Abstract
Rapid detection of antibodies during infection and after vaccination is critical for the control of infectious outbreaks, understanding immune response, and evaluating vaccine efficacy. In this manuscript, we evaluate a simple ultrarapid test for SARS‐CoV‐2 antibodies in COVID‐19 patients, which gives quantitative results (i.e., antibody concentration) in 10–12 s using a previously reported nanomaterial‐based three‐dimensional (3D)‐printed biosensing platform. This platform consists of a micropillar array electrode fabricated via 3D printing of aerosolized gold nanoparticles and coated with nanoflakes of graphene and specific SARS‐CoV‐2 antigens, including spike S1, S1 receptor‐binding domain (RBD) and nucleocapsid (N). The sensor works on the principle of electrochemical transduction, where the change of sensor impedance is realized by the interactions between the viral proteins attached to the sensor electrode surface and the antibodies. The three sensors were used to test samples from 17 COVID‐19 patients and 3 patients without COVID‐19. Unlike other serological tests, the 3D sensors quantitatively detected antibodies at a concentration as low as picomole within 10–12 s in human plasma samples. We found that the studied COVID‐19 patients had higher concentrations of antibodies to spike proteins (RBD and S1) than to the N protein. These results demonstrate the enormous potential of the rapid antibody test platform for understanding patients' immunity, disease epidemiology and vaccine efficacy, and facilitating the control and prevention of infectious epidemics.
Keywords: 3D biosensor, additive manufacturing, COVID‐19 antibodies, graphene oxide, rapid antibody detection, SARS‐CoV‐2
Abbreviations
- ACE2
angiotensin‐converting enzyme 2 receptor
- Ag/AgCl
silver/silver chloride
- AM
additive manufacturing
- BSA
bovine serum albumin
- BSL
biosafety level 3
- CE
counter electrode
- CLIA
chemiluminescence immunoassays
- Cr
chromium
- EDC
1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide hydrochloride
- EIS
electrochemical impedance spectroscopy
- ELISA
enzyme‐linked immunosorbent assays
- IgM
immunoglobulin G (IgG) or M
- IIFT
indirect immunofluorescence tests
- LFAs
lateral flow assays
- LoD
limit‐of‐detection
- MCLIA
magnetic chemiluminescence enzyme immunoassay
- N
nucleocapsid
- NC
nitrocellulose
- NHS
N‐hydroxysuccinimide
- PBS
phosphate‐buffered solution
- PD
peptidase domain
- PDMS
polydimethylsiloxane
- PMMA
polymethylmethacrylate
- RBD
receptor‐binding domain
- R ct
charge transfer resistance
- RE
reference electrode
- rGO
reduced graphene oxide
- rGO‐Au
reduced graphene oxide‐gold
- RT‐qPCR
reverse transcriptional real‐time polymerase chain reaction
- SDS‐PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SEM
scanning electron microscopy
- TBST
tris‐buffered saline with 0.1% tween‐20 solution
- UPMC
University of Pittsburgh Medical Center
- WE
working electrode
1. INTRODUCTION
Understanding the human immune response to pathogens and vaccination is central to the management of infectious epidemics, enabling informed decision‐making in public health policies. 1 , 2 , 3 In COVID‐19, the status of immune response can be revealed through the measurement of antibody titers against SARS‐CoV‐2 infection. 4 , 5 , 6 , 7 , 8 , 9 , 10
The SARS‐CoV‐2 genome consists of several functional genes, of which nucleocapsid (N) and spike (S) proteins are widely used in SARS‐CoV‐2 molecular and serological detection. 11 , 12 , 13 As the structural proteins, S and N, are expected to show high expression levels during infection. The S glycoprotein of SARS‐CoV‐2, which protrudes from the virus surface, plays an important role in viral infection through the specific molecular interaction of its receptor‐binding domain (RBD) with the human angiotensin‐converting enzyme 2 receptor (ACE2). 14 , 15 , 16 Hence, the antibodies to S protein are expected to play an important role in the immunity against SARS‐CoV‐2 infection.
Recent studies have shown that the knowledge gained by antibody tests can help develop preventive and therapeutic approaches against SARS‐CoV‐2. The development of rapid test platforms is expected to facilitate the development of methods to improve the durability of immune responses. 17 , 18 , 19 , 20 The risk of re‐infection is higher when individuals have a lower level of antibodies. For example, severely ill COVID‐19 patients with lower levels of antibodies may have a higher risk of death. 3 , 18 Thus, analyzing the host‐pathogen relationship can improve the disease management for COVID‐19 patients. 21 , 22 , 23
Cheng et al. measured the immunoglobulin G (IgG) or M (IgM) antibody responses to SARS‐CoV‐2 by applying traditional method such as enzyme‐linked immunosorbent assay (ELISA) of a cluster of patients recovered from mild to severe COVID‐19 infection. 24 Each test took about 24 h to generate results. Magnetic chemiluminescence enzyme immunoassay (MCLIA) was used to measure the patient antibody response to SARS‐CoV‐2. 25 This study revealed that the level of S antibody response is 1.5 fold higher than that of an N antibody response. 25 The MCLIA test is quite complex and can take several hours per test. A recent study showed a comprehensive mapping of the immunological response of COVID‐19 patients and investigated the cellular pathways of complex immune networks for patients with severe COVID‐19 infection to understand the correlation between dysregulation of cytokines and immune hyperactivation. 26 Understanding immune response to COVID‐19 infection and/or vaccination is of significant interest and can benefit from the development of new rapid tests that can provide quantitative antibody results, ideally in seconds.
A number of sensing modalities have been explored to detect IgG or IgM responses to SARS‐CoV‐2 infection in different body fluids such as whole blood, serum, and plasma. These include ELISA, 27 microsphere immunoassays, 28 chemiluminescence immunoassays (CLIA), lateral flow assays (LFAs), and indirect immunofluorescence tests (IIFT). 29 , 30 One major concern with these methods (ELISA and LFA) 31 is the relative long detection time (0.3–24 h) and the inability for point‐of‐care use. In addition, a few research reports describe efforts to sense COVID‐19 antibodies using electrochemical tests. 3 , 5 , 32 Their sensing modalities include organic electrochemical transistors, 3 paper‐based analytical devices, 32 gold microcuboids‐based microsensors, 33 microfluidic devices, 34 terahertz plasmonic metasensors, 35 and a set of planar two‐dimensional electrodes. 36 , 37 , 38 None of these methods, however, can provide results in seconds.
To overcome the limitations of the current tests for infections of pathogens, we recently used a completely new jetting‐based microscale additive manufacturing (AM or colloquially called three‐dimensional “[3D] printing”) technique 39 to create a 3D electrochemical sensor device platform that achieves sensitive and rapid detection of biomolecules such as SARS‐CoV‐2 proteins and antibodies in seconds. 40 , 41 Note that 3D printing allows the manufacture of structures via a layer‐by‐layer “addition” of materials, which leads to the realization of structures with complex architectures, controlled microstructures, and material combinations. 42 A large effort in academia as well as in industry is currently devoted to bringing the advantages of AM to biomedical device fabrication, as described in our recent review article. 43 Specifically, we used aerosol‐jet (AJ)‐based 3D printing to develop a fabrication method to make microscale 3D metallic structures such as micropillars and microlattices, without any sacrificial support. 39 , 41 Further, we note that graphene and its derivates are exploited as transducer materials for biosensing of SARS‐CoV‐2 due to their excellent properties. 44 , 45 They have excellent electron transfer characteristics, high surface area, and tunable electrochemical properties, which makes them suitable candidates for biosensing applications. 46 , 47 , 48 , 49 Recently, graphene was also used to develop a capacitance‐based biosensing platform which detects the surface protein of SARS‐CoV‐2 with a limit of detection of 1 fg/ml. 44
Our recent work utilized this capability to build a test platform to quantitatively measure specific antibodies to SARS‐CoV‐2 in seconds. 40 In addition to the incredibly short time, this label‐free platform shows an ultra‐low limit‐of‐detection (LoD) down to the femtomolar level of antibody concentrations. 40 This sensor 40 , 50 demonstrated sensing capability for multiple SARS‐CoV‐2 antibodies to S and N proteins, which is a feature not found in other sensors. The method, however, was successfully tested only on rabbit‐derived antibodies and its validation on human samples remained to be done.
In this study, we use the unique features of the 3D biosensing platform we developed 39 , 40 for the evaluation of antibody responses in COVID‐19 patients. The biosensing platform used in this study allows the detection of antibodies to N, spike 1 (S1), and RBD proteins in seconds via simple direct electrochemical transduction without adding labeling agents. In the sensor structure, the working electrode (WE) is an array of 3D‐printed and sintered 3D gold micropillars coated with reduced graphene oxide (rGO‐Au) and functionalized with S1, RBD, or N proteins (antigens). These proteins are used to analyze the levels of SARS‐CoV‐2 antibodies in plasma samples from COVID‐19 positive (n = 17) and COVID‐19 negative (n = 3) patients confirmed by reverse transcriptional real‐time polymerase chain reaction (RT‐qPCR). The same patient samples were used in three separate antibody tests including antibodies to S1, RBD, and N proteins. We also demonstrate that the sensor has the ability to quantify concentrations of antibodies ranging from femtomolar to nanomolar levels due primarily to the unique 3D micropillar geometry realized via our AJ‐based microscale 3D printing technique.
2. MATERIALS AND METHODS
2.1. Chemicals and sensor materials
Recombinant proteins of SARS‐CoV‐2, namely, RBD‐His protein (50 μg/ml), N‐His protein (50 μg/ml), and spike S1‐His protein (50 μg/ml), were purchased from Sino Biological, Inc. 51 , 52 The S1, RBD, and N antigens were immobilized separately on the sensor surfaces, which acted as a capturing element for corresponding antibodies in an aqueous solution, that is, human plasma samples from SARS‐CoV‐2 patients. These three SARS‐CoV‐2 proteins were prepared in carbonate buffer of pH 9.6 and aliquoted at –80° C before use. For immobilization, the concentrations of all the antigens were set to 5 μg/ml.
Two rabbit IgG antibodies to S1 and RBD, and a mouse monoclonal antibody (IgG) to N protein were purchased from Sino Biological Inc. 50 , 53 For titration measurements, the antibody solutions of S1, RBD, and N were prepared in a phosphate‐buffered solution (PBS) of pH 7.4 (50 mM) containing an equimolar concentration of 5 mM of ferro and ferricyanide. We used the same PBS for serial dilution and spiking antibodies. A set of other chemicals, namely sodium carbonate, sodium bicarbonate, EDC (1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide hydrochloride), and NHS (N‐hydroxysuccinimide) were obtained from Sigma Aldrich. Lyophilized powder of bovine serum albumin (BSA) was also purchased from Sigma Aldrich and prepared in a PBS (50 mM) at a concentration of 2 mg/ml. 54
A commercial ink of gold (Au) nanoparticles (UTDAu40) was purchased from UT Dots Inc. The Au nanoparticle ink has an average particle size of 4 nm and a viscosity of 3 cP. Particle loading in an organic non‐polar solvent was 40 wt% and this ink solution was used to build the 3D sensor using an AJ 3D printer (AJ 300; Optomec Inc.). The SYLGARD™ 184 Silicone Elastomer Kit was purchased from Dow Corning Inc. To create a microfluidic channel in the sensor (Figure 1A), the ratio of polydimethylsiloxane (PDMS) oligomer and linker was set to 10:1 (wt %). The powder form of rGO nanosheets (CAS‐No. 7782‐42‐5) was purchased from ACS Materials LLC. The rGO powder was dispersed in deionized water at a concentration of 0.2mg ml− 1 and sonicated for 2 h before use. A commercial silver/silver chloride (Ag/AgCl) ink was purchased from Ercon, Inc. Silicone oil was purchased from Ease Release™ 205, Reynolds Advanced Materials.
Figure 1.
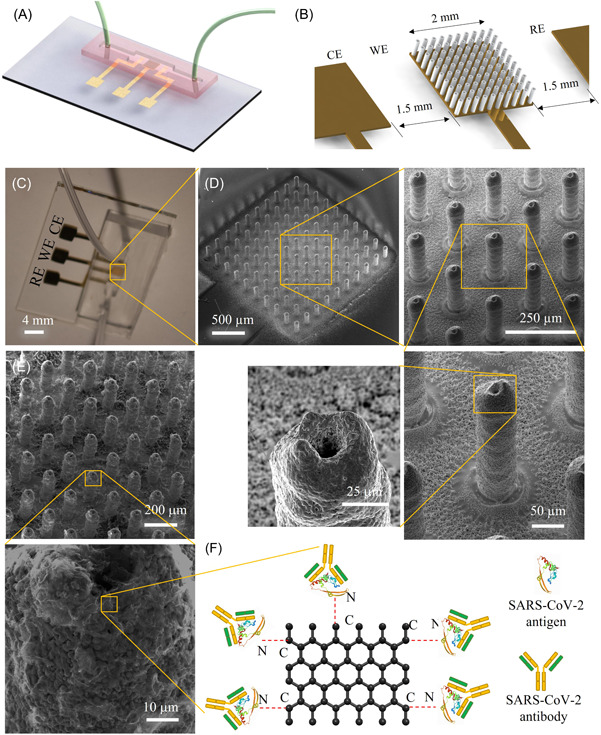
3D‐printed biosensor for in vitro sensing of SARS‐CoV‐2 antibodies in COVID‐19 patients. (A, B) Schematic illustration of a 3D‐printed microfluidic sensor showing that the WE consists of a micropillar array of gold nanoparticles and the WE is situated between the RE and CE (B). (C) An optical image of an actual device. (D) SEM images (tilted view) of the printed micropillar array at different magnifications before coating by graphene and proteins. The zoomed‐in SEM images show the surface microtexture of the sintered gold micropillars, which helps the rGO nanoflakes to bind with this surface. (E) SEM image showing the rGO nanoflakes coated on the array electrode. (F) Schematic illustration depicting the functionalization of the electrode by attaching SARS‐CoV‐2 proteins on rGO sheets via EDC‐NHS conjugation chemistry. In this amidation reaction, EDC activates the COOH groups of rGO, while NHS acts as a stabilizer, resulting in amide bonds between proteins and rGO sheets on the Au pillar surface. Antibodies to SARS‐CoV‐2 proteins attached to the antigens during sensing are also depicted. 3D, three‐dimensional; CE, counter electrode; EDC, 1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide hydrochloride; NHS, N‐hydroxysuccinimide; RE, reference electrode; rGO, reduced graphene oxide; SEM, scanning electron microscopy; WE, working electrode.
2.2. Instruments
A scanning electron microscopy (SEM, Sirion SEM; FEI Inc.) was used to image the sensor array structures. A 3D computer aided design (CAD) software, SolidWorks 2020, was used to create a 3D schematic of the device shown in Figure 1. For antibody sensing, a VersaSTAT 3 Potentiostat/Galvanostat from Princeton Applied Research, with Zview software was used to record the electrochemical impedance spectroscopy (EIS) signals and analyze all the spectra with its in‐built software. The EIS technique was used for the spiking study. This technique measures the change in the electrical impedance at the electrode surface/interface due to protein‐antibody interactions. 55 It is known to be a label‐free, sensitive detection method. 56 In this study, the AC signal during EIS measurement had an amplitude of 1 mV and a frequency range of 10 000–1 Hz. An e‐beam evaporator (Kurt Lesker PVD 75) was utilized to deposit chromium and gold layers on the glass substrates. For the printing of the 3D microelectrode array, the Aerosol Jet nanoprinter (Model AJ‐300; Optomec Inc.) was used. An AutoCAD software (AutoCAD 2021; Autodesk Inc.) was utilized to create the sensor design and CAD programming files for printing. Further, AutoLISP language was used to create a program that was converted to a printing compatible “prg” file to print the sensor structure, similar to that in our previous work. 38 An automated cutter was purchased from Silhouette Curio™, Silhouette America®, Inc., and used to create a Kapton tape‐based shadow mask for the device.
2.3. Details of human COVID‐19 patient samples
A total of 20 human samples were utilized to analyze the presence or absence of COVID‐19 antibodies. The suspected COVID‐19 patients were confirmed by the SARS‐CoV‐2 RT‐qPCR test. Among the 20 samples, 17 samples were from COVID‐19 positive patients and 3 samples were from COVID‐19 negative patients. Each sample, positive and negative, was collected and then divided into three separate tubes, allowing the plasma in each tube to be used for one of the three antibody tests.
The study was approved by the Institutional Review Board (IRB) of the University of Pittsburgh (STUDY20090114). The human plasma samples were collected from the University of Pittsburgh Medical Center (UPMC) hospital from patients who were admitted to an intensive care unit (ICU) with severe COVID‐19. 57 , 58 , 59 We obtained informed consent from the patients or their legally authorized representatives under an IRB‐approved protocol before enrollment and sample collection. Plasma samples used in this study were separated from whole blood samples through centrifugation. These samples were aliquoted and stored at −80°C before the study. These human plasma samples were mixed into PBS at a fixed ratio of 1:250 for this testing. All the testing was conducted in Biosafety Level 2+ (BSL2+) facility.
2.4. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE)
The SARS‐CoV‐2 recombinant proteins were analyzed by SDS‐PAGE. Each recombinant protein at 10 µg/well was loaded into a 12‐well 4%–20% SDS‐PAGE gel and run at 120 V for 80 min. Images of gels were taken with a BIO‐RAD Imagine System following Coomassie brilliant blue staining (Bio‐Rad Laboratories).
2.5. Western‐blotting
For the Western‐blotting analysis, the S1, RBD, and N recombinant proteins were analyzed by SDS‐PAGE gels and then transferred onto nitrocellulose (NC) membranes with a Trans‐Blot Turbo Transfer System (Bio‐Rad). Each membrane with a recombinant protein was blocked by 5.0% nonfat milk for 1 h at room temperature (25°C) and incubated with a specific antibody to the protein diluted in 1.0% BSA in tris‐buffered saline with 0.1% tween‐20 solution (TBST) solution at 4°C overnight. After washing it three times for 5 min each in TBST solution, the membrane was incubated with the respective secondary antibody.
2.6. Sensor construction and functionalization
A schematic diagram of the sensor for human trials is shown in Figure 1. Fabrication details of the 3D‐printed sensor are described in our previous work. 40 In brief, three‐electrodes on a glass substrate were first fabricated by directly depositing a 100 nm‐thick Au layer and 10 nm‐thick Cr layer, which acted as an adhesion layer via an e‐beam evaporator. For this, a shadow mask of a Kapton tape was prepared using an automated cutter with the help of AutoCAD software and mounted on the glass slide before depositing the Au layer. The Au layer was a base connector for the three electrodes, namely the counter electrode (CE), reference electrode (RE), and the WE for the impedance measurements. The Au layer was used to collect the impedance signal from the sensor. The gap of each electrode in the sensor geometry is shown in Figure 1B. Note that the working area was 2 mm × 2 mm where the 3D‐printed structure of the sensor was built. A 10 × 10 array of Au micropillars, made of Au nanoparticles, was manufactured on the evaporated Au layer. The geometrical shape of the micropillar array is shown in Figure 1B.
To obtain this geometry of micropillar electrode, the AJ‐300 nanoparticle printer was used, as described before. The printer is equipped with a programmable motor‐driven stage (X‐Y direction) with a temperature control unit, an ultrasound atomizer, a deposition head containing a nozzle, and a shutter to break the streamline flow of the aerosol ink. During printing, 1 ml of Au nanoparticle ink was loaded into a glass vial and converted into a mist of aerosol droplets (each containing the Au nanoparticles) via the ultrasonic energy of the AJ printer. Note that an external heating chamber was positioned on top of the X‐Y stage to maintain a constant temperature of 150°C for the platen during printing. Nitrogen (N2) gas was used as a carrier gas to transport the aerosol droplets to the deposition head at an optimized pressure of 25 sccm. Further, a sheath gas (also N2) was applied at a pressure of 60 sccm surrounding the printer nozzle to aerodynamically focus the aerosol ink stream before printing. The minimum feature size of Au ink in the study was 15 µm. Before printing, the geometry of the micropillar array was drawn using AutoCAD software and the program was written using AutoLISP software, followed by converting to a prg file which is compatible with the AJ‐300 nanoprinter as described in our previous work. 40 The external heating element helped rapid drying of the binder and solvent from the ink once a layer was printed. Because of its strong surface tension, the dried layer of the ink was able to hold the next layer of printing. Such layer‐by‐layer printing formed the desired micropillar array without any support structures. 60 , 61 Once printed, the dry layer of Au created a surface tension to hold the second printed layer. Au printing was continued until the pillar reached 250 µm in height. The dried micropillar array was then sintered to 400°C for 4 h, wherein the Au nanoparticles formed the micropillars with highly porous surface features (Figure 1D,E). A thin layer (5 µm) of Ag/AgCl ink was coated on top of Au surface of the RE by placing another Kapton tape‐based shadow mask and dried it at 150°C for 2 h. 62 An optical photograph of the complete device is shown in Figure 1B. The diameter of the nozzle was chosen as 150 µm to maintain the uniform pillar diameter (which is known to create a droplet stream of about 15 µm in diameter). The height, pillar‐to‐pillar distance, and diameter of the micropillar were controlled by an AutoCAD program. The micropillar diameter, pillar‐to‐pillar distance, and height were 72, 118, and 250 µm, respectively, as measured by the processing camera of the printer. Sintered pillars formed a micro‐texture of Au nanoparticles (Figure 1D), which is expected to help the adhesion of a coating of rGO sheets during the functionalization step.
Next, the micropillar array was modified with rGO sheets. A PDMS fence was placed surrounding the array, and a 20 µl colloidal suspension of rGO (0.2 mg/ml) was drop‐casted and dried at 80°C for 1 h. This process was repeated for an additional two times, allowing more graphene flakes to adhere on the pillar surface. SEM image of a micropillar array modified with rGO sheets is shown in Figure 1E in which the wrinkled graphene flakes are clearly visible on the pillar surface. After rGO modification, three separate rGO‐Au sensors were made, and each was functionalized with one of three recombinant proteins of SARS‐CoV‐2, either RBD, N, or S1.
The schematic representation of the surface functionalization on the rGO‐Au micropillar array is shown in Figure 1F. For this, three independent rGO‐Au sensors were functionalized with specific proteins, namely S1, RBD, and N proteins. Before applying the proteins, the ─COOH groups present on the rGO‐Au array were first activated by introducing EDC (0.2 M) as a crosslinker and then stabilized with NHS (0.05 M). For a single sensor, a mixture of EDC and NHS solution was prepared first at a ratio of 1:1 (volume). Using a pipette, 30 µl of this mixture solution was applied to the surface of the micropillar. The sensor was then kept in a humid chamber having 100.0% humidity for 4 h and then washed with a buffer solution. A 20 µl RBD protein (5 µg/ml) was spread on the EDC‐NHS modified sensor and kept in the humid chamber for 4 h. The ─NH2 groups of the protein (antigen) bind with ─COOH groups of rGO by creating a C─N amide bond 63 as shown in Figure 1F. In addition to covalent interactions on the sensor surface, we also expected that some of the antigen molecules may interact with the pillar surface via electrostatic interactions. At the end of the functionalization process, a BSA (2 mg/ml) coating on the sensor surface was added to block the nonspecific sites of the sensor. A similar process of functionalization was applied to the N and S1 sensors. For measurements, a set of all three sensors including RBD, N, and S1 were fabricated to complete the tests of the COVID‐19 samples. Note that each sensor is sensitive to specific antibodies to SARS‐CoV‐2 as designed and does not require any external labeling agents. The coated sensors were stored at 4°C until use.
To create a microfluidic channel in the sensor, a soft‐replica method was used and integrated with the sensor containing WE, CE, and RE. 41 The schematic diagram of the PDMS channel that was placed on the glass substrate containing the electrodes is shown in Figure 1A. A polymethylmethacrylate (PMMA) mold containing a channel was fabricated using a high‐precision milling machine. 64 The dimension of the channel was set to 1 mm × 2 mm × 2 mm (depth × width × length). Note that the middle portion (~1 cm) of the channel had a width of 2 mm. A PDMS solution was prepared by mixing PDMS oligomer and crosslinker at a ratio of 10:1 (wt %) and poured into the PMMA channel to get an opposite pattern. Before pouring, the PDMS mixer solution was placed in a degassed chamber to remove bubbles for 1 h at 10− 4Torr. Upon curing at 80°C for 2 h, the opposite pattern of PDMS was peeled off from the PMMA. The resulting channel acted as a mold to make the final microfluidic channel. Silicone oil was sprayed on the top surface of the PDMS mold before applying a new PDMS layer. Finally, a PDMS solution was poured on the PDMS mold and cured at the same temperature (80°C) and peeled off the PDMS mold. The final PDMS channel was utilized as PDMS housing for the biological fluids. At the end of the channel, two holes were created and connected to Tygon tubes and a syringe to handle the aqueous and plasma samples. The resulting PDMS channels were placed manually into the glass substrates containing the functional electrodes for the sensing measurements. The three electrodes of the sensor chip were connected to copper tapes using silver paste which was dried at room temperature. The copper tapes were connected to the potentiostat for the measurements.
2.7. Spiking analysis
Before sensing of antibodies in the plasma samples, the sensor was validated by spiking the specific antibodies to RBD at a final concentration of 10 nM into plasma from a subject without COVID‐19 (Figure 2C–E) at different dilutions in PBS. An RBD sensor was used for this study. As a control, the impedance responses of the RBD sensor were used to measure human plasma at similar dilutions without spiking of RBD antibodies. Specifically, the human plasma was diluted at 1:10, 1:250, 1:500, and 1:1000 in PBS. An AC input signal was applied by setting an amplitude of 1 mV and frequencies of 10 000–1 Hz during all the EIS measurements.
Figure 2.
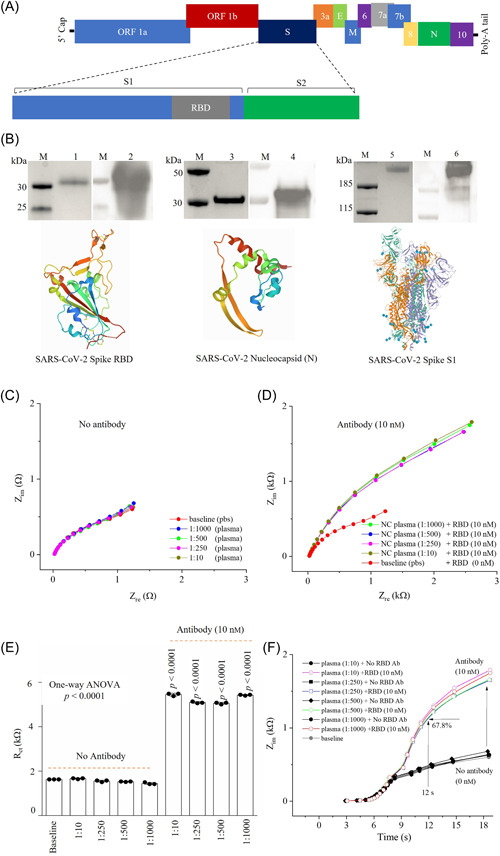
Spiking experiments to validate detection of antibodies to RBD in a plasma with a sensor. (A) Schematic illustration of SARS‐CoV‐2 genome. (B) Detection of SARS‐CoV‐2 recombinant proteins (RBD, N, and S1) used in sensors. Lanes M: protein markers; lanes 1, 3, 5: RBD, N, and S1, respectively, observed by SDS‐PAGE; lanes 2, 4, 6: RBD, N, and S1 detected by Western‐blotting with corresponding specific antibodies, respectively. (C, D) Nyquist plots for the sensor in presence of human plasma at 1:10; 1:250; 1:500, and 1:1000 dilution without (C) and with spiking of 10 nM RBD antibodies (D). (E) R ct values obtained from graphs in (C, D). Three independent experiments were conducted for the plots in (C–E). (F) Impedance, Z, versus detection time(s) without and with spiking RBD antibodies in plasma sample. EIS experiments involved applying an AC signal with an amplitude of 1 mV and a fixed frequency range of 10,000 Hz to 1 Hz. E, envelope; EIS, electrochemical impedance spectroscopy; M, membrane; N, nucleocapsid; ORF1a, open reading frame 1a; ORF1b, open reading frame 1b; RBD, receptor‐binding domain; S, spike; SDS‐PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
2.8. Sensing of antibodies in plasma samples
The sensor with three electrodes (WE, CE, and RE) was connected to an electrochemical analyzer (potentiostat) and used for the EIS experiments to detect the antibodies to SARS‐CoV‐2 proteins. The sensors were tested on 17 samples from COVID‐19 positive patients and 3 samples from COVID‐19 negative patients. These aliquoted samples were mixed with PBS at different dilutions from 1:10 to 1:1000 (plasma: buffer) and injected into sensors via a syringe.
2.9. Statistical analysis
All the samples were tested at least three times per experiment. GraphPad Prism 9.3.1 was used in the present study for plotting figures and for statistical analysis. Sensor testing data were presented as mean ± standard error of the mean (SEM). p Value was calculated by unpaired two‐tailed student's t‐test. p < 0.05 was considered to be statistically significant.
3. RESULTS
3.1. 3D‐printed sensor construction
As discussed in the Methods section, AJ AM method 65 was used to fabricate the 3D geometry of the sensor used in this study. The sensor was constructed to detect antibodies to multiple SARS‐CoV‐2 proteins including S1, RBD, and N in plasma samples. The construction of the sensor device is shown in Figure 1. The details of sensor fabrication is described in our previously reported work. 40 In brief, Figure 1A shows a schematic of the device where a 3D‐printed micropillar array electrode is integrated with a microfluidic channel. The glass substrate in the sensor configuration contains three‐electrodes: WE, CE, and RE, all of which were attached with a PDMS microfluidic channel. The CE is made of Au of thickness ~100 nm. The RE is an Au layer of thickness ~100 nm, which is then coated with a 5 µm thick layer of Ag/AgCl. The WE was made of a 3D‐printed Au micropillar array. A 10 × 10 array was chosen to fit a PDMS (2 × 2 mm2) microchannel. The gaps between RE and CE to WE were set equally to 1.5 mm (Figure 1B). An optical photograph of the sensing device is shown in Figure 1C. The SEM images show the tilted view of the 10 × 10 Au micropillar array at different magnifications (Figure 1D).
3.2. Validation of SARS‐CoV‐2 proteins and specific antibodies
Spike and nucleocapsid proteins are the principal structural proteins of SARS‐CoV‐2 (Figure 2A). In the present study, SARS‐CoV‐2 recombinant S1, RBD, and N proteins were used to treat the sensors for immobilization with which the recombinant protein‐specific antibody was expected to bind to and be detected. SDS‐PAGE analysis with Coomassie blue staining showed that the purities of recombinant S1, RBD, and N proteins are all >95%, which are qualified to be used in sensor functionalization for antibodies testing (Figure 2B, lanes 1, 3, and 5). The protein tertiary structures of S1, RBD, and N proteins are shown in Figure 2B.
Subsequently, Western‐blotting was conducted to detect the antigens with antibodies. The results of Western‐blotting (Figure 2B) show a single band for each protein (S1, RBD, and N). A short exposure time and specific band in Western‐blotting indicated that the specificity of the antibodies.
3.3. Spiking analysis of the sensors
The spiking analysis of the sensors was conducted by EIS. An RBD sensor was used for this study and the human plasma was diluted at 1:10, 1:250, 1:500, and 1:1000. Figure 2C shows the Nyquist plots in the diluted plasma samples without the specific RBD antibodies using the RBD sensor. Compared to the baseline signal, which does not have any human plasma, there was only a minute change of 5.9% the charge transfer resistance (R ct) value observed due to the nonspecific binding of proteins in the human plasma (Figure 2C). This change is insignificant compared to the sensor response with RBD antibodies as shown in Figure 2D. When spiking with RBD antibodies, a drastic change in the signal, 330.7% compared to baseline (p < 0.0001), represents the detection of specific antibodies (Figure 2E). Further, a change of 4.0% was observed with different concentrations of human plasma, which is insignificant when compared to the initial signal. This indicates the reliable sensing of RBD antibodies at different dilutions of human plasma. The sensor impedances (imaginary) are plotted against response time (seconds) with and without RBD antibody spiking to a set of human plasma concentrations (Figure 2F). Upon adding 10 nM of RBD antibodies, the sensor was capable of detecting 67.8% signal within 12 s as compared to the final signal which was found at 18 s. This indicates that the sensor can identify the RBD antibodies within 12 s, which is the fastest readout system compared to any other serological tests.
3.4. Quantitative sensing of SARS‐CoV‐2 antibodies in COVID‐19 positive samples
After the sensor validation, we performed quantitative sensing of the specific SARS‐CoV‐2 antibodies in COVID‐19 patient samples at different dilutions. This experiment allowed the identification of a suitable dilution concentration of human plasma that the sensor can easily detect the SARS‐CoV‐2 antibodies. In addition, this testing establishes the lower limit of concentration of antibodies that the sensor can quantitatively measure. Figure 3 shows the sensing results of the RBD sensor with COVID‐19 patient samples. In this experiment, one COVID‐19 negative sample and two COVID‐19 positive samples were used as negative and positive controls, respectively, at 1:20, 1:40, 1:60, 1:80, 1:100, 1:250, and 1:500 dilutions in PBS. Sensing of these samples was conducted using the EIS method as described above.
Figure 3.
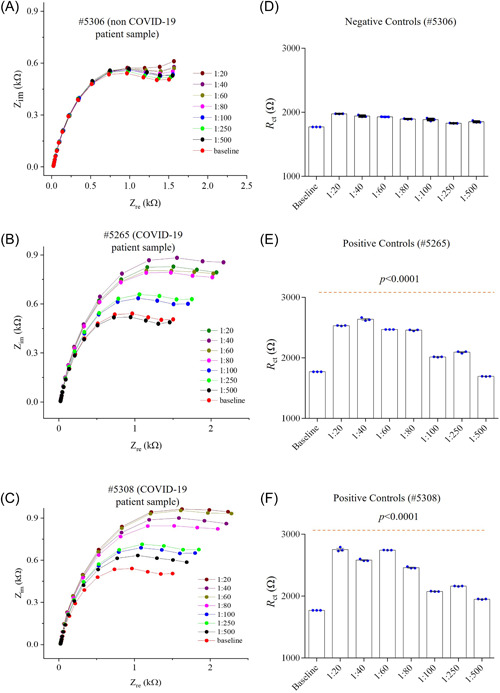
Quantitative detection of COVID‐19 antibodies in plasma samples. (A–C) Response of sensor to a COVID‐19 negative sample (#5306) (A) and two COVID‐19 positive samples #5265 (B) and #5308 (C). The plasma sample was diluted with PBS from 1:20 to 1:500. An RBD sensor was chosen for this study. (D–F) R ct values from the (A–C), respectively. R ct plot shows p values of <0.0001 compared to the baseline for the COVID‐19 positive samples (E, F). EIS, electrochemical impedance spectroscopy; RBD, receptor‐binding domain.
The RBD sensor was first exposed with a negative control human plasma at different dilutions. Testing results of these samples are shown in Figure 3A,D. Compared to the baseline signal (without any human plasma), the RBD sensor signal exhibits a relative standard deviation of 2.5% (Figure 3D) at all the dilutions. Once the positive control (COVID‐19) sample was introduced, the sensor signal shown in Figure 3B,E was increased to a higher value compared to the baseline signal. This is due to the specific interaction of recombinant spike RBD protein on the sensor with specific antibodies in the COVID‐19 plasma, which changes the interfacial impedance of the sensor, resulting in higher impedance.
In the diluted samples or COVID‐19 plasma, the sensor response reduced gradually. The sensor provided an excellent detection capability with a dilution up to 250 times (Figure 3E). Thus, 1:250 was chosen as an initial dilution of COVID‐19 plasma samples for further testing. In the more diluted plasma samples, the reduction of the sensor signal is due to the gradual decrease of antibodies captured by the sensor. However, the sensing impedance is significantly higher with a dilution of 250 times than that of the baseline, indicating that RBD antibodies can be detected at a low concentration of plasma samples (p < 0.0001).
In another COVID‐19 plasma, the sensor shows a similar pattern (Figure 3C,F), but the antibody sensing was realized with a maximum dilution of 500 times. The sensing impedance in Figure 3F is significantly higher compared to the baseline signal (p < 0.0001), indicating that RBD antibodies can also be sensitively detected in this plasma.
3.5. Sensing of RBD antibodies in COVID‐19 patients
The spike protein RBD was used to functionalize the rGO‐Au micropillar array. To measure RBD antibodies, we established a standard curve of the antibodies. A concentration range of RBD antibodies from 10 fM to 50 nM was prepared in PBS and used for the sensor calibration before testing COVID‐19 patient samples. We note that the detection of antibodies within this range of frequencies is high enough compared to the baseline signal. The baseline of the RBD sensor was set with PBS (50 mM; pH ~7.4) containing an equimolar concentration of ferro/ferricyanide (5 mM) without any RBD antibodies (Figure 4A,B). The reason for choosing ferro/ferricyanide as a mediator is to enhance the electron transfer from the electrolyte to the 3D electrode so that a reasonable signal can be detected to differentiate biomolecular events at the electrode surface.
Figure 4.
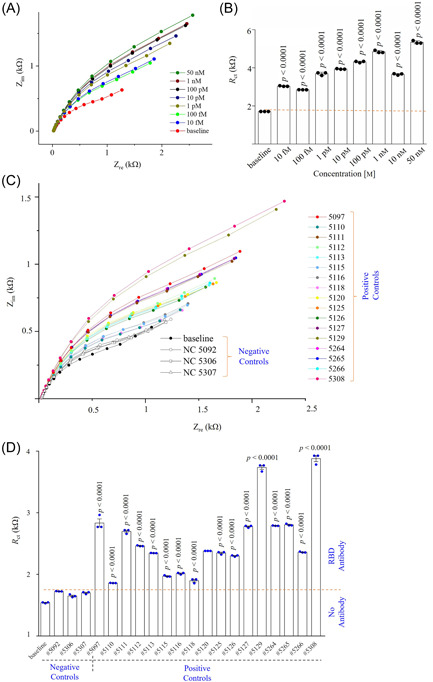
Sensing RBD antibodies in COVID‐19 samples. (A, B) Standard curve of the RBD sensor with Nyquist plots recorded as a function of RBD antibodies from 10 fM to 50 nM (A) and their corresponding charge transfer resistances (R ct) plotted according to the different RBD antibody concentrations (B). (C, D) RBD sensing results of plasma samples showing Nyquist plots of 17 COVID‐19 positive samples and 3 COVID‐19 negative samples (C), and their calculated R ct values plotted as a function of sample numbers (D). The deviation of the fitted semicircle and the raw semicircle of all impedance spectra was less than ±3.0%. All COVID‐19 positive samples have p values of <0.0001 compared to the COVID‐19 negative samples. All EIS experiments for sensing of antibodies were conducted by applying an AC signal with an amplitude of 1 mV and a fixed frequency range of 10 000–1 Hz. PBS (50 mM) mixed with a ferro/ferricyanide (5 mM) mediator was used to dilute plasma samples at 1:250. EIS, electrochemical impedance spectroscopy; PBS, phosphate‐buffered solution; RBD, receptor‐binding domain.
The baseline signal of the RBD sensor exhibited anR ctof 1.7 kΩ. In this study, the R ct values of all EIS signals were calculated by fitting in‐built semicircles in the same instrument (as described in Section 2) to the Nyquist plots (Figure 4A) wherein the diameter of the semicircle is the R ct value. Note that the deviation of fitted semicircles and raw data is from ±0.5% to ±3.9%. Once a 10 fM concentration of RBD antibodies was introduced, the sensor showed a higher R ct of 3 kΩ. This indicates the presence of RBD antibodies in the aqueous solution. Further, the concentration of RBD antibodies increased from 10 fM to 50 nM, and the EIS responses of the RBD sensor were recorded. It is noticed that the semicircle in the Nyquist plot (Figure 4A) increases with increasing concentrations of antibodies. At 50 nM of RBD antibodies, the sensor provided an R ct of 5.43 kΩ. Captured RBD antibodies on the RBD sensor change the interfacial impedance due to hindering of the electron transfer produced from the redox reactions at the electrode/electrolyte. 66 The calibration curve of the RBD sensor is shown in Supporting Information: Figure S1, wherein the R ct value of sensor is directly proportional to the concentration of RBD antibodies. The calibration equation is given as ; r 2 = 0.992. Supporting Information: Figure S2 shows the change of magnitude of impedance (|Z|) with frequency (10 000–1 Hz) for different RBD antibody concentrations. Results indicate that the change of sensor impedance due to protein–antibody interactions are dominant in the range of 10–1 Hz. The analytical sensitivity of the RBD sensor is found to be 10 fM, which is extremely low compared to other serological tests such as ELISA, chemiluminescence, and lateral‐flow immunoassays. 32 Thus, we expect to detect extremely low levels of antibodies.
Next, four RBD sensors were chosen to analyze the 20 plasma samples. These samples were diluted 250 times in PBS before testing via the impedance method. For the sensing measurements, the parameters of EIS measurement were kept the same, with the detection frequency of the input signal at 10 000–1 Hz and its amplitude at 1 mV. Because the sensor surface would be washed with a buffer solution between measurements, the sensor was first exposed to buffer solution without any samples to set a baseline (R ct of 1.55 kΩ). Then, three plasma samples from healthy individuals were injected into the first sensor via the microfluidic channel, and their impedances were recorded.
The testing results of the first sensor for plasma samples are shown in Figure 4C,D. Compared to the baseline signal, a deviation of ±2.5% was found with the three COVID‐19 negative controls. This slight deviation is due to the nonspecific interactions on the sensor with the proteins present in the plasma samples. The negatively charged rGO flakes and BSA molecules on the sensor surface repel the negatively charge proteins (e.g., albumin) at pH 7.4 (due to its low isoelectric point of 5.0) in the plasma, which provides a good specificity of the sensor. 67 Then, 17 COVID‐19 positive samples were introduced into the first sensor one‐by‐one, and the EIS spectra were recorded. The sensor was washed with buffer solution multiple times between sample tests.
The sensor impedances were found to be higher for the COVID‐19 positive human samples due to the capture of the specific spike RBD antibodies on the electrode surface. The R ct values of all samples are different, indicating different levels of RBD antibodies present in the samples. The impedances of COVID‐19 samples are all significantly higher than those of the baseline and the negative controls (p < 0.0001). It is noteworthy that the RBD sensor showed 100% sensitivity toward the detection of COVID‐19 antibodies. To confirm this, we tested three additional RBD sensors with the same samples. The results (R ct values) of the first RBD sensor was compared with the three additional RBD sensors and the correlation co‐efficient (r 2) was evaluated. Supporting Information: Figure S3 shows the EIS spectra of the other three RBD sensors, and the corresponding linear regression plots of R ct values as shown in Figure 5A–C. Almost a perfect corelation (r 2 = 0.905) was found when the same human samples were tested using sensor 2 as shown in Figure 5A. For the third and fourth testing times (sensors 3 and 4), the sensors showed a gradual reduction of R ct values, resulting in decreased r 2 (Figure 5D). The reduction of R ct values in the third and fourth measurements compared to the first measurement may be due to the limited number of RBD antibodies in the samples. However, all sensors showed the presence of RBD antibodies in COVID‐19 samples, indicating that the RBD sensor has a 100% sensitivity without an indication of a false positive signal.
Figure 5.
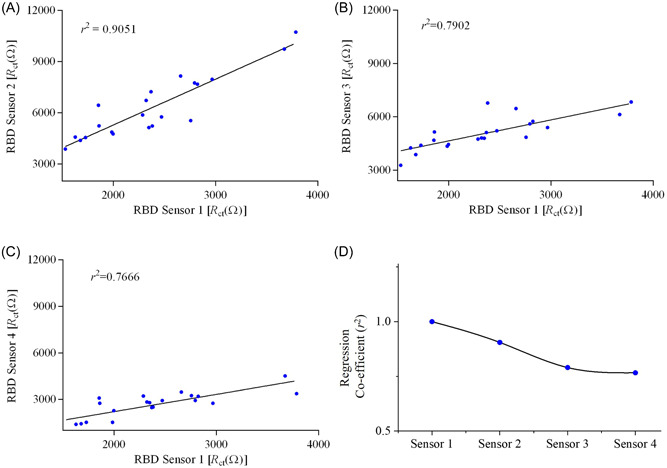
RBD sensor‐to‐sensor variation for detecting SARS‐CoV‐2 antibodies in COVID‐19 samples examined by regression co‐efficient analysis. (A–C) The same samples were tested with four different sensors. Regression co‐efficients (r 2) were calculated for (A) sensor 1 versus sensor 2, (B) sensors 1 versus sensor 3, and (C) sensor 1 versus sensor 4. All responses of sensors are shown in Supporting Information: Figure S3. (D) The values of r 2 are plotted against the sensor number. RBD, receptor‐binding domain.
3.6. Sensing of N antibodies in COVID‐19 samples
In this case, the sensor surface with an rGO‐Au micropillar array was modified with N proteins instead of RBD proteins. Before sensing plasma samples, the sensor was tested with PBS mixed with N antibodies ranging from 10 fM to 50 nM. As in the RBD sensing, impedance spectroscopy was used as the sensing modality, with parameters as previously described.
Figure 6A shows the sensing results with different concentrations of N antibodies to establish the calibration plot. It was expected that when the sensor captured N antibodies from the aqueous solution, the sensor showed a change of impedance signal, which was recorded and analyzed. As we increased the concentration of N antibodies, the sensor showed a significant increase of R ct in their Nyquist plots. Similar to the RBD sensor, a minimum detection time of 12 s was taken to monitor the trace level of N antibodies by the N sensor. The analytical sensitivity of N sensor is found as 1 pm.
Figure 6.

Sensing of N antibodies in COVID‐19 plasma samples. (A) EIS response of N sensor. Nyquist plots of the N sensor were recorded as a function of N antibodies (10 fM to 50 nM). (B, C) N antibody sensing results of COVID‐19 samples with corresponding Nyquist plots of each sensing graph of the samples (B) and calculated R ct values corresponding to samples plotted as a function of the sample number (C). COVID‐19 samples were treated as described in Figure 4. The deviation of fitted semicircle and raw semicircle of all impedance spectra was less than ±3.0%. All COVID‐19 positive samples have p values of <0.0001 compared to the COVID‐19 negative samples except sample #5118, which is not significant. All EIS experiments for sensing of antibodies were conducted by applying an AC signal with an amplitude of 1 mV and a fixed frequency range of 10 000 Hz to 1 Hz. PBS (50 mM) mixed with a ferro/ferricyanide (5 mM) mediator was used to dilute both COVID‐19 negative and positive plasma samples at 1:250. EIS, electrochemical impedance spectroscopy; PBS, phosphate‐buffered saline.
Once the sensor was calibrated with the standard concentrations of N antibodies, we tested the N sensor for the detection of N antibodies in the patient samples. The sensing results are shown in Figure 6B,C. First, the N sensor was set to the baseline by introducing only PBS to the sensor. The R ct value of baseline is calculated as 235 Ω, which is low compared to RBD sensor (1.7 kΩ). This may be due to the low molecular weight of the N proteins attached to the microelectrode array surface. The sensor was then independently tested with three COVID‐19 negative plasma samples. Compared to the baseline signal, the N sensor showed a relative standard deviation of 4.6% (Figure 6C). This indicates that the N sensor has an excellent selectivity because it rejected other proteins that are presented in the plasma samples. The N sensor was used to test the 17 COVID‐19 positive samples. The Nyquist plots and corresponding R ct values of the N sensor are shown in Figure 6B,C, respectively. Once a plasma sample was introduced, the N sensor showed a significant change of R ct values compared to the baseline signal, indicating presence of N antibodies in the patient sample. A statistically significant change in the R ct values was observed for all the plasma samples (p < 0.0001), except for one sample. Thus, the sensor showed a lower, 94.1%, sensitivity toward N antibodies. It is also possible that the one sample may have had an extremely low concentration of N antibodies.
As in the case of the RBD sensor, the N antibodies were evaluated using three additional N sensors. The R ct values of these N sensors were compared with the first N sensors (discussed above) and their correlation co‐efficients (r 2) evaluated. Supporting Information: Figure S4 shows the EIS spectra of the other three RBD sensors, and their corresponding linear regression plots of R ct values are shown in Supporting Information: Figure S5A–C. A correlation co‐efficient (r 2) was obtained as 0.85 when the same samples were tested with sensor 2 (Supporting Information: Figure S5A). Further, when sensor 3 (Supporting Information: Figure S5B) and sensor 4 (Supporting Information: Figure S5C) were tested with the same samples, the sensors showed a gradual reduction of R ct values and their correlation coefficients (Supporting Information: Figure S5D). As described in the RBD sensor, the reduction of R ct values compared to the first measurement may be due to the decreasing number of antibodies that are assessible in each successive measurement. We also note that all sensors showed the presence of N antibodies with COVID‐19‐positive patients. The exact concentration of N antibodies is described in the later section of the study.
3.7. Sensing of spike S1 antibodies in COVID‐19 samples
Similar to the RBD and N sensors, the S1 sensor was used to detect S1 antibodies in the same SARS‐CoV‐2 plasma samples. A total of three independent S1 sensors were chosen to evaluate the sensor‐to‐sensor variation. In this case, the sensor surface of an rGO‐Au micropillar array was modified with S1 proteins instead of RBD or N proteins. The results of the S1 sensor 1 are shown in Figure 7A,B. The Nyquist plots of sensors 1–3 are shown in Figure 7A, and Supporting Information: Figures S6A,B, respectively.
Figure 7.
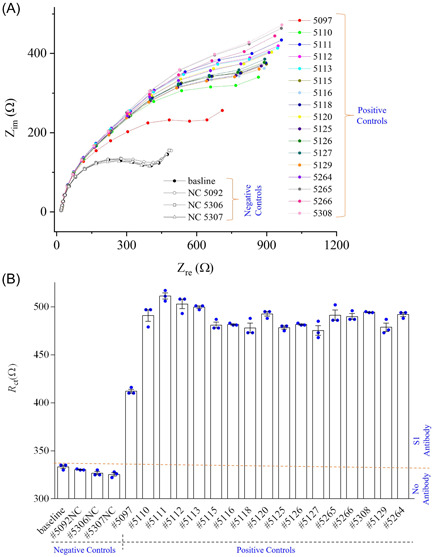
Sensing results of S1 sensor in plasma samples. (A, B) S1 sensor result of COVID‐19 patient samples showing the Nyquist plots for each human samples (A) and the calculated R ct values for each patient samples (B). The deviation of fitted semicircle and raw semicircle of all impedance spectra was less than ±4.0%. All COVID‐19 positive samples have p values of <0.0001 compared to the COVID‐19 negative samples. All EIS experiments for the sensing of antibodies were conducted by applying an AC signal with an amplitude of 1 mV and a fixed frequency range of 10 000–1 Hz. The PBS (50 mM) mixed with a ferro/ferricyanide (5 mM) mediator was used to dilute both negative and positive COVID‐19 human plasma (ratio of buffer to human plasma set to 250:1). EIS, electrochemical impedance spectroscopy; PBS, phosphate‐buffered saline.
The S1 sensor 1 was set to the baseline by exposing it to PBS. The R ct value of the baseline with PBS was calculated as 335 Ω (Figure 7B). When the sensor was exposed to COVID‐19 negative samples, a standard deviation 1.4% was calculated. This indicates that the S1 sensor was not sensitive to the proteins and other antibodies present in the samples. Once a COVID‐19 sample was introduced, the S1 sensor showed a significant change of R ct (416 Ω) compared to the baseline signal (335 Ω), indicating the presence of S1 antibodies (Figure 7B). Similarly, the impedance signals of the S1 sensor were recorded for other COVID‐19 positive samples. The significant changes of R ct values are observed for all 17 COVID‐19 samples (p < 0.0001). The S1 sensor showed a sensitivity of 100% towards S1 antibodies. Further, the same testing was performed with the other two sensors (sensors 2 and 3) with the same COVID‐19 samples (Supporting Information: Figure S6). These two sensors also showed a similar trend with a 100% sensitivity. Based on their R ct values, the sensor‐to‐sensor variation is evaluated (Supporting Information: Figure S7).
Before testing COVID‐19 samples, the S1 sensor was also tested with S1 antibodies to establish a standard curve. The S1 sensor showed a significant increase of R ct with increasing concentrations of S1 antibodies. This is due to the capture of S1 antibodies by the specific S1 proteins on the sensor surface. The analytical sensitivity of the S1 sensor was found to be 0.1 pm. To evaluate the correlation coefficients, the R ct values of S1 sensor 1 were compared with sensor 2 (Supporting Information: Figure S7A) and sensor 3 (Supporting Information: Figure S7B). A correlation co‐efficient was obtained as 0.924 for sensor 1 as compared to sensor 2 (Supporting Information: Figure S7A). When the R ct values of sensor 1 were compared to sensor 3, the correlation co‐efficient was found to be 0.727 (Supporting Information: Figure S7C). Similar to the RBD and N sensors, the gradual reduction of correlation coefficients in the sensor testing is primarily due to the repeated exposing of the same samples to multiple sensors. Supporting Information: Figure S7C shows the change of correlation co‐efficient with the sensors.
3.8. Comparative results of analysis of antibodies with different sensors in COVID‐19 samples
Table 1 summarizes the qualitative sensing results of SARS‐CoV‐2 antibodies in the patient samples detected by the RBD, N, and S1 sensors. Except N‐sensor, which failed to detect 1 of 17 (5.9%) COVID‐19 samples, the other two sensors had a 100% sensitivity. Figure 8 shows the detected concentrations of RBD, N, and S1 antibodies in samples. These results were obtained by comparing the R ct values for RBD, N, and S1 antibodies (Figures 4, 6, and 7B, respectively) with the respective sensor calibrations. The concentrations of RBD antibodies are plotted in Figure 8A. This figure shows that all COVID‐19 samples have detectable RBD antibodies ranging from >1 fM to <1 nM. Similarly, the concentrations of N antibodies were calculated from Figure 6C using standard curve (Figure 6A) and plotted against the patient samples (Figure 8B). The N sensor detected antibodies in 94.0% of the COVID‐19 patients. The COVID‐19 sample that was negative for N antibodies were positive for spike antibodies (RBD and S1). The highest concentration of N antibodies detected was 0.21 nM. Similarly, the concentrations of S1 antibodies are plotted against the patient samples (Figure 8C). The values of S1 concentrations are calculated from Figure 8B and standard curve (not shown). The S1 sensor shows a sensitivity of 100%. Note that the concentrations of S1 antibodies were found to be the highest compared to spike RBD and N antibodies. The highest of S1 antibodies detected was 6.3 nM. It is noteworthy that we observed variations of RBD, N, and S1 antibodies in COVID‐19 samples, which might reflect the different infection kinetics of these subjects, which could also be impacted by individual genetics, health history, and general immune responses, an area that is worth future investigation.
Table 1.
Comparative analysis of the detection of RBD, N, and S1 antibodies
| Samples/sensors | RBD | N | S1 | PCR test | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | ||
| 5092 | + | + | + | + | + | + | + | + | + | + |
| 5097 | − | − | − | − | − | − | − | − | − | − |
| 5110 | + | + | + | + | + | + | + | + | + | + |
| 5111 | + | + | + | + | + | + | + | + | + | + |
| 5112 | + | + | + | + | + | + | + | + | + | + |
| 5113 | + | + | + | + | + | + | + | + | + | + |
| 5115 | + | + | + | + | + | + | + | + | + | + |
| 5116 | + | + | + | + | + | + | + | + | + | + |
| 5118 | + | + | + | − | − | − | + | + | + | + |
| 5120 | + | + | + | + | + | + | + | + | + | + |
| 5125 | + | + | + | + | + | + | + | + | + | + |
| 5126 | + | + | + | + | + | + | + | + | + | + |
| 5127 | + | + | + | + | + | + | + | + | + | + |
| 5129 | + | + | + | + | + | + | + | + | + | + |
| 5264 | + | + | + | + | + | + | + | + | + | + |
| 5265 | + | + | + | + | + | + | + | + | + | + |
| 5266 | + | + | + | + | + | + | + | + | + | + |
| 5306 | − | − | − | − | − | − | − | − | − | − |
| 5307 | − | − | − | − | − | − | − | − | − | − |
| 5308 | + | + | + | + | + | + | + | + | + | + |
Note: The COVID‐19 status of the patients was confirmed by RT‐qPCR test.
Abbreviations: RBD, receptor‐binding domain; RT‐qPCR, reverse transcriptional real‐time polymerase chain reaction.
Figure 8.
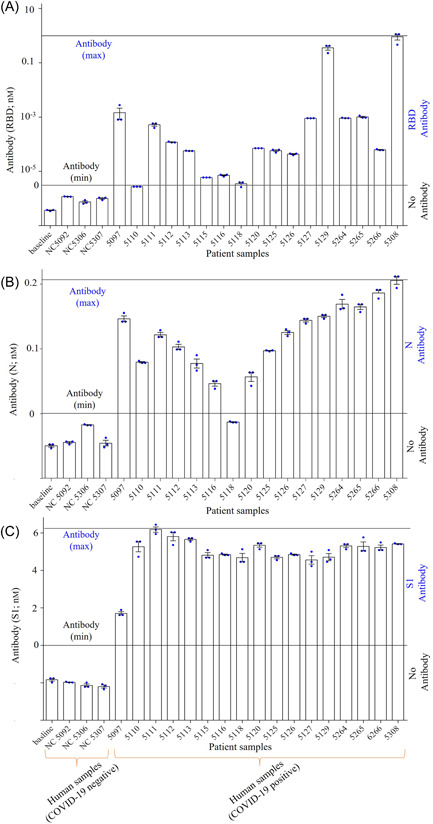
Quantitative examination of COVID‐19 antibodies using RBD, N and S1 sensors. The concentrations of RBD, N and S1 antibodies were calculated using the respective standard curves in Figures 4D, 6C, and 7B. (A) Concentration of RBD antibodies in positive and negative COVID‐19 samples. The maximum concentration of RBD antibodies detected was 1 nM. (B) Concentration of N antibodies in positive and negative COVID‐19 samples. The maximum concentration of N antibodies detected was 0.21 nM. (C) Concentration of spike S1 antibodies for the positive and negative COVID‐19 samples. The maximum concentration of S1 antibodies detected was 6.3 nM. Black lines indicate the maximum and minimum antibodies concentrations detected. RBD, receptor‐binding domain.
4. DISCUSSION
The results presented in this study provide a validation of our ultrarapid and ultrasensitive detection platform for pathogen biomarkers 40 , 41 in clinical samples. We achieve a label‐free detection of pathogen biomarkers in seconds using this platform. The speed of antibody detection demonstrated in this study is the fastest yet reported.
We note that in the test we conducted (e.g., Figure 6C), the sensor could not detect N‐antibodies for one of the COVID‐19 positive patients. The same patient, however, showed RBD and S1 antibodies. Thus, compared to the 100.0% (17/17) sensitivities of RBD and S1 sensors, N sensor showed a sensitivity of 94.0% (16/17). A number of serological studies demonstrate that the concentrations of S antibodies are usually higher than those of N antibodies in infected individuals. 68 , 69 Future studies with larger sample sizes could further compare the sensitivities and specificities of these sensors.
Our platform provides a means of quantifying antibodies. As different SARS‐CoV‐2 variants of concern (VOCs) have spread worldwide causing numerous re‐infection waves asynchronously in various regions, 70 , 71 obtaining such quantitative results can be of significant value for analyzing the efficacies of vaccines to SARS‐CoV‐2, 72 especially, the quantification of RBD and S1 antibodies. Similarly, detection of N antibodies to SARS‐CoV‐2 could help determine the status of infection, particularly in vaccinated subjects. Quantification of the exact concentrations of SARS‐CoV‐2 antibodies in plasma or whole blood could reveal the infection statuses and vaccine effectiveness. 73 The sensor has a high specificity because of the negatively charged graphene layer 74 on the array electrode repels the negatively charged albumin molecules (isoelectric point ~5) 75 and other proteins present in human samples. Finally, the short detection time and the ability to detect low concentrations is likely due to the enhanced interaction of the analyte molecules (antibodies in our case) within the micropillar electrode. 41
It is noted that the viral load determines the severity of the disease for COVID patients. 76 , 77 A recent study that correlates the viral load with pathogenesis has shown the kinetics of viral load and antibody response for COVID‐19 patients with different severities. 77 It was found that for most of the severely ill COVID‐19 patients (66.7%) in the ICU, the viral shedding in a variety of tissues was within ~20–40 days of the disease onset. For a majority of mildly ill COVID‐19 patients (81.8%), however, the viral shedding was limited to the respiratory tract ~10 days after the onset of the disease. 77 A low level of antibody response was noticed in mildly ill patients compared to severely ill patients. 77 From this perspective, a rapid point‐of‐care antibody test can aid in the understanding of the kinetics and tissue distribution of SARS‐CoV‐2 viral load, as well as neutralization of antibodies in patients, which could further help disease management in a personalized manner. 78
The results of this study demonstrate that the 3D‐printed ultrarapid sensing platform we developed can be used in the diagnosis of biomarkers for other pathogens. We can potentially also attach antibodies to the electrodes to detect antigens or pathogen proteins to detect antibodies. The developed sensors use spike S1 and RBD in addition to N protein to detect antibodies. It is noted that VOCs have a high mutation rate in the spike protein. 79 The use of specific antibodies to individual VOCs could potentially detect the different variants. 80 , 81 , 82 In fact, our unpublished research shows that we can attach SARS‐CoV‐2 antibodies to the electrode surface and detect corresponding SARS‐CoV‐2 antigens and SARS‐CoV‐2 pseudoviruses. We can also integrate multiple working electrodes in the device to create a multiplexed system that detects different antibodies to the same pathogen or different pathogen biomarkers. Finally, pathogen‐related ailments such as Sepsis, a leading cause of death in the United States 83 , 84 would greatly benefit from the timely detection offered by our sensing platform. Future research efforts will be focused on these areas.
5. CONCLUSION
In this manuscript, we have demonstrated the feasibility of rapid detection of SARS‐CoV‐2 antibodies in human plasma samples within 10–12 s by using a 3D‐printed sensor platform. The concentrations of antibodies to SARS‐CoV‐2 RBD, N, and S1 proteins were detected in 17 COVID‐19 positive samples but not in 3 COVID‐19 negative samples. The sensing was achieved by modifying the surfaces of each sensor with rGO and one of the viral proteins of SARS‐CoV‐2 (RBD, N, and S1). We conclude that:
(a) We have validated the 3D‐printed rapid biosensing platform (Ali et al. 40 ) with human plasma samples with detection times for RBD, N, and S1 antibodies within 12 s. This represents by far the fastest detection platform of antibodies for any pathogens.
(b) In addition to the detection of antibodies, the rapid platform can quantify the actual concentrations of different antibodies in the plasma samples. The results show that the concentrations of S1 antibodies are much higher than those of RBD and N antibodies in COVID‐19 samples. The rapid sensing of antibodies to viral proteins is achieved based on the novel 3D microneedle array geometry of the sensor.
(c) Direct simultaneous detection of multiple antibodies, specifically N and S (RBD and S1) antibodies, in the same COVID‐19 sample can provide a complete spectrum of antibodies in the tested human subjects.
AUTHOR CONTRIBUTIONS
Rahul Panat and Shou‐Jiang Gao developed with the concept and directed the research. Md. Azahar Ali built the device. Md. Azahar Ali and George Fei Zhang carried out all the antibodies detection tests. Chunshan Hu and Bin Yuan carried out the AJ printing of the micropillar electrodes. Sanjida Jahan carried out the SEM imaging of sensor geometries. Chunshan Hu, Bin Yuan, and Sanjida Jahan gave inputs to the manuscript. Georgios D. Kitsios, and Alison Morris collected and labeled human plasma samples. Md. Azahar Ali, Shou‐Jiang Gao, Rahul Panat, and George Fei Zhang processed the data and co‐wrote the manuscript. All the authors read and edited the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
The authors gratefully acknowledge partial financial support from the NIH R01 award #RF1NS110483 and NSF award #CMMI‐1757117 to Rahul Panat, and UPMC Hillman Cancer Center Startup Fund and Pittsburgh Foundation Endowed Chair in Drug Development for Immunotherapy to Shou‐Jiang Gao. The authors acknowledge the use of the Materials Characterization Facility at Carnegie Mellon University, supported by grant MCF‐677785.
Ali MA, Zhang GF, Hu C, et al. Ultrarapid and ultrasensitive detection of SARS‐CoV‐2 antibodies in COVID‐19 patients via a 3D‐printed nanomaterial‐based biosensing platform. J Med Virol. 2022;1‐19. 10.1002/jmv.28075
Md. Azahar Ali and George Fei Zhang authors contributed equally to this work.
Contributor Information
Shou‐Jiang Gao, Email: gaos8@upmc.edu.
Rahul Panat, Email: rpanat@andrew.cmu.edu.
DATA AVAILABILITY STATEMENT
All relevant data that support the findings of this study are presented in the manuscript and Supporting Information file. Data are available from the corresponding authors upon reasonable request.
REFERENCES
- 1. Brodin P. Immune determinants of COVID‐19 disease presentation and severity. Nature Med. 2021;27(1):28‐33. [DOI] [PubMed] [Google Scholar]
- 2. Ravindran R, McReynolds C, Yang J, et al. Immune response dynamics in COVID‐19 patients to SARS‐CoV‐2 and other human coronaviruses. PLoS One. 2021;16(7):e0254367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu H, Yang A, Song J, et al. Ultrafast, sensitive, and portable detection of COVID‐19 IgG using flexible organic electrochemical transistors. Sci Adv. 2021;7(38):eabg8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. West R, Kobokovich A, Connell N, Gronvall GK. COVID‐19 antibody tests: a valuable public health tool with limited relevance to individuals. Trends Microbiol. 2021;29(3):214‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu G, Rusling JF. COVID‐19 antibody tests and their limitations. ACS Sens. 2021;6(3):593‐612. [DOI] [PubMed] [Google Scholar]
- 6. Shuren J, Stenzel T. The FDA's experience with covid‐19 antibody tests. N Engl J Med. 2021;384(7):592‐594. [DOI] [PubMed] [Google Scholar]
- 7. Baraniuk C. Covid‐19 antibody tests: a briefing. BMJ. 2020;369:m2284. [DOI] [PubMed] [Google Scholar]
- 8. Cele S, Gazy I, Jackson L, et al. Escape of SARS‐CoV‐2 501Y. V2 from neutralization by convalescent plasma. Nature. 2021;593(7857):142‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barda N, Dagan N, Cohen C, et al. Effectiveness of a third dose of the BNT162b2 mRNA COVID‐19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet. 2021;398(10316):2093‐2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao C, Li X, Liu S, Sang Y, Gao S‐J, Gao F. HIV‐1 did not contribute to the 2019‐nCoV genome. Emerg Microbes Infect. 2020;9(1):378‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol. 2021;19(3):141‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramos da Silva S, Ju E, Meng W, et al. Broad severe acute respiratory syndrome coronavirus 2 cell tropism and immunopathology in lung tissues from fatal coronavirus disease 2019. J Infect Dis. 2021;223(11):1842‐1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H, Fischer DK, Shuda M, et al. Construction and characterization of two SARS‐CoV‐2 minigenome replicon systems. J Med Virol. 2022;94(6):2438‐2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS‐CoV‐2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike‐based vaccine immunogens. Front Immunol. 2020;11:2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang J, Petitjean SJL, Koehler M, et al. Molecular interaction and inhibition of SARS‐CoV‐2 binding to the ACE2 receptor. Nat Commun. 2020;11(1):4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A. 2020;117(21):11727‐11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garcia‐Beltran WF, Lam EC, Astudillo MG, et al. COVID‐19‐neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muruato AE, Fontes‐Garfias CR, Ren P, et al. A high‐throughput neutralizing antibody assay for COVID‐19 diagnosis and vaccine evaluation. Nat Commun. 2020;11(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fenwick C, Turelli P, Pellaton C, et al. A high‐throughput cell‐and virus‐free assay shows reduced neutralization of SARS‐CoV‐2 variants by COVID‐19 convalescent plasma. Sci Transl Med. 2021;13(605):eabi8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang F, Li W, Feng J, et al. SARS‐CoV‐2 pseudovirus infectivity and expression of viral entry‐related factors ACE2, TMPRSS2, Kim‐1, and NRP‐1 in human cells from the respiratory, urinary, digestive, reproductive, and immune systems. J Med Virol. 2021;93:6671‐6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varghese PM, Tsolaki AG, Yasmin H, et al. Host‐pathogen interaction in COVID‐19: pathogenesis, potential therapeutics and vaccination strategies. Immunobiology. 2020;225:152008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laing AG, Lorenc A, Del Molino Del Barrio I, et al. A dynamic COVID‐19 immune signature includes associations with poor prognosis. Nature Med. 2020;26(10):1623‐1635. [DOI] [PubMed] [Google Scholar]
- 23. Li K, Huang B, Wu M, et al. Dynamic changes in anti‐SARS‐CoV‐2 antibodies during SARS‐CoV‐2 infection and recovery from COVID‐19. Nat Commun. 2020;11(1):6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng M‐L, Liu H‐Y, Zhao H, et al. Longitudinal dynamics of antibody responses in recovered COVID‐19 patients. Signal Transduct Target Ther. 2021;6(1):1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li K, Huang B, Wu M, et al. Dynamic changes in anti‐SARS‐CoV‐2 antibodies during SARS‐CoV‐2 infection and recovery from COVID‐19. Nat Commun. 2020;11(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koutsakos M, Rowntree LC, Hensen L, et al. Integrated immune dynamics define correlates of COVID‐19 severity and antibody responses. Cell Rep Med. 2021;2(3):100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nature Med. 2020;26(7):1033‐1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ayouba A, Thaurignac G, Morquin D, et al. Multiplex detection and dynamics of IgG antibodies to SARS‐CoV2 and the highly pathogenic human coronaviruses SARS‐CoV and MERS‐CoV. J Clin Virol. 2020;129:104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deeks JJ, Dinnes J, Takwoingi Y, et al. Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Database Syst Rev. 2020;6(6):013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kubina R, Dziedzic A. Molecular and serological tests for COVID‐19. A comparative review of SARS‐CoV‐2 coronavirus laboratory and point‐of‐care diagnostics. Diagnostics. 2020;10(6):434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Elslande J, Houben E, Depypere M, et al. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID‐19 patients. Clin Microbiol Infect. 2020;26(8):1082‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yakoh A, Pimpitak U, Rengpipat S, Hirankarn N, Chailapakul O, Chaiyo S. based electrochemical biosensor for diagnosing COVID‐19: detection of SARS‐CoV‐2 antibodies and antigen. Biosens Bioelectron. 2021;176:112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El‐Said WA, Al‐Bogami AS, Alshitari W, et al. Electrochemical microbiosensor for detecting COVID‐19 in a patient sample based on gold microcuboids pattern. Biochip J. 2021;15(3):287‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ko DH, Hosseini A, Karaosmanoglu H, Taredun K, Jones L, Partridge A. Microfluidic separation of capture from detection and its application for determination of COVID‐19 antibodies. Sens. Actuators B Chem. 2022;351:130918. [Google Scholar]
- 35. Ahmadivand A, Gerislioglu B, Ramezani Z, Kaushik A, Manickam P, Ghoreishi SA. Functionalized terahertz plasmonic metasensors: femtomolar‐level detection of SARS‐CoV‐2 spike proteins. Biosens Bioelectron. 2021;177:112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, Qin Z, Fu H, et al. Enhancing the performance of paper‐based electrochemical impedance spectroscopy nanobiosensors: an experimental approach. Biosens Bioelectron. 2021;177:112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Torrente‐Rodríguez RM, Lukas H, Tu J, et al. SARS‐CoV‐2 RapidPlex: a graphene‐based multiplexed telemedicine platform for rapid and low‐cost COVID‐19 diagnosis and monitoring. Matter. 2020;3(6):1981‐1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lukas H, Xu C, Yu Y, Gao W. Emerging telemedicine tools for remote COVID‐19 diagnosis, monitoring, and management. ACS Nano. 2020;14(12):16180‐16193. [DOI] [PubMed] [Google Scholar]
- 39. Saleh MS, Hu C, Panat R. Three‐dimensional microarchitected materials and devices using nanoparticle assembly by pointwise spatial printing. Sci Adv. 2017;3(3):e1601986. 10.1126/sciadv.1601986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ali MA, Hu C, Jahan S, et al. Sensing of COVID‐19 antibodies in seconds via aerosol jet nanoprinted reduced‐graphene‐oxide‐coated 3D electrodes. Adv Mater. 2021;33(7):2006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ali MA, Hu C, Yuan B, et al. Breaking the barrier to biomolecule limit‐of‐detection via 3D printed multi‐length‐scale graphene‐coated electrodes. Nat Commun. 2021;12(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gibson I, Rosen DW, Stucker B, et al. Additive Manufacturing Technologies. Vol 17. Springer; 2021. [Google Scholar]
- 43. Ali MA, Hu C, Yttri EA, Panat R. Recent advances in 3D printing of biomedical sensing devices. Adv Funct Mater. 2021;32(9):2107671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharma PK, Kim E‐S, Mishra S, et al. Ultrasensitive and reusable graphene oxide‐modified double‐interdigitated capacitive (DIDC) sensing chip for detecting SARS‐CoV‐2. ACS Sens. 2021;6(9):3468‐3476. [DOI] [PubMed] [Google Scholar]
- 45. Mujawar MA, Gohel H, Bhardwaj SK, Srinivasan S, Hickman N, Kaushik A. Nano‐enabled biosensing systems for intelligent healthcare: towards COVID‐19 management. Mater Today Chem. 2020;17:100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sengupta J, Hussain CM. Graphene‐based field‐effect transistor biosensors for the rapid detection and analysis of viruses: a perspective in view of COVID‐19. Carbon Trends. 2021;2:100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palmieri V, Papi M. Can graphene take part in the fight against COVID‐19? Nano Today. 2020;33:100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Afroj S, Britnell L, Hasan T, Andreeva DV, Novoselov KS, Karim N. Graphene‐based technologies for tackling COVID‐19 and future pandemics. Adv Funct Mater. 2021;31(52):2107407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beduk T, Beduk D, de Oliveira Filho JI, et al. Rapid point‐of‐care COVID‐19 diagnosis with a gold‐nanoarchitecture‐assisted laser‐scribed graphene biosensor. Anal Chem. 2021;93(24):8585‐8594. [DOI] [PubMed] [Google Scholar]
- 50. Ali MA, Hu C, Zhang F, et al. N protein‐based ultrasensitive SARS‐CoV‐2 antibody detection in seconds via 3D nanoprinted, microarchitected array electrodes. J Med Virol. 2022;94:2067‐2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shen S, Tan TH, Tan Y‐J. Expression, glycosylation, and modification of the spike (S) glycoprotein of SARS CoV. Glycovirology Protocols. 2007;379:127‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiao X, Dimitrov D. The SARS‐CoV S glycoprotein. Cell Mol Life Sci. 2004;61(19):2428‐2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bisht H, Roberts A, Vogel L, et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci U S A. 2004;101(17):6641‐6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ali MA, Mondal K, Singh C, Malhotra BD, Sharma A. Anti‐epidermal growth factor receptor conjugated mesoporous zinc oxide nanofibers for breast cancer diagnostics. Nanoscale. 2015;7(16):7234‐7245. [DOI] [PubMed] [Google Scholar]
- 55. Xu Q, Davis JJ. The diagnostic utility of electrochemical impedance. Electroanalysis. 2014;26(6):1249‐1258. [Google Scholar]
- 56. Suni II. Impedance methods for electrochemical sensors using nanomaterials. TrAC Trends Anal Chem. 2008;27(7):604‐611. [Google Scholar]
- 57. Jacobs JL, Bain W, Naqvi A, et al. Severe acute respiratory syndrome coronavirus 2 viremia is associated with coronavirus disease 2019 severity and predicts clinical outcomes. Clin Infect Dis. 2022;74(9):1525‐1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jacobs JL, Naqvi A, Shah FA, et al. Plasma SARS‐CoV‐2 RNA levels as a biomarker of lower respiratory tract SARS‐CoV‐2 infection in critically ill patients with COVID‐19. J Infect Dis . Published online May 2, 2022;jiac157. 10.1093/infdis/jiac157 [DOI] [PMC free article] [PubMed]
- 59. Bain W, Yang H, Shah FA, et al. COVID‐19 versus Non‐COVID‐19 acute respiratory distress syndrome: comparison of demographics, physiologic parameters, inflammatory biomarkers, and clinical outcomes. Ann Am Thorac Soc. 2021;18(7):1202‐1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saleh MS, Li J, Park J, Panat R. 3D printed hierarchically‐porous microlattice electrode materials for exceptionally high specific capacity and areal capacity lithium ion batteries. Addit Manuf. 2018;23:70‐78. [Google Scholar]
- 61. Saleh MS, Hu C, Panat R. Three‐dimensional microarchitected materials and devices using nanoparticle assembly by pointwise spatial printing. Sci Adv. 2017;3(3):e1601986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ali MA, Wang X, Chen Y, et al. Continuous monitoring of soil nitrate using a miniature sensor with poly (3‐octyl‐thiophene) and molybdenum disulfide nanocomposite. ACS Appl Mater Interfaces. 2019;11(32):29195‐29206. [DOI] [PubMed] [Google Scholar]
- 63. Liu EY, Jung S, Weitz DA, Yi H, Choi C‐H. High‐throughput double emulsion‐based microfluidic production of hydrogel microspheres with tunable chemical functionalities toward biomolecular conjugation. Lab Chip. 2018;18(2):323‐334. [DOI] [PubMed] [Google Scholar]
- 64. Ali MA, Mondal K, Jiao Y, et al. Microfluidic immuno‐biochip for detection of breast cancer biomarkers using hierarchical composite of porous graphene and titanium dioxide nanofibers. ACS Appl Mater Interfaces. 2016;8(32):20570‐20582. [DOI] [PubMed] [Google Scholar]
- 65. Secor EB. Principles of aerosol jet printing. Flex Print Electron. 2018;3(3):035002. [Google Scholar]
- 66. Heine V, Kremers T, Menzel N, Schnakenberg U, Elling L. Electrochemical impedance spectroscopy biosensor enabling kinetic monitoring of fucosyltransferase activity. ACS Sens. 2021;6(3):1003‐1011. [DOI] [PubMed] [Google Scholar]
- 67. Jansen RW, Schols D, Pauwels R, De Clercq E, Meijer D. Novel, negatively charged, human serum albumins display potent and selective in vitro anti‐human immunodeficiency virus type 1 activity. Mol Pharmacol. 1993;44(5):1003‐1007. [PubMed] [Google Scholar]
- 68. Dodd RY, Notari EP, Brodsky JP, et al. Patterns of antibody response to severe acute respiratory syndrome coronavirus 2 among 1.6 million blood donors: impact of vaccination, United States, December 2020‐June 2021. J Infect Dis. 2022;225(1):5‐9. [DOI] [PubMed] [Google Scholar]
- 69. Murhekar MV, Bhatnagar T, Thangaraj JWV, et al. SARS‐CoV‐2 seroprevalence among the general population and healthcare workers in India, December 2020‐January 2021. Int J Infect Dis. 2021;108:145‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS‐CoV‐2 Omicron variant in southern Africa. Nature. 2022;603(7902):679‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cheng VC, Ip JD, Chu AW, et al. Rapid spread of SARS‐CoV‐2 Omicron subvariant BA.2 in a single‐source community outbreak. Clin Infect Dis . 2022;ciac203. 10.1093/cid/ciac203 [DOI] [PMC free article] [PubMed]
- 72. Yuce M, Filiztekin E, Ozkaya KG. COVID‐19 diagnosis—a review of current methods. Biosens Bioelectron. 2021;172:112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hwang Y‐C, Lu R‐M, Su S‐C, et al. Monoclonal antibodies for COVID‐19 therapy and SARS‐CoV‐2 detection. J Biomed Sci. 2022;29(1):1‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vallés C, Drummond C, Saadaoui H, et al. Solutions of negatively charged graphene sheets and ribbons. J Am Chem Soc. 2008;130(47):15802‐15804. [DOI] [PubMed] [Google Scholar]
- 75. Wiig H, Kolmannskog O, Tenstad O, et al. Effect of charge on interstitial distribution of albumin in rat dermis in vitro. J Physiol. 2003;550(2):505‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kaushik AK, Dhau JS, Gohel H, et al. Electrochemical SARS‐CoV‐2 sensing at point‐of‐care and artificial intelligence for intelligent COVID‐19 management. ACS Appl Bio Mater. 2020;3(11):7306‐7325. [DOI] [PubMed] [Google Scholar]
- 77. Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID‐19 severity. J Clin Invest. 2020;130(10):5235‐5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kaushik AK, Dhau JS, Gohel H, et al. Electrochemical SARS‐CoV‐2 sensing at Point‐of‐Care and artificial intelligence for intelligent COVID‐19 management. ACS Appl Bio Mater. 2020;3(11):7306‐7325. [DOI] [PubMed] [Google Scholar]
- 79. Zhang GF, Meng W, Chen L, et al. Neutralizing antibodies to SARS‐CoV‐2 variants of concern including Delta and Omicron in subjects receiving mRNA‐1273, BNT162b2 and Ad26.COV2.S vaccines. J Med Virol . Published online Jul 28, 2022. 10.1002/jmv.28032 [DOI] [PMC free article] [PubMed]
- 80. Mostafavi E, Dubey AK, Teodori L, Ramakrishna S, Kaushik A. SARS‐CoV‐2 Omicron variant: a next phase of the COVID‐19 pandemic and a call to arms for system sciences and precision medicine. MedComm. 2022;3(1):e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kujawska M, Mostafavi E, Kaushik A. SARS‐CoV‐2 getting into the brain; neurological phenotype of COVID‐19, and management by nano‐biotechnology. Neural Regen Res. 2022;18(2023):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gao SJ, Guo H, Luo G. Omicron variant (B. 1.1. 529) of SARS‐CoV‐2, a global urgent public health alert. J Med Virol. 2022;94(4):1255‐1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. 2010;14(1):R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sehgal M, Ladd HJ, Totapally B. Trends in epidemiology and microbiology of severe sepsis and septic shock in children. Hosp Pediatr. 2020;10(12):1021‐1030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
All relevant data that support the findings of this study are presented in the manuscript and Supporting Information file. Data are available from the corresponding authors upon reasonable request.


