGCTATCTGCCCTAACCATGGCC-3′). The hheAAD2 gene was cloned behind the T7 promoter in the NcoI site, and hheC and hheBGP1 were cloned between the NcoI and BamHI sites of the expression vector pGEF+ (25).
Nucleotide sequencing.
Sequencing on double-stranded DNA was performed with the Amersham Thermo Sequenase cycle sequencing kit (Amersham BV, Roosendaal, The Netherlands), with 7-deaza-dGTP and 5′ Cy5 fluorescent primers. Sequence reactions were run on the Pharmacia ALF-Express automatic sequencing machine (Uppsala, Sweden) at the BioMedical Technology Centre (Academic Hospital, Groningen, The Netherlands). Both strands were sequenced to ensure accuracy.
Homology searches and structure prediction.
The BLAST program was used to screen DNA and protein databases for similar proteins. Multiple sequence alignments were made in ClustalW v1.7. Secondary structures were predicted with the programs SopM (10) and SSP (26). Tertiary structure modeling was done by comparative protein modeling to known three-dimensional structures of members of the SDR protein family with the program SWISS-MODEL (12) by using the structures of 2HSD, 1AE1, 2AE1, 1AHH, and 1A4U (Protein Data Base codes) as templates.
Overexpression and purification of the halohydrin dehalogenases.
Both wild-type and mutant halohydrin dehalogenase genes were expressed in E. coli BL21(DE3) as described before (23). A 1-liter culture of E. coli BL21(DE3) (pGEFHheC) was harvested by centrifugation for purification of HheC. Cells were resuspended in 10 mM Tris-sulfate buffer (pH 7.5), and all further steps were carried out at 0 to 4°C. Cells were washed twice with this buffer before they were resuspended in 10 mM Tris-sulfate buffer containing 1 mM EDTA and 1 mM β-mercaptoethanol (TEM buffer) or TEM buffer containing 3 mM NaN3 (TEMA buffer). After sonication, a crude extract was obtained by centrifugation (200,000 × g, 60 min).
The crude extract was applied to a Resource Q anion-exchange column (6 ml; Pharmacia Biotech, Uppsala, Sweden) that was connected to an LCC500 type fast protein liquid chromatography system (Pharmacia Biotech). The buffer system consisted of TEMA buffer (buffer A) and TEMA buffer with 0.45 M (NH4)2SO4 (buffer B). Retained protein was eluted with a three-step increasing linear gradient: 0 to 5% buffer B in 15 ml, 15 to 45% buffer B in 100 ml, and 45 to 100% buffer B in 35 ml (flow rate, 5 ml min−1; fraction volume, 5 ml). The dehalogenase eluted at 110 to 150 mM (NH4)2SO4, and active fractions were pooled.
Ammonium sulfate was added to a concentration of 1.5 M, and the protein was applied to a Resource Phe column (1 ml; Pharmacia Biotech). Retained protein was eluted with a 20-ml decreasing linear gradient of 1.5 to 0 M ammonium sulfate in buffer A (flow rate, 0.5 ml min−1; fraction volume, 0.5 ml). The dehalogenase eluted at 1.0 to 0.8 M (NH4)2SO4, and active fractions were pooled, yielding a pure protein as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was dialyzed against TEM buffer to remove azide, filtered with a 0.2-μm-pore-diameter filter, and stored at 4°C.
Construction of HheC mutants.
Site-directed mutagenesis was done by using the Quickchange site-directed mutagenesis kit of Stratagene (La Jolla, Calif.) with pGEFHheC as a template. Ser132 was mutated to Cys with the primer set PCODS132C (5′-GGACATATTATCTTTATTACCTGTGCAACGCCCTTCGGGCCTTGG-3′) and PNONS132C (5′-CCAAGGCCCGAAGGGCGTTGCACAGGTAATAAAGATAATATGTCC-3′) (codon of substituted residue shown in boldface, substituted nucleotides shown in italics). Ser132 was mutated to Ala with the primer set PCODS132A (5′-GGACATATTATCTTTATTACCGCTGCAACGCCCTTCGGGCCTTGG-3′) and PNONS132A (5′-CCAAGGCCCGAAGGGCGTTGCAGCGGTAATAAAGATAATATGTCC-3′). Tyr145 was mutated to Phe with the primer set PCODY145F (5′-CCTTGGAAGGAACTTTCTACCTTCACGTCAGCCCGAGCAGGTGC-3′) and PNONY145F (5′-GCACCTGCTCGGGCTGACGTGAAGGTAGAAAGTTCCTTCCAAGG-3′). Arg149 was mutated to Lys with the primer set PCODR149K (5′-CCTACACGTCAGCCAAAGCAGGTGCATGCACCTTGGC-3′) and PNONR149K (5′-GCCAAGGTGCATGCACCTGCTTTGGCTGACGTGTAGG-3′). Arg149 was mutated to Gln with the primer set PCODR149Q (5′-CCTACACGTCAGCCCAGGCAGGTGCATGCACCTTGGC-3′) and PNONR149Q (5′-GCCAAGGTGCATGCACCTGCCTGGGCTGACGTGTAGG-3′). Arg149 was mutated to Glu with the primer set PCODR149E (5′-CCTACACGTCAGCCGAAGCAGGTGCATGCACCTTGGC-3′) and PNONR149E (5′-GCCAAGGTGCATGCACCTGCTTCGGCTGACGTGTAGG-3′).
CD spectroscopy.
Circular dichroism (CD) spectra were recorded with an AVIV 62A DS spectrometer. The far-UV spectra of both the wild type and the HheC variants were recorded at 25°C from 190 to 250 nm with a 0.1-cm cuvette containing 0.1 mg of halohydrin dehalogenase (5 mM potassium phosphate [pH 7.5]) per ml.
Enzyme assays.
Halohydrin dehalogenase activities were assayed at 30°C in 50 mM Tris-sulfate buffer (pH 8.0) containing 5 mM substrate by monitoring halide liberation or epoxide formation or with a colorimetric assay using the chromogenic substrate p-nitro-2-bromo-1-phenylethanol. Protein concentration and halide liberation were determined as described before (30). Halohydrins and epoxides were analyzed by gas chromatography. Samples (1.5 ml) were extracted with 1.5 ml of diethyl ether containing 0.05 mM 1-chlorohexane, 1-bromohexane, or mesitylene as an internal standard. Extracts were analyzed by split injection of 2 or 4 μl on an HP5 column (model HP 19091J-413; Hewlett-Packard) with helium as a carrier gas. Separation of enantiomers of chiral compounds was carried out with chiral gas chromatography as described before (16).
Nucleotide sequence accession numbers.
The nucleotide sequences described in this article have been deposited in the EMBL/DDBJ/GenBank database under the following accession numbers: HheC, AF397296; HheAAD2, AF397297; HheBGP1, AY044094.
RESULTS
Cloning and sequence analysis of the halohydrin dehalogenase gene from Agrobacterium radiobacter strain AD1.
The gram-negative bacterium A. radiobacter strain AD1 was isolated for its capability to use chloropropanols and epichlorohydrin as growth substrates (30). The organism utilizes (R)-2,3-dichloro-1-propanol, whereas the (S)-enantiomer is not degraded (4). This is caused by the high enantioselectivity of the first enzyme in the degradation pathway, the halohydrin dehalogenase (16). To clone the gene encoding the halohydrin dehalogenase, a gene bank of strain AD1 was constructed in E. coli with the cosmid vector pLAFR3. Three active clones were identified when 1,000 cosmid clones were screened for dehalogenase activity with 1,3-dichloro-2-propanol. Sequence analysis of one of the clones, pAD1–9B2, showed the presence of a complete open reading frame of 765 nucleotides, designated hheC. The first 104 bp were identical to a fragment of a putative halohydrin dehalogenase gene that was located on a genomic DNA segment of A. radiobacter strain CFZ11 that carried the epoxide hydrolase gene (echA) (23). The deduced protein, HheC, has a predicted molecular mass of 27,954 Da and is highly similar to HalB of Agrobacterium tumefaciens (Table 1), for which no biochemical characterization has been published. The hheC gene was amplified by PCR, and the start codon was fused into the NcoI site of pGEF+. The resulting expression vector was designated pGEFHheC.
TABLE 1.
Pairwise sequence identities of halohydrin dehalogenases and SDRs
| Enzyme acronym and function | Organism | Source or reference | Identity (%)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HheA | HHeAAD2 | HheB | HheBGP1 | HheC | HalB | 1AE1 | 2AE1 | 1AHH | 2HSD | 1AU4 | |||
| Halohydrin dehalogenases | |||||||||||||
| Group A | |||||||||||||
| HheA | Corynebacterium sp. strain N-1074 | 36 | 100 | 97.1 | 24.1 | 24.0 | 32.0 | 29.2 | 24.3 | 24.1 | 25.5 | 24.2 | 19.9 |
| HheAAD2 | Arthrobacter sp. strain AD2 | This study | 100 | 24.5 | 24.2 | 32.4 | 29.6 | 24.6 | 24.1 | 25.1 | 23.5 | 20.2 | |
| Group B | |||||||||||||
| HheB | Corynebacterium sp. strain N-1074 | 36 | 100 | 98.4 | 25.4 | 23.9 | 20.9 | 18.7 | 22.2 | 24.2 | 18.0 | ||
| HheBGP1 | Mycobacterium sp. strain GP1 | This study | 100 | 25.8 | 23.9 | 20.2 | 18.7 | 21.5 | 23.8 | 18.2 | |||
| Group C | |||||||||||||
| HheC | A. radiobacter AD1 | This study | 100 | 80.7 | 22.2 | 23.1 | 25.5 | 23.7 | 20.4 | ||||
| HalB | A. tumefaciens | —a | 100 | 17.6 | 20.6 | 22.6 | 21.7 | 20.2 | |||||
| SDR oxidoreductases | |||||||||||||
| 1AE1 (tropinone reductase I) | Datura stramonium | 8 | 100 | 61.5 | 29.7 | 27.4 | 21.9 | ||||||
| 2AE1 (propinone reductase II) | D. stramonium | 8 | 100 | 32.4 | 27.6 | 20.4 | |||||||
| 1AHH (7α-hydroxysteroid dehydrogenase) | E. coli | 29 | 100 | 32.2 | 19.7 | ||||||||
| 2HSD (20-β-hydroxysteroid dehydrogenase) | Streptomyces exfoliatus | 11 | 100 | 22.6 | |||||||||
| 1A4U (alcohol dehydrogenase) | Drosophila lebanonensis | 3 | 100 | ||||||||||
—, unpublished (accession no. AAD34609).
Cloning and sequence analysis of the halohydrin dehalogenase gene of Arthrobacter sp. strain AD2.
Previously, Van den Wijngaard et al. (31) purified the halohydrin dehalogenase of the 3-chloro-1,2-propanediol-degrading Arthrobacter sp. strain AD2. Out of the 34 N-terminal residues, 32 were identical to the halohydrin dehalogenase HheA of Corynebacterium sp. strain N-1074 (36), which suggests that the halohydrin dehalogenase genes of both strains are highly similar. Therefore, primers designed for the hheA gene were used to amplify the halohydrin dehalogenase gene of strain AD2. Three independent clones were sequenced and found to have identical DNA sequences. The gene encodes a 244-amino-acid protein that was designated HheAAD2, since it was identical to HheA except for 7 amino acid substitutions (Table 1). The gene was fused into the start codon of pGEF+, and the resulting plasmid was designated pGEFHheAAD2.
Cloning and sequence analysis of the halohydrin dehalogenase gene of Mycobacterium sp. strain GP1.
At least two dehalogenases are produced by Mycobacterium sp. strain GP1 when 1,2-dibromoethane serves as a growth substrate (22). A haloalkane dehalogenase, encoded by dhaAf, catalyzes the hydrolytic dehalogenation of 1,2-dibromoethane to 2-bromoethanol, which is subsequently dehalogenated by a halohydrin dehalogenase to epoxyethane. No halohydrin dehalogenase-producing clones were identified when a gene library of strain GP1 in E. coli was screened for dehalogenase activity with 1,3-dichloro-2-propanol, suggesting that transcription or translation signals were not recognized in E. coli (22). However, when cosmid pGP1–4B5, which carries dhaAf, was transferred to Pseudomonas sp. strain GJ1 or Burkholderia cepacia G4, the resulting transconjugants rapidly dehalogenated 1,3-dichloro-2-propanol, indicating that active halohydrin dehalogenase was produced. Hence, pGP1–4B5 also harbors the gene encoding the halohydrin dehalogenase of strain GP1. Sequencing showed that the halohydrin dehalogenase gene is located 2,637 bp downstream of dhaAf and encodes a 235-amino-acid protein. The gene was identical to dehalogenase gene hheB of Corynebacterium sp. strain N-1074, except for 4 nucleotide substitutions that result in 4 amino acid substitutions in the encoded protein (Table 1). As in strain N-1074, a duplication of a 27-nucleotide region (36) has resulted in a duplicate set of AGGA ribosome binding sites, each located 12 nucleotides upstream of an ATG start codon. Hence, two polypeptides can be produced, which differ in the presence or absence of a MANGRKRE amino acid sequence extension at the N terminus, by translation starting from the first or the second start codon, respectively. Previously, Yu et al. (36) have shown that HheB is active as a tetramer and that all possible combinations of the two slightly different subunits occur. However, this subunit composition had little effect on substrate specificity, since enzyme variants exclusively composed of either the long or the short protein were kinetically indistinguishable. The hheBGP1 gene was amplified by PCR, and the second possible start codon was fused into the NcoI site of pGEF+ to yield the expression vector pGEFHheBGP1. In this way, a homotetrameric protein consisting only of the shorter 245-amino-acid polypeptide was produced.
Substrate range of halohydrin dehalogenases.
All proteins were expressed in a soluble and active form up to 15 to 25% of the total cellular protein content of E. coli BL21(DE3) as judged by SDS-PAGE. Further experiments focused on HheC, because it is enantioselective with various valuable halohydrins (16). Moreover, its sequence is very different from that of halohydrin dehalogenases that have previously been characterized. HheC was purified by anion-exchange chromatography followed by hydrophobic interaction chromatography as described in Materials and Methods. Purified HheC displayed optimal activity at around pH 8.0 to 9.0, and the temperature optimum for activity was 50°C.
Purified HheC and crude extracts of E. coli BL21(DE3) overexpressing HheAAD2 or HheBGP1 were used to study the substrate range of the three halohydrin dehalogenases (Table 2). Similar to the known HheA and HheB halohydrin dehalogenases, all three enzymes were active with all chlorinated and brominated C2 and C3 vicinal halohydrins tested. The only exception was HheAAD2, for which no activity was detected with 2,3-dichloro-1-propanol. The activities with brominated substrates were in most cases higher than with their chlorinated analogs. The substrate range of recombinant HheAAD2 was in agreement with that of the enzyme isolated from strain AD2 (31). The substrate range of HheBGP1 was similar to those reported for HheB (20, 21) (Table 2) and DehA (2). Halohydrin dehalogenase HheC displayed a relatively high level of activity with chloroacetone, which clearly distinguishes this enzyme from HheAAD2 and HheBGP1. The kinetics of 1,3-dichloro-2-propanol conversion by HheC were evaluated by measuring initial degradation rates at various substrate concentrations (Table 3). The enzyme followed Michaelis-Menten kinetics with a kcat value of 37 s−1 and a Km value of 0.010 mM. The specific activity of HheC with this substrate is similar to that reported for HheAAD2 (31). However, the Km value of HheC for 1,3-dichloro-2-propanol is 2 to 3 orders of magnitude lower than those of HheA, HheAAD2, and HheB (17, 21, 31).
TABLE 2.
Substrate range of halohydrin dehalogenases HheC, HheAAD2, and HheBGP1
| Substrate | % of activitya
|
||||
|---|---|---|---|---|---|
| HheC | HheAAD2 | HheA | HheBGP1 | HheB | |
| 1,3-Dichloro-2-propanol | 100 | 100 | 100 | 100 | 100 |
| (R,S)-2,3-Dichloro-1-propanol | 10.1 | <0.05 | 0.15 | 0.33 | 0.12 |
| 3-Chloro-1,2-propanediol | 18.8 | 9.30 | 35.1 | 1.76 | 0.98 |
| 2-Chloroethanol | 5.30 | 0.31 | 0.19 | 0.73 | 0.10 |
| 1,3-Dibromo-2-propanol | 80.2 | 6,680 | 20,000 | 76.3 | 166 |
| 2-Bromoethanol | 128 | 70.6 | 5.8 | 71.8 | 9.15 |
| Chloroacetone | 165 | 7.89 | 1.78 | ||
| (R)-2-Chloro-1-phenylethanol | 13.2 | 3.62 | 1.73 | ||
| (S)-2-Chloro-1-phenylethanol | 24.2 | 15.5 | 16.9 | ||
Activity was measured with purified HheC and crude extracts of E. coli BL21(DE3) overexpressing HheAAD2 or HheBGP1. With purified HheC, a rate of 100% corresponds to 20.7 μmol of halide released min−1 mg of protein−1 with 1,3-dichloro-2-propanol as the substrate. With crude extracts of E. coli BL21(DE3) overexpressing HheAAD2 or HheBGP1, these values were 0.480 and 8.52 μmol min−1 mg of protein−1, respectively. Relative activities of HheA and HheB were obtained from reference 21.
TABLE 3.
Steady-state kinetic parameters of purified HheC
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) |
|---|---|---|---|
| 1,3-Dichloro-2-propanol | 37 | 0.010 | 3.7 × 106 |
| (R,S)-2,3-Dichloro-1-propanol | 6.5 | 0.82 | 7.9 × 103 |
| 2-Chloroethanol | 3.9 | 0.84 | 4.6 × 103 |
| Chloroacetone | 235 | 2.4 | 9.8 × 104 |
| 2-Bromoethanol | 26.5 | <0.2 | >1.3 × 105 |
| (R)-2-Chloro-1-phenylethanol | 48.5 | 0.37 | 1.3 × 105 |
| (S)-2-Chloro-1-phenylethanol | 8.9 | 4.2 | 2.2 × 103 |
| (R)-p-Nitro-2-bromo-1-phenylethanol | 75 | <0.01 | >7.5 × 106 |
Recently, it was shown that aromatic halohydrins can also be dehalogenated by HheC (16). To explore the substrate specificity of HheC, the steady-state kinetic parameters of HheC with a range of aliphatic and aromatic substrates were determined (Table 3). It was found that HheC can efficiently convert aliphatic and aromatic halohydrins. Purified HheC has kcat and Km values for (R)-2-chloro-1-phenylethanol of 48.5 s−1 and 0.37 mM, respectively, and for the (S)-enantiomer, these values are 8.9 s−1 and 4.2 mM (Table 3). From these steady-state kinetic data, it can be calculated that the enantioselectivity (E-value) of HheC for this substrate is 73, which is in close agreement with the value calculated from kinetic resolutions by Lutje Spelberg et al. (16). We tested whether HheAAD2 and HheBGP1 could also convert 2-chloro-1-phenylethanol, and found that both enzymes were active with both enantiomers. The highest activities were observed with the (S)-enantiomer (Table 2). The enantioselectivity of 2-chloro-1-phenylethanol conversion by HheBGP1 was evaluated by means of kinetic resolution (data not shown), which showed that the (S)-enantiomer was preferentially converted with an E-value of 8. Hence, HheC and HheBGP1 exhibit opposite enantioselectivities for 2-chloro-1-phenylethanol conversion.
Sequence similarities of halohydrin dehalogenases with SDRs.
The pairwise sequence identities of the three halohydrin dehalogenases described in this paper are between 24.2 and 32.4% (Table 1). Interestingly, similarity searches with the amino acid sequences of HheAAD2, HheBGP1, and HheC in various protein and DNA databases showed that these enzymes are similar to proteins belonging to the family of SDR enzymes. Sequence similarities of up to 25.5% were observed with well-characterized SDR family members (Table 1). The SDR family constitutes a large number of proteins that catalyze oxidation or reduction reactions with NAD(H) or NADP(H) as a cofactor (13). They are active as dimers or tetramers, where each monomer consists of approximately 250 residues. Halohydrin dehalogenases consist of subunits with similar sizes, and HheA (17) and HheB (20) also appear to be active as tetramers.
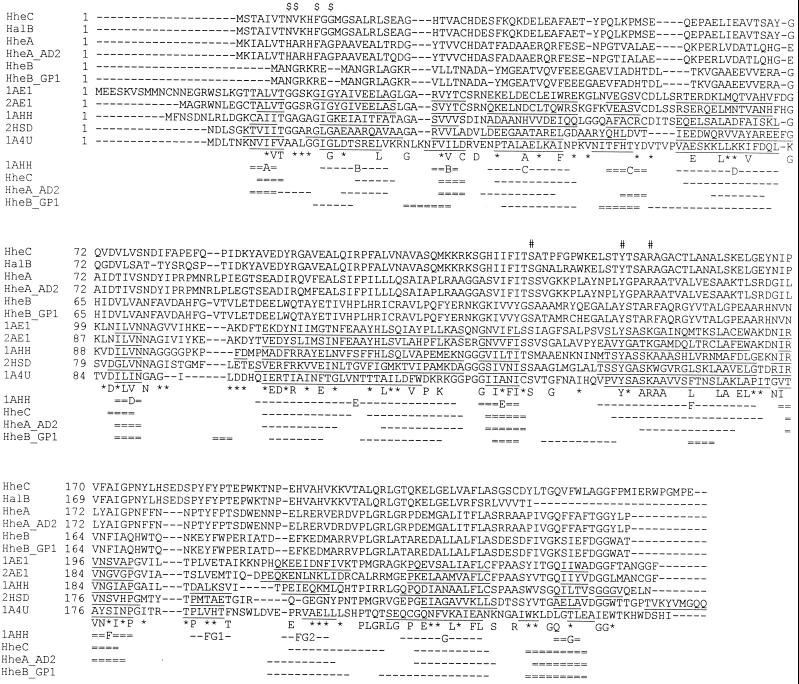
In Fig. 1 is depicted an alignment of six halohydrin dehalogenase sequences together with five members of the SDR protein family for which the three-dimensional structure is known. The highest degree of conservation between members of the SDR protein family is observed within the N-terminal part of these proteins, which contains a typical (G/A)-(G/A)-X-X-(G/A)-X-G fingerprint. This fingerprint is characteristic for the Rossman fold of the cofactor binding site (13), but it is not conserved in the halohydrin dehalogenases. The absence of a cofactor-binding motif in halohydrin dehalogenases is in agreement with the fact that the dehalogenation reaction that is catalyzed by these enzymes is not a redox reaction.
FIG. 1.
Sequence alignment of halohydrin dehalogenases with SDRs with known three-dimensional structures. The sequences were aligned by using the multiple alignment program ClustalW and are shown in decreasing sequence similarity to HheC. Amino acids that are identical in six or more sequences are depicted below the sequence. Positions at which six or more similar residues occur are indicated by an asterisk. The position of the G/A-G/A-X-X-G/A-X-G/A fingerprint typical of the Rossman fold in the SDR family is indicated by $ on top of the alignment. The proposed active site residues in halohydrin dehalogenases are indicated by #. The structural elements identified in three-dimensional structures of SDR family members are underlined. The first line below the sequence alignment shows the secondary structure elements and the nomenclature of 7α-hydroxysteroid dehydrogenase (29). β-Strands are indicated as double broken lines, and α-helices are indicated as single broken lines. The second, third, and fourth lines show the predicted secondary structure elements for HheC, HheAAD2, and HheBGP1, respectively. The nomenclature of the sequences is explained in Table 1.
The C-terminal part of members of the SDR family is involved in substrate binding and is much less conserved within the SDR protein family. However, it contains three highly conserved residues (a Ser, Tyr, and Lys) that play a critical role in catalysis (11, 13, 29). The conserved serine and tyrosine residues can also be identified in the halohydrin dehalogenase sequences (Ser132 and Tyr145 in HheC). However, halohydrin dehalogenases differ from members of the SDR family by the presence of an arginine residue at a position at which a lysine is conserved in the SDR family (Fig. 1). The implications of the partial conservation of the active site residues for the catalytic mechanism of halohydrin dehalogenases will be discussed below.
Conservation in several other sequence regions also suggests a structural relationship between SDR oxidoreductases and halohydrin dehalogenases. For example, Jörnvall et al. (13) have noticed that specific glycine and proline residues are typically conserved in more than 90% of the SDR family. Both residues are also present in four out of six halohydrin dehalogenases (Gly125 and Pro175 in HheC). Furthermore, two residues (Arg98 and Glu102 in HheC) that form salt bridges that stabilize dimers in some SDR family members (11, 29) are also present in the halohydrin dehalogenases HheA, HheAAD2, HheC, and HalB.
Structure determination by X-ray crystallography of several members of the SDR protein family has shown that the subunits share a common fold that is highly conserved despite the low overall sequence identity of 15 to 25% (3, 11, 18, 29). Each subunit has an α/β doubly wound structure in which seven parallel β-strands form a core β-sheet that is sandwiched between two arrays of three α-helices (3, 11, 18, 29). We compared the available X-ray structures with predicted secondary structure elements of HheC, HheAAD2, and HheBGP1 (Fig. 1). All β-strands and α-helices were predicted at the same position at which these elements are found in the structures of SDR family members. These results strongly suggest that the overall folding of halohydrin dehalogenases closely resembles that of SDR oxidoreductases. This includes a conserved topology for the cofactor binding domain, which consists of a βA-αB-βB-αC-βC-αD-βD-αE motif, despite the fact that the characteristic Rossman fold fingerprint for this motif in the SDR family (Fig. 1) (described above) is not conserved in the dehalogenases. With HheBGP1, some differences were observed, since the prediction of α-helix F is somewhat shifted and an extra β-strand is predicted between α-helix F and β-strand F (Fig. 1).
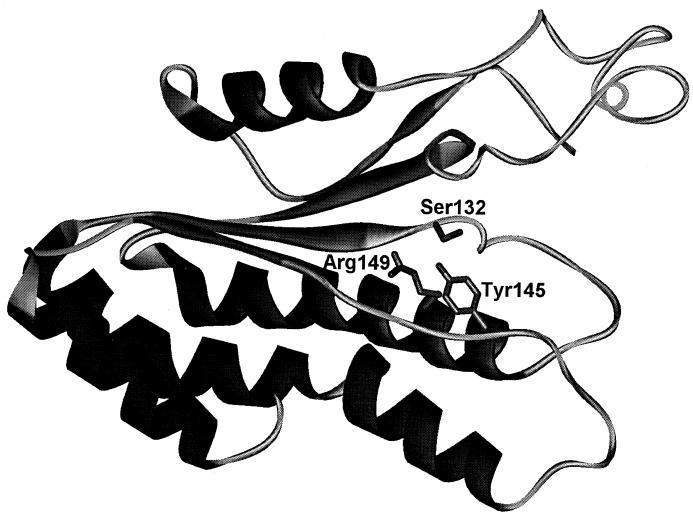
The sequence similarity and presence of conserved residues allowed us to generate a model of halohydrin dehalogenases by using the coordinates of SDR family members as a template (Fig. 2). The predicted model of α-helix D to the C terminus of HheC closely resembles that of the X-ray structure of 7α-hydroxysteroid dehydrogenase (29). Moreover, the proposed catalytic residues of HheC are found at the same positions as their counterparts in 7α-hydroxysteroid dehydrogenase. Similar results were obtained with HheAAD2, whereas with HheBGP1, a smaller portion of the protein could be predicted.
FIG. 2.
Proposed tertiary structure of halohydrin dehalogenase (HheC) of A. radiobacter AD1. A ribbon representation of residues Lys52 to Ile246 of HheC is shown. The active site residues that are critical for catalysis are indicated. Due to low sequence homology, the N-terminal fraction of HheC could not be predicted by using SDR proteins as a template.
Catalytic mechanism of halohydrin dehalogenases.
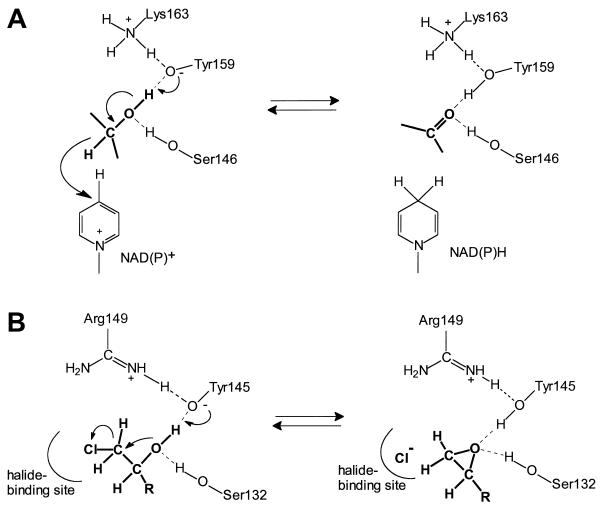
The strict conservation of Ser132, Tyr145, and Arg149 (HheC numbering) in halohydrin dehalogenases suggests a role of these residues similar to that of their counterparts in members of the SDR protein family. Studies of various SDR family members have shown that the active site residues play a role in catalysis, as depicted in Fig. 3A (3, 13). In this mechanism, the tyrosine acts as a catalytic base to extract a proton from the substrate, and simultaneously NAD(P)+ accepts a hydride from the carbon atom. Throughout the catalytic cycle, the conserved serine is hydrogen bonded with the substrate oxygen, thus facilitating proper positioning of the substrate and stabilization of the reaction intermediate (29). The conserved Lys149 may be involved in lowering the pKa of the tyrosine as well as positioning of the ribose moiety of the cofactor via interaction with its 2′- and 3′-hydroxyl groups (29).
FIG. 3.
Proposed reaction mechanism of halohydrin dehalogenase. (A) Reaction mechanism and active site residues of SDRs exemplified by 7α-hydroxysteroid dehydrogenase of E. coli (adapted from references 13 and 29). Interactions of the conserved lysine residue with the ribose moiety of the cofactor are not included. (B) Proposed reaction mechanism and active site residues of halohydrin dehalogenase exemplified by HheC of A. radiobacter (see text for explanation).
The role of the conserved Ser132, Tyr145, and Arg149 in the dehalogenation catalyzed by HheC was investigated by site-directed mutagenesis. All HheC mutants (Ser132Ala, Ser132Cys, Tyr145Phe, Arg149Lys, Arg149Glu, and Arg149Gln) were expressed as soluble proteins at room temperature at levels similar to that of overexpressed wild-type enzyme, as judged by SDS-PAGE (data not shown). Crude extracts of E. coli BL21(DE3) expressing mutant or wild-type enzyme were tested for activity with 2-bromoethanol and chloroacetone. While chloroacetone was shown to be a very good substrate for wild-type HheC, all mutant enzymes were inactive with this compound. The extracts containing the Ser132 and Tyr145 mutants also did not show significant activity with 2-bromoethanol, indicating the critical role of these residues. Only when Arg149 was replaced by a basic lysine residue, as is found in the SDR family at this position, was some activity still observed for 2-bromoethanol. For a more detailed kinetic analysis, several HheC mutants (Ser132Ala, Tyr145Phe, Arg149Lys, and Arg149Gln) were purified. Proper folding of these HheC mutants was verified by CD spectroscopy. All mutants displayed highly similar CD spectra compared with wild-type HheC, confirming proper folding of these HheC variants (data not shown). A kinetic analysis of the purified mutants revealed that the Ser132Ala mutant was totally inactive, while all other mutant enzymes analyzed displayed strongly decreased kcat/Km values for p-nitro-2-bromo-1-phenylethanol (Table 4).
TABLE 4.
Steady-state kinetic parameters of purified wild-type and mutant HheC with p-nitro-2-bromo-1-phenylethanol
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|
| Wild type | <0.010 | 75 | >7.5 × 106 |
| Tyr145Phe | 0.13 | 0.072 | 5.5 × 102 |
| Arg149Lys | 0.056 | 0.33 | 5.9 × 103 |
| Arg149Gln | >0.40 | >0.15 | 3.8 × 102 |
These data support the proposed mechanism shown in Fig. 3B in which the residues Ser132, Tyr145, and Arg149 have a similar function to their counterparts in SDR family members. In the first step, Tyr145 abstracts a proton from the hydroxyl group in the substrate, and concomitantly the substrate oxygen performs a nucleophilic attack on the neighboring halogen-substituted carbon atom, resulting in ring closure and liberation of halide. The conserved Ser132 could be hydrogen bonded, thus ensuring proper positioning of the substrate as in the SDR family, whereas Arg149 may play a critical role by lowering the pKa of Tyr145.
DISCUSSION
Three genes encoding different halohydrin dehalogenases were cloned, sequenced, and overexpressed in E. coli. The enzymes have significant sequence similarity to proteins of the SDR family, and several residues that are highly conserved in SDR enzymes are also present in halohydrin dehalogenases. The typical catalytic triad (Ser-Tyr-Lys) observed in SDR enzymes (11, 13, 29) was partially conserved in halohydrin dehalogenases. In the halohydrin dehalogenases, an arginine is observed at the position at which a lysine is found in the SDR family. However, replacement of the catalytic lysine by an arginine is not unique for halohydrin dehalogenases, since it has also occasionally been observed in other members of the SDR family (e.g., in sorbitol-6-phosphate 2-dehydrogenase of Streptococcus mutans [5] and 1-cyclohexenylcarbonyl coenzyme A reductase from Streptomyces collinus [34]). The observed sequence similarities between halohydrin dehalogenases and proteins of the SDR family strongly point to a common evolutionary origin. Furthermore, kinetic analysis of several HheC mutants revealed that the conserved Ser-Tyr-Arg triad in halohydrin dehalogenases is crucial for efficient catalysis. These results indicate that the dehalogenation reaction catalyzed by halohydrin dehalogenases also shares interesting similarities with the redox reaction that is catalyzed by members of the SDR family of proteins (Fig. 3). These data also suggest that the dehalogenation reaction catalyzed by halohydrin dehalogenases is fundamentally different from that of hydrolytic dehalogenases, since it does not involve a covalent intermediate.
Including the three new sequences presented in this paper, six complete sequences of halohydrin dehalogenases are now available in the databases. Based on sequence identities, they can be divided into three distinct groups that share 23.9 to 32.4% sequence identity (Table 1). Each group is represented by two sequences, but there are some biochemical and immunological data that allow a preliminary assignment of enzymes of which no complete sequence is known. Group A halohydrin dehalogenases consist of the closely related halohydrin dehalogenases HheA and HheAAD2. The dehalogenase of the unidentified chloropropanol-utilizing strain AD3 and the DehC produced by Arthrobacter erithrii H10a may also belong to this group (2, 31). Group B enzymes are constituted of HheB and HheBGP1. The DehA of A. erithrii H10a probably also belongs to this group, since sequencing of peptides of a tryptic digest showed 100% homology to the sequence of HheB (2). Finally, group C halohydrin dehalogenases consist of HheC and halohydrin dehalogenase B of Agrobacterium tumefaciens. A recently isolated halohydrin dehalogenase from an Agrobacterium strain probably also belongs to this group, because it exhibits a similar substrate specificity (9).
The halohydrin dehalogenases HheAAD2, HheBGP1, and HheC were active with C2, C3, and aromatic halohydrins and exhibit some remarkable differences in substrate range. The enantioselectivity of halohydrin dehalogenases can be useful for the preparation of optically active compounds. Previously, the epoxide hydrolase of Agrobacterium radiobacter sp. strain AD1 has been cloned, overexpressed, and used for the preparation of optically pure epoxides (15, 23). HheC is the second enzyme in the degradation pathway of chloropropanols in this organism that exhibits high enantioselectivity to chiral substrates. Some promising examples of the application of HheC (16) and other halohydrin dehalogenases for the preparation of optically active aliphatic or aromatic epoxides or halohydrins have been described (1, 14). Interestingly, HheBGP1 exhibits the opposite enantioselectivity for 2-chloro-1-phenylethanol. Thus, several members of the SDR family and the halohydrin dehalogenases display a high enantioselectivity to many chiral substrates. The stereoselectivity may be determined by only a small number of residues that constitute the substrate binding site (18). These residues are located in the substrate binding loop or in the loops linking β-strand D with α-helix E and β-strand E with α-helix F. In two highly identical tropinone reductases (Table 1 and Fig. 1) that have opposite enantioselectivities, stereoselectivity is determined by only 5 amino acid residues (19).
The present study provides insight in the structure and catalytic mechanism of halohydrin dehalogenases, but some important mechanistic questions remain. The reverse reaction, the halogenation of epoxides, is also efficiently catalyzed by these enzymes (1, 31). This probably requires the activation and proper positioning of the halide ion that acts as a nucleophile. From sequence alignments, we were not able to identify a halide-binding site, but it may be formed by residues located at the position at which the cofactor is bound in the SDR family. Tryptophan or arginine residues are involved in halide binding in hydrolytic dehalogenases. Three of the four tryptophan residues of HheC are located in the part of the protein that is predicted in the structure model in Fig. 3. Comparisons with structures of proteins belonging to the SDR family suggest these three residues could interact with the substrate. Future experiments will aim at identifying a halide binding site and elucidating the kinetic properties.
ACKNOWLEDGMENTS
This research was financially supported by the EU Environment and Climate Program (grant ENV5-CT95-0086) and the Innovation Oriented Research Program of the Dutch Ministry of Economic Affairs.
REFERENCES
- 1.Assis H M S, Bull A T, Hardman D J. Synthesis of chiral epihalohydrins using haloalcohol dehalogenase A from Arthrobacter erithriiH10a. Enzyme Microb Technol. 1998;22:545–551. doi: 10.1016/s0141-0229(97)00254-8. [DOI] [PubMed] [Google Scholar]
- 2.Assis H M S, Sallis P J, Bull A T, Hardman D J. Biochemical characterization of a haloalcohol dehalogenase from Arthrobacter erithriiH10a. Enzyme Microb Technol. 1998;22:568–574. doi: 10.1016/s0141-0229(97)00254-8. [DOI] [PubMed] [Google Scholar]
- 3.Benach J, Atrian S, Gonzalez-Duarte R, Ladenstein R. The refined crystal structure of Drosophila lebanonensisalcohol dehydrogenase at 1.9 A resolution. J Mol Biol. 1998;282:383–399. doi: 10.1006/jmbi.1998.2015. [DOI] [PubMed] [Google Scholar]
- 4.Bosma T, Kruizinga E, de Bruin E J, Poelarends G J, Janssen D B. Utilization of trihalogenated propanes by Agrobacterium radiobacter AD1 through heterologous expression of the haloalkane dehalogenase from Rhodococcussp. strain m15-3. Appl Environ Microbiol. 1999;65:4575–4581. doi: 10.1128/aem.65.10.4575-4581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd D A, Thevenot T, Gumbmann M, Honeyman A L, Hamilton I R. Identification of the operon for the sorbitol (glucitol) phosphoenolpyruvate:sugar phosphotransferase system in Streptococcus mutans. Infect Immun. 2000;68:925–930. doi: 10.1128/iai.68.2.925-930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Castro C E, Bartnicki E W. Epoxidation of halohydrins, epoxide opening and transhalogenation by a Flavobacteriumsp. Biochemistry. 1968;7:3213–3218. doi: 10.1021/bi00849a025. [DOI] [PubMed] [Google Scholar]
- 8.Copley S D. Microbial dehalogenases: enzymes recruited to convert xenobiotic substrates. Curr Opin Chem Biol. 1998;2:613–617. doi: 10.1016/s1367-5931(98)80092-6. [DOI] [PubMed] [Google Scholar]
- 9.Effendi A J, Greenaway S D, Dancer B N. Isolation and characterization of 2,3-dichloro-1-propanol-degrading rhizobia. Appl Environ Microbiol. 2000;66:2882–2887. doi: 10.1128/aem.66.7.2882-2887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh D, Weeks C M, Grochulski P, Duax W L, Erman M, Rimsay R L, Orr J C. Three-dimensional structure of holo 3α,20β-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci USA. 1991;88:10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 13.Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 14.Kasai N, Suzuki T, Furukawa Y. Chiral C3 epoxides and halohydrins: their preparation and synthetic application. J Mol Catal. 1998;4:237–252. [Google Scholar]
- 15.Lutje Spelberg J H, Rink R, Kellogg R M, Janssen D B. Enantioselectivity of a recombinant epoxide hydrolase from Agrobacterium radiobacter. Tetrahedron Asymmetry. 1998;9:459–466. [Google Scholar]
- 16.Lutje Spelberg J H, van Hylckama Vlieg J E T, Bosma T, Kellogg R M, Janssen D B. A tandem enzyme reaction to produce optically active halohydrins, epoxides and diols. Tetrahedron Asymmetry. 1999;10:2863–2870. [Google Scholar]
- 17.Nagasawa T, Nakamura T, Yu F, Watanabe I, Yamada H. Purification and characterization of halohydrin hydrogen-halide lyase from recombinant Escherichia coli containing the gene from a Corynebacteriumsp. Appl Microbiol Biotechnol. 1992;36:478–484. [Google Scholar]
- 18.Nakajima K, Yamashita A, Akama H, Nakatsu T, Kato H, Hashimoto T, Oda J, Yamada Y. Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold. Proc Natl Acad Sci USA. 1998;95:4876–4881. doi: 10.1073/pnas.95.9.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima K, Kato H, Oda J, Yamada Y, Hashimoto T. Site-directed mutagenesis of putative substrate-binding residues reveals a mechanism controlling the different stereospecificities of two tropinone reductases. J Biol Chem. 1999;274:16563–16568. doi: 10.1074/jbc.274.23.16563. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Nagasawa T, Yu F, Watanabe I, Yamada H. Characterization of a novel enantioselective halohydrin-hydrogen-halide lyase. Appl Environ Microbiol. 1994;60:1297–1301. doi: 10.1128/aem.60.4.1297-1301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, Nagasawa T, Yu F, Watanabe I, Yamada H. Resolution and some properties of enzymes involved in enantioselective transformation of 1,3-dichloro-2-propanol to (R)-3-chloro-1,2-propanediol by Corynebacteriumsp. strain N-1074. J Bacteriol. 1992;174:7613–7619. doi: 10.1128/jb.174.23.7613-7619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poelarends G J, van Hylckama Vlieg J E T, Marchesi J R, Freitas dos Santos L M, Janssen D B. Degradation of 1,2-dibromoethane by Mycobacteriumsp. strain GP1. J Bacteriol. 1999;181:2050–2058. doi: 10.1128/jb.181.7.2050-2058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of epoxide hydrolase from Agrobacterium radiobacterAD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Schanstra J P, Rink R, Pries F, Janssen D B. Construction of an expression and site-directed mutagenesis system of haloalkane dehalogenase in Escherichia coli. Protein Expr Purif. 1993;4:479–489. doi: 10.1006/prep.1993.1063. [DOI] [PubMed] [Google Scholar]
- 26.Solovyev V V, Salamov A A. Secondary structure prediction based on discriminant analysis. In: Kolchanov N A, Lim H A, editors. Computer analysis of genetic macromolecules. Singapore: World Scientific; 1994. pp. 352–364. [Google Scholar]
- 27.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringaepv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. Crystal structures of the binary and ternary complexes of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry. 1996;35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 30.Van den Wijngaard A J, Janssen D B, Witholt B. Degradation of epichlorohydrin and halohydrins by bacteria isolated from freshwater sediment. J Gen Microbiol. 1989;135:2199–2208. [Google Scholar]
- 31.Van den Wijngaard A J, Reuvekamp P T W, Janssen D B. Purification and characterization of haloalcohol dehalogenase from Arthrobactersp. strain AD2. J Bacteriol. 1991;173:124–129. doi: 10.1128/jb.173.1.124-129.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Hylckama Vlieg J E T, Leemhuis H, Janssen D B. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcussp. strain AD45. J Bacteriol. 2000;182:1956–1963. doi: 10.1128/jb.182.7.1956-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verschueren K H, Seljee F, Rozeboom H J, Kalk K H, Dijkstra B W. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- 34.Wang P, Denoya C D, Morgenstern M R, Skinner D D, Wallace K K, Digate R, Patton S, Banavali N, Schuler G, Speedie M K, Reynolds K A. Cloning and characterization of the gene encoding 1-cyclohexenylcarbonyl coenzyme A reductase from Streptomyces collinus. J Bacteriol. 1996;178:6873–6881. doi: 10.1128/jb.178.23.6873-6881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 36.Yu F, Nakamura T, Mizunashi W, Watanabe I. Cloning of two halohydrin hydrogen-halide-lyase genes of Corynebacteriumsp. strain N-1074 and structural comparison of the genes and gene products. Biosci Biotechnol Biochem. 1994;58:1451–1457. doi: 10.1271/bbb.58.1451. [DOI] [PubMed] [Google Scholar]