Abstract
The aminophenol (AP) catabolic operon in Pseudomonas putida HS12 mineralizing nitrobenzene was found to contain all the enzymes responsible for the conversion of AP to pyruvate and acetyl coenzyme A via extradiol meta cleavage of 2-aminophenol. The sequence and functional analyses of the corresponding genes of the operon revealed that the AP catabolic operon consists of one regulatory gene, nbzR, and the following nine structural genes, nbzJCaCbDGFEIH, which encode catabolic enzymes. The NbzR protein, which is divergently transcribed with respect to the structural genes, possesses a leucine zipper motif and a MarR homologous domain. It was also found that NbzR functions as a repressor for the AP catabolic operon through binding to the promoter region of the gene cluster in its dimeric form. A comparative study of the AP catabolic operon with other meta cleavage operons led us to suggest that the regulatory unit (nbzR) was derived from the MarR family and that the structural unit (nbzJCaCbDGFEIH) has evolved from the ancestral meta cleavage gene cluster. It is also proposed that these two functional units assembled through a modular type gene transfer and then have evolved divergently to acquire specialized substrate specificities (NbzCaCb and NbzD) and catalytic function (NbzE), resulting in the creation of the AP catabolic operon. The evolutionary process of the AP operon suggests how bacteria have efficiently acquired genetic diversity and expanded their metabolic capabilities by modular type gene transfer.
Nitroaromatic compounds are widely used in the manufacture of dyes, drugs, explosives, and solvents, and massive amounts of their discharge have had detrimental effects on the environment. Much attention has been paid to the bioremediation of these compounds, and both aerobic and anaerobic degradation by microorganisms have been reported (13, 16, 45). As a typical recalcitrant nitroaromatic compound, nitrobenzene (NB) is known to be metabolized by aerobic bacteria through either an oxidative pathway (35) or a partial reductive pathway (20, 22, 34, 38). In the partial reductive pathway, NB is converted into 2-aminophenol (AP), which is subsequently metabolized via a meta-like cleavage pathway. Several enzymes involved in NB catabolism of Pseudomonas pseudoalcaligenes JS45 were purified and characterized (21, 23, 24, 31, 44). However, despite intensive studies on the catabolic pathway of NB and characterization of the relevant enzymes, a detailed molecular basis of and a regulatory mechanism for NB catabolism remain yet to be elucidated. Recently, we reported the novel genetic organization of the NB catabolic gene clusters that are on the catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12 (37). All the genes except for that of mutase, which was found on pNB2, were clustered on pNB1. Of the nbz (for nitrobenzene degradation) genes, the AP gene cluster was revealed to be a tightly regulated one.
It has been generally accepted that horizontal gene transfer (HGT) has played an integral role in the dissemination of antibiotic resistance genes, biodegradative genes, and pathogenicity-conferring genes in bacteria (7, 10, 27, 32, 36). Recently, it was reported that HGT is also responsible for bacterial speciation (11, 29). HGT is known to be mediated dominantly by three mobile genetic elements: conjugative plasmids (for cell contact-dependent gene transfer), transposons (for Tn-mediated gene transfer), and bacteriophages (which act as the vector for gene transfer) (2, 10, 11). In the gene transfer event, most of mobilized genes cannot be stably maintained in the recipient cells without appropriate selection. Moreover, in order for the transferred genes to confer stable phenotypes (antibiotic resistance, catabolic capability, and virulence), they have to be assembled into a single functional unit; otherwise, the whole functional operon needs to be transferred (29). Lawrence and Roth recently proposed the selfish operon model, in which the gradual formation of gene cluster and their subsequent integration into an operon is facilitated by HGT (30). In this way, HGT has played a major role in the diversification and evolution of microorganisms.
In this paper, to further elucidate the genetic origin and evolutionary process of the AP catabolic operon, we carried out a genetic and biochemical characterization of the AP catabolic operon and investigated its regulatory mechanism. A comparative study of the AP gene cluster with other well-studied meta cleavage operons was also conducted. Based on the results, we propose that the AP operon has evolved through modular type gene transfer, a specific type of HGT. The modular type gene transfer, in which functional gene units (regulatory and structural gene clusters originated from different sources), not arbitrary genes, are transferred and assembled into a new operon with specific function, is considered a fast and efficient mechanism for bacteria to acquire genetic diversity and expand their metabolic capabilities.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Cells of P. putida strains were grown in defined mineral medium at 30°C as described elsewhere (37). Cells of Escherichia coli strains were cultured in Luria-Bertani medium at 37°C. pBluescript SK/KS (Stratagene) and pUCP22 (a gift from G. J. Zylstra; GenBank accession no. U07166) vectors were used as cloning vehicles for E. coli and P. putida strains, respectively. When necessary, 100 μg of ampicillin/ml was added to E. coli cultures and 40 μg of gentamicin/ml was used for P. putida cultures.
DNA manipulation and sequencing.
DNA manipulations, such as subcloning and transformation, were performed in accordance with the standard procedure described by Sambrook et al. (41). Plasmids of P. putida HS12 were isolated as previously described (37). DNA fragments from pNB1 and pNB2 were cloned into pBluescript SK/KS and pUCP22, a broad-host-range vector for gram-negative bacteria. Expression profiles of the constructed plasmid in E. coli JM109 and P. putida strains were analyzed by assaying the corresponding enzymes.
For DNA sequencing, physical mapping of pNB1 was followed by subcloning into pBluescript SK/KS. Both strands of the 11.1-kb DNA fragment from pNB1 were sequenced by the dideoxy-chain termination method, using double-stranded DNA as the template (42). Sequencing was carried out by a combination of manual sequencing and automated sequencing with an API 377 sequencer (Applied Biosystems Inc.).
Enzyme assays.
Cell extracts were prepared as previously described (37). The analysis of AP dioxygenase, 2-aminomuconic semialdehyde (AMS) dehydrogenase, and 2-aminomuconate (AM) deaminase activities was performed by using previously determined methods (37). 4-Oxalocrotonate (OC) decarboxylase activity was analyzed spectrophotometrically by measuring the decrease of OC at 236 nm (18). 2-Oxopent-4-enoate (OP) hydratase activity was determined by the method described by Collinsworth et al. (8), with some modifications. One unit of activity was defined as the amount of enzyme required to cause a decrease in A265 of 1.0 per minute. 4-Hydroxy-2-oxovalerate (HOV) aldolase was assayed by measuring the oxidation of NADH at A340 in the presence of excess l-lactate dehydrogenase (40). To measure acetaldehyde (ACT) dehydrogenase activity, the coenzyme A-stimulated reduction of NAD+ was monitored at 340 nm (40).
Chemicals.
2-Hydroxymuconic semialdehyde (HMS) and AM for AMS dehydrogenase and AM deaminase were synthesized and purified by using established methods (37). OC was synthesized as described elsewhere (28, 53). OP was prepared enzymatically from l-allylglycine as reported by Collinsworth et al. (8). HOV was prepared by mild alkaline hydrolysis of 4-methyl-2-oxobutyrolactone (40).
RT-PCR.
Total RNA was isolated from HS12 cells grown on NB or succinate by using an RNeasy total RNA kit (Qiagen). Purified RNA was treated with RNase-free DNase (Promega) to remove DNA contaminants and then was concentrated by ethanol precipitation. Reverse transcription (RT)-PCR was carried out with an Access RT-PCR kit (Promega). The following primer pairs were utilized: P2281 and P3721R for the nbzCaCbD region, P4801 and P6001R for the nbzDGF region, and P6361 and P8281R for the nbzFEIH region (Fig. 1A). The sequences of the primers are available through personal communication.
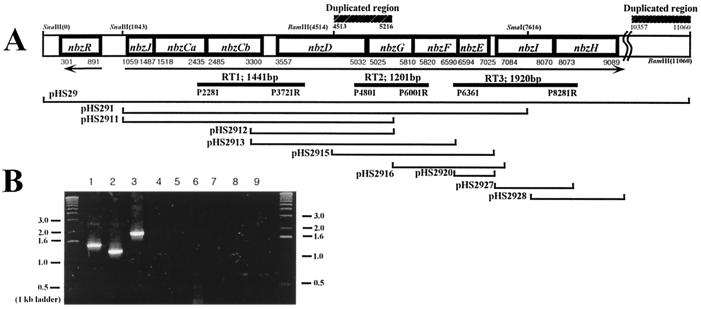
FIG. 1.
(A) The genetic organization of the AP catabolic operon on the pNB1 plasmid in P. putida HS12. Arrows indicate the direction of transcription. Duplicated DNA regions are shown above the physical map. DNA fragments subcloned for functional analysis are indicated by thin lines below the map. The locations of the primers and the amplified DNA fragments for RT-PCR are shown by thick lines. (B) RT-PCR amplification of the AP gene clusters. Lanes 1, 2, and 3, RT-PCR products with total RNA from NB-grown HS12; lanes 4, 5, and 6, total RNA from succinate-grown HS12; lanes 7, 8, and 9, PCR products with total RNA from NB-grown HS12 without RT. Amplifications were performed with the RT1 set (lanes 1, 4, and 7) the RT2 set (lanes 2, 5, and 8), and the RT3 set (lanes 3, 6, and 9).
Purification of NbzR and DNA binding assay.
The nbzR gene was amplified using PCR with a PN primer for the N-terminal region and a PC primer for the C-terminal region. The amplified DNA fragment was digested with EcoRI (partial) and HindIII and then was inserted into pMAL-c2X (New England BioLabs). The recombinant plasmid, designated pMR1, was expressed in E. coli BL21(DE3) cells under the control of isopropyl-β-d-thiogalactopyranoside (IPTG). The overexpressed NbzR protein was cleaved from the maltose binding protein and purified according to the manufacturer's instructions.
DNA binding experiments were performed by Fried and Crothers' method (14), with slight modifications. Two sets of primers, P793-P1157R and P1300-P1583, were used to amplify the intergenic regions of nbzR-nbzJ and nbzJ-nbzCa, respectively. The PCR-amplified PI (365 bp from the P793-P1157R set) and PII (284 bp from the P1300-P1583 set) fragments (50 ng) were incubated with purified NbzR protein (0 to 300 ng) in 20 μl of binding buffer (10 mM Tris-HCl [pH 7.2], 10 mM β-mercaptoethanol, 1 mM EDTA, 0.1% Triton X-100, 4% glycerol, 50 mM KCl, 5 mM MgCl2) for 10 min at 4°C. The reaction mixture was subjected to electrophoresis on 5% native polyacrylamide gels in TBE buffer (41).
Cross-linking experiments.
Cross-linking of NbzR was carried out as follows. Purified NbzR (1 mg/ml) was first activated by adding 2 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and 5 mM N-hydroxysuccinimide (NHS) in 20 μl of 10 mM MES (morpholineethanesulfonic acid) (pH 6.0), and then β-mercaptoethanol (final concentration, 20 mM) and 20 μl of 50 mM HEPES (pH 8.0) were added to quench the EDC and raise the pH to 7.5. After incubation at 4°C for 1 h, the reaction was stopped by adding hydroxylamine (final concentration, 10 mM) and analyzed with sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (41).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in GenBank under the accession no. AF319593.
RESULTS AND DISCUSSION
Sequence and functional analysis of the AP catabolic gene cluster.
We previously found that the AP catabolic gene cluster might constitute a single operonic structure in P. putida HS12 (37). To further characterize the AP catabolic genes, the nucleotide sequence of a 11.1-kb DNA fragment of the pNB1 plasmid was determined. Analysis of the sequence identified 10 open reading frames (ORFs), all of which were organized in the same direction with the exception of the nbzR gene, a putative regulatory gene (Fig. 1A). Databases in GenBank were searched for proteins having a high degree of similarity to the deduced amino acid sequences of the AP catabolic gene products by using the BLAST program. Sequences were then retrieved and compared with the deduced amino acid sequences by using the ClustalX program. The location of each gene and the predicted molecular masses of the gene products are listed in Table 1, along with details of proteins bearing significant similarity to the predicted gene products.
TABLE 1.
Comparison of NbzR and NbzJCaCbDEFGHI with related proteins
| Gene | Position in sequence (nt) | Protein function | Calculated mol mass (Da) (no. of aa) | Protein with homologous sequence | Source | % Identitya/ no. of aa | Accession no. |
|---|---|---|---|---|---|---|---|
| nbzR | 301–891 | Regulatory factor | 21,886 (196) | MarR | E. coli | 26/144 | D90797 |
| nbzJ | 1,059–1,487 | Putative ferredoxin | 15,686 (142) | XylT | P. putida mt-2 | 20/112 | M64747 |
| nbzCa | 1,518–2,435 | β-Subunit of aminophenol | 54,483 (305) | AmnB | P. pseudoalcaligenes JS45 | 98/305 | U93363 |
| 1, 6-dioxygenase | AmnB | Pseudomonas sp. strain AP-3 | 84/305 | D89855 | |||
| HpcB | E. coli | 19/276 | O05353 | ||||
| nbzCb | 2,485–3,300 | α-Subunit of aminophenol 1,6-dioxygenase | 29,475 (271) | AmnA | P. pseudoalcaligenes JS45 | 99/271 | U93363 |
| AmnA | Pseudomonas sp. strain AP-3 | 67/271 | AB020521 | ||||
| CarBb | Pseudomonas sp. | 22/269 | D89064 | ||||
| nbzD | 3,557–5,032 | Aminomuconic semialdehyde dehydrogenase | 53,294 (491) | AmnC | P. pseudoalcaligenes JS45 | 66/542 | U93363 |
| AnmC | Pseudomonas sp. strain AP-3 | 85/491 | AB020521 | ||||
| XylG | P. putida mt-2 | 55/486 | M64747 | ||||
| nbzG | 5,025–5,810 | 2-Oxopent-4-enoate hydratase | 27,834 (261) | AmnF | Pseudomonas sp. strain AP-3 | 73/261 | AB020521 |
| XylJ | P. putida mt-2 | 35/222 | M64747 | ||||
| nbzF | 5,820–6,590 | 4-Oxalocrotonate decarboxylase | 27,258 (256) | AmnD | Pseudomonas sp. strain AP-3 | 77/256 | AB020521 |
| XylI | P. putida mt-2 | 38/254 | M94186 | ||||
| nbzE | 6,594–7,025 | Aminomuconate deaminase | 15,793 (143) | AmD | P. pseudoalcaligenes JS45 | 100/54b | P81593 |
| AmnE | Pseudomonas sp. strain AP-3 | 76/142 | AB020521 | ||||
| XylH | P. putida mt-2 | 29/63 | M94186 | ||||
| nbzI | 7,084–8,070 | Acetaldehyde dehydrogenase | 36,283 (338) | AmnH | Pseudomonas sp. strain AP-3 | 71/316 | AB020521 |
| XylQ | P. putida mt-2 | 50/312 | M94186 | ||||
| nbzH | 8,073–9,089 | 4-hydroxy-2-oxovalerate aldolase | 34,905 (328) | AmnG | Pseudomonas sp. strain AP-3 | 85/343 | AB020521 |
| XylK | P. putida mt-2 | 78/345 | M94186 |
Percent identity of partial amino acid sequence of NbzR with that of MarR (residues 28 to 144).
Only a partial peptide sequence is available.
The deduced amino acid sequences of NbzCa, NbzCb, NbzD, and NbzE were found to have extremely high degrees of identity (98 to 99%) with the β-subunit and α-subunit of AP dioxygenase, the N-terminal region of AMS dehydrogenase (residues 1 to 325), and a partial peptide sequence of AM deaminase (residues 1 to 54) from P. pseudoalcaligenes JS45 (GenBank accession no. U93363 and P81593). Despite the fact that HS12 and JS45 were separately isolated from geographically remote areas (34, 38), DNA and amino acid analyses of the above enzymes imply that these strains are closely related to each other. Recently, Takenaka et al. reported the nucleotide sequence of the AP catabolic genes in Pseudomonas sp. strain AP-3 (49). Interestingly, AP structural gene products from Pseudomonas sp. strain AP-3 revealed high identity levels with those from HS12, as shown in Table 1. Pseudomonas sp. strain AP-3 was isolated on the basis of growth ability on AP, but HS12 uses NB, not AP, as a growth substrate and inducer (37) and contains all the catabolic genes for NB degradation. Nonetheless, AP gene clusters in AP-3 and HS12 seem to have a similar physiological role in AP catabolism. NbzJ has 20% amino acid identity with chloroplast-type ferredoxin (XylT) in the TOL operon that mediates the reductive reactivation of catechol 2,3-dioxygenase (25). Lendenmann and Spain reported that purified AP dioxygenase from JS45 was severely inactivated by catechol (31), as was found to be the case for the inactivation of catechol 2,3-dioxgenase by 4-methylcatechol (25). This suggests the possible participation of NbzJ in the protection and reactivation of NbzCaCb. However, because only two of four cystein residues presumed to serve as ligands for the [2Fe-2S] cluster in XylT were conserved in NbzJ (data not shown), it is not clear yet whether NbzJ is involved in AP catabolism. When compared with other meta cleavage gene clusters, NbzD showed 55% amino acid identity with HMS dehydrogenase (XylG) from P. putida mt-2, but NbzCa and NbzCb had relatively low identities with other extradiol dioxygenases, such as HpcB from Escherichia coli and CarBb from Pseudomonas sp. (Table 1). As for NbzE, no significant level of similarity with other deaminases has been found. Instead, part of NbzE (residues 44 to 111) possessed an identity of 25% with 4-oxalocrotonate tautomerase (XylH) from P. putida mt-2 (Table 1). Interestingly, the gene products of nbzF, nbzG, nbzH, and nbzI exhibited relatively high levels of identity with the well-characterized meta cleavage enzymes (XylI, XylJ, XylK, and XylQ) of P. putida mt-2 (Table 1). In order to confirm the predicted catalytic function of the AP catabolic gene products, a functional analysis of the subclones containing the ORFs was attempted with E. coli JM109 cells (Fig. 1A and Table 2). All the subclones carrying part or all of the catabolic genes exhibited the corresponding catalytic activities, as predicted by sequence comparison analysis. Based on the above results, the AP catabolic gene products are as follows: NbzCaCb, β- and α-subunits of AP dioxygenase; NbzD, AMS dehydrogenase; NbzE, AM deaminase; NbzF, OC decarboxylase; NbzG, OP hydratase; NbzH, HOV aldolase; and NbzI, ACT dehydrogenase.
TABLE 2.
Enzyme activities of AP catabolic enzymes in various plasmids
| Plasmidb | Enzyme sp act (nmol/min/mg of protein)a
|
||||||
|---|---|---|---|---|---|---|---|
| NbzC (dioxygenase) | NbzD (dehydrogenase) | NbzE (deaminase) | NbzF (decarboxylase) | NbzG (hydratase) | NbzH (aldolase) | NbzI (dehydrogenase) | |
| pHS29 | 69 ± 3 | 46 ± 3 | 314 ± 25 | 2,104 ± 23 | 80 ± 4 | 171 ± 12 | 106 ± 15 |
| pHS291 | 175 ± 20 | 74 ± 5 | 164 ± 15 | 3,510 ± 29 | 113 ± 6 | — | — |
| pHS2911 | 333 ± 21 | 208 ± 16 | — | — | — | — | — |
| pHS2912 | —c | 304 ± 13 | — | — | — | — | — |
| pHS2913 | — | 262 ± 28 | — | — | 209 ± 19 | — | — |
| pHS2915 | — | — | 480 ± 27 | 7,326 ± 542 | 365 ± 32 | — | — |
| pHS2916 | — | — | 725 ± 46 | 4,534 ± 359 | — | — | — |
| pHS2920 | — | — | 613 ± 35 | — | — | — | — |
| pHS2927 | — | — | — | — | — | — | 359 ± 58 |
| pHS2928 | — | — | — | — | — | 113 ± 25 | — |
Enzyme assays were performed with cell extracts prepared as described in Materials and Methods and each enzyme activity is a mean value of three independent measurements.
All E. coli plasmids were not induced.
—, not detected.
In order to determine whether the AP catabolic genes are cotranscribed, three sets of primers were designed (Fig. 1A) and used for PCR amplification with total RNAs from NB-grown and succinate-grown P. putida HS12. The RT1 set (spanning the nbzCaCbD gene region), RT2 set (spanning the nbzDGF gene region), and RT3 set (spanning the nbzFEIH gene region) resulted in the amplification of 1.4-, 1.2-, and 1.9-kb DNA fragments, respectively, only from a reverse-transcribed RNA sample of HS12 grown on NB (Fig. 1B). This result indicates that the nbzCaCbDGFEIH genes are transcribed as a single transcript and their expressions are coordinately regulated as a whole, thus confirming the conclusion that these genes constitute a single operon. Although Takenaka et al. reported a structural gene organization of amn genes (amnBACFDEHG) that is identical to that of nbz genes of the AP operon (49), all the structural gene products are not functionally identified, and whether these genes are clustered in a single operon was not investigated. ORF1 in P. pseudoalcaligenes JS45 was classified as a putative member of the YjgF protein family and found to be cotranscribed with the amnBAC genes (9). High DNA identity (98%) between the NbzJ-nbzCaCb region in HS12 and the orf1-amnBA region in JS45 implies that, as in JS45, NbzJ in the AP gene cluster of HS12 might be cotranscribed with the other structural genes, nbzCaCbDGFEIH. Therefore, the overall genetic organization of the AP catabolic gene cluster is probably nbzJCaCbDGFEIH. A truncated duplicate of nbzDG genes was found in the downstream of the structural genes. But, the possible involvement of this region in the catabolism is not clear yet.
Regulation of the AP operon and NbzR.
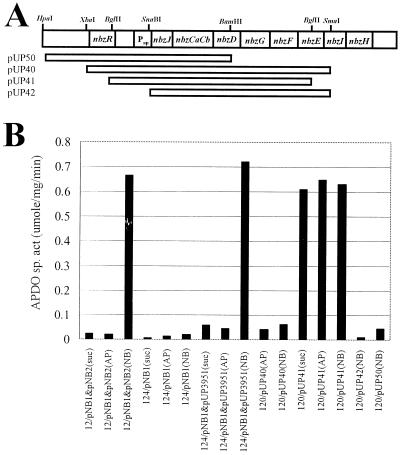
The catabolic genes and their products of the AP operon were studied extensively (21, 23, 31, 37, 48), but the regulatory mechanism of the operon has not been reported so far. It was previously observed that the AP-degrading pathway is inducible (37). When pUP3951, containing only the nbzB gene (1.0 kb) from the pNB2 plasmid, was expressed in P. putida HS124 harboring a pNB1 plasmid with the nbzA and nbzJCaCbDGFEIH genes, the induction profile of the AP catabolic genes and the ability of P. putida HS124 (pNB1 + pUP3951) to grow on NB were similar to those of the wild-type HS12 (pNB1 + pNB2) (Fig. 2B). Only NbzB from pNB2 is required both for the growth of HS12 on NB and for the full induction of the AP catabolic genes by NB. Therefore, contrary to a previous report (37) suggesting the possible existence of a regulatory factor in pNB2, we reasoned that a potential regulatory factor(s) controlling the expression of the AP catabolic genes might be located on the pNB1 plasmid based on the above results. The presence of mutase from pNB2 only contributes to the conversion of hydroxylaminobenzene to AP (37). To get some insights into the regulatory mechanism of the AP operon, several plasmids were constructed, and the expression profile was analyzed by assaying AP dioxygenase activity in HS120, a derivative of HS12 cured of pNB1 and pNB2. As shown in Fig. 2, unlike deletion of both the nbzR gene and putative promoter region on the AP operon in HS120 (pUP42), only deletion of the nbzR gene in HS120 (pUP41) led to the constitutive expression of the AP-encoded pathway regardless of the growth substrates utilized. This result indicates that NbzR is a negative regulator and that the promoter of the AP operon is located upstream of the nbzJ gene. Although only NB was found to be an inducer for the AP catabolic operon in wild-type P. putida HS12 (pNB1 plus pNB2) and P. putida HS124 (pNB1 plus pUP3951), NB could not induce the expression of the AP operon significantly in P. putida HS120 (pUP40 or pUP50) containing only the nbzR-NbzJCaCb or the nbzR-NbzJCaCbDGFE region, which suggests the requirement of secondary trans-acting factor(s) for the regular induction of the AP operon.
FIG. 2.
(A) Construction of plasmids containing various regions of the AP operon. DNA fragments used for subcloning are indicated by open bars below the physical map. (B) Expression profile of the AP dioxygenase from strains with different plasmids. The carbon source used as a growth substrate or an inducer is indicated in parentheses (NB, nitrobenzene; AP, 2-aminophenol; suc, succinate). 12, HS12; 124, HS124; 120, HS120. Plasmid pUP3951 was constructed by cloning a 1.0-kb SalI-SphI DNA fragment (the nbzB gene) from pNB2 (37) into pUCP22.
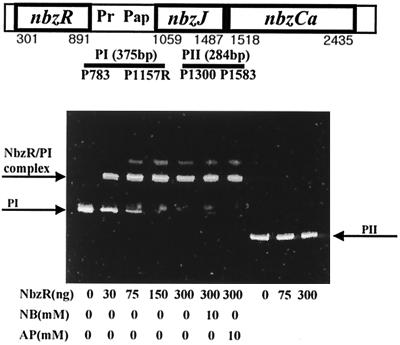
To determine whether the putative regulator NbzR is involved in the expression of the AP operon through direct contact with the promoter region, a gel mobility shift assay with the purified NbzR and the presumed promoter region was carried out. Figure 3 shows a clear mobility shift of the PI fragment containing the putative promoter region (Pap) of the AP operon by NbzR, whereas no retardation was observed with the PII fragment containing an intergenic region of the nbzJ and the nbzCa genes. As expected from the induction profile of HS120 (pUP40) in Fig. 2B, neither NB nor AP affected the binding of NbzR to the promoter region. From the gel mobility assay (Fig. 3) and the induction profile of HS120 (Fig. 2), the NbzR binding site and the transcription initiation site of the AP operon were presumed to be located upstream of the nbzJ gene. We carried out a footprinting analysis and a primer extension assay to determine the precise NbzR binding site and transcription start site, but we could not identify the exact sites under our experimental conditions.
FIG. 3.
Specific binding of NbzR to nbzR-nbzJ intergenic region. DNA fragments (PI and PII) used for the binding assay are indicated by bars below the physical map. For the gel mobility shift assay, various amounts of purified NbzR (0 to 300 ng) were incubated with a PI or PII DNA fragment. NB or AP was added optionally at a final concentration of 10 mM. The PI-NbzR complex, PI, and PII are indicated by arrows.
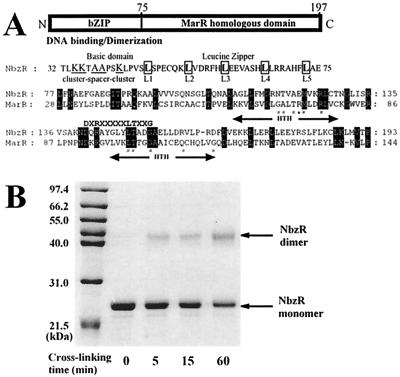
The C-terminal region of the predicted NbzR (residues 77 to 193) shared a modest identity with a group of negative transcription regulators belonging to the MarR family (47). In addition, NbzR possessed a putative basic leucine zipper motif with a basic region and an imperfect heptad leucine repeat (6) in the N-terminal region (residues 34 to 73), which is expected to be responsible for the dimerization and DNA binding of NbzR (Fig. 4A and B). Based on these observations, it is concluded that the NbzR protein acts as a dimeric repressor for the AP catabolic operon through specific binding to the divergently located promoter regions of the nbzR gene and the AP structural genes.
FIG. 4.
(A) Schematic domain structure of NbzR. The typical characteristics of bZIP, repeating leucine residues and a preceding basic region, and an alignment of NbzR (residues 77 to 193) with MarR from E. coli K-12 (GenBank accession no. AE000250) are shown below. A preliminary consensus sequence (DXRXXXXXLTXXG) of the MarR family (47) is shown above the alignment. The putative HTH regions in MarR are indicated below the alignment. Amino acids highlighted in black boxes are identical residues. #, Residues of MarR causing negative complementary trans-dominant mutations; ∗, residue of MarR making a specific operator contact. (B) Chemical cross-linking of NbzR by EDC. NbzR (1 mg/ml) was reacted with 2 mM EDC and 5 mM NHS at 4°C for different times as described in Materials and Methods. Arrows indicate monomeric and dimeric forms of NbzR.
Comparative study of AP operon and its origin.
A comparative study of the catabolic pathway and the genetic organization of the AP operon with other well-studied meta cleavage gene clusters provides an intriguing insight into the evolution of the AP catabolic operon. In general, microbial catabolic routes of various aromatic compounds converge at the ring cleavage steps, that is, at the ortho and meta cleavage pathways which are responsible for the conversion of catechol derivatives into Krebs cycle intermediates, such as pyruvate and acetyl coenzyme A (52). The NB catabolic pathway also follows this general route. NB is first transformed to AP, a catechol analogue, by a partial reductive pathway involving NB nitroreductase (NbzA) and HAB mutase (NbzB), and then AP is further metabolized via a meta-like cleavage pathway.
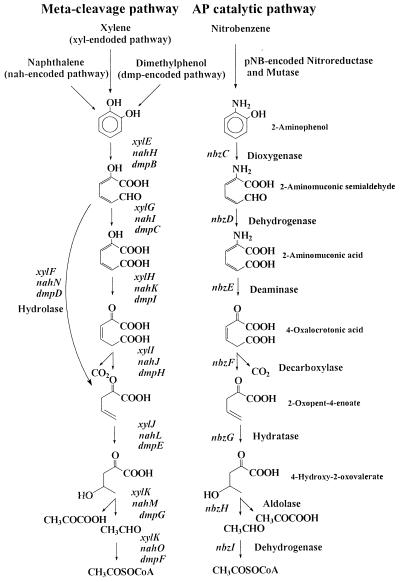
In most of the meta cleavage pathways, two main different catabolic branches exist, the oxalocrotonate pathway (central meta pathway) and the hydrolytic route, which is signified by the presence of hydrolase (Fig. 5). These two branches have different substrate preferences in the TOL pathway. Catechol and 4-methylcatechol are catabolized via the oxalocrotonate pathway, but 3-methylcatechol is dissimilated via the hydrolytic branch (19). The AP catabolic pathway has a distinct ring cleavage pathway as depicted in Fig. 5. It has only the central meta cleavage pathway and unique catabolic enzymes that differentiate the AP catabolic pathway from common meta cleavage pathways, especially when it comes to ring cleavage (AP dioxygenase), dehydration (AMS dehydrogenase), and deamination (AM deaminase) reactions. It has been proposed that the hydrolytic branch in the meta cleavage pathway has been recruited from cellular hydrolase to degrade keto ring products (26). The absence of hydrolase in the AP catabolic operon suggests that the recruitment event of the AP structural genes may have preceded the incorporation of hydrolytic branch in ancestral meta cleavage operon (see below).
FIG. 5.
Comparison of the AP catabolic pathway with other meta cleavage pathways encoded on the TOL, NAH, and DMP operons.
A widespread distribution of common meta cleavage routes in different catabolic pathways provides a typical clue to horizontal transfer of catabolic gene clusters in the course of adaptation to organic compounds available in the natural environment (52). The well-conserved genetic organization of the meta cleavage pathway has been reported for the toluene degradation pathway encoded in the TOL plasmid of P. putida mt-2 (17), the dimethylphenol operon on plasmid pVI150 of Pseudomonas sp. CF600 (43), and the naphthalene operon on the chromosome of Pseudomonas stutzeri AN10 (4). Comparison of the AP catabolic operon with the meta cleavage operons revealed some interesting facts. While the gene products of nbzF, nbzG, nbzH, and nbzI, which encode the same catabolic enzymes of the meta cleavage operons, showed relatively high amino acid identities (40 to 78%), the gene products of nbzCaCb and nbzE, which encode enzymes with different specificities and distinctive catalytic functions, shared low levels of identity with the counterparts of meta cleavage operons (15 to 25%).
NbzCaCb, as in the case of AmnBA in Pseudomonas sp. strain AP-3 (48) and P. pseudoalcaligenes JS45 (9), consists of a β-subunit containing conserved histidine residues (His-14, His-65, His-141, His-205), which seem to constitute a heme iron cofactor coordinate site found in class III extradiol ring cleavage enzymes proposed by Spence et al. (46), and an α-subunit possibly contributing to the stabilization of the heterotetrameric structure (48). High substrate specificity towards AP, rather than catechol (31), as well as the above result support the hypothesis that NbzCaCb would have divergently evolved from an ancestral gene for class III ring cleavage enzymes through a gene duplication mechanism (50) to acquire the specific ring cleavage function toward AP. Despite a high degree of identity between NbzD and HMS dehydrogenase of the meta cleavage pathway and the well-conserved putative NAD+ binding site in NbzD (data not shown), the substrate binding affinity of NbzD towards AMS is three times higher than that of HMS (23), which also implies the occurrence of divergent evolution of NbzD from HMS dehydrogenase of the ancestral meta cleavage pathway. AM deaminase, which is a unique enzyme found only in the AP catabolic pathway, was reported to have biochemical characteristics similar to those of 4-oxalocrotonate tautomerase (XylH) from the TOL plasmid. Both of these enzymes have hexameric structures and share tautomerization reactions in catalysis (21). Analyzing the amino acid sequence of NbzE, we could not find any similarity with other deaminases reported so far, and NbzE seems not to possess any cofactor binding site commonly found in several types of deamination-catalyzing enzymes, such as PLP in aminotransferase (12), NAD+ in glutamate dehydrogenase (51), and a Zn+ coordinate site in adenosine deaminases (3). Instead, we observed a modest level of amino acid identity between part of NbzE and XylH, and the nbzE gene has identity as high as 54% with the xylH gene, which implies a close evolutionary relationship between these genes. Based on the above observations, it is proposed that NbzE might have evolved from 4-oxalocrotonate tautomerase of the ancestral meta cleavage operon to transform an amino group to a keto one, leading to the subsequent degradation in the meta cleavage pathway. Further biochemical study including site-directed mutagenesis would be necessary to elucidate a more detailed catalytic mechanism and the genetic origin of AM deaminase. Thus, it can be concluded that the structural genes of the AP operon might have originated from ancestral meta cleavage gene cluster and have divergently evolved for the catabolism of AP.
The AP operon has a different regulatory system from that of the meta cleavage operons that are regulated by one of the following regulatory systems: LysR (52), AraC/XylS (15), and a ς54-dependent regulator (5). NbzR, which acts as a repressor for the expression of the AP operon, contains two distinct motifs, a putative basic leucine zipper motif and a MarR homologous domain. Recently, it was shown that the MarR protein has two helix-turn-helix (HTH) DNA binding motifs (1), as shown in Fig. 4A. Although the MarR homologous domain contains the consensus sequence (DXRXXXXLTXXG) as proposed by Sulavik et al. (47) and some conserved sequences in HTH regions, many of the residues (Glu-69, Ala-70, The-72, Arg-73, Gln-110, and Gly-116) critical for DNA binding were not conserved in NbzR. In particular, Arg-73, which makes a specific contact with the operator sequence, was replaced with Glu, a negatively charged amino acid. In addition to the incongruity in critical amino acid residues in HTH motifs and operator DNA sequences (1) (Fig. 3B), NbzR was observed to bind to the promoter region probably as a dimer, in contrast to oligomeric binding of the MarR protein (33). Many of the proteins belonging to the MarR family are known to interact with phenolic compounds, whereas NbzR did not interact with AP, as observed in Fig. 2 and 3. These results suggest that the MarR homologous domain of NbzR do not contribute to DNA binding, and a putative basic leucine zipper motif in the N-terminal region may lead to a dimerization and the consequent DNA binding of NbzR to the Pap promoter region. The elucidation of derepression and the DNA binding mechanism of NbzR in vivo should provide a more detailed regulatory system of the AP catabolic operon. From the above analyses, it is reasonable to suggest that the regulatory unit in the AP operon might have originated from the Mar gene cluster and has become involved in the regulation of the AP structural gene expression.
Evolution of AP operon in a modular fashion.
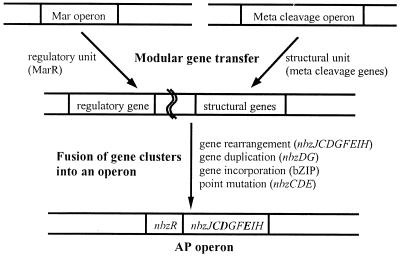
Based on previous results (37) and a comparative study of the AP catabolic genes, it is inferred that P. putida HS12 has acquired nbzA, nbzB, and the AP catabolic operon from different sources in order to adapt to an NB-contaminated habitat. Unlike nbzA and nbzB genes, which are scattered and free of regulation to some extent, the AP catabolic gene cluster was revealed to form a well-defined operonic structure under tight regulation. Thus, the evolutionary process of the AP operon is expected to provide a critical insight into the gene transfer mechanism. In general, HGT in bacteria is mediated by transformation, conjugation, and transduction (2, 10, 11). It has been known that bacterial evolution towards the acquisition of antibiotic resistance and pathogenicity-conferring genes, expansion of xenobiotics degradation capabilities, and speciation has proceeded via the horizontal transfer of gene(s) or whole operon, followed by modification of the respective gene(s) (7, 10, 27, 32, 36). However, it is unlikely that the HGT model can fully explain the mechanism by which bacteria have efficiently acquired genetic diversity and metabolic capabilities. In this regard, the present study on the organization and origin of the regulatory and structural units of the AP catabolic operon led us to propose that the AP catabolic operon has evolved in a modular fashion. Herein, we term it modular type gene transfer, which refers to a specific kind of HGT comprising fusion of functional gene units (regulatory and structural gene clusters) originating from different sources and their subsequent arrangement, resulting in the rapid generation of genetic diversity in bacteria. In the creation of the AP operon on plasmid pNB1, both the regulatory unit (nbzR), which seems to have been derived from the Mar operon, and the structural unit (nbzJCaCbDFGEIH), from the ancestral meta cleavage operon, assembled through modular type gene transfer and then rearranged for adaptation, as depicted in Fig 6. HGT through a conjugative catabolic plasmid to indigenous bacteria for efficient xenobiotics utilization was observed in the natural environment (39), and we also found a Tn5501-like transposon in the catabolic plasmid of HS12 (37). Maybe the mobile genetic elements, such as conjugative plasmids and transposons, might have been involved in the gene transfer. During the fusion of gene clusters into an operon, directional mutations, such as gene rearrangement (differential genetic organization of the structural genes; nbzJCaCbDGFEIH), gene duplication (truncated genes found downstream of the operon; nbzDG gene segment), gene incorporation (DNA binding domain of bZIP; nbzR), and point mutations (generation of new catalytic functions; nbzC, nbzD, and nbzE), must have occurred for the creation of the AP operon. Therefore, the evolutionary process of the AP operon provides evidence that modular type gene transfer is considered an efficient mechanism for bacteria to create genetic diversities and to expand metabolic capabilities.
FIG. 6.
Proposed mechanism for the creation of the AP operon. Modular type gene transfer of regulatory and structural units was followed by fusion of these gene clusters into an operon. Boldtype ORFs in the AP operon represent enzymes with different substrate specificities and catalytic function from meta cleavage operons.
ACKNOWLEDGMENTS
We are grateful to J. C. Spain, P. R. Reeves, J. Davison, A. M. Campbell, and A. Salyers for critical comments on the manuscript.
REFERENCES
- 1.Alekshun M N, Kim Y S, Levy S B. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 2.Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik D, Medin J, Gathy K, Coleman M S. Mutational analysis of active site residues of human adenosine deaminase. J Biol Chem. 1993;268:5464–5470. [PubMed] [Google Scholar]
- 4.Bosch R, Garcia-Valdes E, Moore E R B. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene. 2000;245:65–74. doi: 10.1016/s0378-1119(00)00038-x. [DOI] [PubMed] [Google Scholar]
- 5.Buck M, Gallegos M T, Studholme D J, Guo Y, Gralla J D. The bacterial enhancer-dependent ς54 (ςN) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch S J, Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- 7.Campbell A M. Lateral gene transfer in prokaryotes. Theor Popul Biol. 2000;57:71–77. doi: 10.1006/tpbi.2000.1454. [DOI] [PubMed] [Google Scholar]
- 8.Collinsworth W L, Chapman P J, Dagley S. Stereospecific enzymes in the degradation of aromatic compounds by Pseudomonas putida. J Bacteriol. 1973;113:922–931. doi: 10.1128/jb.113.2.922-931.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis J K, He Z, Somerville C C, Spain J C. Genetic and biochemical comparison of 2-aminophenol 1,6-dioxygenase of Pseudomonas pseudoalcaligenes JS45 to meta-cleavage dioxygenases: divergent evolution of 2-aminophenol meta-cleavage pathway. Arch Microbiol. 1999;172:330–339. doi: 10.1007/s002030050787. [DOI] [PubMed] [Google Scholar]
- 10.Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 11.de la Cruz F, Davies J. Horizontal gene transfers and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 12.Denessiouk K A, Denesyuk A I, Lehtonen J V, Korpela T, Johnson M S. Common structural elements in the architecture of the cofactor-binding domains in unrelated families of pyridoxal phosphate-dependent enzymes. Proteins. 1999;35:250–261. doi: 10.1002/(sici)1097-0134(19990501)35:2<250::aid-prot10>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Esteve-Nunez A, Lucchesi G, Philipp B, Schink B, Ramos J L. Respiration of 2,4,6-trinitrotoluene by Pseudomonas sp. strain JLR11. J Bacteriol. 2000;182:1352–1355. doi: 10.1128/jb.182.5.1352-1355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6526. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos M T, Schleif R, Bairoch A, Hofmann K, Ramos J L. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haidour A, Ramos J L. Identification of products resulting from the biological reduction of 2,4,6-trinitrotoluene, 2,4-dinitrotoluene, and 2,6-dinitrotoluene by Pseudomonas sp. Environ Sci Technol. 1996;30:2365–2370. [Google Scholar]
- 17.Harayama S, Rekik M. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol Gen Genet. 1990;221:113–120. doi: 10.1007/BF00280375. [DOI] [PubMed] [Google Scholar]
- 18.Harayama S, Rekik M, Ngai K-L, Ornston N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989;171:6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harayama S, Mermod N, Rekik M, Lehrhach P R, Timmis K N. Roles of the divergent branches of the meta-cleavage pathway in the degradation of benzoate and substituted benzoates. J Bacteriol. 1987;169:558–564. doi: 10.1128/jb.169.2.558-564.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Z, Spain J C. Studies of the catabolic pathway of degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45: removal of the amino group from 2-aminomuconic semialdehyde. Appl Environ Microbiol. 1997;63:4839–4853. doi: 10.1128/aem.63.12.4839-4843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z, Spain J C. A novel 2-aminomuconate deaminase in the nitrobenzene degradation pathway of Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1998;180:2502–2506. doi: 10.1128/jb.180.9.2502-2506.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Z, Spain J C. Comparison of the downstream pathways for degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45 (2-aminophenol pathway) and by Comamonas sp. JS765 (catechol pathway) Arch Microbiol. 1999;171:309–316. doi: 10.1007/s002030050715. [DOI] [PubMed] [Google Scholar]
- 23.He Z, Davis J K, Spain J C. Purification, characterization, and sequence analysis of 2-aminomuconic 6-semialdehyde dehydrogenase from Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1998;180:4591–4595. doi: 10.1128/jb.180.17.4591-4595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Z, Nadeau L J, Spain J C. Characterization of hydroxylaminobenzene mutase from pNBZ139 cloned from Pseudomonas pseudoalcaligenes JS45. A highly associated SDS-stable enzyme catalyzing an intramolecular transfer of hydroxy groups. Eur J Biochem. 2000;267:1110–1116. doi: 10.1046/j.1432-1327.2000.01107.x. [DOI] [PubMed] [Google Scholar]
- 25.Hogu N, Armengaud J, Gaillard J, Timmis K N, Jouanneau Y. A novel -2Fe-2S- ferredoxin from Pseudomonas putida mt-2 promotes the reductive reactivation of catechol 2,3-dioxygenase. J Biol Chem. 1998;273:9622–9629. doi: 10.1074/jbc.273.16.9622. [DOI] [PubMed] [Google Scholar]
- 26.Horn J M, Harayama S, Timmis K N. DNA sequence determination of the TOL plasmid (pWW0) xylGFJ genes of Pseudomonas putida: implications for the evolution of aromatic catabolism. Mol Microbiol. 1991;5:2459–2474. doi: 10.1111/j.1365-2958.1991.tb02091.x. [DOI] [PubMed] [Google Scholar]
- 27.Lan R, Reeves P R. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- 28.Lapworth A. The form of change in organic compounds and the function of the alpha-meta-orientating groups. J Chem Soc. 1901;79:1265–1284. [Google Scholar]
- 29.Lawrence J G. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;142:11–24. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lendenmann U, Spain J C. 2-aminophenol 1,6-dioxygenase: a novel aromatic ring cleavage enzyme purified from Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1996;178:6227–6232. doi: 10.1128/jb.178.21.6227-6232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Pro Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishino S F, Spain J C. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl Environ Microbiol. 1993;59:2520–2525. doi: 10.1128/aem.59.8.2520-2525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishino S F, Spain J C. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl Environ Microbiol. 1995;61:2308–2313. doi: 10.1128/aem.61.6.2308-2313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 37.Park H-S, Kim H-S. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J Bacteriol. 2000;182:573–580. doi: 10.1128/jb.182.3.573-580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park H-S, Lim S-J, Chang Y-K, Livingston A G, Kim H-S. Degradation of chloronitrobenzenes by a coculture of Pseudomonas putida and Rhodococcus sp. Appl Environ Microbiol. 1999;65:1083–1091. doi: 10.1128/aem.65.3.1083-1091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters M, Heinaru E, Talpsep E, Wand H, Stottmeister U, Heinaru A, Nurk A. Acquisition of a deliberately introduced phenol degradation operon. pheBA, by different indigenous Pseudomonas species. Appl Environ Microbiol. 1997;63:4899–4906. doi: 10.1128/aem.63.12.4899-4906.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platt A, Shingler V, Taylor S C, Williams P A. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology. 1995;141:2223–2233. doi: 10.1099/13500872-141-9-2223. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somerville C C, Nishino S F, Spain J C. Purification and characterization of nitrobenzene nitroreductase from Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1995;177:3837–3842. doi: 10.1128/jb.177.13.3837-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spain J C. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol. 1995;49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 46.Spence E L, Kawamukai M, Sanmoisin J, Braven H, Bugg T D H. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J Bacteriol. 1996;178:5249–5256. doi: 10.1128/jb.178.17.5249-5256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 48.Takenaka S, Murakami S, Shinke R, Hatakeyama S, Yukawa H, Aoki K. Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and catalytic properties of the purified enzyme. J Biol Chem. 1997;272:14727–14732. doi: 10.1074/jbc.272.23.14727. [DOI] [PubMed] [Google Scholar]
- 49.Takenaka S, Murakami S, Kim Y-J, Aoki K. Complete nucleotide sequence and functional analysis of the genes for 2-aminophenol metabolism from Pseudomonas sp. AP-3. Arch Microbiol. 2000;174:265–272. doi: 10.1007/s002030000203. [DOI] [PubMed] [Google Scholar]
- 50.Todd A E, Orengo C A, Thornton J M. Evolution of protein function, from a structural perspective. Curr Opin Chem Biol. 1999;3:548–556. doi: 10.1016/s1367-5931(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 51.Todone F, Vanoni M A, Mozzarelli A, Bolognesi M, Coda A, Curti B, Mattevi A. Active site plasticity in D-amino acid oxidase: a crystallographic analysis. Biochemistry. 1997;36:5853–5860. doi: 10.1021/bi9630570. [DOI] [PubMed] [Google Scholar]
- 52.van der Meer J R, De Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiley R H, Hart A J. 2-Pyrones. IX. 2-Pyrone-6-carboxylic acid and its derivatives. J Am Chem Soc. 1954;76:1942–1944. [Google Scholar]