Dear Editor,
1.
Due to the continuing evolution of SARS‐CoV‐2, the formation of new variants is not a novelty or a sporadic case, but it is a certainty that periodically recurs. 1 One of the most recent variants is represented by the subvariant BA.2.75, also known as Centaurus. In comparison to the BA.2 variant, BA.2.75 carries nine additional mutations in the sequence of the spike protein genes: K147E, W152R, F157L, I210V, G257S G339H, G446S, N460K, and R493Q. The site G446S has been indicated as the site of potential escape from antibodies evoked by current vaccines that are capable of neutralizing Omicron 5 2 and it has been hypothesized that the BA.2.75 spike significantly reduces sensitivity toward therapeutic monoclonal antibodies than BA.5 (and thus than BA.2 and BA.4). 3
Here below, you can find a genetic and structural point of view:
1.1.
The phylogenomic reconstruction, available at https://gisaid.org/phylodynamics/global/nextstrain/, indicates that the evolutionary condition of BA.2.75 seems to be very similar to the sublineage BA.2.12.1, which represents an evolutionary blind background with no further descendant. Indeed, the branches' length in the phylogenetic tree suggested that BA.2.75 lacks the rapid diversification which is typical of a dangerous lineage at the beginning of its evolutionary path. Bayesian Skyline Plot (BSP) reconstruction (Figure 1A), estimated on 270 whole genomes of BA.2.75 (2022/05/26–2022/07/20), indicates a constant trend after a brief expansion with a substantial lack of variation in genetic variability during the time. Indeed, according to BSP, the plateau seems to be already achieved and started about June 23 (27 days before July 20). This is not the trend of a lineage that is about to explode in terms of population size and, accordingly, in terms of contagiousness as shown at the beginning of the pandemic when variability increased very quickly with a very vertical curve. 4 Conversely, this condition is quite typical of an evolutionary lineage that presents new features in comparison to its direct progenitor (BA.2) but these new features, at the present stage, do not represent a further boost able to promote an abnormal expansion. Indeed, the reconstruction of lineages through time (Figure 1B) further confirms the lack of an increase in the number of haplotypes, not even in recent times. Reconstruction of BSP for BA.5 variant, performed on 430 whole genomes (2022/05/08–2022/07/30), shows a similar trend but with a more vertical increase of the population size during the peak‐reaching phase (Figure 1C) as well as occurred in the number of lineages (Figure 1D). Evolutionary rate estimated for BA.2.75 amount to 1.619e−4 [6.5554e−8−4.3236e−4] subs/site/year. It represents a low level of genetic variation with poor capability for demographic expansion. Variant BA.5, nowadays presents a similar and slightly higher evolutionary rate of about 7e−4 [1.072e−3−3.494e−4] subs/site/year. Considering that BA.5 has been circulating for several months, the evolutionary rate of BA.2.75, this last one, should be greater if it were dangerous with a highly contagious capability lineage. Indeed, at the beginning of the current pandemic, the evolutionary rate of the first lineage of SARS‐CoV‐2 was about 6.58e−3 subs/site/year, 5 which means that BA.2.75 presents a factor of 10−1 more slow than Wuhan‐Hu‐1 variant.
Figure 1.
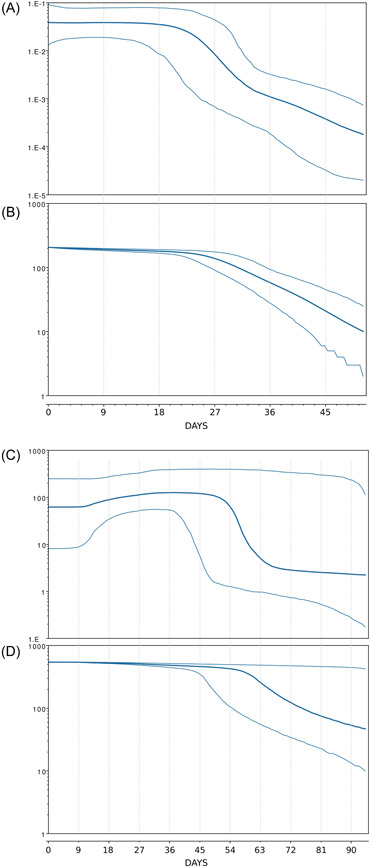
(A) Bayesian skyline plot of SARS‐CoV‐2 BA.2.75 variant. (B) SARS‐CoV‐2 BA.2.75 variant lineages through time. (C) Bayesian skyline plot of SARS‐CoV‐2 BA.5 variant. (D) SARS‐CoV‐2 BA.5 variant lineages through time. The viral effective population size and the number of lineages (y‐axis) are shown as a function of days (x‐axis). Thin lines represent the 95% high posterior density (HPD) region. These plots have been reconstructed by using the software Beast 1.10.4 10 following Mugosa et al. 11
Five of the additional nine specific mutations (K147E, W152R, F157L, I210V, G257S) of the BA.2.75 Spike occur in the N‐terminal domain (NTD), while the other four (G339H, G446S, N460K, R493Q) are found in the receptor binding domain (RBD). Among the structural properties that may be altered by residue mutations, the surface electrostatic potential is interesting as it guides the electrostatic interactions between macromolecules and molecules. Calculation performed following Pascarella et al., 6 indicates that the BA.2.75 net charge of NTD is similar to that of BA.2 (+0.38 and +0.35, respectively) but lower than that predicted for BA.5 NTD (about + 1.4). However, it should be noted that the BA.2.75 mutations K147E, W152R, and G257S occur at the putative site involved in the interaction with the AXL receptor. 7 At this stage, it is difficult to predict the effect of these mutations on the NTD affinity for AXL, although modifications of the interaction NTD‐AXL are likely. BA.2.75 RBD displays a net charge more positive than that of BA.2 and BA.5 (+6.6, +5.2, and +5.2, respectively). However, the increase of the positive charge does not involve directly the RBD interface to the ACE2 receptor. Indeed, mapping of the surface electrostatic potential shows that the interface is more positive in BA.2 than in BA.2.75 and BA.5 (Figure 2). The local increase of the positive potential is mainly caused by the presence of the positively charged R493 that in BA.2.75 is reverted to the neutral Q493. BA.5 conserves the Q493 while R493 is present in BA.1 and BA.3. R493 may interact with ACE2 E35 through a salt bridge that is absent in BA.5 and in BA.2.75 where R is replaced by Q. This may suggest that BA.2.75 has an interface interaction to ACE2 similar to BA.5 but different from BA.1, BA.2 and BA.3.
Figure 2.
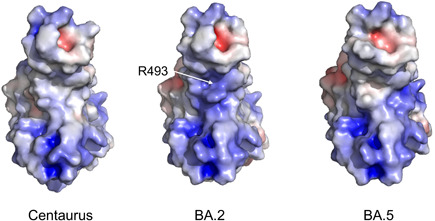
Comparison of the electrostatic potential surface of the Centaurus and BA.2 Spike receptor binding domains (RBDs). Red and blue colors indicate negative and positive potential, respectively. The color scale ranges from −5.0 to +5.0 kT/e. The RBD is oriented with the ACE2 interface in the front. Position of the R493 in BA.2 is marked by the white arrow. Calculations have been carried out with the software PROPKA3 12 while in‐silico mutagenesis by means of PyMOL Molecular Graphics System v.2 (available at https://pymol.org/2/). The conformation of the side chains of the mutagenized structures was optimized by the application of the software FoldX 5.0. 13
In conclusion, genetic and structural data on SARS‐CoV‐2 BA.2.75 suggest that this new variant currently does not show evidence of its particular dangerous or high expansion capability. On the contrary, as of now, it appears even more slowly than the last variant that became dominant at the commencement of the year, the BA.5 one. Of course, this does not imply that BA.2.75 cannot become more dangerous or generate new subvariants following new mutations going forward. 8 , 9 However, based on current data, it does not appear to present an alarming situation and phylogenomic reconstruction shows that BA.5 and BA.4 Omicron descendent lineages continued to be dominant globally. Of course, the monitoring on BA.2.75 must continue uninterrupted (as well as the monitoring of all other lineages) to identify and/or predict the occurrence of an expansion in population size flaked by an increase of genetic variability, if any.
AUTHOR CONTRIBUTIONS
Conceptualization: Fabio Scarpa, Daria Sanna, Marco Casu, Pier Luigi Fiori, and Massimo Ciccozzi. Data Analyses: Fabio Scarpa and Stefano Pascarella. Writing‐original draft preparation: Fabio Scarpa, Daria Sanna, and Marco Casu. Writing‐review and editing: Fabio Scarpa, Daria Sanna, Ilenia Azzena, Marta Giovanetti, Domenico Benvenuto, Silvia Angeletti, Giancarlo Ceccarelli, Stefano Pascarella, Marco Casu, Pier Luigi Fiori, and Massimo Ciccozzi. Resources: Fabio Scarpa, Daria Sanna, and Marco Casu.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We thank all the authors who have kindly deposited and shared genomes on GSAID. This research was funded from by FONDAZIONE DI SARDEGNA bando 2022–2023 for the Dipartimento di Scienze Biomediche—UNISS (to Daria Sanna and Fabio Scarpa). Marta Giovanetti is funded by PON “Ricerca e Innovazione” 2014–2020.
Fabio Scarpa and Daria Sanna contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the GSAID database at https://gisaid.org/
REFERENCES
- 1. Zella D, Giovanetti M, Benedetti F, et al. The variants question: what is the problem. J Med Virol. 2021;93:6479‐6485. 10.1002/jmv.27196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greaney AJ, Starr TN, Bloom JD. An antibody‐escape estimator for mutations to the SARS‐CoV‐2 receptor‐binding domain. Virus Evol. 2022;8(1):veac021. 10.1093/ve/veac021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamasoba D, Yosugi Y, Kimura I, et al. Neutralisation sensitivity of SARS‐CoV‐2 omicron subvariants to therapeutic monoclonal antibodies. The Lancet Infectious Diseases. 2022;22(7):942‐943. 10.1016/S1473-3099(22)00365-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lai A, Bergna A, Acciarri C, Galli M, Zehender G. Early phylogenetic estimate of the effective reproduction number of SARS‐CoV‐2. J Med Virol. 2020;92:675‐679. 10.1002/jmv.25723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benvenuto D, Giovanetti M, Salemi M, et al. The global spread of 2019‐nCoV: a molecular evolutionary analysis. Pathog Glob Health. 2020;114(2):64‐67. 10.1080/20477724.2020.1725339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pascarella S, Ciccozzi M, Bianchi M, Benvenuto D, Cauda R, Cassone A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta‐plus variants: a hint to higher transmissibility? J Med Virol. 2022;94(4):1277‐1280. 10.1002/jmv.27528 [DOI] [PubMed] [Google Scholar]
- 7. Wang S, Qiu Z, Hou Y, et al. AXL is a candidate receptor for SARS‐CoV‐2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126‐140. 10.1038/s41422-020-00460-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borsetti A, Scarpa F, Maruotti A, et al. The unresolved question on COVID‐19 virus origin: the three cards game. J Med Virol. 2022;94:1257‐1260. 10.1002/jmv.27519 [DOI] [PubMed] [Google Scholar]
- 9. Scarpa F, Casu M, Sanna D. Evolutionary and conservation genetics. Life. 2021;11(11):1160. 10.3390/life11111160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:e214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mugosa B, Vujosevic D, Ciccozzi M, et al. Genetic diversity of the haemagglutinin (HA) of human influenza A (H1N1) virus in Montenegro: focus on its origin and evolution. J Med Virol. 2016;88:1905‐1913. 10.1002/jmv.24552 [DOI] [PubMed] [Google Scholar]
- 12. Olsson MH, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: consistent treatment of internal and surface residues in empirical pK a predictions. J Chem Theory Comput. 2011;7(2):525‐537. 10.1021/ct100578z [DOI] [PubMed] [Google Scholar]
- 13. Delgado J, Radusky LG, Cianferoni D, Serrano L. FoldX 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics. 2019;35(20):4168‐4169. 10.1093/bioinformatics/btz184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in the GSAID database at https://gisaid.org/


