Abstract
Our previous studies revealed that lysine is synthesized through α-aminoadipate in an extremely thermophilic bacterium, Thermus thermophilus HB27. Sequence analysis of a gene cluster involved in the lysine biosynthesis of this microorganism suggested that the conversion from α-aminoadipate to lysine proceeds in a way similar to that of arginine biosynthesis. In the present study, we cloned an argD homolog of T. thermophilus HB27 which was not included in the previously cloned lysine biosynthetic gene cluster and determined the nucleotide sequence. A knockout of the argD-like gene, now termed lysJ, in T. thermophilus HB27 showed that this gene is essential for lysine biosynthesis in this bacterium. The lysJ gene was cloned into a plasmid and overexpressed in Escherichia coli, and the LysJ protein was purified to homogeneity. When the catalytic activity of LysJ was analyzed in a reverse reaction in the putative pathway, LysJ was found to transfer the ɛ-amino group of N2-acetyllysine, a putative intermediate in lysine biosynthesis, to 2-oxoglutarate. When N2-acetylornithine, a substrate for arginine biosynthesis, was used as the substrate for the reaction, LysJ transferred the δ-amino group of N2-acetylornithine to 2-oxoglutarate 16 times more efficiently than when N2-acetyllysine was the amino donor. All these results suggest that lysine biosynthesis in T. thermophilus HB27 is functionally and evolutionarily related to arginine biosynthesis.
Two pathways have been described for lysine biosynthesis in prokaryotes and eukaryotes: the diaminopimelate (DAP) pathway and the α-aminoadipate (AAA) pathway (Fig. 1). In the former pathway, found in most bacteria and plants, lysine is synthesized from aspartate via DAP, while in the latter pathway, found in yeast (2) and fungi (11, 35), lysine is synthesized from 2-oxoglutarate through AAA. Recently, however, we found that an extreme thermophile, Thermus thermophilus HB27, which belongs to the domain Bacteria, synthesized lysine through the AAA pathway (16). We also cloned a gene cluster involved in lysine biosynthesis. Sequence analysis of the components in the cluster indicates that the Thermus lysine biosynthetic enzyme gene involved in the conversion of 2-oxoglutarate into AAA is homologous to the corresponding genes of fungi and yeast. It was also suggested that the pathway from AAA to lysine is dissimilar to those found in fungi and yeasts but that it resembles the pathway from glutamate to ornithine in bacterial arginine biosynthesis (6, 24). To establish lysine biosynthesis in T. thermophilus HB27 in detail, characterization of the gene products is necessary. However, we have not yet succeeded in enzymatic characterization of these gene products because of their low level of production in Escherichia coli and the difficulty of preparing the putative substrates in several reactions.
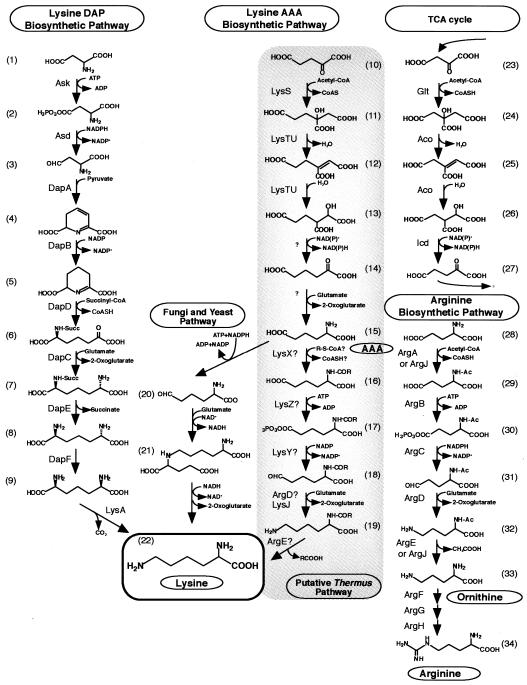
FIG. 1.
Proposed lysine AAA biosynthetic pathway, aligned with the lysine DAP biosynthetic pathway, the arginine biosynthetic pathway, and corresponding portions of the tricarboxylic acid cycle. 1, l-Aspartate; 2, l-aspartyl-γ-phosphate; 3, l-aspartate semialdehyde; 4, l-dihydrodipicolinate; 5, l-tetrahydrodipicolinate; 6, N2-succinyl-l-2-amino-6-oxopimelate; 7, N2-succinyl-l,l-DAP; 8, l,l-DAP; 9, d,l-DAP; 10, 2-oxoglutarate; 11, homocitrate; 12, homoaconitate; 13, homoisocitrate; 14, 2-oxoadipate; 15, AAA; 16, N2-acetyl-l-aminoadipate; 17, N2-acetyl-l-aminoadipyl-δ-phosphate; 18, N2-acetyl-l-aminoadipate semialdehyde; 19, N2-acetyl-l-lysine; 20, AAA semialdehyde; 21, l-saccharopine; 22, l-lysine; 23, 2-oxaloacetate; 24, citrate; 25, aconitate; 26, isocitrate; 27, 2-oxoglutarate; 28, l-glutamate; 29, N2-acetyl-l-glutamate; 30, N2-acetyl-l-glutamyl-γ-phosphate; 31, N2-acetyl-l-glutamate semialdehyde; 32, N2-acetyl-l-ornithine; 33, l-ornithine; 34, l-arginine. CoASH, coenzyme A; Succ, succinyl moiety; Ac, acetyl moiety.
The gene cluster from T. thermophilus HB27 contains several genes encoding enzymes involved in the reactions related to arginine biosynthesis. The cluster, however, lacks two genes corresponding to the argD and argE homologs, which are probably involved in the last two reactions of the putative lysine biosynthetic pathway in T. thermophilus HB27. In this report, we describe the cloning of an argD homolog, termed lysJ, that is essential for lysine biosynthesis in T. thermophilus HB27. We also report the kinetic properties of LysJ, which uses N2-acetylornithine, a precursor of ornithine in arginine biosynthesis, more efficiently than N2-acetyllysine, a putative natural precursor of lysine in T. thermophilus HB27. The evolutionary relationship between arginine and the newly identified biosynthesis of lysine is also discussed.
MATERIALS AND METHODS
Strains, media, and chemicals.
The extreme thermophile T. thermophilus HB27 was cultivated as described previously (16, 17, 31). E. coli DH5α and JM105 (28) were used for DNA manipulation, and E. coli BL21-CodonPlus(DE3)-RIL [F− ompT hsdS (rB− mB−) dcm Tetr gal λ(DE3) endA Hte (argU ileY leuW Camr)] (Stratagene, La Jolla, Calif.) was used as the host for gene expression. A medium, 2× YT (28), was generally used for cultivation of E. coli cells.
All the chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) or Kanto Chemicals (Tokyo, Japan). NAD+-dependent glutamate dehydrogenase was purchased from TOYOBO (Osaka, Japan). Enzymes for DNA manipulation were purchased from TAKARA Shuzo (Kyoto, Japan).
Molecular cloning and sequencing.
DNA manipulation was performed according to the methods in reference 28. Based on amino acid sequence alignment among N2-acetylornithine aminotransferases from various sources, oligonucleotides, 5′-GAGGC(G/C)GC(G/C)CT(G/C)AAGTTCGC(G/C)-3′ (ARD1), 5′-GCA(G/C)GC(G/C)AG(G/C)GGGTT(G/C)CC(G/C)CCGAA(G/C) GT-3′ (ARD2), and 5′-(G/C)CC(G/C)GTCTG(G/C)ACCTCGTC-3′ (ARD3), were designed and used as degenerate primers for PCR. The following thermal cycle was used: (step 1) 94°C for 2 min, (step 2) 94°C for 1 min, (step 3) 57°C for 1 min, (step 4) 72°C for 2 min, and (step 5) 72°C for 5 min; steps 2 to 4 were repeated 30 times. An amplified 515-bp fragment was cloned into the pT7Blue vector by using a Perfectly Blunt Cloning kit (Novagen, Madison, Wis.) and used as a probe for Southern hybridization. Southern hybridization against chromosomal DNA of T. thermophilus HB27 was carried out by using a Random Primer Fluorescein Labeling kit (New England Nuclear, Boston, Mass.). A BamHI fragment of about 3.2 kb that was positive in the hybridization assay against the 515-bp probe was ligated into pUC18 digested with BamHI and then introduced into E. coli DH5α. A colony that was positive in the colony hybridization assay using the same probe was selected. A plasmid was recovered from the colony and named pRDBamL. Its nucleotide sequence was determined by the method of Sanger et al. (29).
Disruption of lysJ in T. thermophilus HB27.
Disruption of the chromosomal copy of lysJ was performed as described previously (10, 17) with minor modifications. The plasmid, pUC39-442KmR (22), was digested with HindIII and blunt ended with T4 DNA polymerase, and the 1.4-kb fragment which contained the kanamycin nucleotidyltransferase (KNT) gene (20) was inserted into pRDBamL at the Aor51HI site, which is present in the middle of the lysJ gene. The resulting plasmid was named pRDKmR. T. thermophilus HB27 was cultured in TM medium (17), and when the turbidity (the optical density at 600 nm) reached 0.6, pRDKmR was added to the culture. After 2 h of cultivation, the cells were spread on TM plates containing 50 μg of kanamycin per ml and incubated at 65°C for 2 days. Colonies that grew on these plates were picked up as putative strains with a knockout in the lysJ gene. Disruption was confirmed by Southern hybridization.
Auxotrophic complementation test.
Each lysJ mutant was cultured in 1 ml of TM medium overnight. After centrifugation of the culture, the precipitate was washed with minimal medium (MP medium) (16, 31) four times and resuspended in 1 ml of MP medium. Cells (1 ml of the resuspension) were pipetted on an MP plate supplemented with 0.1 mM lysine, 0.1 mM ornithine, or a 0.1 mM concentration of both lysine and ornithine and incubated at 65°C for 2 days.
Expression of the argD homolog from T. thermophilus HB27 in E. coli
NdeI and EcoRI recognition sites were introduced around the start codon and the termination codon of the lysJ gene from T. thermophilus HB27, respectively, by PCR using the synthetic oligonucleotides 5′-AAAAAACATATGGAGACGAGAACCCTGGAAGAC-3′ and 5′-AAAGAATTCCTATGCTAGCACCGCCCGCACCGC-3′. The following program was used: (step 1) 95°C for 2 min, (step 2) 95°C for 1 min, (step 3) 68°C for 1 min, (step 4) 72°C for 2 min, and (step 5) 72°C for 7 min; steps 2 to 4 were repeated 30 times. An amplified fragment was digested with NdeI and EcoRI and cloned into pET26b (+) (Novagen). The resulting plasmid, pETTRDNE, was used for expression of the lysJ gene. E. coli BL21-CodonPlus(DE3)-RIL cells harboring pETTRDNE were cultured in 2 liters of 2× YT medium containing 50 μg of kanamycin per ml and 30 μg of chloramphenicol per ml. When the E. coli cells were grown to an optical density at 600 nm of 0.5, isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 1 mM) was added. The culture was continued for an additional 12 h after the induction.
Purification of the recombinant LysJ.
E. coli (pETTRDNE) cells (12 g) collected from the 2-liter culture were suspended in 24 ml of buffer I (20 mM potassium phosphate buffer [pH 6.5], 0.5 mM EDTA) and disrupted by sonication. The supernatant prepared by centrifugation at 40,000 × g for 20 min was heated at 80°C for 20 min, and denatured proteins from E. coli cells were removed by centrifugation as described above. Supernatant fractions were applied onto an anion-exchange column (DE-52; Whatman, Tokyo, Japan), preequilibrated with buffer I, and eluted with buffer I containing 0.1 M NaCl. The fractions showing LysJ activity were collected and pooled. After addition of sodium sulfate to a final concentration of 45%, the resultant precipitate was collected by centrifugation at 40,000 × g for 30 min. The precipitated proteins were solubilized with buffer I, dialyzed against buffer I containing 1 M sodium sulfate, and loaded onto a Phenyl Superose 5/5 column (Pharmacia Biotech, Tokyo, Japan) equilibrated with buffer I containing 1 M sodium sulfate. Proteins absorbed were eluted with a linear gradient of 1.0 to 0 M sodium sulfate. Active fractions were pooled, concentrated, and purified using a Hi-load 26/60 Superdex 200 prep-grade column (Pharmacia Biotech) equilibrated with buffer I containing 0.2 M NaCl. The purity of the recombinant enzyme was verified by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS-PAGE). The protein amount was determined by the method of Bradford (1) using a Bio-Rad protein assay kit (Nippon Bio-Rad, Tokyo, Japan). Molecular size was estimated by gel filtration using Superose 12 (Pharmacia Biotech) at a 0.5-ml/min flow rate.
LysJ assay.
Ten microliters of the enzyme solution (0.5 mg/ml) was added to the reaction buffer (50 mM N-cyclohexyl-2-aminoethanesulfonic acid [CHES; pH 8.9], 100 mM KCl, 5 mM 2-oxoglutarate, 10 mM pyridoxal-5′-phosphate, 0.15 mM NAD+, 5 to 25 mM N2-acetyllysine, and 2.37 U of glutamate dehydrogenase per ml), which was preincubated at 45°C for 5 min. For measuring the activity of N2-acetylornithine, 0.5 to 10 mM N2-acetylornithine was added to the reaction buffer instead of N2-acetyllysine. The reaction was monitored at 45°C by monitoring the increase in absorption at 340 nm. Kinetic parameters were calculated by using an initial velocity program of Cleland (4) with the equation for a steady-state ping-pong bi-bi mechanism, v = VAB/(KaB + AKb + AB), where v is velocity, V is maximum velocity, A is the concentration of substrate A, B is the concentration of substrate B, Ka is the Michaelis constant for substrate A, and Kb is the Michaelis constant for substrate B.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ sequence database under accession no. AB055203.
RESULTS
Cloning and sequencing of lysJ, an argD gene homolog.
Sequence analysis of each component in the cloned major lysine biosynthetic gene cluster of the extreme thermophilic bacterium T. thermophilus HB27 revealed that the lysY and lysZ genes have high identity to the argC and argB genes, respectively, which are involved in arginine biosynthesis, suggesting that the conversion of AAA to lysine proceeds in a manner similar to that in arginine biosynthesis. The T. thermophilus major lysine biosynthetic gene cluster contained several genes probably involved in the process but obviously lacked two genes which catalyzed the last two steps of the reactions of the putative lysine biosynthetic pathway. To elucidate the whole lysine biosynthetic pathway in T. thermophilus HB27, we tried to clone an argD homolog from T. thermophilus HB27 using three degenerate primers. DNA fragments of 515 and 340 bp were amplified by PCR with two combinations of degenerate primers, ARD1-ARD2 and ARD1-ARD3, using the genomic DNA of T. thermophilus HB27 as the template. Since both the amplified fragments were confirmed to have sequences similar to that of the argD gene by sequencing and the sequence of the smaller fragment was entirely contained in the longer one, Southern hybridization was carried out using the 515-bp fragment as the probe. A 3.2-kb hybridization-positive band was detected when the chromosomal DNA was digested with BamHI. The DNA fragment of 3.2 kb was recovered, ligated into pUC18 previously digested with BamHI, and introduced into E. coli DH5α cells. A hybridization-positive clone was isolated by colony hybridization. The plasmid contained in the cells was named pRDBamL. A faint but obvious band of about 5 kb was also detected in the Southern hybridization when the chromosomal DNA was digested with BamHI, suggesting the presence of an additional argD homolog in T. thermophilus. Cloning and characterization of the homolog is under way and will be described elsewhere in the near future.
Of the five open reading frames (ORFs) found in this fragment, one contained an argD homolog, but there was no argE homolog, unlike in Pyrococcus horikoshii, which possesses argDE homologs (PH1716 and PH1715) in a tandem manner at a position distal to the putative lysine biosynthetic gene cluster of this organism (14, 24). The lysJ gene encodes a 395-residue polypeptide. The deduced amino acid sequence of LysJ showed considerable identity to that of the DR0794 gene product from Deinococcus radiodurans (61% identity) (36), which is consistent with the taxonomically close relationship between T. thermophilus HB27 and D. radiodurans. Like many genes contained in the major lysine biosynthetic gene cluster of T. thermophilus HB27, LysJ also exhibited significant identity (44%) in amino acid sequence to a putative Pyrococcus homolog, PH1716. It should be noted that LysJ is more closely related to ArgD homologs from archaea than to those from bacteria (37). Although four additional ORFs, tentatively named orfA, orfB, orfC, and orfD, were founds in this fragment, those ORFs did not show amino acid sequence similarity to other proteins whose functions are identified.
Disruption of lysJ in T. thermophilus HB27.
We next investigated the role of the lysJ gene in T. thermophilus HB27. For this purpose, we constructed a mutant of T. thermophilus HB27 with a disruption in lysJ as described in Materials and Methods. The lysJ disruptant, RV4, could not grow on a minimal medium. However, addition of lysine restored the growth of the disruptant on minimal medium. On the other hand, the addition of ornithine, a precursor of arginine, had no effect on the growth of the disruptant. Thus, the lysJ gene was shown to be essential only for lysine biosynthesis in T. thermophilus HB27.
Expression of lysJ in E. coli.
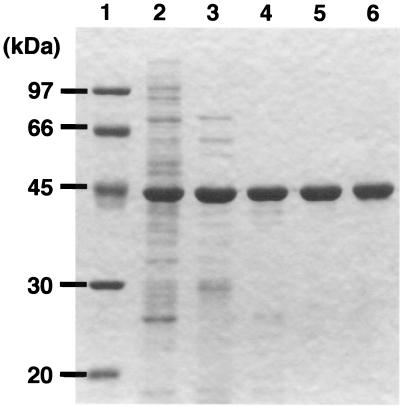
LysJ was purified to homogeneity by SDS-PAGE (Fig. 2), and the apparent molecular weight of 43,000 on SDS-PAGE coincided well with the molecular weight (43,503) calculated from the amino acid sequence. Through these five steps, 47 mg of purified LysJ protein was isolated from E. coli cells in a 2-liter culture.
FIG. 2.
Purification of the LysJ protein of T. thermophilus from recombinant E. coli cells. Lane 1, molecular mass markers; lane 2, supernatant of the sonicate of E. coli cells; lane 3, supernatant of the sonicate after heat treatment at 80°C for 20 min; lane 4, active fraction after DE52 anion-exchange column chromatography; lane 5, active fraction after Phenyl Superose 5/5 column chromatography; lane 6, purified LysJ protein after gel filtration using Hi-load 26–60 Superdex 200. Molecular size markers used were phosphorylase b (97 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (30 kDa), and soybean trypsin inhibitor (20 kDa).
The molecular size of this enzyme was estimated to be 80 kDa by gel filtration using Superose 12. This size indicated that LysJ is a dimeric enzyme composed of two identical subunits (data not shown).
Kinetic properties of LysJ from T. thermophilus HB27.
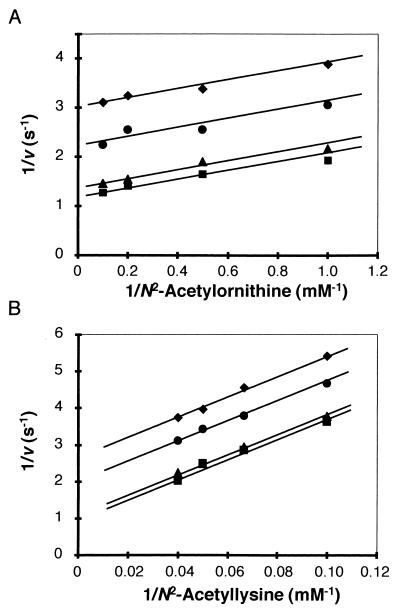
We next determined the catalytic activity in the reverse reaction using, for convenience, N2-acetyllysine and 2-oxoglutarate as the amino donor and amino acceptor, respectively. As shown in Fig. 3, the reaction catalyzed by LysJ proceeded through a ping-pong bi-bi mechanism, similar to results obtained with other aminotransferases. Kinetic parameters indicated that the catalytic efficiency, kcat/km, of LysJ using N2-acetyllysine was low, due to the high km value for N2-acetyllysine (Table 1). When similar steady-state kinetic assays were done with N2-acetylornithine, an intermediate of arginine biosynthesis, as a substrate, the catalytic efficiency was much higher (16-fold) than that obtained for N2-acetyllysine. Comparison of kinetic parameters for both the substrates revealed that the high catalytic efficiency with N2-acetylornithine was attributed to the low km value for the substrate. The kcat values were almost the same for both reactions.
FIG. 3.
Lineweaver-Burk plots of LysJ activity. (A) N2-Acetylornithine; (B) N2-acetyllysine. Diamonds, circles, triangles, and squares indicate results with 1, 2, 5, and 10 mM 2-oxoglutarate, respectively.
TABLE 1.
Kinetic parameters of LysJ
| Substrate | km (mM) | kcat (S−1) | kcat/km (S−1 M−1) |
|---|---|---|---|
| N2-Acetyllysine | 10.0 ± 1.6 | 0.8 ± 0.1 | 78.6 |
| N2-Acetylornithine | 0.8 ± 0.1 | 1.0 ± 0.0 | 1,266.1 |
DISCUSSION
Our previous study suggested that in T. thermophilus HB27 lysine is biosynthesized through the AAA pathway, which contains AAA as a biosynthetic intermediate of lysine, as in the fungal AAA pathway (16). However, that study also suggested that the Thermus AAA pathway is different from the fungal pathway in that the conversion of AAA to lysine proceeds in a manner similar to that of ornithine synthesis from glutamate in arginine biosynthesis (6). In the present study, we demonstrated that the argD homolog lysJ is essential for lysine biosynthesis in T. thermophilus HB27. This finding indicated that a homolog of a gene involved in arginine biosynthesis is actually responsible for lysine biosynthesis in T. thermophilus HB27 and that the Thermus AAA pathway is related evolutionarily to the arginine biosynthetic pathway. This result is further supported by our recent detection of the activity converting N2-acetyllysine to lysine in the crude extract of T. thermophilus (unpublished result).
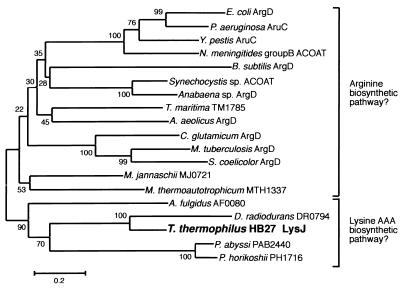
When the evolutionary relationships between LysJ, ArgDs, and their homologs were phylogenetically analyzed, LysJ was found to be closely related to DR0794 of D. radiodurans (61% identity), which is a bacterium closely related taxonomically to T. thermophilus HB27 (36) (Fig. 4). The phylogenetic tree also shows that LysJ is grouped with ArgD homologs of two archaea, Pyrococcus horikoshii (PH1716) (14) and Pyrococcus abyssi (PAB2440), and ArgD-1 of another archaeon, Archaeglobus fulgidus (15). Both Pyrococcus strains have a gene cluster similar to that for lysine biosynthesis in T. thermophilus HB27 (24). Our previous study also showed that each component of the cluster of T. thermophilus HB27 is related evolutionarily to each counterpart in P. horikoshii. That study therefore suggested that, in Pyrococcus, lysine is synthesized through the bacterial AAA pathway found in T. thermophilus HB27. The Thermus lysine biosynthetic gene cluster lacks two genes corresponding to PH1716 and PH1715 in the putative lysine biosynthetic gene cluster of P. horikoshii. Based on homology and phylogenetic analysis, we concluded that the lysJ gene cloned in this study corresponds to PH1716 and PAB2440 of P. horikoshii and P. abyssi, respectively.
FIG. 4.
Phylogenetic tree of various N2-acetylornithine aminotransferases, LysJ, and related transferases. A phylogenetic tree was constructed by using maximum-parsimony and neighbor-joining methods. Sequence data for the analysis were obtained from the GenBank and Protein Information Resource databases. The amino acid sequences were aligned by using CLASTAL W (33). Using this aligned data, a phylogenetic tree was constructed by the computer program Mega (18). Numbers on selected nodes indicate bootstrap values. This figure includes LysJ and related transferases from E. coli (9), Pseudomonas aeruginosa (12), Yersinia pestis (GenBank accession no. GI 4106567), Neisseria meningitidis group B (32), Bacillus subtilis (25), Synechocystis sp. strain PCC6803 (13), Anabaena sp. strain PCC7120 (8), Thermotoga maritima (23), Aquifex aeolicus (7), Corynebacterium glutamicum (27), Mycobacterium tuberculosis (5), Streptomyces coelicolor A3 (2) (GenBank accession no. GI 7106695), Methanococcus jannaschii (3), Methanobacterium thermoautotrophicum (30), Archaeoglobus fulgidus (15), D. radiodurans (36), P. abyssi (GenBank accession no. GI 5457730), and P. horikoshii (14).
In addition to lysJ, genes corresponding to the components for lysine biosynthesis in T. thermophilus HB27 are all present in D. radiodurans, suggesting that this bacterium also synthesizes lysine through the bacterial AAA pathway. Interestingly, the corresponding genes are not clustered but are spread over the genome of D. radiodurans, which is in contrast to what occurs in Thermus and Pyrococcus. In a recent review, Makarova and coworkers indicate that the absence of all key enzymes for lysine biosynthesis through the DAP pathway is a puzzling feature of Deinococcus metabolism since it does not require lysine for growth (21). The absence of typical prokaryotic lysine biosynthetic enzymes may be compensated for by the presence of all the enzyme homologs for prokaryotic lysine biosynthesis through AAA in the D. radiodurans genome. It should be noted that although D. radiodurans possesses all the components for the bacterial AAA pathway, the microorganism also has a homolog (DR1758) of lysA which is possibly involved in the decarboxylation of DAP to produce lysine using the typical DAP pathway for lysine biosynthesis. Therefore, lysine biosynthesis in D. radiodurans may be a new target for elucidating the evolutionary relationship between the DAP pathway and the prokaryotic AAA pathway for lysine biosynthesis.
The lysJ mutant of T. thermophilus HB27 showed only a lysine-auxotrophic phenotype. This result may indicate the presence of other argD homologs that play a role primarily in arginine biosynthesis in T. thermophilus HB27. On the other hand, kinetic analysis for LysJ revealed that LysJ preferred N2-acetylornithine to N2-acetyllysine as the substrate. The kinetic data suggest that LysJ may function in supporting arginine biosynthesis when the activity of the ArgD homolog responsible for arginine biosynthesis is lost. Thermus species possess ornithine in place of DAP as a cell wall component, which may confer an advantage for growth at high temperatures and render dispensable the synthesis of DAP acid for growth (26). Furthermore, ornithine is a precursor for not only arginine but also polyamines, which are involved in several cellular processes, such as the stabilization of a ternary complex consisting of a ribosome, mRNA, and aminoacyl-tRNA in T. thermophilus cells (34). Thus, species of the genus Thermus are able to grow at an extremely elevated temperature by producing a large amount of polyamines to protect their own machinery. These observations suggest the importance of ornithine-synthesizing activity for the growth of the microorganism at an elevated temperature and therefore may explain the presence of an isozyme(s) having ornithine-synthesizing activity. Recently, E. coli ArgD was shown to catalyze the N2-succinyl-l,l-DAP-dependent transamination of 2-oxoglutarate, which is the sixth reaction in the DAP pathway for lysine biosynthesis (19). In that study, Ledwidge and Blanchard suggested that E. coli ArgD has key functions in the biosynthetic pathways for both arginine and lysine in E. coli. Their study as well as ours demonstrates that arginine and lysine biosyntheses functionally correlate with each other, although both lysine biosynthetic pathways are totally different from each other.
N2-Acetyllysine and N2-acetylornithine are compounds structurally related to each other, which may explain the dual functions of LysJ and ArgD. Based on homology in amino acid sequence and by analogy to reactions mediated by two enzymes, it is evident that LysJ shares an ancestor with ArgD. Similarly, other members of lysine biosynthesis in T. thermophilus HB27 share common ancestors with counterparts in arginine biosynthesis (6). In addition, several reactions, from citrate to 2-oxoglutarate, in the tricarboxylic acid cycle are related evolutionarily to those of the first half of the lysine AAA biosynthetic pathway (3, 6).
LysJ has become the first well-characterized enzyme in the lysine AAA pathway in T. thermophilus HB27. We have shown that the lysine biosynthetic pathway is clearly related to the arginine biosynthetic pathway. In consideration of the fact that the corresponding enzymes in both pathways have evolved from a single common ancestral enzyme, these two pathways have probably diverged from a common ancestral pathway. Through further detailed studies of the lysine AAA biosynthetic pathway in T. thermophilus HB27, for example, structural and biochemical analyses, we expect to reveal principles for the evolution of the enzyme along with its amino acid biosynthesis.
ACKNOWLEDGMENTS
We thank Hiromi Nishida (The University of Tokyo) for his help in phylogenetic analysis. We also thank Michael E. P. Murphy (University of British Columbia) for providing helpful comments for completing the draft.
REFERENCES
- 1.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Broquist H P. Lysine biosynthesis (yeast) Methods Enzymol. 1971;17:112–129. [Google Scholar]
- 3.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J D, Geoghagen N S, Weidman J F, Fuhrmann J L, Nguyen D T, Utterback T, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B B, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 4.Cleland W W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;78:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 5.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Sqares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Cunin R, Glandsorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 8.Floriano B, Herrero A, Flores E. Analysis of expression of the argC and argD genes in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1994;176:6397–6401. doi: 10.1128/jb.176.20.6397-6401.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heimberg H, Boyen A H, Crabeel M, Glansdorff N. Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferases: evolutionary relationship with ornithine aminotransferases. Gene. 1990;90:69–78. doi: 10.1016/0378-1119(90)90440-3. [DOI] [PubMed] [Google Scholar]
- 10.Hidaka Y, Hasegawa M, Nakahara T, Hoshino T. The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci Biotechnol Biochem. 1994;58:1338–1339. doi: 10.1271/bbb.58.1338. [DOI] [PubMed] [Google Scholar]
- 11.Irvin S D, Bhattacharjee J K. A unique fungal lysine biosynthesis enzyme shares a common ancestor with tricarboxylic acid cycle and leucine biosynthetic enzymes found in diverse organisms. J Mol Evol. 1998;46:401–408. doi: 10.1007/pl00006319. [DOI] [PubMed] [Google Scholar]
- 12.Itoh Y. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7280–7290. doi: 10.1128/jb.179.23.7280-7290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 14.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Nakamura Y, Robb T F, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 15.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D'Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 16.Kobashi N, Nishiyama M, Tanokura M. Aspartate kinase-independent lysine synthesis in an extremely thermophilic bacterium, Thermus thermophilus: lysine is synthesized via α-aminoadipic acid, not via diaminopimeric acid. J Bacteriol. 1999;181:1713–1718. doi: 10.1128/jb.181.6.1713-1718.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumer S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park, Pa: Pennsylvania State University; 1993. [Google Scholar]
- 19.Ledwidge R, Blanchard J S. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynthesis. Biochemistry. 1999;38:3019–3024. doi: 10.1021/bi982574a. [DOI] [PubMed] [Google Scholar]
- 20.Liao H, McKenzie T, Hageman R. Isolation of a thermostable enzyme variant by cloning and selection in thermophile. Proc Natl Acad Sci USA. 1986;83:576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova K S, Aravind L, Wolf Y I, Tatusov R L, Minton K W, Koonin E V, Daly M J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maseda H, Hoshino T. Development of expression vector for Thermus thermophilus. J Ferment Bioeng. 1998;86:121–124. [Google Scholar]
- 23.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, White O, Salzberg S L, Smith H O, Venter J C, Fraser C M. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 24.Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res. 1999;9:1175–1183. doi: 10.1101/gr.9.12.1175. [DOI] [PubMed] [Google Scholar]
- 25.O'Reilly M, Devine K M. Sequence and analysis of the citrulline biosynthetic operon argC-F from Bacillus subtilis. Microbiology. 1994;140:1023–1025. doi: 10.1099/13500872-140-5-1023. [DOI] [PubMed] [Google Scholar]
- 26.Quintela J C, Pittenauer E, Allmaier G, Aran V, de Pedro M A. Structure of peptidoglycan from Thermus thermophilus HB8. J Bacteriol. 1995;177:4947–4962. doi: 10.1128/jb.177.17.4947-4962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakanyan V, Petrosyan P, Lecocq M, Boyen A, Legrain C, Demarez M, Hallet J N, Glansdorff N. Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacterium glutamicum: enzyme evolution in the early steps of the arginine pathway. Microbiology. 1996;142:99–108. doi: 10.1099/13500872-142-1-99. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Nicklen E F, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J, Rice P, Noelling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka T, Kawano N, Oshima T. Cloning of 3-isopropylmalate dehydrogenase gene of an extreme thermophile and partial purification the gene products. J Biochem (Tokyo) 1981;89:677–682. doi: 10.1093/oxfordjournals.jbchem.a133245. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Venter J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 33.Tompson J D, Higgins D G, Gilson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4553–4559. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzawa T, Hamasaki N, Oshima T. Effects of novel polyamines on cell-free polypeptide synthesis catalyzed by T. thermophilus HB8 extract. J Biochem (Tokyo) 1993;114:478–486. doi: 10.1093/oxfordjournals.jbchem.a124203. [DOI] [PubMed] [Google Scholar]
- 35.Vogel H J. Distribution of lysine pathways among fungi: evolutionary implications. Am Natl. 1964;98:446–455. [Google Scholar]
- 36.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffat K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Minton K W, Fleischmann R D, Ketchum K A, Nelson K E, Salzberg S, Smith H O, Venter J C, Fraser C M. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]