Abstract
The newly emerged severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants with high transmission rates and striking immune evasion have posed a serious challenge to the application of current first‐generation SARS‐CoV‐2 vaccines. Other sarbecoviruses, such as SARS‐CoV and SARS‐related coronaviruses (SARSr‐CoVs), have the potential to cause outbreaks in the future. These facts call for the development of variant‐proof SARS‐CoV‐2, pan‐sarbecovirus or pan‐β‐CoV vaccines. Several novel vaccine platforms have been used to develop vaccines with broad‐spectrum neutralizing antibody responses and protective immunity to combat the current SARS‐CoV‐2 and its variants, other sarbecoviruses, as well as other β‐CoVs, in the future. In this review, we discussed the major target antigens and protective efficacy of current SARS‐CoV‐2 vaccines and summarized recent advances in broad‐spectrum vaccines against sarbecoviruses and β‐CoVs.
Keywords: COVID‐19, pan‐sarbecovirus, pan‐β‐coronavirus, RBD, SARS‐CoV‐2, vaccines
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has severely threatened global public health, as well as social and economic development. Substantial strides have been made in the development of first‐generation SARS‐CoV‐2 vaccines during the past 2 years. Moreover, authorized SARS‐CoV‐2 vaccines, such as BNT162b2, mRNA‐1273, and BBIBP‐CorV, have played a critical role in controlling the COVID‐19 pandemic.
However, amid the circulation of SARS‐CoV‐2, several SARS‐CoV‐2 variants have emerged under immune pressure. For example, SARS‐CoV‐2 variants of concern (VOCs), including B.1.1.7 (Alpha), 1 B.1.351 (Beta), 2 P.1 (Gamma), 3 B.1.617.2 (Delta), 4 and B.1.1529 (Omicron), 5 have posed a serious challenge for the protective efficacy of first‐generation SARS‐CoV‐2 vaccines. 5 , 6 The recently emerged Omicron and its subvariants, such as BA.2, BA.2.12.1, and BA.4/BA.5, have rapidly replaced other VOCs and become the dominant strain around the world. 7 Especially, striking immune evasion from the current SARS‐CoV‐2 vaccines has been found for Omicron, reducing overall protective efficiency. 8 Titers of neutralizing antibodies (nAbs) induced by two doses of mRNA vaccines against the Omicron variant are significantly decreased. 9 Similarly, the Omicron variant shows a significantly decreased sensitivity to neutralizing antibodies induced by ChAdOx1 nCoV‐19 (AstraZeneca) in vaccinees. 8 To address the poor protective efficiency of first‐generation vaccines against Omicron, many vaccine makers have already started the development of Omicron variant‐specific COVID‐19 vaccines. 10 However, given the continuing pandemic of SARS‐CoV‐2 and increased selection pressure of antibodies in COVID‐19 convalescents and vaccinees, new variants may also be on the cusp of emerging and very likely further evade the Omicron variant‐specific COVID‐19 vaccines. Therefore, developing the next generation of variant‐proof SARS‐CoV‐2 vaccines against current and future emerging SARS‐CoV‐2 variants is urgently needed.
Apart from SARS‐CoV‐2, the other two highly pathogenic β‐CoVs, including SARS‐CoV and Middle East respiratory syndrome coronavirus (MERS‐CoV), have caused severe pneumonia in humans over the past 20 years. 11 , 12 , 13 SARS‐CoV has caused 8096 reported cases, with about a 10% fatality rate, whereas MERS‐CoV has caused 1728 confirmed cases with about a 34.4% fatality rate in humans. 14 , 15 Studies have shown 75.9% S protein amino acid sequence identity between SARS‐CoV‐2 and SARS‐CoV, 16 and they use the same human angiotensin‐converting enzyme 2 (hACE2) as their receptor to infect target cells. 17 , 18 Moreover, similar to SARS‐CoV, most SARS‐related CoVs (SARSr‐CoVs) from bats, such as WIV1 and RsSHC014, show a higher amino acid identity (~88%) with SARS‐CoV‐2. 19 , 20 , 21 From a historical perspective, SARS‐CoV or bat‐SARSr‐CoVs can utilize hACE2 as their common receptor to infect primary airway epithelial cells, posing potential threats to humans. 19 , 22 , 23 , 24 In another scenario, SARS‐CoV‐2 and MERS‐CoV are less homologous, but they still infect the same type‐II alveolar cells and use identical transcription regulatory sequences. 25 , 26 Saudi Arabia has reported several cases infected with MERS‐CoV and SARS‐CoV‐2 simultaneously, further leading to the concern that new coronaviruses with high transmission and fatality rates might emerge when recombination occurs between MERS‐CoV and SARS‐CoV‐2 in the same host. 27 , 28 Although a large number of vaccines against SARS‐CoV and MERS‐CoV have been developed, 29 , 30 , 31 none of them has been approved for the clinical prevention or treatment of highly pathogenic CoV infections in humans.
Taken together, these facts call for the urgent development of broad‐spectrum anti‐COVID‐19, anti‐sarbecovirus or even anti‐coronavirus vaccines with strong efficacy and safety to combat the current SARS‐CoV‐2 and its variants, as well as the expected emergence of novel coronaviruses.
2. MAJOR ANTIGEN TARGETS OF CURRENT SARS‐COV‐2 VACCINES
SARS‐CoV‐2 contains four major structural proteins, three important structural proteins embedded in the viral surface envelope, including spike protein (S), envelope protein (E), and membrane protein (M), and one in the ribonucleoprotein termed nucleocapsid (N) protein. 32 , 33 Among them, S protein plays an important role in the processes of receptor recognition, binding, and membrane fusion. 34 Therefore, it serves as the major target for most SARS‐CoV‐2 vaccines. 35
The S protein is a 180–200 kDa type I transmembrane glycoprotein, consisting of S1 and S2 subunits (Figure 1). 36 The S1 subunit contains the N‐terminal domain (NTD) and the C‐terminal domain (CTD). CTD contains an important receptor‐binding domain (RBD), which is a key functional domain in S protein, consisting of a ring structure named receptor‐binding motif (RBM) that contacts host receptors directly. 37 , 38 Heptad repeat 1 (HR1) and heptad repeat 2 (HR2) in the S2 subunit form a six‐helix bundle (6‐HB) core structure during the fusion process, leading to viral entry. 39 S proteins harbor multiple B cell epitopes and T cell epitopes that elicit immune responses against virus infection, enabling a more rational approach to design effective vaccine antigen. 40 , 41 , 42 Prefusion‐stabilized S, achieved by using two consecutive proline substitutions (S‐2P) in the S2 subunit between the central helix and HR1, has been widely used as the immunogen for the development of anti‐SARS‐CoV‐2, anti‐SARS‐CoV, and anti‐MERS‐CoV vaccines since it could increase the quality and quantity of antibodies compared with their wild‐type (WT) counterparts. 43 , 44 Currently, antigen S‐2P, comprising proline substitutions at residues K986 and V987, is being used in several SARS‐CoV‐2 vaccine candidates (BNT16B2/mRNA‐1273/Ad26. COV2‐S/NVX‐CoV2373) (Table 1).
Figure 1.
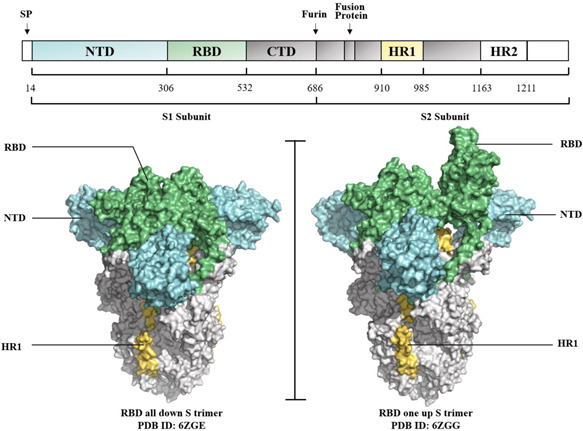
Spike protein of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Sequence diagram and structure diagram of S protein. S protein contains S1 and S2 subunits. The left (three RBDs, down S trimer) and the right (one RBD, up S trimer) were shown as surface in Cryo‐EM resolved structures (modified from 6ZGE and 6ZGG). CTD, C terminal domain; HR, heptad repeat; NTD, N terminal domain; RBD, receptor binding domain.
Table 1.
WHO Emergency Use Listing and Prequalification (EUL/PQ)‐approved vaccines and their antigens
| Vaccine | Manufacturer | Antigen | Type of vaccine | Reference |
|---|---|---|---|---|
| CoronaVac/PiCoVacc | Sinovac, National Institute for Communicable Disease Control and Prevention, China | Whole virus (CN2 strain) | Inactivated virus | [160] |
| BBIBP‐CorV | Beijing Institute of Biological Products, Sinopharm and Institute of Viral Disease Control and Prevention, China | Whole virus (HB02 strain) | Inactivated virus | [161] |
| Covaxin/BBV152 | Bharat Biotech and Indian Council of Medical Research | Whole virus (NIV‐2020‐770) | Inactivated virus | [90] |
| BNT16B2 | BioNTech, Fosun Pharma, and Pfizer, Germany | S‐2P | LNP‐mRNA | [162] |
| mRNA‐1273 | Moderna, National Institute of Allergy and Infectious Diseases, USA | S‐2P | LNP‐mRNA | [163, 164] |
| ChAdOx1 nCoV‐19/AZD1222 | The University of Oxford, and AstraZeneca, UK | Full‐length S | Nonreplicating viral vector | [165, 166] |
| Convidecia™ Ad5‐nCoV | CanSino Biological Inc. and Beijing Institute of Biotechnology, China | Full‐length S | Nonreplicating viral vector | [167, 168] |
| Ad26. COV2‐S | Johnson & Johnson, USA | S‐2P | Nonreplicating viral vector | [169, 170] |
| NVX‐CoV2373 | Novavax, USA | S‐2P | Protein‐based | [171] |
RBD is another important target site for developing anti‐coronavirus vaccines. 45 , 46 We have previously convincingly demonstrated that RBD‐based vaccines could induce highly protective immunity against SARS‐CoV or MERS‐CoV. 47 , 48 , 49 , 50 For example, we reported that a recombinant fusion protein containing SARS‐CoV RBD and the fragment‐crystallizable (Fc) region of human IgG (RBD‐Fc) could induce highly potent nAbs against SARS‐CoV in immunized rabbits and mice. 49 RBD‐Fc could induce potent and durable S‐specific antibodies able to maintain high titers for 12 months after immunization and protect most vaccinated mice against SARS‐CoV challenge. 51 Also, a recombinant RBD protein expressed in 293T cells with excellent conformation and good antigenicity could elicit highly effective nAbs that completely protected immunized mice from SARS‐CoV challenge. 52 Furthermore, several subunit vaccines against MERS‐CoV have been designed based on RBD. RBD‐immunized mice or nonhuman primates (NHPs) exhibited potent humoral and cellular immune responses to effectively neutralize MERS‐CoV infection and provide protection against MERS‐CoV challenge. 53 , 54
As for SARS‐CoV‐2, neutralizing antibodies from COVID‐19 convalescent sera mainly target the RBD. 45 , 55 , 56 , 57 Currently, a large number of RBD‐based SARS‐CoV‐2 vaccines have been reported. 58 For instance, we have developed a SARS‐CoV‐2 vaccine candidate, consisting of SARS‐CoV‐2 RBD‐Fc protein and Freund's adjuvant. 59 The vaccine could produce high titers of SARS‐CoV‐2 RBD‐specific antibodies, neutralize SARS‐CoV‐2 and its variants, and show cross‐neutralization activity against SARS‐CoV and SARSr‐CoVs. Yang et al. have reported a recombinant RBD vaccine that could induce strong virus‐neutralizing activities in mice and protect NHPs against live SARS‐CoV‐2 challenge. Importantly, they compared four structure‐based SARS‐CoV‐2 vaccine candidates, including RBD, extracellular domain (ECD), S1 subunit and S2 subunit, and found that RBD elicited a much higher viral neutralization activity than that achieved with ECD or S1 subunit, and no viral neutralization activity of S2 subunit was detected. 60 Similarly, in a recent study, we evaluated the immunogenicity of RBD‐Fc, RBD, or S‐trimer with different adjuvants. Compared with RBD and S‐trimer, we found that RBD‐Fc homodimer effectively elicited more potent RBD‐specific IgG and neutralizing antibodies against SARS‐CoV‐2 and its Delta variant in mice. 61 Moreover, a COVID‐19 RBD‐dimer‐based protein subunit vaccine candidate, ZF2001, 62 exhibited high protective efficacy (more than 80%) in preventing symptomatic COVID‐19 in phase 3 clinical trials. 63 Recent studies have demonstrated that it could induce increased titers of nAbs against Omicron after administration of multiple booster doses. 64 , 65 In line with our conclusion, compared with the monomeric RBD, they found that the RBD‐dimer significantly enhanced SARS‐CoV‐2 neutralizing antibodies in immunized mice. 66 Together, these results demonstrated that the RBD, especially the RBD homodimer, is a promising immunogen for the development of SARS‐CoV‐2 vaccines.
The reported neutralizing epitopes of SARS‐CoV‐2 RBD in recent studies rationally explain why RBD is an excellent antigen. Located in the S trimer, RBD possesses flexibility and undergoes structural fluctuation between a “down” and “up” conformation, 67 , 68 and both conformations could induce antibodies targeting different epitopes when being used as immunogens. 69 We previously isolated 48 monoclonal antibodies (mAbs) from memory B cells in a convalescent individual and found four nonoverlapping epitopes on the RBD. 57 Among the 48 mAbs, RBD‐targeted mAbs, such as XG014, could exert highly cross‐neutralizing activity against SARS‐CoV‐2, SARS‐CoV‐2 variants, and SARS‐CoV. 70 Based on antigenic mapping analyses and functional characteristics, anti‐RBD antibodies have been grouped into four classes (Classes 1–4), 57 , 71 , 72 , 73 and we have listed the representative antibodies in Figure 2. RBM‐targeting antibodies in Class 1, such as REGN10933 and CB6, block hACE2 binding and only access their epitopes in the RBD “up” conformation. 56 , 74 , 75 Other antibodies in Class 2 bind both RBD “up” and “down” conformations and contact‐adjacent RBDs. For example, LY‐CoV555 could render complete steric hindrance interference to limit RBD binding with ACE2. 73 , 76 , 77 Some non‐ACE2‐competing antibodies, such as DH1047, have been identified to target the Class 3 epitope which is outside the RBM and could interact with an “up” RBD conformation from an adjacent protomer. 67 , 78 Class 4 antibodies, such as S309, could target a conserved proteoglycan epitope distal of the RBM, neutralize the virus by one or more IgG‐specific bivalent mechanisms and have broad cross‐reactivity against related zoonotic coronaviruses. 79 Among the group of four epitopes, Classes 1 and 2 were highly variable, whereas the epitopes of Classes 3 and 4 were more conserved, but less accessible (Figure 2). 72 Therefore, eliciting neutralizing antibodies targeting Classes 3 and 4 epitopes are desirable for enhanced broad‐spectrum protective immunity to combat current and future SARS‐CoV‐2 variants and emerging zoonotic sarbecoviruses. 80 , 81 , 82
Figure 2.
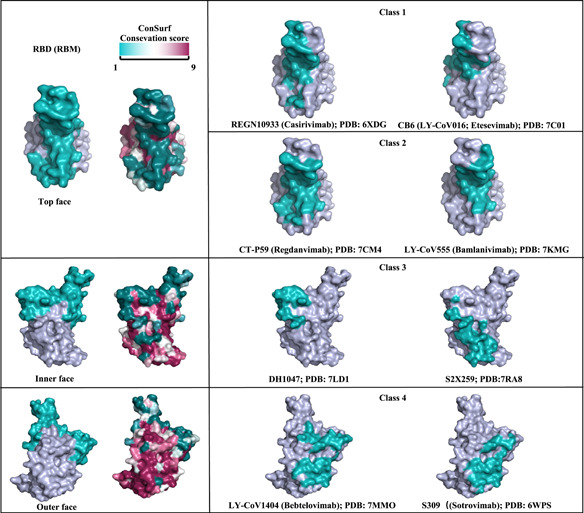
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RBD‐specific binding footprints of neutralizing antibodies. RBD (RBM) top face, inner face and outer face were shown in the left with teal RBM. Sequence conservation was calculated by the ConSurf Database. RBD‐specific binding interface footprints of four classifications of neutralizing antibody regions were colored in teal in the right. For each class of antibody binding region (RBD1‐4), the footprints of two representative antibodies on the RBD are shown. RBD, receptor binding domain; RBM, receptor binding motif.
3. EFFICACY OF CURRENTLY APPROVED SARS‐COV‐2 VACCINES
As of July 2022, more than 360 SARS‐CoV‐2 vaccines were under development at different stages, and about 160 vaccines are now in the clinical development stage (SARS‐CoV‐2 vaccine and therapeutics tracker) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines). In this part, we mainly summarize and discuss the authorized SARS‐CoV‐2 vaccines shown in Table 1, and they could be generally grouped into three categories (Figure 3). 83 , 84
Figure 3.
Currently utilized severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) vaccine types. Six types of currently used vaccines against the COVID‐19 pandemic. Illustration created by the authors using BioRender (http://www.biorender.com).
The first category includes protein‐based vaccines, such as inactivated virus vaccines and subunit vaccines. Inactivated vaccines can induce a wider range of immune responses compared with other vaccines by using the entire virus as an immunogen. 84 Three inactivated vaccine candidates have been approved by WHO, BBIBP‐CorV, CoronaVac, and BBV152. Among them, two inactivated vaccines from Sinopharm (BBIBP‐CorV) and Sinovac (CoronaVac) were developed in China (Table 1). BBIBP‐CorV showed 78.1% efficacy, 85 and CoronaVac showed 50.7% efficacy for preventing symptomatic COVID‐19 in the phase 3 trial. 86 However, the neutralizing activities of sera from individuals immunized with BBIBP‐CorV or CoronaVac against Omicron were limited. The neutralizing activities of BBIBP‐CorV vaccinee serum showed a 20.1‐fold reduction against the Omicron variant relative to the WT strain. 87 Cohorts receiving two doses of CoronaVac vaccine showed a 6.5‐fold reduction in anti‐Omicron antibody titers. 88 Similarly, BBV152, also known as Covaxin, which has been approved for restricted emergency use in India, 89 is an inactivated virus‐based vaccine adjuvanted with a TLR7/8 agonist adsorbed on Algel. 90 The phase 3 trial showed that BBV152 conferred 65.2% protection against the Delta variant. 91 Further studies demonstrated that vaccination with BBV152 induced a 26.6‐fold reduction in neutralization activity against the Omicron variant. 92
Subunit vaccines include protein‐based vaccines and virus‐like particle (VLP) vaccines. The protein‐based vaccine NVX‐CoV2373 is constructed from the full‐length SARS‐CoV‐2 trimeric S protein stabilized in the prefusion conformation based on a recombinant nanoparticle platform. Phase 3 trials have shown that this vaccine in a two‐dose regimen conferred 89.7% protection against SARS‐CoV‐2 infection. 93 Six VLP vaccine candidates are in clinical development. A candidate vaccine named coronavirus‐like particles (CoVLP) has been tested in a phase 3 trial and showed 69.5% efficacy at preventing symptomatic COVID‐19 infection and 78.8% efficacy against moderate‐to‐severe disease. 94
The second category includes gene‐based vaccines, such as virus‐vectored vaccines, DNA vaccines, and mRNA vaccines. Virus‐vectored vaccines using safe viral vectors can stimulate potent humoral and cellular immune responses, and viral vectors might be an efficient strategy for delivering genes encoding key antigens of targeted pathogens. 95 WHO has approved three virus‐vectored vaccines, including ChAdOx1 nCoV‐19, Ad26. COV‐2‐S and Ad5‐nCoV. ChAdOx1 nCoV‐19, known as AZD1222, is a chimpanzee adenovirus‐based non‐replicating vector vaccine. It conferred 81.3% protection against symptomatic COVID‐19 in participants who received two doses. 96 Studies have shown that it confers 74.5%, 67%, and 10.4% protection against Alpha, Delta, and Beta VOCs, respectively. 97 , 98 However, compared to the ancestral variant, the neutralizing titers were 14‐ to 21‐fold lower against the Omicron variant. 99 , 100 Ad26.COV‐2‐S is an adenovirus serotype 26‐based non‐replicating vector vaccine encoding a prefusion‐stabilized SARS‐CoV‐2 S glycoprotein, which conferred 66% protection against symptomatic disease in the clinical trial, 101 , 102 but plasma specimens from recipients with a single‐dose of Ad26.COV‐2‐S vaccine lacked detectable neutralization activity against the Omicron variant. 103 Ad5‐nCoV, also known as Convidecia, is also a non‐replicating adenovirus‐vectored vaccine that expresses optimized full‐length SARS‐CoV‐2 S glycoprotein with the TPA signal peptide gene. The phase 3 trial indicated that one dose of Ad5‐nCoV exhibited over 90% efficacy at preventing severe disease and death. 104 In addition, these virus‐vectored vaccines can induce Th‐1 cell responses, thus inducing strong cellular immunity.
DNA and mRNA vaccines are promising platforms for SARS‐CoV‐2 vaccines owing to their fast and flexible design and production. At present, INO‐4800, ZyCoV‐D, AG0302‐COVID‐19, and GX‐19N are the four DNA vaccines progressing through phase 3 clinical trials (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines). As for mRNA vaccines, two COVID‐19 LNP‐encapsulated mRNA vaccines (mRNA‐1273 and BNT162b2) have now been approved for commercial use. 105 , 106 Clinical trials showed that mRNA‐1273 had 94.1% efficacy 107 and BNT162b2 had 95% efficacy in preventing symptomatic SARS‐CoV‐2 infections. 108 However, the effectiveness of both vaccines against symptomatic disease caused by Omicron dropped to 75.1% and 65.5% by 2–4 weeks post the second dose. 8
The last category comprises a combination of both protein‐based and gene‐based approaches represented as live‐attenuated virus vaccines. 83 , 84 Live‐attenuated virus vaccines take advantage of mimicking natural infection to stimulate humoral and cellular immune responses that provide long‐lasting protection in the host. 109 However, viruses can regain their toxicity owing to mutations after vaccination, raising public concerns about the safety of live‐attenuated SARS‐CoV‐2 vaccines. 110 Thus, the development of widely‐used COVID‐19 live‐attenuated vaccines remains a challenge. So far, only two candidate COVID‐19 live‐attenuated vaccines have been approved for clinical trials as of August 8, 2022 (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines).
4. BROAD‐SPECTRUM ANTI‐CORONAVIRUS VACCINES
Coronavirus is a family of enveloped viruses with a positive‐sense, single‐stranded RNA, comprised of alpha, beta, gamma, and delta genera. 111 β‐CoVs can be further divided into A, B, C, and D lineage. 112 Developing broad‐spectrum vaccines against β‐CoV lineage B (sarbecovirus), including SARS‐CoV‐2, SARS‐CoV, and bat‐SARSr‐CoV, is essential. 18 Developing an effective and broad‐spectrum next‐generation vaccine involves three main strategies. First, multiple antigens from different coronavirus subgroups are displayed simultaneously onto a carrier (e.g., nanoparticles) to elicit protective immunity against multiple coronaviruses. 113 Second, multiplexed‐chimeric spike, which contains mixtures, such as RBD, NTD, or S2 from different CoVs, is designed. 114 Third, a novel and potent adjuvant is developed to boost the immune response of conserved epitope from the proper antigen. 115 The currently developed coronavirus vaccines could be classified into three types according to their protective breadth: variant‐proof SARS‐CoV‐2 vaccines, pan‐sarbecovirus vaccines, and pan‐β‐CoVs vaccines.
4.1. Variant‐proof SARS‐CoV‐2 vaccines
Some vaccines have been reported as having broad protective efficacy against SARS‐CoV‐2 variants (Table 2). For instance, a multivalent COVID‐19 inactivated trivalent vaccine based on the original strain (HB02), Delta, and Omicron provided broad‐spectrum protection against SARS‐CoV‐2 HB02, Beta, Delta, and Omicron variants in humoral immunity. The multivalent vaccine could enhance the immune responses of virus‐specific T cells in immunized mice. The authors clarified that cellular immune responses could work in their inactivated vaccines, but the mechanism remained to be studied. 116 Moreover, the RBD‐dimer protein subunit vaccine ZF2001 could exhibit a broadly neutralizing antibody response against five VOCs (Alpha, Beta, Gamma, Delta, and Omicron) and three other variants of interest (VOIs) (Epsilon, Eta, and Kappa). 65 , 117 , 118 At present, ZF2001 has been used in China and several other countries. 119 Furthermore, nanoparticle‐based protein vaccines could elicit broad immunity. For example, the vaccine RBD‐NP utilized a two‐component icosahedral protein nanoparticle (I53‐50) to display the SARS‐CoV‐2 RBD protein. 113 The vaccine's efficacy in different animal models was confirmed (Table 2). And RBD‐NP could elicit potent neutralizing antibody responses against Alpha, Beta, and Gamma variants, as well as 12 single‐residue SARS‐CoV‐2 RBD mutants. Simultaneously, they found that RBD‐NP‐induced polyclonal Abs weakly neutralized SARS‐CoV pseudovirus. Also, Joyce et al. have designed a ferritin nanoparticle immunogen named SpFN that recapitulates the structural and antigenic property of the prefusion SARS‐CoV‐2 S‐domain with two stabilizing prolines (K986P, V987P). 120 , 121 SpFN formulated with a liposome‐based adjuvant, ALFQ, could induce Th1‐biased immune responses and elicit potent binding and neutralizing antibodies against SARS‐CoV‐2 VOCs and SARS‐CoV in immunized mice.
Table 2.
Variant‐proof COVID‐19 vaccines
| Variant‐proof COVID‐19 vaccines | Properties | Species immunized | Dose | Neutralization assay method | Neutralizing and cross‐neutralizing activity for SARS‐CoV‐2 and its variants | Reference |
|---|---|---|---|---|---|---|
| Multivalent COVID‐19 inactivated vaccine | HB02 + Delta + Omicron trivalent inactivated vaccine | Mice |
2 doses Day 0‐prime Day 21‐boost |
Microplate CPE (micro‐cytopathogenic efficiency) assay |
WT(HB02): GM NT50 up to ~3271 Beta: GM NT50 up to ~1753 Delta: GM NT50 up to ~1544 Omicron (B.1): GM NT50 up to ~2072 |
[116] |
| ZF2001 | Tandem‐repeat dimeric RBD (Wuhan‐Hu‐1) with alum‐based adjuvant | Human (in clinical phase 3) |
3 doses Month 0‐prime Month 1‐boost Month 4–6‐boost |
VSV‐ΔG‐GFP based SARS‐CoV‐2 pseudovirus neutralization assay |
WT: pVNT50 ~390 Alpha: pVNT50 ~536 Beta: pVNT50 ~323 Gamma: pVNT50 ~418 Delta: pVNT50 ~407 Omicron: (BA.2: pVNT50 ~629 BA.2.12.1: pVNT50 ~457 BA.4/BA.5: pVNT50 ~270) Epsilon: pVNT50 ~529 Eta: pVNT50 ~310 Kappa: pVNT50 ~262 |
[65, 117, 118] |
| RBD‐NP | Multivalent SARS‐CoV‐2 RBD nanoparticle (RBD‐NP) with AS03‐adjuvanted |
Rhesus macaques Pigtail macaques |
2 doses for Rhesus macaques Day 0‐prime Day 21‐boost 3 doses for Pigtail macaques Day 0‐prime Day 28‐boost Day 168‐boost |
HIV‐based SARS‐CoV‐2 pseudovirus neutralization assay |
Rhesus macaques WT(D614G): GM NT50 up to ~103 Alpha: GM NT50 up to ~103 Beta: GM NT50 up to ~102 Gamma: GM NT50 up to ~102 Pigtail macaques: WT(D614G): GM NT50 up to ~2 × 104 Alpha: GM NT50 up to ~2 × 104 Beta: GM NT50 up to ~8 × 103 |
[113, 172] |
| SpFN | A stabilized S trimer with stabilizing prolines (K986P, V987P), removal of the furin cleavage site (RRAS to GSAS) ormation on the ferritin scaffold with ALFQ‐adjuvanted | Mice |
3 doses Day 0‐prime Day 21‐boost Day 42‐boost |
HIV‐based SARS‐CoV‐2 pseudovirus neutralization assay |
WT(Wuhan): GM NT50 up to ~104 Alpha: GM NT50 up to ~2 × 103 Beta: GM NT50 up to ~2 × 103 |
[120, 173] |
|
circRNARBD‐Delta (circular RNA) |
A covalently closed ring RNA molecules encoding trimeric SARS‐CoV‐2 Delta‐RBD | Mice |
2 doses Day 0‐prime Day 14‐boost |
VSV‐based SARS‐CoV‐2 pseudovirus neutralization assay |
WT: GM NT50 up to ~104 Alpha: GM NT50 up to ~104 Beta: GM NT50 up to ~103 Delta: GM NT50 up to ~1.4 × 105 Omicron: GM NT50 up to ~ 4.7 × 103 |
[122] |
| VFLIP‐X | A circRNA encoding the full‐length engineered VFLIP S protein possessing native‐like glycosylation with substitutions of six amino acids (D614G, K417N, L452R, T478K, E484K, and N501Y). | Mice |
2 doses Day 0‐prime Day 21‐boost |
Microneutralization assay of infectious SARS‐CoV‐2 variant Lentivirus‐based pseudovirus neutralization assay |
Infectious SARS‐CoV‐2 variants: WT: NT50 up to ~102 Alpha: NT50 up to ~103 Beta: NT50 up to ~103 Delta: NT50 up to ~102 Omicron: NT50 up to ~102 Pseudovirus: B.1: pVNT50 up to ~103 Epsilon: pVNT50 up to ~102 Mu: pVNT50 up to ~102 Lambda: pVNT50 up to ~102 Omicron: pVNT50 up to ~103 |
[123] |
Abbreviations: CTD, C terminal domain; HR, heptad repeat; NTD, N terminal domain; RBD, receptor binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Apart from these protein‐based subunit vaccines, a circular RNA (circRNA) vaccine that expresses trimeric RBD has been reported. 122 The circRNARBD‐Delta vaccine elicited high levels of broad‐spectrum neutralizing antibodies against four VOCs (Table 2). The NT50 against Omicron variants elicited by circRNARBD‐Delta was about 4.7 × 103. After that, Seephetdee et al. reported a circular mRNA vaccine that produces a VFLIP‐X S protein that contains six substituted amino acids, including K417N, L452R, T478K, E484K, N501Y, and D614G. It conferred broad neutralization against SARS‐CoV‐2 VOCs and VOIs in immunized mice. A two‐dose immunization of VFLIP‐X induced strong Omicron S‐specific Th1‐biased cellular immune response in mice. 123 They demonstrated that circRNAs exhibit superior stability to their linear counterparts owing to the covalently closed structure, which could prevent exonuclease degradation and thus prolong the half‐life of RNA. In conclusion, these different designs and technology routes provide a broader understanding and framework for the development and advancement of ongoing and future SARS‐CoV‐2 vaccines, as well as pan‐coronavirus vaccines.
4.2. Pan‐sarbecovirus vaccines
Our group has developed a pan‐sarbecovirus vaccine, CF501/RBD‐Fc, containing an RBD of original SARS‐CoV‐2 strain conjugated with a human IgG1 Fc fragment and a stimulator of interferon genes (STING) agonist CF501 as an adjuvant (Figure 4). STING agonist was found to be a new candidate adjuvant to promote the transcription of type I interferons (IFNs) and other pro‐inflammatory cytokines for modulating antigen presentation and immune responses. 124 Cyclic GMP‐AMP (cGAMP) is a natural agonist of STING. 125 We previously reported a pulmonary surfactant–biomimetic liposome encapsulating cGAMP (PS‐cGAMP), which could be used as an adjuvant for influenza H1N1 vaccine and elicited a strong and durable heterosubtypic immunity response. 126 However, several limitations have arisen when using cGAMP as an adjuvant since it showed high sensitivity to phosphodiesterases and did not easily accumulate in the cell cytoplasm. 125 , 127 Based on these findings, we further designed and synthesized several small‐molecule STING agonists and found that one of these compounds, CF501, could effectively activate the STING‐TBK1‐IRF3 signaling pathway in vitro and potently, if only transiently, activate innate immunity in vivo. 115 , 128 Furthermore, when CF501 was used as the adjuvant for original SARS‐CoV‐2 RBD‐Fc vaccines, CF501/RBD‐Fc could potently enhance both humoral and T‐cell immune responses, as well as significantly increase Th‐1 immune responses. It also elicited potent cross‐nAb responses against SARS‐CoV‐2 and its variants, including 41 S‐mutants, five VOCs (Alpha, Beta, Gamma, Delta, and Omicron), and five previously identified VOIs (Epsilon, Zeta, Eta, Iota, and Kappa), as well as SARS‐CoV and SARS‐related coronaviruses (SARSr‐CoVs). Notably, CF501/RBD‐Fc‐elicited antibodies in the sera of immunized macaques could potently neutralize Omicron. 128 The animal challenge test indicated that CF501/RBD‐Fc almost completely protected immunized human ACE2 transgenic mice (hACE2‐Tg mice) from SARS‐CoV‐2 challenge, even at 6 months post the first immunization. Similarly, CF501/RBD‐Fc could also elicit durable protective immunity in NHPs since the nAb against SARS‐CoV‐2 remained at high titer for 6 months, and the viral loads in the upper and lower airways were remarkably reduced when macaques were challenged at 108 days post the final immunization. The potent, durable, and broad protective immunity elicited by CF501/RBD‐Fc is promising to combat the current and future sarbecoviruses.
Figure 4.
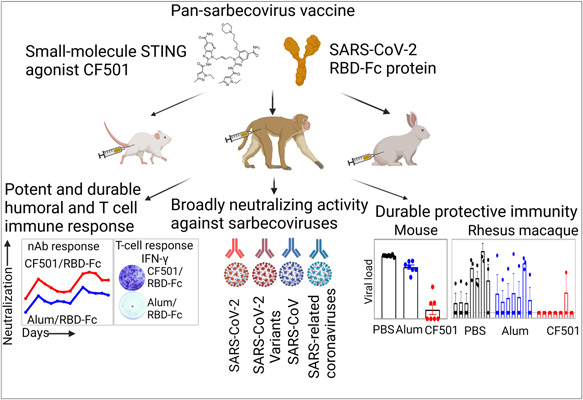
Potent efficacy of pan‐sarbecovirus vaccine CF501/RBD‐Fc. CF501 is a small‐molecule STING agonist. severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) receptor binding domain (RBD)‐Fc adjuvanted with CF501 elicited potent and durable neutralizing antibody and T cell responses, broad neutralizing activity against sarbecoviruses, and durable protective immunity in mice, rabbits, and nonhuman primates.
Simultaneous co‐display of a variety of different β‐coronavirus RBDs onto nanoparticles, such as Spycather003‐mi3, I53‐50A/I53‐50B, or ferritin, provides another strategy to develop pan‐sarbecovirus vaccines (Table 3, Figure 5A). For instance, based on the design of a multivalent subunit vaccine (RBD‐NP) displaying the SARS‐CoV‐2 RBD, Walls et al. further designed mosaic (mRBD‐NP) and cocktail (cRBD‐NP) nanoparticle immunogens displaying four RBDs from SARS‐CoV‐2, SARS‐CoV, and the bat coronaviruses WIV1 and RaTG13. These RBDs were genetically fused to the trimeric I53‐50A and then mixed with the I53‐50B pentamer as shown in Figure 5A. They found that both mRBD‐NP and cRBD‐NP elicited broad cross‐reactive neutralizing sarbecovirus Abs against SARS‐CoV‐2, SARS‐CoV, and RsSHC014 pseudoviruses and protected immunized mice against heterotypic SARS‐CoV MA15 challenge. 113 Beyond recognizing strains displayed on the nanoparticles, mosaic nanoparticles elicited antibodies that also recognized mismatched strains. Similarly, SpyTagged RBDs from SARS‐CoV‐2‐Beta and seven sarbecoviruses were simultaneously displayed on SpyCatcher003‐mi3 nanoparticles, which were termed as mosaic‐8b. 80 , 129 Mosaic‐8b elicited significantly higher neutralizing titers of antisera than homotypic SARS‐CoV‐2 RBD nanoparticles against WIV1 and RsSHC014 and mismatched strains, such as SARS‐CoV. After three doses of mosaic‐8b immunization, the polyclonal antisera in NHPs recognized the RBDs and neutralized pseudoviruses from different sarbecoviruses, including the matched SARS‐CoV‐2 Beta, WIV1, RsSHC014, as well as some mismatched viruses (SARS‐CoV, LYRa3, RshSTT200, BM48–31, BtKY72, Khosta‐2, and Yun11). Researchers observed that mosaic‐8b elicited strong immune responses and protected K18‐hACE2 mice from SARS‐CoV‐2 and SARS‐CoV challenges. In a similar approach, a homotypic SARS‐CoV‐2 RBD‐based sortase A‐conjugated ferritin nanoparticle, 130 named RBD‐scNP, could induce neutralizing antibodies against SARS‐CoV‐2 and three pre‐emergent sarbecoviruses, SARS‐CoV, WIV1, and RsSHC014. 131 RBD‐scNP protected immunized NHPs from SARS‐CoV‐2 WA‐1, Beta, and Delta variant challenges and protected aged mice from challenges of SARS‐CoV‐2 Beta variant, SARS‐CoV and RsSHC014. In general, the three nanoparticles resulted in high neutralizing antibody levels against not only SARS‐CoV‐2 variants, but also different sarbecoviruses. These results suggested that RBD immunogenicity can be augmented by arraying multiple copies on nanoparticles.
Table 3.
Pan‐sarbecovirus (β‐CoV‐B) vaccines
| Pan‐sarbecovirus vaccines | Properties | Animal immunized | Dose | Analysis method | Neutralizing and cross‐neutralizing activity against sarbecovirus | Reference | |
|---|---|---|---|---|---|---|---|
| CF501/RBD‐Fc | SARS‐CoV‐2 RBD conjugated with a human IgG Fc fragment (RBD‐Fc) and a STING agonist CF501 as an adjuvant |
Mice Rabbits Rhesus macaques |
3 doses for mice Day 0‐prime Day 14‐boost Day 28‐boost 4 doses for rabbits Day 0‐prime Day 14‐boost Day 28‐boost Day 42‐boost 3 doses for NHPs Day 0‐prime Day 21‐boost Day 115‐boost |
HIV‐based pseudovirus neutralization assay Cell‐cell fusion assay |
Mice: SARS‐CoV‐2: GM NT50 up to ~104 SARS‐CoV: GM NT50 up to ~102 Rs3367: GM NT50 up to ~103 WIV1: GM NT50 up to ~102 RsSHC014: IT50 up to ~103 Rabbit SARS‐CoV‐2: GM NT50 up to ~103 SARS‐CoV: GM NT50 up to ~103 Rs3367: GM NT50 up to ~104 WIV1: GM NT50 up to ~103 RsSHC014: IT50 up to ~103 Rhesus macaques SARS‐CoV: GM NT50 up to ~103 Rs3367: GM NT50 up to ~104 WIV1: GM NT50 up to ~104 |
115 | |
| Mosaic‐sarbecovirus RBD‐NPs | Equimolar mixture of RBD of SARS‐CoV‐2, SARS‐CoV, WIV1, and RaTG13 were added to nanoparticles | Mice |
2 doses Day 0‐prime Day 21‐boost |
MLV‐based SARS‐CoV‐2 and SARS‐CoV pseudovirus neutralization assay VZV‐based RsSHC014 pseudovirus neutralization assay |
SARS‐CoV‐2(D614G): GM NT50 up to ~103 SARS‐CoV: GM NT50 up to ~103 RsSHC014: GM NT50 up to ~102 |
113 | |
| Cocktail‐sarbecovirus RBD‐NPs | Independently assembled nanoparticles, each displaying a single RBD from SARS‐CoV‐2, SARS‐CoV, WIV1, or RaTG13 | Mice |
2 doses Day 0‐prime Day 21‐boost |
MLV‐based SARS‐CoV‐2 and SARS‐CoV pseudovirus neutralization assay VZV‐based SHC014 pseudovirus neutralization assay |
SARS‐CoV‐2(D614G): GM NT50 up to ~103 SARS‐CoV: GM NT50 up to ~103 RsSHC014: GM NT50 up to ~102 |
113 | |
| Mosaic‐8b | Nanoparticle presenting the SARS‐2 Beta RBD and seven other sarbecovirus RBDs (RaTG13, RsSHC014, and Rs4081 pang17, RmYN02, Rf1, and WIV1) |
Mice cynomolgus macaques |
2 doses for mice Day 0‐prime Day 28‐boost 3 doses for NHPs Day 0‐prime Day 28‐boost Day 92‐boost |
HIV‐based pseudovirus neutralization assay |
Mice: SARS‐CoV‐2(Wuhan‐hu‐1): GM NT50 up to ~102 SARS‐CoV: GM NT50 up to ~103 WIV1: GM NT50 up to ~104 RsSHC014: GM NT50 up to ~104 Cynomolgus macaques: SARS‐CoV: ID50 up to ~103 WIV1: ID50 up to ~103 RsSHC014: ID50 up to ~104 |
80 | |
| SARS‐CoV‐2 RBD‐scNP | SARS‐CoV‐2 RBD sortase A‐conjugated ferritin nanoparticle formulated with the TLR7/8 agonist 3M‐052‐aqueous formulation (3M‐052‐AF) plus Alum | Cynomolgus macaque |
3 doses Day 0‐prime Day 28‐boost Day 56‐boost |
Live virus neutralization assays. |
SARS‐CoV‐2: ID50 up to ~104 SARS‐CoV: ID50 up to ~103 WIV1: ID50 up to ~103 RsSHC014: ID50 up to ~102 |
130, 131 | |
| Chimeric spike mRNA vaccines | Nucleoside‐modified mRNA‐lipid nanoparticle (LNP) vaccines expressing chimeric spikes that contain admixtures of different RBD, NTD, and S2 modular domains from HKU3‐1, SARS‐CoV, SARS‐CoV‐2, or RsSHC014 | Mice |
2 doses Day 0‐prime Day 21‐boost |
Nanoluciferase‐expressing‐based recombinant live viruses neutralization assay. |
SARS‐CoV‐2: logID50 up ~102 SARS‐CoV: logID50 up ~104 WIV1: logID50 up ~103 RsSHC014: logID50 up ~103 |
114 | |
Abbreviations: CTD, C terminal domain; HR, heptad repeat; NTD, N terminal domain; RBD, receptor binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Figure 5.
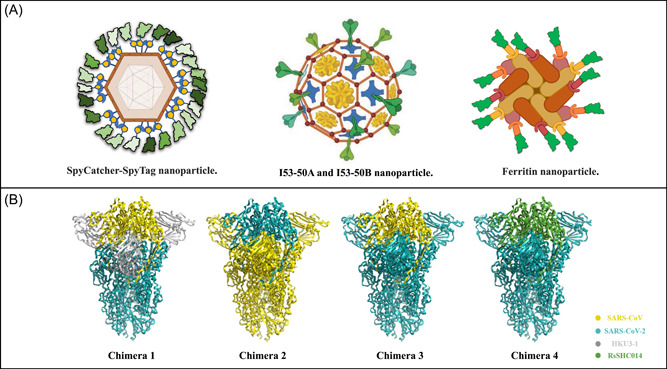
Strategies of protein nanoparticle vaccines and antigen chimera vaccines for broad‐spectrum neutralization. (A) Three representative types of selfassembled nanoparticles for antigen delivery. Spy‐tagged antigen was conjugated with a Spycather003‐mi3, forming nanoparticles. I53‐50A and I53‐50B could assemble into virus‐like particle (VLP) nanoparticles after mixing. Helicobacter pylori‐derived ferritin conjugated with antigens could form 24‐mer or 60‐mer nanoparticles. (B) Four types of genetically fused S chimera trimer were shown above, and S protein was generally divided into 3 parts, N terminal domain (NTD), receptor binding domain (RBD), and others (CTD and S2). For each part, one type of coronavirus was filled in with specific colors. Yellow stands for SARS‐CoV, teal for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), gray for HKU3‐1 and green for RsSHC014. Chimera 1 contained SARS‐CoV (RBD)—HKU3‐1 (NTD)—SARS‐CoV‐2 (Others); chimera 2 contained SARS‐CoV‐2 (RBD)—SARS‐CoV (NTD)—SARS‐CoV (Others); chimera 3 contained SARS‐CoV (RBD)—SARS‐CoV‐2 (NTD)—SARS‐CoV‐2 (Others) and chimera 4 contained RsSHC014 (RBD)—SARS‐CoV‐2 (NTD)—SARS‐CoV‐2 (Others).
The above protein‐based vaccines show promise for the development of β‐CoV vaccines against sarbecoviruses. A novel mRNA chimeric vaccine could also perform broad immune responses against sarbecoviruses. Nucleoside‐modified mRNA‐lipid nanoparticle (LNP) vaccines were designed by splicing different RBD, NTD, and S2 domains from zoonotic and pandemic Hong Kong University 3‐1(HKU3‐1), SARS‐CoV, SARS‐CoV‐2, and RsSHC014 into chimeric spikes (Figure 5B). For example, one chimera consists of HKU3‐1 NTD, SARS‐CoV RBD, and SARS‐CoV‐2 S2. The authors have verified that mRNA‐LNP could stimulate robust germinal center (GC) B cell responses, durable long‐lived plasma cells, as well as T follicular helper cell activity. The vaccines elicited neutralizing antibody responses against live SARS‐CoV, RsSHC014, WIV1, SARS‐CoV‐2 infection, infection from SARS‐CoV‐2 VOCs, and protected aged mice from SARS‐CoV, SARS‐CoV‐2 and RsSHC014 challenges. Mice immunized with chimeric spike mRNA vaccines fully prevented heterologous WIV1 challenge, whereas mice that received the monovalent SARS‐CoV‐2 mRNA vaccines had breakthrough replication in their lung. 114
4.3. Research and development of pan‐β‐CoV vaccines
To date, the three life‐threatening CoVs identified all belong to β‐CoV, so it is essential to develop vaccines against CoVs in these genera. Development of pan‐β‐CoV vaccines is difficult, and only few studies have reported on pan‐β‐CoV. The United Kingdom government is now funding clinical trials for a new vaccine candidate called DIOS‐CoVax2, which was developed by researchers from DIOSynVax. They used 3D computer modeling to design Vaccine Antigen Payloads (the immune instructions of a vaccine) from multiple synthetic antigens. Vaccine Antigen Payloads can be deployed in the form of vaccine vectors, such as nucleic acid‐based, virus vectored‐based, and protein‐based platforms. DIOS‐CoVax2 was designed to provide broad protection against multiple β‐CoVs, such as SARS‐CoV and MERS‐CoV, that bear rapid transmission and high pathogenicity among humans (https://www.diosvax.com/technology). Yet, so far, the clinical data of this pan‐β‐CoV vaccine have not been reported.
Although few pan‐β‐CoV vaccines have been reported, it is worth noting that many cross‐reactive epitopes in CoVs have recently been described. 56 , 81 , 132 , 133 Pinto et al. recently described some mAbs isolated from COVID‐19 convalescent individuals, which exhibited cross‐reactivity targeting the stem helix of multiple β‐CoV spike glycoproteins. 134 Among these mAbs, S2P6 exhibited the broadest neutralizing activity. S2P6 showed similar potency against SARS‐CoV‐2 VOCs, including Alpha, Beta, Gamma, and Kappa, compared with parental SARS‐CoV‐2 D614G. Moreover, S2P6 could broadly neutralize SARS‐CoV, Pangolin Guangdong 2019 (PANG/GD), MERS‐CoV, and OC43 S pseudoviruses with median inhibitory concentration (IC50) values ranging from 0.02 to 17 μg/ml. S2P6 is therefore a broad‐spectrum anti‐coronavirus neutralizing mAb. Simultaneously, Sun et al. isolated a human antibody named 76E1, which has extremely broad neutralizing activity against multiple α‐ and β‐coronaviruses, including SARS‐CoV‐2 and its variants. The antibody targets a highly conserved S2' cleavage site and fusion peptide epitope within the S protein, and the crucial epitope is exposed when the S protein binds to ACE2. This observation suggests that 76E1 blocks membrane fusion and viral entry by binding to the epitope at an intermediate conformation of S during the transition process from the prefusion to the post‐fusion state. 135 These data, along with the structural and functional characteristics of conservative epitopes in different β‐CoVs, provide insights for the rational design of pan‐β‐CoV vaccines.
5. CONCLUSION AND PERSPECTIVES
In this review, we first discussed the main targets of current SARS‐CoV‐2 vaccines that produce broad‐spectrum protection. Then, we summarized the protective efficacy of currently approved vaccines. Finally, we mainly reviewed the broad‐spectrum anti‐coronavirus vaccines, including variant‐proof SARS‐CoV‐2 vaccines, pan‐sarbecovirus vaccines, and pan‐β‐CoV vaccines, currently in development.
As the COVID‐19 pandemic continues, the effective global control of COVID‐19 via immunization with first‐generation vaccines is threatened by VOCs and waning vaccine‐induced antibody immunity. 9 , 136 , 137 Nowadays, Omicron has outcompeted the previous VOCs and exhibits the largest prevalence in the world. It is worth noting that Omicron infections have been mainly in the upper respiratory tract, thriving in the throat and nose, but not in the lung within the lower airway system, making it easy for virus particles to be exhaled as aerosols from the nose or mouth and leading to new host infections. 138 However, while the current SARS‐CoV‐2 vaccines might reduce disease severity, they fail to prevent infection of SARS‐CoV‐2 VOCs, particularly Omicron variant, since Omicron breakthrough infection has occurred in many vaccinated individuals. 138 , 139 , 140 , 141 In addition, Zhou et al. have described robust SARS‐CoV‐2 infection in nasal turbinates of golden Syrian hamsters challenged with SARS‐CoV‐2, despite the presence of potent systemic neutralizing antibodies. 142 Therefore, given the transmission characteristics of this virus and the complexity of adaptive host immunity, both systemic and mucosal protective immunity need to be taken into consideration when developing next‐generation vaccines (Figure 6). Currently, most vaccines in clinical trials are administered intramuscularly or intradermally to elicit activated T and B cell responses, but these routes of vaccination are unable to induce effective mucosal IgA antibodies or tissue‐resident T cells (TRM) in bodies. 143 Notably, intranasal (IN) immunization effectively induces the establishment of IgA‐secreting cells, mucosal IgA antibodies, and TRM in the lung (Figure 6). 144 , 145 , 146 Therefore, mucosal vaccination should be emphasized as it would be beneficial to induce robust mucosal immune responses against SARS‐CoV‐2 transmission through the respiratory tract, rather than only curtail infection or prevent the development of disease symptoms. Accordingly, an aerosolized Ad5‐nCoV vaccine has been developed by CanSino Biologics. This vaccine against COVID‐19 in an aerosol route with an equivalent to one‐ or two‐fifths of an intramuscular (IM) dose was well tolerated in healthy adults in clinical phase 1 trial. 147 This aerosol vaccination could trigger not only broad and polyfunctional T cell responses, but also a higher ratio of neutralizing antibodies to total antibodies compared with intramuscular vaccination. Their recent clinical results showed that a booster vaccination with aerosolized Ad5‐nCoV induced an effective serum nAbs response against SARS‐CoV‐2 in adults who had already received two doses of CoronaVac. 148 The increased knowledge of mucosal antigen‐presenting cells and resident memory cells in mucosal sites allows for the design of antigens and the development of effective mucosal adjuvants, thus facilitating the rational design of next‐generation mucosal vaccines. 149
Figure 6.
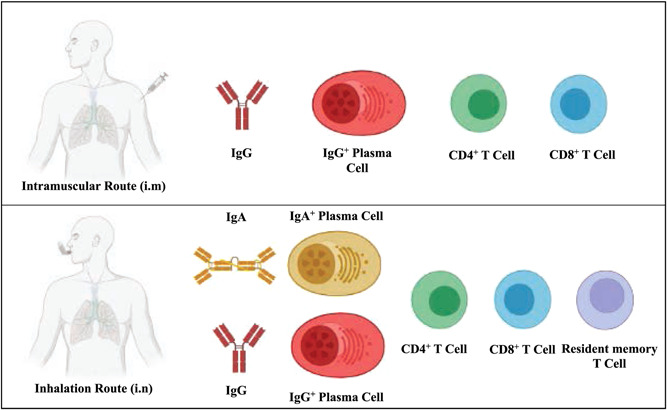
Corresponding differences in immunization reactions between inhalation and intramuscular route. Intramuscular immunization induces antigen‐directed innate and adaptive responses and activates T and B cell responses. IgG+ plasma cells are generated, differentiated and maturated to create anti‐antigen neutralization antibodies. Apart from these immunological responses, inhalation immunization also induces tissue‐resident T cells and IgA+ B cells for circulating mucosal IgA antibodies.
Recently, some studies have noted that the S2 subunit is an alternative target that harbors promising cross‐reactive antibodies and CD4+T cell epitopes. 150 , 151 , 152 Identification of conserved epitopes in S2 may hold promise for developing a universal pan‐coronavirus vaccine. 153 Accordingly, Ma et al. developed a combination of nanoparticle vaccines by displaying SARS‐CoV‐2 RBD and/or HR1 to HR2 fragment from S2. They found that RBD‐HR nanoparticle vaccines could induce nAbs against pseudotyped SARS‐CoV, MERS‐CoV, HCoV‐229E, HCoV‐OC43, and RaTG13 and elicit T cell responses potentially cross‐reactivated with β‐coronavirus HCoV‐OC43 and α‐coronavirus HCoV‐229E. 154 More recently, Ng et al. observed that two doses of a SARS‐CoV‐2 S2‐targeted DNA vaccine could confer protection against infection with distinct coronaviruses. S2‐targeted vaccination could produce relatively weak neutralizing activities against diverse α‐coronaviruses and β‐CoVs, including SARS‐CoV‐2 Wuhan strain and its two VOCs (Alpha and Beta), HCoV‐HKU1, HCoV‐OC43, HCoV‐229E, HCoV‐NL63, WIV1, and RaTG13. Nevertheless, the relatively low immunogenicity of S2 has been a matter of debate. It has been explained that some neutralizing epitopes on S2 subunit are shielded by N‐glycan or other parts in trimeric S, making S2 less accessible for immune recognition than the S1 subunit. 155 , 156 Therefore, the addition of novel adjuvants or new antigen presentation strategies to stimulate these concealed epitopes on the S2 subunit may be a viable option for improving S2 immunogenicity and developing broad‐spectrum protective vaccines.
In general, the first‐generation SARS‐CoV‐2 vaccines will ultimately be replaced by second‐generation vaccines with broader protective efficiency and more durable immunity. Meanwhile, as a continuing risk, animal coronaviruses may spill over from animals to humans and generate novel viruses by recombination, considering the continuous and frequent contact between humans and wild animals. 157 , 158 We must therefore accelerate our efforts in the development of broadly protective vaccines. 159 However, there remain some challenges in the development of broad‐spectrum sarbecovirus and β‐coronavirus vaccines. For instance, it is difficult to construct a vaccine consisting of the highly conserved neutralizing epitopes in coronaviruses, such as those in S2 that we mentioned above, in appropriate conformation. An approved novel adjuvant that can strongly enhance the immunogenicity of vaccine to elicit potent and long‐lasting neutralizing antibody and T cell immune responses is still not available. These suggest that it is not easy to promptly develop a broad‐spectrum coronavirus vaccine, but the pan‐sarbecovirus and pan‐β‐coronavirus vaccines are still urgently needed for combating the pandemics or epidemics that will be caused by emerging or reemerging coronaviruses in the foreseeable future.
AUTHOR CONTRIBUTIONS
Shibo Jiang, Lu Lu, and Zezhong Liu conceived the idea and planned the study. Jie Zhou, Zezhong Liu, Wei Xu, and Lixiao Xing collected the data and devised the initial draft. Guangxu Zhang and Zezhong Liu modified figures and performed critical reading. Shibo Jiang, Qian Wang, and Lu Lu reviewed, finalized, and approved the final version of the manuscript for submission.
CONFLICTS OF INTEREST
Shibo Jiang, Lu Lu, Zezhong Liu, Jie Zhou, Wei Xu, and Qian Wang are inventors of the patent application related to the pan‐sarbecovirus vaccines described in this review, while other authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (92169112 and 82161138002 to Shibo Jiang), the National Key Research and Development Program of China (2021YFC2300703 to Lu Lu), and the Shanghai Municipal Science and Technology Major Project (ZD2021CY001 to Shibo Jiang and Lu Lu).
Zhou J, Liu Z, Zhang G, et al. Development of variant‐proof severe acute respiratory syndrome coronavirus 2, pan‐sarbecovirus, and pan‐β‐coronavirus vaccines. J Med Virol. 2022;1‐19. 10.1002/jmv.28172
Jie Zhou and Zezhong Liu contributed equally to this study.
Contributor Information
Qian Wang, Email: wang_qian@fudan.edu.cn.
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Volz E, Mishra S, Chand M, et al. Assessing transmissibility of SARS‐CoV‐2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266‐269. [DOI] [PubMed] [Google Scholar]
- 2. Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS‐CoV‐2 variant of concern in South Africa. Nature. 2021;592(7854):438‐443. [DOI] [PubMed] [Google Scholar]
- 3. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science. 2021;372:815‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS‐CoV‐2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220‐4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carreño JM, Alshammary H, Tcheou J, et al. Activity of convalescent and vaccine serum against SARS‐CoV‐2 omicron. Nature. 2022;602(7898):682‐688. [DOI] [PubMed] [Google Scholar]
- 6. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130‐135. [DOI] [PubMed] [Google Scholar]
- 7. Shrestha LB, Foster C, Rawlinson W, Tedla N, Bull RA. Evolution of the SARS‐CoV‐2 omicron variants BA.1 to BA.5: implications for immune escape and transmission. Rev Med Virol. 2022;32:e2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrews N, Stowe J, Kirsebom F, et al. Covid‐19 vaccine effectiveness against the omicron (B.1.1.529) variant. N Engl J Med. 2022;386(16):1532‐1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann M, Krüger N, Schulz S, et al. The omicron variant is highly resistant against antibody‐mediated neutralization: implications for control of the COVID‐19 pandemic. Cell. 2022;185(3):447‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma KA, Kumar A. Molecular aspects of omicron, vaccine development, and recombinant strain XE: a review. J Med Virol. 2022;94:4628‐4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353‐1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang N, Shang J, Jiang S, Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 16. Liu Z, Xia S, Wang X, et al. Inefficiency of sera from mice treated with pseudotyped SARS‐CoV to neutralize 2019‐nCoV infection. Virol Sin. 2020;35(3):340‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;588:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Menachery VD, Yount BL Jr., Debbink K, et al. A SARS‐like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21(12):1508‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol. 2021;19(3):141‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Menachery VD, Yount BL Jr., Sims AC, et al. SARS‐like WIV1‐CoV poised for human emergence. Proc Natl Acad Sci USA. 2016;113(11):3048‐3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X, Zhang L, Chen S, Ouyang H, Ren L. Possible targets of Pan‐Coronavirus antiviral strategies for emerging or Re‐Emerging coronaviruses. Microorganisms. 2021;9(7):1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Boheemen S, de Graaf M, Lauber C, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3(6): e00473‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elhazmi A, Al‐Tawfiq JA, Sallam H, et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and middle east respiratory syndrome coronavirus (MERS‐CoV) coinfection: a unique case series. Travel Med Infect Dis. 2021;41:102026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su S, Li W, Jiang S. Developing pan‐β‐coronavirus vaccines against emerging SARS‐CoV‐2 variants of concern. Trends Immunol. 2022;43(3):170‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang S, He Y, Liu S. SARS vaccine development. Emerging Infect Dis. 2005;11(7):1016‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS‐CoV‐‐a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du L, Tai W, Zhou Y, Jiang S. Vaccines for the prevention against the threat of MERS‐CoV. Expert Rev Vaccines. 2016;15(9):1123‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srinivasan S, Cui H, Gao Z, et al. Structural genomics of SARS‐CoV‐2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus‐2 (SARS CoV‐2 or n‐COV19), the cause of COVID‐19. Protein J. 2020;39(3):198‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295(37):12910‐12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jadaan SA, Khan AW. Recent update of COVID‐19 vaccines. Adv Pharm Bull. 2022;12(2):219‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science. 2005;309(5742):1864‐1868. [DOI] [PubMed] [Google Scholar]
- 38. Yi C, Sun X, Ye J, et al. Key residues of the receptor binding motif in the spike protein of SARS‐CoV‐2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol. 2020;17(6):621‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou J, Xu W, Liu Z, et al. A highly potent and stable pan‐coronavirus fusion inhibitor as a candidate prophylactic and therapeutic for COVID‐19 and other coronavirus diseases. Acta Pharm Sin B. 2022;12(4):1652‐1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang S, Lu L, Liu Q, Xu W, Du L. Receptor‐binding domains of spike proteins of emerging or re‐emerging viruses as targets for development of antiviral vaccines. Emerg Microbes Infect. 2012;1(8):e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Z, Ruan P, Wang L, Nie X, Ma X, Tan YT. B cell epitope analysis of SARS‐CoV‐2 S protein based on immunoinformatics and experimental research. J Cell Mol Med. 2021;25(2):1274‐1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID‐19 coronavirus (SARS‐CoV‐2) based on SARS‐CoV immunological studies. Viruses. 2020;12(3):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li T, Zheng Q, Yu H, et al. SARS‐CoV‐2 spike produced in insect cells elicits high neutralization titres in non‐human primates. Emerg Microbes Infect. 2020;9(1):2076‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pallesen J, Wang N, Corbett KS, et al. Immunogenicity and structures of a rationally designed prefusion MERS‐CoV spike antigen. Proc Natl Acad Sci USA. 2017;114(35):E7348‐E7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Premkumar L, Segovia‐Chumbez B, Jadi R, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS‐CoV‐2 patients. Sci Immunol. 2020;5(48):eabc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Du L, He Y, Jiang S, Zheng BJ. Development of subunit vaccines against severe acute respiratory syndrome. Drugs Today (Barc). 2008;44(1):63‐73. [DOI] [PubMed] [Google Scholar]
- 47. Jiang S, Bottazzi ME, Du L, et al. Roadmap to developing a recombinant coronavirus S protein receptor‐binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccines. 2012;11(12):1405‐1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou Y, Yang Y, Huang J, Jiang S, Du L. Advances in MERS‐CoV vaccines and therapeutics based on the receptor‐binding domain. Viruses. 2019;11(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. He Y, Zhou Y, Liu S, et al. Receptor‐binding domain of SARS‐CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He Y, Lu H, Siddiqui P, Zhou Y, Jiang S. Receptor‐binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation‐dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908‐4915. [DOI] [PubMed] [Google Scholar]
- 51. Du L, Zhao G, He Y, et al. Receptor‐binding domain of SARS‐CoV spike protein induces long‐term protective immunity in an animal model. Vaccine. 2007;25(15):2832‐2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Du L, Zhao G, Chan CC, et al. Recombinant receptor‐binding domain of SARS‐CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393(1):144‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Du L, Zhao G, Kou Z, et al. Identification of a receptor‐binding domain in the S protein of the novel human coronavirus middle east respiratory syndrome coronavirus as an essential target for vaccine development. J Virol. 2013;87(17):9939‐9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lan J, Deng Y, Chen H, et al. Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the middle east respiratory coronavirus (MERS‐CoV) receptor‐binding domain as an antigen. PLoS One. 2014;9(11):e112602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ni L, Ye F, Cheng ML, et al. Detection of SARS‐CoV‐2‐specific humoral and cellular immunity in COVID‐19 convalescent individuals. Immunity. 2020;52(6):971‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barnes CO, West AP Jr., Huey‐Tubman KE, et al. Structures of human antibodies bound to SARS‐CoV‐2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828‐842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou Y, Liu Z, Li S, et al. Enhancement versus neutralization by SARS‐CoV‐2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021;34(5):108699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kleanthous H, Silverman JM, Makar KW, Yoon IK, Jackson N, Vaughn DW. Scientific rationale for developing potent RBD‐based vaccines targeting COVID‐19. NPJ Vaccines. 2021;6(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Z, Xu W, Xia S, et al. RBD‐Fc‐based COVID‐19 vaccine candidate induces highly potent SARS‐CoV‐2 neutralizing antibody response. Signal Transduct Target Ther. 2020;5(1):282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang J, Wang W, Chen Z, et al. A vaccine targeting the RBD of the S protein of SARS‐CoV‐2 induces protective immunity. Nature. 2020;586(7830):572‐577. [DOI] [PubMed] [Google Scholar]
- 61. Zhang N, Ji Q, Liu Z, et al. Effect of different adjuvants on immune responses elicited by protein‐based subunit vaccines against SARS‐CoV‐2 and its delta variant. Viruses. 2022;14(3):501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang S, Li Y, Dai L, et al. Safety and immunogenicity of a recombinant tandem‐repeat dimeric RBD‐based protein subunit vaccine (ZF2001) against COVID‐19 in adults: two randomised, double‐blind, placebo‐controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1107‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dai L, Gao L, Tao L, et al. Efficacy and safety of the RBD‐dimer‐based Covid‐19 vaccine ZF2001 in adults. N Engl J Med. 2022;386(22):2097‐2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang B, Dai L, Wang H, et al. Serum sample neutralisation of BBIBP‐CorV and ZF2001 vaccines to SARS‐CoV‐2 501Y.V2. Lancet Microbe. 2021;2(7):e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao X, Zhang R, Qiao S, et al. Omicron SARS‐CoV‐2 neutralization from inactivated and ZF2001 vaccines. N Engl J Med. 2022;387:277‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dai L, Zheng T, Xu K, et al. A universal design of betacoronavirus vaccines against COVID‐19, MERS, and SARS. Cell. 2020;182(3):722‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Qi H, Liu B, Wang X, Zhang L. The humoral response and antibodies against SARS‐CoV‐2 infection. Nat Immunol. 2022;23:1008‐1020. [DOI] [PubMed] [Google Scholar]
- 68. Liu Z, Lu L, Jiang S. Up or down: where comes omicron? Cell Res. 2022;32(7):601‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ke Z, Oton J, Qu K, et al. Structures and distributions of SARS‐CoV‐2 spike proteins on intact virions. Nature. 2020;588(7838):498‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu Z, Xu W, Chen Z, et al. An ultrapotent pan‐β‐coronavirus lineage B (β‐CoV‐B) neutralizing antibody locks the receptor‐binding domain in closed conformation by targeting its conserved epitope. Protein Cell. 2022;13(9):655‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hastie KM, Li H, Bedinger D, et al. Defining variant‐resistant epitopes targeted by SARS‐CoV‐2 antibodies: a global consortium study. Science. 2021;374(6566):472‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gruell H, Vanshylla K, Weber T, Barnes CO, Kreer C, Klein F. Antibody‐mediated neutralization of SARS‐CoV‐2. Immunity. 2022;55(6):925‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Barnes CO, Jette CA, Abernathy ME, et al. SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS‐CoV‐2 antibody cocktail. Science. 2020;369(6506):1010‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor‐binding site of SARS‐CoV‐2. Nature. 2020;584(7819):120‐124. [DOI] [PubMed] [Google Scholar]
- 76. Lempp FA, Soriaga LB, Montiel‐Ruiz M, et al. Lectins enhance SARS‐CoV‐2 infection and influence neutralizing antibodies. Nature. 2021;598(7880):342‐347. [DOI] [PubMed] [Google Scholar]
- 77. Jones BE, Brown‐Augsburger PL, Corbett KS, et al. The neutralizing antibody, LY‐CoV555, protects against SARS‐CoV‐2 infection in nonhuman primates. Sci Transl Med. 2021;13(593):eabf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li D, Edwards RJ, Manne K, et al. In vitro and in vivo functions of SARS‐CoV‐2 infection‐enhancing and neutralizing antibodies. Cell. 2021;184(16):4203‐4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pinto D, Park YJ, Beltramello M, et al. Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature. 2020;583(7815):290‐295. [DOI] [PubMed] [Google Scholar]
- 80. Cohen AA, van Doremalen N, Greaney AJ, et al. Mosaic RBD nanoparticles protect against challenge by diversesarbecoviruses in animal models. Science. 2022;377(6606):eabq0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu H, Wu NC, Yuan M, et al. Cross‐neutralization of a SARS‐CoV‐2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53(6):1272‐1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Burnett DL, Jackson KJL, Langley DB, et al. Immunizations with diverse sarbecovirus receptor‐binding domains elicit SARS‐CoV‐2 neutralizing antibodies against a conserved site of vulnerability. Immunity. 2021;54(12):2908‐2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Graham BS. Rapid COVID‐19 vaccine development. Science. 2020;368(6494):945‐946. [DOI] [PubMed] [Google Scholar]
- 84. Dai L, Gao GF. Viral targets for vaccines against COVID‐19. Nat Rev Immunol. 2021;21(2):73‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Al Kaabi N, Zhang Y, Xia S, et al. Effect of 2 inactivated SARS‐CoV‐2 vaccines on symptomatic COVID‐19 infection in adults: a randomized clinical trial. JAMA. 2021;326(1):35‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Palacios R, Patiño EG, de Oliveira Piorelli R, et al. Double‐blind, randomized, placebo‐controlled phase III clinical trial to evaluate the efficacy and safety of treating healthcare professionals with the adsorbed COVID‐19 (inactivated) vaccine manufactured by sinovac ‐ PROFISCOV: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yu X, Wei D, Xu W, et al. Reduced sensitivity of SARS‐CoV‐2 omicron variant to antibody neutralization elicited by booster vaccination. Cell Discov. 2022;8(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cheng SMS, Mok CKP, Leung YWY, et al. Neutralizing antibodies against the SARS‐CoV‐2 omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022;28(3):486‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Raina SK, Kumar R. “Covishield and covaxin” ‐ India's contribution to global COVID‐19 pandemic. J Family Med Prim Care. 2021;10(7):2433‐2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ella R, Vadrevu KM, Jogdand H, et al. Safety and immunogenicity of an inactivated SARS‐CoV‐2 vaccine, BBV152: a double‐blind, randomised, phase 1 trial. Lancet Infect Dis. 2021;21(5):637‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ella R, Reddy S, Blackwelder W, et al. Efficacy, safety, and lot‐to‐lot immunogenicity of an inactivated SARS‐CoV‐2 vaccine (BBV152): interim results of a randomised, double‐blind, controlled, phase 3 trial. Lancet. 2021;398(10317):2173‐2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Medigeshi GR, Batra G, Murugesan DR, et al. Sub‐optimal neutralisation of omicron (B.1.1.529) variant by antibodies induced by vaccine alone or SARS‐CoV‐2 infection plus vaccine (hybrid immunity) post 6‐months. EBioMedicine. 2022;78:103938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Heath PT, Galiza EP, Baxter DN, et al. Safety and efficacy of NVX‐CoV2373 Covid‐19 vaccine. N Engl J Med. 2021;385(13):1172‐1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hager KJ, Pérez Marc G, Gobeil P, et al. Efficacy and safety of a recombinant plant‐based adjuvanted Covid‐19 vaccine. N Engl J Med. 2022;386(22):2084‐2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Deng S, Liang H, Chen P, et al. Viral vector vaccine development and application during the COVID‐19 pandemic. Microorganisms. 2022;10(7):1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Voysey M, Costa Clemens SA, Madhi SA, et al. Single‐dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV‐19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397(10277):881‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid‐19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385(7):585‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV‐19 Covid‐19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(20):1885‐1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cameroni E, Bowen JE, Rosen LE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 omicron antigenic shift. Nature. 2022;602(7898):664‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Willett BJ, Grove J, MacLean OA, et al. SARS‐CoV‐2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022;7 (8):1161‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Logunov DY, Dolzhikova IV, Zubkova OV, et al. Safety and immunogenicity of an rAd26 and rAd5 vector‐based heterologous prime‐boost COVID‐19 vaccine in two formulations: two open, non‐randomised phase 1/2 studies from Russia. Lancet. 2020;396(10255):887‐897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single‐dose Ad26.COV2.S vaccine against Covid‐19. N Engl J Med. 2021;384(23):2187‐2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 omicron to antibody neutralization. Nature. 2022;602(7898):671‐675. [DOI] [PubMed] [Google Scholar]
- 104. Halperin SA, Ye L, MacKinnon‐Cameron D, et al. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: an international, multicentre, randomised, double‐blinded, placebo‐controlled phase 3 trial. Lancet. 2022;399(10321):237‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Anderson EJ, Rouphael NG, Widge AT, et al. Safety and immunogenicity of SARS‐CoV‐2 mRNA‐1273 vaccine in older adults. N Engl J Med. 2020;383(25):2427‐2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sahin U, Muik A, Derhovanessian E, et al. Concurrent human antibody and TH1 type T‐cell responses elicited by a COVID‐19 RNA vaccine. medRxiv. 2020;2020:2007.2017.20140533. [Google Scholar]
- 107. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Vergnes JN. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2021;384(16):1577. [DOI] [PubMed] [Google Scholar]
- 109. McCullers JA, Dunn JD. Advances in vaccine technology and their impact on managed care. P T. 2008;33(1):35‐41. [PMC free article] [PubMed] [Google Scholar]
- 110. Okamura S, Ebina H. Could live attenuated vaccines better control COVID‐19? Vaccine. 2021;39(39):5719‐5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Xia S, Yan L, Xu W, et al. A pan‐coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4):eaav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995‐4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Walls AC, Miranda MC, Schäfer A, et al. Elicitation of broadly protective sarbecovirus immunity by receptor‐binding domain nanoparticle vaccines. Cell. 2021;184(21):5432‐5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Martinez DR, Schäfer A, Leist SR, et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373(6558):991‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu Z, Zhou J, Xu W, et al. A novel STING agonist‐adjuvanted pan‐sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res. 2022;32(3):269‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang Y, Tan W, Lou Z, et al. Immunogenicity evaluating of the multivalent COVID‐19 inactivated vaccine against the SARS‐CoV‐2 variants. Vaccines (Basel). 2022;10(6):956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhao X, Zheng A, Li D, et al. Neutralisation of ZF2001‐elicited antisera to SARS‐CoV‐2 variants. Lancet Microbe. 2021;2(10):e494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. An Y, Li S, Jin X, et al. A tandem‐repeat dimeric RBD protein‐based covid‐19 vaccine zf2001 protects mice and nonhuman primates. Emerg Microbes Infect. 2022;11(1):1058‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhao Y, Zhao X, Zhang R, et al. Heterologous BBIBP‐CorV/ZF2001 vaccination augments neutralization against SARS‐CoV‐2 variants: a preliminary observation. Lancet Reg Health West Pac. 2022;21:100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Joyce MG, Chen WH, Sankhala RS, et al. SARS‐CoV‐2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021;37(12):110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. King HAD, Joyce MG, Lakhal‐Naouar I, et al. Efficacy and breadth of adjuvanted SARS‐CoV−2 receptor‐binding domain nanoparticle vaccine in macaques. Proc Natl Acad Sci USA. 2021;118(38):e2106433118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Qu L, Yi Z, Shen Y, et al. Circular RNA vaccines against SARS‐CoV‐2 and emerging variants. Cell. 2022;185(10):1728‐1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Seephetdee C, Bhukhai K, Buasri N, et al. A circular mRNA vaccine prototype producing VFLIP‐X spike confers a broad neutralization of SARS‐CoV‐2 variants by mouse sera. Antiviral Res. 2022;204:105370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wang J, Li P, Wu MX. Natural STING agonist as an “ideal” adjuvant for cutaneous vaccination. J Invest Dermatol. 2016;136(11):2183‐2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chauveau L, Bridgeman A, Tan TK, et al. Inclusion of cGAMP within virus‐like particle vaccines enhances their immunogenicity. EMBO Rep. 2021;22(8):e52447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang J, Li P, Yu Y, et al. Pulmonary surfactant‐biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science. 2020;367(6480):eaau0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li L, Yin Q, Kuss P, et al. Hydrolysis of 2'3’‐cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat Chem Biol. 2014;10(12):1043‐1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Liu Z, Chan JF, Zhou J, et al. A pan‐sarbecovirus vaccine induces highly potent and durable neutralizing antibody responses in non‐human primates against SARS‐CoV‐2 omicron variant. Cell Res. 2022;32(5):495‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cohen AA, Gnanapragasam PNP, Lee YE, et al. Mosaic nanoparticles elicit cross‐reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371(6530):735‐741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Saunders KO, Lee E, Parks R, et al. Neutralizing antibody vaccine for pandemic and pre‐emergent coronaviruses. Nature. 2021;594(7864):553‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Li D, Martinez DR, Schäfer A, et al. Breadth of SARS‐CoV‐2 neutralization and protection induced by a nanoparticle vaccine. bioRxiv. 2022:477915. 10.1101/2022.03.04.479488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wrapp D, De Vlieger D, Corbett KS, et al. Structural basis for potent neutralization of betacoronaviruses by single‐domain camelid antibodies. Cell. 2020;181(6):1436‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sauer MM, Tortorici MA, Park YJ, et al. Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol. 2021;28(6):478‐486. [DOI] [PubMed] [Google Scholar]
- 134. Pinto D, Sauer MM, Czudnochowski N, et al. Broad betacoronavirus neutralization by a stem helix‐specific human antibody. Science. 2021;373(6559):1109‐1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sun X, Yi C, Zhu Y, et al. Neutralization mechanism of a human antibody with pan‐coronavirus reactivity including SARS‐CoV‐2. Nat Microbiol. 2022;7(7):1063‐1074. [DOI] [PubMed] [Google Scholar]
- 136. Lyke KE, Atmar RL, Islas CD, et al. Rapid decline in vaccine‐boosted neutralizing antibodies against SARS‐CoV‐2 omicron variant. Cell Rep Med. 2022;3(7):100679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. VanBlargan L, Errico J, Halfmann P, et al. An infectious SARS‐CoV‐2 B.1.1.529 omicron virus escapes neutralization by therapeutic monoclonal antibodies. Res Sq . 2021. [DOI] [PMC free article] [PubMed]
- 138. Zhou H, Møhlenberg M, Thakor JC, et al. Sensitivity to vaccines, therapeutic antibodies, and viral entry inhibitors and advances to counter the SARS‐CoV‐2 omicron variant. Clin Microbiol Rev. 2022;35(3):e00014‐e00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Helmsdal G, Hansen OK, Møller LF, Christiansen DH, Petersen MS, Kristiansen MF. Omicron outbreak at a private gathering in the Faroe Islands, infecting 21 of 33 triple‐vaccinated healthcare workers. Clin Infect Dis. 2022;75:893‐896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Scott J, Richterman A, Cevik M. Covid‐19 vaccination: evidence of waning immunity is overstated. BMJ. 2021;374:n2320. [DOI] [PubMed] [Google Scholar]
- 141. Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS‐CoV‐2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021;2021:2012.2007.21267432. [Google Scholar]
- 142. Zhou D, Chan JF, Zhou B, et al. Robust SARS‐CoV‐2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021;29:551‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Xu H, Cai L, Hufnagel S, Cui Z. Intranasal vaccine: factors to consider in research and development. Int J Pharm. 2021;609:121180. [DOI] [PubMed] [Google Scholar]
- 144. Oh JE, Song E, Moriyama M, et al. Intranasal priming induces local lung‐resident B cell populations that secrete protective mucosal antiviral IgA. Sci Immunol. 2021;6(66):eabj5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID‐19 vaccine strategies. Nat Rev Immunol. 2020;20(10):615‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Afkhami S, D'agostino MR, Zhang A, et al. Respiratory mucosal delivery of next‐generation COVID‐19 vaccine provides robust protection against both ancestral and variant strains of SARS‐CoV‐2. Cell. 2022;185(5):896‐915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wu S, Huang J, Zhang Z, et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type‐5 vector‐based COVID‐19 vaccine (Ad5‐nCoV) in adults: preliminary report of an open‐label and randomised phase 1 clinical trial. Lancet Infect Dis. 2021;21(12):1654‐1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Li JX, Wu SP, Guo XL, et al. Safety and immunogenicity of heterologous boost immunisation with an orally administered aerosolised Ad5‐nCoV after two‐dose priming with an inactivated SARS‐CoV‐2 vaccine in Chinese adults: a randomised, open‐label, single‐centre trial. Lancet Respir Med. 2022;10:739‐748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Lavelle EC, Ward RW. Mucosal vaccines: fortifying the frontiers. Nat Rev Immunol. 2022;22(4):236‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in coronavirus disease patients. Emerging Infect Dis. 2020;26(7):1478‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wec AZ, Wrapp D, Herbert AS, et al. Broad neutralization of SARS‐related viruses by human monoclonal antibodies. Science. 2020;369(6504):731‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhou P, Song G, He WT, et al. Broadly neutralizing anti‐S2 antibodies protect against all three human betacoronaviruses that cause severe disease. bioRxiv. 2022:479488. 10.1101/2022.03.04.479488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ng KW, Faulkner N, Cornish GH, et al. Preexisting and de novo humoral immunity to SARS‐CoV‐2 in humans. Science. 2020;370(6522):1339‐1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ma X, Zou F, Yu F, et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS‐CoV‐2 elicit robust protective immune responses. Immunity. 2020;53(6):1315‐1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science. 2020;369(6501):330‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li Y, Lai DY, Zhang HN, et al. Linear epitopes of SARS‐CoV‐2 spike protein elicit neutralizing antibodies in COVID‐19 patients. Cell Mol Immunol. 2020;17(10):1095‐1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Koff WC, Berkley SF. A universal coronavirus vaccine. Science. 2021;371(6531):759. [DOI] [PubMed] [Google Scholar]
- 158. Wang N, Li SY, Yang XL, et al. Serological evidence of bat SARS‐Related coronavirus infection in humans, China. Virol Sin. 2018;33(1):104‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Burton DR, Topol EJ. Variant‐proof vaccines ‐ invest now for the next pandemic. Nature. 2021;590(7846):386‐388. [DOI] [PubMed] [Google Scholar]
- 160. Gao Q, Bao L, Mao H, et al. Development of an inactivated vaccine candidate for SARS‐CoV‐2. Science. 2020;369(6499):77‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Wang H, Zhang Y, Huang B, et al. Development of an inactivated vaccine candidate, BBIBP‐CorV, with potent protection against SARS‐CoV‐2. Cell. 2020;182(3):713‐721.e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Walsh EE, Frenck RW Jr., Falsey AR, et al. Safety and immunogenicity of two RNA‐Based Covid‐19 vaccine candidates. N Engl J Med. 2020;383(25):2439‐2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Corbett KS, Flynn B, Foulds KE, et al. Evaluation of the mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. N Engl J Med. 2020;383(16):1544‐1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Corbett KS, Edwards DK, Leist SR, et al. SARS‐CoV‐2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV‐19 vaccine prevents SARS‐CoV‐2 pneumonia in rhesus macaques. Nature. 2020;586(7830):578‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Folegatti PM, Ewer KJ, Aley PK, et al. Safety and immunogenicity of the ChAdOx1 nCoV‐19 vaccine against SARS‐CoV‐2: a preliminary report of a phase 1/2, single‐blind, randomised controlled trial. Lancet. 2020;396(10249):467‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zhu FC, Guan XH, Li YH, et al. Immunogenicity and safety of a recombinant adenovirus type‐5‐vectored COVID‐19 vaccine in healthy adults aged 18 years or older: a randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet. 2020;396(10249):479‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zhu FC, Li YH, Guan XH, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type‐5 vectored COVID‐19 vaccine: a dose‐escalation, open‐label, non‐randomised, first‐in‐human trial. Lancet. 2020;395(10240):1845‐1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Bos R, Rutten L, van der Lubbe JEM, et al. Ad26 vector‐based COVID‐19 vaccine encoding a prefusion‐stabilized SARS‐CoV‐2 spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines. 2020;5:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sadoff J, Le Gars M, Shukarev G, et al. Interim results of a phase 1‐2a trial of Ad26.COV2.S Covid‐19 vaccine. N Engl J Med. 2021;384(19):1824‐1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Keech C, Albert G, Cho I, et al. Phase 1‐2 trial of a SARS‐CoV‐2 recombinant spike protein nanoparticle vaccine. N Engl J Med. 2020;383(24):2320‐2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Walls AC, Fiala B, Schäfer A, et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS‐CoV‐2. Cell. 2020;183(5):1367‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Wuertz KM, Barkei EK, Chen WH, et al. A SARS‐CoV‐2 spike ferritin nanoparticle vaccine protects hamsters against alpha and beta virus variant challenge. NPJ Vaccines. 2021;6(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


