Abstract
Tagged mutants affected in the degradation of hydrophobic compounds (HC) were generated by insertion of a zeta-URA3 mutagenesis cassette (MTC) into the genome of a zeta-free and ura3 deletion-containing strain of Yarrowia lipolytica. MTC integration occurred predominantly at random by nonhomologous recombination. A total of 8,600 Ura+ transformants were tested by replica plating for (i) growth on minimal media with alkanes of different chain lengths (decane, dodecane, and hexadecane), oleic acid, tributyrin, or ethanol as the C source and (ii) colonial defects on different glucose-containing media (YPD, YNBD, and YNBcas). A total of 257 mutants were obtained, of which about 70 were affected in HC degradation, representing different types of non-alkane-utilizing (Alk−) mutants (phenotypic classes alkA to alkE) and tributyrin degradation mutants. Among Alk− mutants, growth defects depending on the alkane chain length were observed (alkAa to alkAc). Furthermore, mutants defective in yeast-hypha transition and ethanol utilization and selected auxotrophic mutants were isolated. Flanking borders of the integrated MTC were sequenced to identify the disrupted genes. Sequence analysis indicated that the MTC was integrated in the LEU1 locus in N083, a leucine-auxotrophic mutant, in the isocitrate dehydrogenase gene of N156 (alkE leaky), in the thioredoxin reductase gene in N040 (alkAc), and in a peroxine gene (PEX14) in N078 (alkD). This indicates that MTC integration is a powerful tool for generating and analyzing tagged mutants in Y. lipolytica.
About 20% of all yeast species have been recorded as being able to utilize as carbon substrates hydrophobic compounds (HC) like n-alkanes, fatty acids, and triglycerides (for reviews see references 15 and 36). This list of species was significantly extended by studies on single-cell protein production in the mid-1960s (30). Among these yeasts, however, genetic and genetic-engineering techniques were widely developed only for the dimorphic fungus Yarrowia (formerly Candida, Endomycopsis, or Saccharomycopsis) lipolytica (2).
Y. lipolytica very efficiently utilizes long- and short-chain triglycerides, such as olive oil and tributyrin, fatty acids, and the corresponding n-alkanes, from decane (C10) to octadecane (C18) and longer chains (2). Screening of mutants affected in HC utilization was initially performed by R. K. Mortimer's group (5) using n-decane as the substrate. Based on the relative frequencies of alkane and auxotrophic mutants, 80 to 90 genes appeared to be involved in n-alkane assimilation. The Alk− mutants were classified into five phenotypic classes (alkA to alkE) depending on their use of intermediates of the alkane degradation pathway (fatty alcohol, fatty aldehyde, fatty acid, and acetate) (5, 6, 14, 17). Among at least 26 loci affecting uptake and primary alkane oxidation, 16 were required for efficient n-alkane uptake (6). Mauersberger et al. isolated Alk− mutants of Y. lipolytica and Candida maltosa using C10, C12, and C16 as substrates and subdivided alkA mutants into alkAa (unable to use any n-alkane), alkAb (no growth on C8 to C10), and alkAc (no growth on C16) (17). This, and studies on cytochrome P450 regulation in Y. lipolytica and C. maltosa, suggested the existence of several chain length-specific alkane uptake or cytochrome P450 monooxygenase systems catalyzing the primary terminal hydroxylation (17). This was proven for C. maltosa (23, 37) and more recently for Y. lipolytica (12), by identifying up to eight cytochrome P450 genes (ALK1 to ALK8) in this species, all belonging to the CYP52 gene family. A Y. lipolytica strain with ALK1 deleted grew very poorly on C10 but almost normally on C12 or C16 (11).
Other groups isolated mutants unable to utilize short-chain triglycerides after nystatin enrichment using tributyrin as a carbon source (19, 20) or unable to utilize the long-chain oleic acid (C18) in a screen of genes involved in peroxisome biogenesis using a rapid immunofluorescence assay (22). Some of the Pex mutants exhibited pleiotropic phenotypes affecting peroxisome biogenesis, secretion, and morphology (32). Several PEX genes were isolated, and their functions were analyzed (32, 33).
Through both reverse and classical genetics, we identified multigene families involved in these metabolic pathways, such as those encoding acyl-coenzyme A oxidases of the peroxisomal β-oxidation (POX1 to POX5 genes) (35) or lipases (LIP genes) (24), and genes impairing the anaplerotic glyoxylate cycle and its regulation during metabolism of alkanes, ethanol, or acetate (ICL1, ACS1, and GPR1) (4, 14, 34).
Several of the previously isolated Alk− mutants were simultaneously affected in mating and/or sporulation, in colonial and cellular morphology, and/or in transformation ability (5, 6). In addition, for several chemically induced alkE mutants, revertants with new phenotypes occurred at high frequencies (16). To circumvent these difficulties, we first developed a transposon tagging approach to identify genes involved in HC utilization (18), leading to the characterization of PEX10 (J.-M. Nicaud, unpublished data), which is involved in peroxisome biogenesis. However, identification of the tagged genes was plagued by a high level of nonhomologous integration (26).
We recently developed new integrative vectors (mono- and multicopy) for gene expression in Y. lipolytica (25), carrying the zeta long terminal repeat of the Y. lipolytica retrotransposon Ylt1 (29). We observed that this long terminal repeat directed random integration of the transforming DNA into the genome of strains devoid of Ylt1. Here, we report on the use of a short zeta-based mutagenesis cassette (MTC) for generating tagged mutants. We demonstrate that this MTC inserts at random by nonhomologous recombination, that mutant phenotypes are due to cassette integration, and that tagged genes are easily identified. This provides a powerful tool for the identification of genes involved in different pathways, as demonstrated here for HC utilization, morphogenesis, and auxotrophic mutants.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The Escherichia coli DH5α strain was used for transformation and amplification of recombinant plasmids DNA. Cells were grown in Luria-Bertani medium (27) supplemented with ampicillin (100 μg/ml) or kanamycin (40 μg/ml) for plasmid selection. The Y. lipolytica strains used in this study are listed in Table 1. They were grown at 28°C in complete media, YPD (3), and YNBcas (YNBD with 0.2% Casamino Acids) (35) or in minimal media derived from YNB (35) or M (a slightly modified YNB medium) (17) containing the following carbon sources: glucose (1 or 2%; YNBD), oleic acid (1% in 0.05% Tween 80, added as 20-fold sonicated stock emulsion; YNBO), tributyrin (1% in 0.05% Tween 80, added as 20-fold sonicated emulsion; YNBT), alkanes (1 or 2%) of different chain lengths (YNBC10, decane; YNBC12, dodecane; YNBC16, hexadecane). For solid media, 20 g of agar per liter was added. For alkane growth test on plates, alkanes were supplied as vapor phase by placing 200 μl of the n-alkane on a sterile filter paper in the lid of the petri dish (16, 17). Amino acids and uracil were supplied when necessary.
TABLE 1.
Y. lipolytica strains used in this study
| Name | Genotype | References |
|---|---|---|
| W29 | MATA, wild-type | 3 |
| PO1d | MATA leu2-270 ura3-302 xpr2-322 SUC2 | 3 |
| E129 | MATA leu2-270 lys11-23 ura3-302 xpr2-322 SUC2 | 3 |
| E150 | MATB leu2-270 his1 ura3-302 xpr2-322 SUC2 | 3 |
| YB423-12 | MATA, wild-type | 3 |
| CX161-1B | MATA ade1 | 5 |
| CXAU1 | MATA ade1, ura3 | 11 |
| H222 | MATA, wild-type | 3 |
| H222-67 | MATA ura3-67 | 16 |
| H222-41 (JMY322) | MATA ura3-41 | This work |
| H222-S4 (JMY323) | MATA ura3-302 SUC2 | This work |
| B204-12C-20 | MATA leu2-20 met6-1 spo1-1 | 14 |
| B204-67 (JMY324) | MATA leu2-20 met6-1 spo1-1 ura3-674 | This work |
| T1 to T20 | H222-41 transformed with JMP5 digested by Notl, randomly selected Ura+ | |
| P1 to P20 | H222-41 transformed with JMP5 digested by Notl, with phenotype | |
| N001 to N257 | H222-41 transformed with JMP5 digested by Notl with selected phenotypes |
Cultivation in liquid media was performed with 100 or 200 ml of minimal YNB or M medium in 500-ml Erlenmeyer shaking flasks; baffled flasks were used to improve dispersion of alkanes and oxygen supply. Cells from overnight YPD cultures were centrifuged, washed twice with minimal medium without a carbon source, and used to inoculate the culture at an initial optical density at 600 nm (OD600) of 0.4 to 0.6. Growth was followed by measuring the OD600 or alkali (2.5 N NaOH) consumption used for maintaining pH at 5.3 to 5.5 in minimal medium (10).
Plasmid constructions.
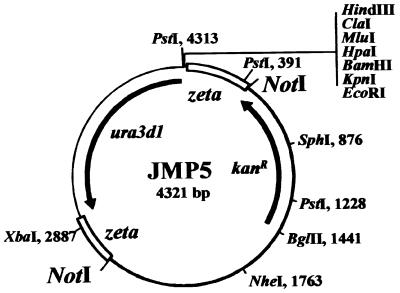
All basic DNA manipulation procedures were performed according to reference 27. The construction of plasmids JMP5 (Fig. 1) and pINA302 was described previously (21, 25); the construction of pCR4 is described below.
FIG. 1.
Schematic map of plasmid JMP5. For insertion mutagenesis, plasmid JMP5 (35) was digested by NotI prior transformation to eliminate the bacterial pHSS6 region (thick line) and to liberate the MTC containing only the nondefective Y. lipolytica ura3d1 allele (arrow), flanked by two inverted partial zeta regions of 401 and 312 bp (open boxes). The gene conferring kanamycin resistance (kanR) in E. coli is also shown by an arrow.
Sequencing of the URA3 locus, construction of pCR4, and isolation of strains carrying nonreverting ura3 alleles.
To increase the upstream and downstream sequence information about the URA3 gene locus (U40564), we sequenced over 4,844 bp for this locus (AJ306421) by primer walking using plasmid pLD55, containing a 4.6-kb Sau3A Y. lipolytica DNA insert (obtained from L. S. Davidow, Pfizer Inc. [8a]), and plasmid AWOAA010FO3, from a library of 2,284 plasmids used for generating 4,940 random sequence tags (RSTs) from strain W29 (8).
URA3 deletions in the recipient strains were constructed by transformation using either pINA302 (containing a ura3::SUC2 construct [21]) or a new URA3 disruption plasmid, pCR4, containing larger flanking regions. Plasmid pCR4 was constructed by PCR amplification of URA3 promoter (620 bp) and terminator regions (2.6 kb) from plasmid pLD55 and ligation as HindIII/blunt-end and blunt-end/SstI fragments into pUC18. It contains a nearly complete deletion (bp 1206 to 2019) of the URA3 open reading frame (bp 1195 to 2049). After transformation of strains H222, B204-12C, and B204-12C-20 with plasmid pCR4 (digested with HindIII and SstI) and subsequent 5-fluoroorotic acid selection, we isolated several stable Ura− strains. Southern blotting and PCR analysis of 15 Ura− clones from each strain revealed that several harbored URA3 deletions, but never of the expected type. The URA3 deletions were mapped by sequencing after PCR amplification using the primer pair URA3-dis1 (GGGGTGACACTGCACTATTGGTTTG) and URA3-dis2 (CATGTACTCTGCCTCTCAG AACGC). The coordinates, corresponding to the known 4,844-bp sequence (AJ306421) of the Y. lipolytica URA3 locus are bp 1195 to 2049 for the URA3 open reading frame, bp 1804 to 1814 for a ura3-41 10-bp deletion in strain H222-41, bp 1211 to 2152 for the ura3-302 deletion in strain H222-S4, and bp 1167 to 3385 for the ura3-67 deletion in strain B204-67.
Transformation of Y. lipolytica.
Transformation was performed by the lithium acetate method as previously described (3). Plasmid JMP5 (Fig. 1) was digested by NotI prior to transformation. For each transformation assay, 0.5 to 1 μg of plasmid DNA was used, yielding about 1,000 to 1,500 transformants per μg of DNA. Ura+ transformants were selected on YNBcas.
Chromosomal DNA preparation and Southern blot analysis.
Genomic DNA was prepared as described previously (3). Probes were prepared by PCR or as DNA fragments from plasmids and labeled with the Gene Images random prime labeling and detection system (Amersham Life Science).
Amplification of the MTC borders and sequences analysis.
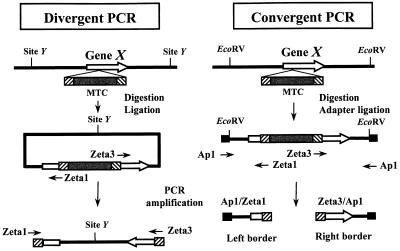
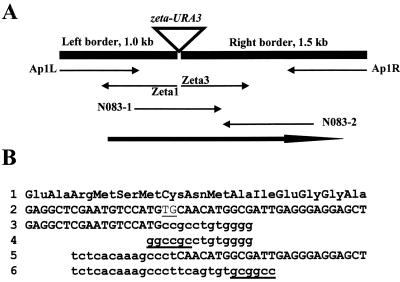
The MTC borders were amplified either by divergent PCR (Fig. 2) as described previously (26) or by convergent PCR walking (31) (Fig. 2). Primers Ad1, Ad2, and Ap1 are described in references 9 and 31, and the MTC-specific primers are Zeta1 (CCCCACTATGAATACATCAG) and Zeta3 (CACTACCGAGGTTACTAGAG). For PCR walking or convergent PCR (Fig. 2), left and right borders were amplified with the primer pairs Ap1-Zeta1 and Ap1-Zeta3, respectively. All amplifications were performed on a Perkin-Elmer thermal cycler 2400 with the Expand long-template PCR system (Roche Diagnostics GmbH) as previously described (26). PCR fragments were sequenced directly after gel purification on an ABI373 DNA sequencer according to the manufacturer's instructions (Perkin-Elmer Biosystems). Sequence comparisons were done by BLAST search (1) against a Y. lipolytica RST database (8) and at http://www.ncbi.nlm.nih.gov/blast/blast.cgi. The LEU1 gene was amplified with primers N083-1 (TCAAGGACTTTGGCGTG) and N083-2 (GAAAAAGAGACCCGAGG).
FIG. 2.
Strategies for sequencing of the zeta-URA3 MTC insertion sites in the tagged mutants. Divergent and convergent PCR methods were used for amplification of the MTC (grey box, ura3d1; flanking hatched boxes, zeta fragments) insertion site borders. (Left) Divergent PCR method to rescue the genomic sequences flanking the MTC inserted into gene X (white arrows, borders after insertion). Genomic DNA (black line) of MTC mutants was digested with an enzyme which does not cut in the MTC (site Y) and circularized by ligation. PCR amplification was performed with oligonucleotide primers (thin arrows) specific for the MTC (Zeta1 and Zeta3). Alternatively, a restriction enzyme was used that cuts in the polylinker of the MTC (Fig. 1, HindIII to EcoRI), followed by amplification of the left and right borders as described previously for Tn3 insertion in Y. lipolytica (26). The PCR product was sequenced using the same primers. (Right) Convergent PCR method (PCR walking). Genomic DNA of selected MTC mutants was digested with enzymes giving blunt ends, such as EcoRV or EcoRV plus StuI plus PvuII), and ligated with the specific adapter oligonucleotides Ad1 and Ad2 (black boxes) as previously described (9). PCR was performed with a primer specific to the adapter (Ap1) and with a primer specific to the MTC, like Zeta1 and Zeta3, generating left and right border fragments, respectively. Border fragments were sequenced with the same primers.
Nucleotide sequence accession numbers.
DNA sequences were deposited in the EMBL database under the accession numbers AJ306421 for URA3 and AJ278693 for LEU1.
RESULTS AND DISCUSSION
Choice and construction of recipient strains.
Natural isolates of Y. lipolytica appear to have widely divergent genetic structures, as indicated by chromosome length polymorphism (7) and by the presence or absence of the retrotransposon Ylt1 (8). For this study, we required a strain that (i) was devoid of Ylt1 in order to obtain dispersed integration of a zeta-URA3 cassette, (ii) grew efficiently on hydrophobic substrates, and (iii) exhibited a clear dimorphic switch. We therefore tested for the presence of Ylt1 in various strains, like the wild-type strain YB423, its derivatives CX161-1B and CXAU1 (American strain series), or the wild-type strain H222 from Germany, all previously used for isolation of non-alkane-utilizing (Alk−) mutants (5, 6, 16) and cytochrome P450 studies (11, 12). We also tested inbred German strains of G. Barth's laboratory, such as B204-12C and its derivatives, inbred French strains such as E129 and E150, used for the isolation of peroxisomal mutants (22), and the French wild-type strain W29, used in the recent Y. lipolytica sequencing projects (8). The American strains and all inbred strains derived from them contain the Ylt1 retrotransposon, while the French (W29) and German (H222) wild-type strains were devoid of it (data not shown; see also reference 13).
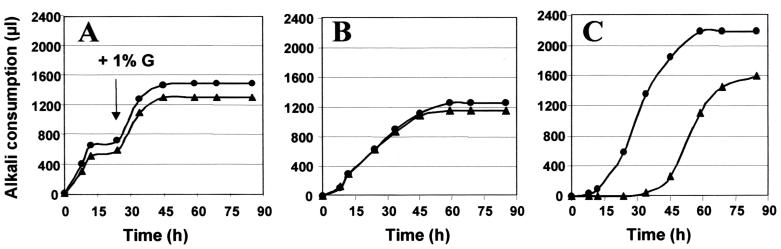
Both W29 and H222 exhibit similar morphology and growth kinetics on the various media tested, except for n-alkanes, where W29 exhibits a very long lag phase or no growth at all when replica plated from a glucose to an alkane medium. Similar growth kinetics are observed in liquid medium on glucose (Fig. 3A) and oleic acid (Fig. 3B), while W29 exhibits a 30- to 35-h lag phase on alkanes of different chain lengths (C10 to C16), as shown for hexadecane (Fig. 3C).
FIG. 3.
Growth comparison of Y. lipolytica wild-type strains. Growth of the strains H222 (●) and W29 (▴) in minimal medium M with 1% glucose (arrow, additional 1% glucose supplied at 24 h) (A), 1% oleic acid (B), and 2% hexadecane (baffled shaking flasks) (C) as carbon sources. Cultures in 100 ml of M medium in 500-ml shaking flasks were inoculated with washed cells from overnight YPD precultures at 2 × 106 cells/ml (OD600 = 0.4). Growth was monitored by measuring the amount of alkali (2.5 N NaOH) consumption used for pH titration to 5.5 as described previously (10).
In addition, we observed that amino acid-auxotrophic strains (Leu−, Met−, Lys−, or His−) exhibited a reduced growth rate in minimal medium on HC, but also on glucose, glycerol, or ethanol, when ammonium salts were given as nitrogen source (35; T. Juretzek and S. Mauersberger, unpublished results). Thus, only a ura3 derivative of the wild-type strain H222 exhibited the required characteristics for this study.
Existing UV-induced mutants like H222-67 (ura3-67) were found to revert at too high a frequency for tagged mutagenesis. We therefore constructed H222 derivatives with nonreverting ura3 alleles (see Materials and Methods).
We used here strain H222-41, which contains a nonreverting 10-bp deletion (allele ura3-41), although strains with larger deletions obtained later (see Materials and Methods) might be less prone to URA3 conversion events during transformation (see below).
Isolation of tagged mutants with a zeta-URA3 MTC.
During analysis of Y. lipolytica Tn3-tagged mutants (18), we frequently observed unexpected events such as nonhomologous recombination, which complicated identification of the insertion site (26; J.-M. Nicaud et al., unpublished data).
We therefore wanted to test if a simpler MTC, containing only a selectable marker flanked by zeta regions for promoting random integration into the genome of Ylt1-free strains, could be used to tag Y. lipolytica genes. For this purpose the strain Y. lipolytica H222-41 (ura3-41) was transformed with NotI-digested JMP5 (Fig. 1), thus liberating the MTC from the vector. Ura+ transformant colonies were selected on YNBcas at a frequency of 1,000 to 1,500 transformants per μg of DNA. A total of approximately 8,600 transformants were isolated and tested for phenotypes (see below).
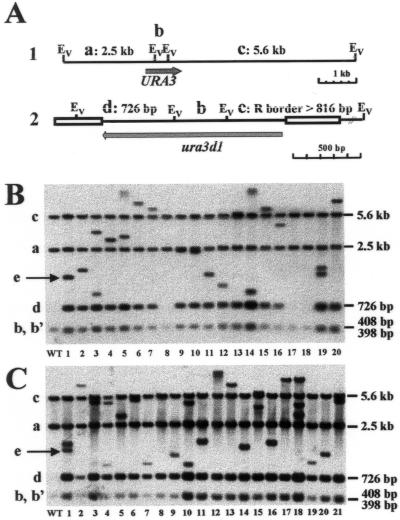
The integration of the MTC into the genome was studied by Southern blot analysis of 20 randomly selected Ura+ transformants (T clones) (Fig. 4B) and of 21 Ura+ transformants exhibiting specific phenotypes (P clones) (Fig. 4C). When genomic DNA was digested by EcoRV and hybridized with a URA3 probe, several bands were expected (Fig. 4A). For the genomic ura3-41 locus, three bands of 5.6 kb (band a), 2,507 bp (band b), and 398 bp (band c) were expected, the latter being replaced by a 408-bp band in the case of the wild-type locus or of the ura3d1 allele (Fig. 4A, construct 1). This was indeed observed in strain H222 and in all transformants (Fig. 4B and C, lanes 1, 2 to 20, and 2 to 21, respectively). When the MTC inserted randomly into genomic DNA, three bands were predicted: a new 726-bp band (band d), an internal 408-bp fragment (band b), and one larger than 816 bp and reflecting the location of the closest genomic EcoRV site in the right border (band e) (Fig. 4A, construct 2).
FIG. 4.
Southern blot analysis of MTC transformants revealing random cassette integration. (A) Schematic representation of the genomic URA3 locus (construct 1) and of a locus where a zeta-URA3 MTC was inserted (construct 2). The expected fragments (a to e) and their sizes are indicated. Abbreviations: Ev, EcoRV; R, right. (B) MTC integration into the genome was determined by Southern blot analysis of 20 randomly selected Ura+ transformants (T clones) of strain H222-41 with the MTC. Genomic DNA of the wild-type strain H222 (lane WT) and 20 T clones (lanes 1 to 20) from one transformation plate was digested with EcoRV and probed with the entire URA3 open reading frame. (C) Southern blot hybridization of 21 selected transformants of strain H222-41 with the MTC, showing phenotypes in the first screen (P clones, lanes 1 to 21) and of the wild-type strain H222 (WT). Conditions were as described for panel B. (B and C) Band names and sizes correspond to those in panel A. The variable right border (band e) is indicated by an arrow for the first transformants, T1 (B) and P1 (C).
In the transformants (Fig. 4B and C), three types of patterns could be observed. A wild-type pattern (type I, bands a, b, and c), identical to that of strain H222, was observed for the randomly selected T clones 8, 17, and 18 (Fig. 4B and C, lanes 1), reflecting ura3-41 conversion in 15% of the transformants. No type I event was observed among P clones (Fig. 4C). The expected profile after MTC integration was observed in most of the transformants, resulting in five bands (type II), as predicted above (the two smallest bands, bands b and b′, comigrate and show doubled intensity [Fig. 4B and C]). This type II pattern was observed in 13 out of 20 T clones (65%) (Fig. 4B) and in 18 out of 21 P clones (86%) (Fig. 4C). Nonexpected patterns suggesting multiple insertion of the MTC (more than five bands and higher intensity of band d, type III) were observed for four T clones (Fig. 4B, clones 3, 19, and probably 5 and 14) and for three P clones (Fig. 4C, clones 1, 10, and 13). This indicates that most events resulted from single MTC integration at different loci.
Phenotypic analysis of tagged mutants.
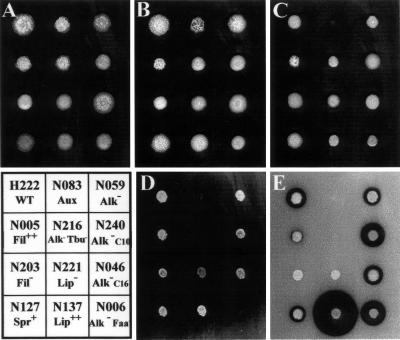
To assess the efficiency of MTC for isolating mutants affected in HC utilization, we screened approximately 8,600 Ura+ transformants for their ability to grow on nine media, including HC (alkanes, fatty acid, and triglyceride), ethanol, and glucose, and on media inducing hyphal growth. Transformants were first transferred onto YNBcas and then replica plated to avoid colony size effects. YPD, YNBcas, and YNBG were used for morphology and auxotrophy testing. An amino acid-auxotrophic strain such as N083 (Fig. 5) will grow only on YPD and YNBcas. HC utilization was analyzed on n-alkanes of different chain lengths (YNBC10, YNBC12, or YNBC16), oleic acid (YNBO), and tributyrin (YNBT) and compared to the growth on the hydrophilic substrates ethanol (YNBE) and glucose (YNBD). The YNBT medium also revealed halo formation due to extracellular lipase or esterase production.
FIG. 5.
Phenotypes of selected zeta-URA3 MTC insertion mutants. Wild-type strain H222 (WT) and selected MTC mutants were pregrown on YNBcas, transferred as suspensions onto YPD (A), YNBcas (B), YNBD (C), YNBO (D), and YNBT (E) plates, and incubated at 28°C for 2 to 5 days. Mutants and their phenotypes: auxotrophic mutant N083 (Aux, later shown to be leu1 [Fig. 7]); morphology mutants N005 (Fil++ [very rough hyperfilamentous colonies] Alk+ Faa+ Tbu+ Lip− [no halo formation due to extracellular lipase activity] Glu+), N203 (Fil− [smooth colonies, only yeast form] Alk+ Faa+ Tbu+ Lip− Glu+), and N127 (Spr+ [spreading and filamenting] Alk+ Faa+ Tbu+ Lip+ Glu+); and HC degradation mutants N006 (alkD/E: Alk− [C10− C12− C16−] Faa− Tbu+ Lip+ [normal halo formation] Eth+/− Glu+ Fil−), N046 (alkAc: Alk− C16 [C10+ C12+/− C16−] Faa+ Tbu+ Lip+ Glu+ Fil− [Fig. 6]), N059 (Alk− [growth delay] [C10+/− C12+/− C16+/−] Faa+ Tbu+ Lip+ Glu+), N137 (Lip++ [large halo formation on YNBT] Alk+), N216 (alkE: all HC− Alk− [C10− C12− C16−] Faa− Tbu− Eth− Glu+ Fil−), N221 (Alk+/− Faa+/− Tbu+ Lip− [no halo] Glu+ Fil+/−), and N240 (alkAb: Alk− C10 [C10− C12+/− C16+/−] Faa+ Tbu+ Lip+ Glu+).
Thus, 257 mutants were isolated out of 8,600 transformants, purified, and retested on the same media in 96-well microtiter plates as described previously (18). Clear phenotypes were confirmed for about 170 of these primary clones. This mutant frequency of 2% is close to the 2.5% frequency previously obtained with Tn3 insertion (18). Selected mutants were subsequently tested in liquid cultures. Examples of mutant phenotypes are shown in Fig. 5 and 6.
FIG. 6.
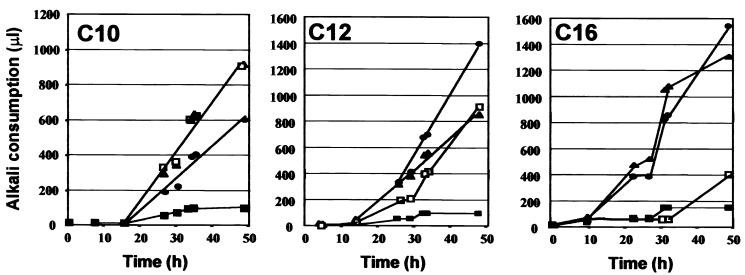
Growth of selected Alk− mutants on alkanes of different chain lengths. Growth of the insertion mutant was tested in shaking flasks with 1% alkane medium (YNBC10, YNBC12, or YNBC16). ▪, N002 (alkD: Alk− [C10− C12− C16−] Faa− Tbu+ Eth+ Glu+ Fil−); ●, N032 (alkAb or alkA leaky: Alk+/− [C10+/− C12+/− C16+] Faa+ Tbu+ Eth+ Glu+); □, N046 (alkAc: Alk− C16 [C10+ C12+/− C16−] Faa+ Tbu+ Glu+ Fil−); ▴, N233 (alkA leaky: Alk+/− [C10+ C12+/− C16+] Faa+ Eth+ Glu+). Mutants were cultivated, and growth was monitored as for Fig. 3.
About 70 mutants were affected with regard to growth on at least one of the three types of hydrophobic substrates (alkanes [Alk−], the fatty acid oleate [Faa−], and the triglyceride tributyrin [Tbu−]). Other carbon source utilization phenotypes, such as no growth on ethanol only (Alk+ Eth− Glu+, similar to N004), and four yellowish or brownish mutants were also obtained. Other less clear phenotypes, such as reduced growth on glucose, which are not easily distinguished from leaky auxotrophy were eliminated.
Among these HC− mutants, about 45 exhibited a phenotype involved in the utilization of alkanes and fatty acid, such as alkA (Alk− Faa+ Eth+ Glu+), alkD (Alk− Faa− Eth+ Glu+), and alkE (Alk− Faa− Eth+ Glu+). Examples of these mutants are shown in Fig. 5 and 6. The alkD clones N002 (Fig. 6) and N040 (data not shown) did not grow on any alkane or on oleic acid but did grow on ethanol and glucose and showed a smooth (Fil−) colony morphology. The alkE clones N216 (Fig. 5), N078, and N156 (data not shown) did not grow on any alkane, on fatty acid, or on ethanol but did grow on glucose and also showed a smooth (Fil−) colony morphology. The alkA (Alk− Faa+) mutants exhibited growth defects depending on alkane chain length. They either did not grow on any alkane (alkAa [C10− C12− C16−]), like N029 (data not shown), or exhibited chain length preferences; for example, N032 grew with a lag on C10 but well on C16 (alkAb), whereas N046 grew on C10 but not on C16 (alkAc) (Fig. 6), representing the first stable alkAc Y. lipolytica mutants isolated. Other Alk mutants appeared to be leaky (delayed growth on alkanes) or unstable after replica plating or in drop tests (not shown), indicating frequent occurrence of spontaneous suppressor mutations.
Several Alk− and Faa− mutants were also affected in tributyrin utilization (Tbu−), although this was not the case for all Alk− mutants, like N006 (Alk− Faa− Tbu+ Lip+ Glu+) (Fig. 5). On YNBT medium, 36 mutants appeared to be affected with regard to extracellular lipase activities, as observed by halo formation. Besides normal halo formation (Lip+, wild-type) observed for several mutants showing other phenotypes, we observed both Lip− phenotypes (no halo formation at all, as for N047, N203, and N221) and Lip++ phenotypes (larger halo, like N137, N185, N235, and N256) (Fig. 5D).
In the primary screening, about 50 auxotrophic mutants were obtained. Strain N083 was found to be auxotrophic for leucine and shown to be interrupted in the LEU1 gene (see below).
We also observed about 90 mutants with morphologies that were abnormal compared to that of the wild-type strain. Although colonial morphology varied on different media, about 25 mutants were hyperfilamentous on various media (Fil++, like N005), a phenotype not reported previously. A majority of mutants produced smooth, yeast-cell-only-containing colonies (Fil−, like N002, N006, and N203), and eight mutants with larger and flat colonies were called spreading (Spr+, like N127). Some examples of these phenotypes are shown in Fig. 5. Interestingly, most Fil− mutants were also affected in alkane and fatty acid utilization, presenting mostly an alkD or alkE phenotype, like N002, N006, or N216. They might be due to MTC insertion in a PEX gene, since mutants affected in both morphology and fatty acid utilization were previously described as being affected in peroxisome biogenesis (32), like mutant JMY226 (18), in which PEX10 is interrupted.
These results show that MTC is highly efficient for the induction of mutants affected in different pathways and can be used for creating various mutant libraries in Y. lipolytica.
Analysis of the insertion event and identification of the interrupted gene.
To analyze the insertion event and identify the interrupted gene in tagged mutants, the MTC borders were amplified by either divergent or convergent PCR (26, 31) (Fig. 2) as described in Materials and Methods.
Results for the leucine-auxotrophic mutant N083 are presented in Fig. 7. Borders of the MTC were amplified by PCR walking and sequenced with the specific MTC primers Zeta1 and Zeta3 and with the adapter-specific primer Ap1. BLAST analysis revealed sequences similar to part of the LEU1 gene, encoding 3-isopropylmalate dehydratase, of Saccharomyces cerevisiae (YGL009c). Within the Zeta1 and Zeta3 sequences (Fig. 7B, lines 3 and 5, respectively), the left and right parts of the MTC sequences (Fig. 7B, lines 4 and 6, respectively) were followed by LEU1 sequences (Fig. 7B, line 2). To identify the genomic sequence at the insertion site, part of the LEU1 gene was amplified with primer pair N083-1–N083-2 using H222-41 DNA as the template, and the PCR fragment was sequenced. Comparison of the genomic DNA sequence with the left and right border sequences revealed that the MTC was partially degraded (deletion of 2 and 14 bp from the left and right ends, respectively) (Table 2). Additionally, no sequence similar to the NotI site could be observed at the insertion site, indicating that insertion was not site specific or mediated by a restriction enzyme-mediated integration-type mechanism into NotI-like GC rich regions (28) and that MTC integrates by nonhomologous recombination.
FIG. 7.
Sequencing of the insertion site in mutant N083. (A) Schematic representation of the zeta-URA3 MTC insertion in mutant N083. The amplified border fragments obtained by convergent PCR (black boxes; PCR walking as described in reference 9) and the sequence determined (arrows) using the MTC-specific primers Zeta1 and Zeta3, the adapter-specific primer Ap1 for the left (Ap1L) and the right (Ap1R) border fragments, and the gene-specific primers N083-1 and N083-2 are shown. The bold arrow indicates the location of the LEU1 gene (AJ278693). (B) Partial amino acid (line 1) and corresponding nucleic (line 2) sequences of the wild-type LEU1 gene (amplified with the primers N083-1 and N083-1 from the genomic locus) and the sequences obtained with Zeta1 (line 3) and Zeta3 (line 5), compared with the left (line 4) and right (line 6) borders of the NotI zeta-URA3 cassette (in lowercase letters). The underlined lowercase letters correspond to the NotI site of the cassette borders (lines 4 and 6). The sequence underlined in line 2 corresponds to the 2-bp deletion at the site of MTC insertion into the LEU1 gene.
TABLE 2.
MTC insertion characteristics
| Mutant | Sequence origin | Deletion size (bp)a
|
Size (kb) of amplified fragmentb
|
||
|---|---|---|---|---|---|
| Left | Right | Left | Right | ||
| N083 | PCR fragment | 2 | 14 | 0.9 | 1.6 |
| N156 | PCR fragment | 2 | 0 | 0.5 | 1.5 |
| N216 | AWOAA019F02T1c | 6 | 13 | 0.7 | 0.8 |
| N222 | AWOAA012C01D1c | 7 | 0 | 1.4 | 1.2 |
| N225 | AWOAA017A04D1c | —d | 1 | 1.1 | 1.7 |
Deletion observed on both sides of the MTC after insertion.
The left and right borders were amplified by the convergent method using Ap1-Zeta 1 and Ap1-Zeta3 primer pairs, respectively.
RSTs overlapping the insertion site, from reference 8.
Addition of 50 bp.
A similar approach was used for 64 MTC insertion mutants. Results are presented in Table 2 for clones N156, N216, N222, and N225. For the last three clones, RSTs from Y. lipolytica (8) overlap the two borders, allowing identification of the wild-type sequence at the insertion site of the MTC. This comparison shows that MTC ends were trimmed over a few base pairs on both sides and integrated by nonhomologous recombination with a few base pairs modified at the insertion site.
A total of 64 mutants were tested to determine the MTC insertion site by convergent PCR amplification after EcoRV restriction and PCR walking (Fig. 2). Both borders were obtained for 32 clones, one border was obtained only for 25 clones, and none was obtained for 7 clones. A new PCR walking was performed with genomic DNA digested by three restriction enzymes, EcoRV, StuI, and PvuII. This increased the number of borders that could be amplified. However, the sizes of the amplified fragments were smaller, decreasing the number of significant BLAST results (data not shown).
For 15 clones, sequence analysis revealed atypical integration events. We obtained seven clones with the insertion of two MTCs either head to tail, head to head, or tail to tail. For example, in the alkD mutant N002 (Alk− Faa− Eth+ Glu+), two copies of the MTC integrated in tandem and in the same orientation. For five clones, we observed integration of JMP5, resulting from a single NotI digestion of the vector. For three clones, we observed the insertion of one MTC flanked by two copies of the vectors or insertion of the vector flanked by two MTCs. These events probably reflect partial digestion of JMP5 and/or in vivo ligation of the MTCs prior integration.
To identify the disrupted genes, we sequenced the borders of the MTC using the PCR walking method as shown above for N083 (Fig. 7). Sequence analysis revealed that the insertion occurred in the isocitrate dehydrogenase gene for mutant N156 (alkE leaky: Alk− Faa− Eth+/− Glu+), in the thioredoxin reductase gene for N040 (alkD: Alk− Faa− Eth− Glu+ Fil− and yellowish), and in the peroxine 14 gene (PEX14 [unpublished data]) for mutant N078 (alkD: Alk− Faa− Eth− Glu+ Fil−). A thorough analysis of the disrupted genes will be presented elsewhere.
Taken together, these results demonstrate that amplification and sequencing of the MTC insertion sites permit efficient and unambiguous identification of the interrupted genes and suggest that this method should be generally useful to identify genes in any pathway.
ACKNOWLEDGMENTS
This work was supported by the Institut National de la Recherche Agronomique and by the Centre National de la Recherche Scientifique (France), and it benefited from the France-Germany exchange program PROCOPE for 1998–2000 (MAE—A.P.A.P.E. no. 98185; DAAD PKZ 9723054).
The technical assistance of Susann Berthold is gratefully acknowledged. We thank Claudia Rentsch for the construction of plasmid pCR4, Fanny Aubertin for participation in mutant screening, and Antje Augstein for sharing results on the presence of Ylt1 in different Y. lipolytica strains.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth G, Gaillardin C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev. 1997;19:219–237. doi: 10.1111/j.1574-6976.1997.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 3.Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Nonconventional yeasts in biotechnology. Berlin, Germany: Springer-Verlag; 1996. pp. 313–388. [Google Scholar]
- 4.Barth G, Scheuber T. Cloning of the isocitrate lyase gene (ICL1) from Yarrowia lipolytica and characterization of the deduced protein. Mol Gen Genet. 1993;241:422–430. doi: 10.1007/BF00284696. [DOI] [PubMed] [Google Scholar]
- 5.Bassel J B, Mortimer R K. Genetic and biochemical studies of n-alkane non-utilizing mutants of Saccharomycopsis lipolytica. Curr Genet. 1982;5:77–88. doi: 10.1007/BF00365697. [DOI] [PubMed] [Google Scholar]
- 6.Bassel J B, Mortimer R K. Identification of mutations preventing n-hexadecane uptake among 26 n-alkane non-utilizing mutants of Yarrowia lipolytica. Curr Genet. 1985;9:579–586. [Google Scholar]
- 7.Casaregola S, Feynerol C, Diez M, Fournier P, Gaillardin C. Genomic organization of the yeast Yarrowia lipolytica. Chromosoma. 1997;106:380–390. doi: 10.1007/s004120050259. [DOI] [PubMed] [Google Scholar]
- 8.Casaregola S, Neuveglise C, Lepingle A, Bon E, Feynerol C, Artiguenave F, Wincker P, Gaillardin C. Genomic exploration of the hemiascomycetous yeasts: 17. Yarrowia lipolytica. FEBS Lett. 2000;487:95–100. doi: 10.1016/s0014-5793(00)02288-2. [DOI] [PubMed] [Google Scholar]
- 8a.Davidow, L. S., and J. R. Dezeeuw. 10 May 1983. Process for transforming Yarrowia lipolytica. U.S. patent US539591.
- 9.Devic M, Albert S, Delseny M, Roscoe T. Efficient PCR walking on plant genomic DNA. Plant Physiol Biochem. 1997;35:331–339. [Google Scholar]
- 10.Huth J, Werner S, Müller H-G. The proton extrusion of growing yeast cultures as an on-line parameter in fermentation processes: quantitative determination of growth from milligram amounts of substrate in a minimized fed-batch fermentation apparatus. J Basic Microbiol. 1990;7:489–497. [Google Scholar]
- 11.Iida T, Ohta A, Takagi M. Cloning and characterization of an n-alkane-inducible cytochrome P450 gene essential for n-decane assimilation by Yarrowia lipolytica. Yeast. 1998;14:1387–1397. doi: 10.1002/(SICI)1097-0061(199811)14:15<1387::AID-YEA333>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Iida T, Sumita T, Ohta A, Takagi M. The cytochrome P450ALK multigene family of an n-alkane-assimilating yeast, Yarrowia lipolytica: cloning and characterization of genes coding for new CYP52 family members. Yeast. 2000;16:1077–1087. doi: 10.1002/1097-0061(20000915)16:12<1077::AID-YEA601>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Juretzek T, Le Dall M T, Mauersberger S, Gaillardin C, Barth G, Nicaud J-M. Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast. 2001;18:97–113. doi: 10.1002/1097-0061(20010130)18:2<97::AID-YEA652>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Kujau M, Weber H, Barth G. Characterization of mutants of the yeast Yarrowia lipolytica defective in acetyl-coenzyme A synthetase. Yeast. 1992;8:193–203. doi: 10.1002/yea.320080305. [DOI] [PubMed] [Google Scholar]
- 15.Lindley N D. Bioconversion and biodegradation of aliphatic hydrocarbons. Can J Bot. 1995;73(Suppl. 1):S1034–S1042. [Google Scholar]
- 16.Mauersberger S. Mutants of alkane oxidation in the yeasts Yarrowia lipolytica and Candida maltosa. In: Sharyshev A A, Finogenova T V, editors. Alkane metabolism and oversynthesis of metabolites by microorganisms. Pushchino, USSR: Center for Biological Research, USSR Academy of Sciences; 1991. pp. 59–78. [Google Scholar]
- 17.Mauersberger S, Ohkuma M, Schunck W-H, Takagi M. Candida maltosa. In: Wolf K, editor. Nonconventional yeasts in biotechnology. Berlin, Germany: Springer-Verlag; 1996. pp. 411–580. [Google Scholar]
- 18.Neuveglise C, Nicaud J-M, Ross-Macdonald P, Gaillardin C. A shuttle mutagenesis system for tagging genes in the yeast Yarrowia lipolytica. Gene. 1998;213:37–46. doi: 10.1016/s0378-1119(98)00205-4. [DOI] [PubMed] [Google Scholar]
- 19.Nga B H, Gaillardin C M, Fournier P, Heslot H. Genetic analysis of lipase low-producing mutants of Yarrowia lipolytica. J Gen Microbiol. 1989;135:2439–2444. [Google Scholar]
- 20.Nga B H, Heslot H, Gaillardin C M, Fournier P, Chan K, Chan Y N, Lim E W, Nai P C. Use of nystatin for selection of tributyrin non-utilizing mutants in Yarrowia lipolytica. J Biotechnol. 1988;7:83–86. [Google Scholar]
- 21.Nicaud J-M, Fabre E, Gaillardin C. Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Curr Genet. 1989;16:253–260. doi: 10.1007/BF00422111. [DOI] [PubMed] [Google Scholar]
- 22.Nuttley W M, Brade A M, Gaillardin C, Eitzen G A, Glover J R, Aitchison J D, Rachubinski R A. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- 23.Ohkuma M, Zimmer T, Iida T, Schunck W-H, Ohta A, Takagi M. Isozyme function of n-alkane-inducible cytochromes P450 in Candida maltosa revealed by sequential gene disruption. J Biol Chem. 1998;273:3948–3953. doi: 10.1074/jbc.273.7.3948. [DOI] [PubMed] [Google Scholar]
- 24.Pignède G, Wang H J, Fudalej F, Gaillardin C, Seman M, Nicaud J-M. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol. 2000;182:2802–2810. doi: 10.1128/jb.182.10.2802-2810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pignède G, Wang H-J, Fudalej F, Seman M, Gaillardin C, Nicaud J-M. Autocloning and amplification of LIP2 in Yarrowia lipolytica. Appl Environ Microbiol. 2000;66:3283–3289. doi: 10.1128/aem.66.8.3283-3289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richard M, Quijano R, Bezzate S, Bordon-Pallier F, Gaillardin C. Tagging morphogenetic genes by insertional mutagenesis in the yeast Yarrowia lipolytica. J Bacteriol. 2001;183:3098–3107. doi: 10.1128/JB.183.10.3098-3107.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sanchez O, Navarro R E, Aguirre J. Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI) Mol Gen Genet. 1998;258:89–94. doi: 10.1007/s004380050710. [DOI] [PubMed] [Google Scholar]
- 29.Schmid-Berger N, Schmid B, Barth G. Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J Bacteriol. 1994;176:2477–2482. doi: 10.1128/jb.176.9.2477-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shennan J L, Levi J D. The growth of yeast on hydrocarbons. Prog Ind Microbiol. 1974;13:1–57. [PubMed] [Google Scholar]
- 31.Siebert P, Chenchik A, Kellogg D, Lukyanov K, Lukyanov S. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Titorenko V I, Ogrydziak D M, Rachubinski R A. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol. 1997;17:5210–5226. doi: 10.1128/mcb.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Titorenko V I, Rachubinski R A. Dynamics of peroxisome assembly and function. Trends Cell Biol. 2001;11:22–29. doi: 10.1016/s0962-8924(00)01865-1. [DOI] [PubMed] [Google Scholar]
- 34.Tzschoppe K, Augstein A, Bauer R, Kohlwein S D, Barth G. Trans-dominant mutations in the GPR1 gene cause high sensitivity to acetic acid and ethanol in the yeast Yarrowia lipolytica. Yeast. 1999;15:1645–1656. doi: 10.1002/(SICI)1097-0061(199911)15:15<1645::AID-YEA491>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 35.Wang H J, Le Dall M-T, Wach Y, Laroche C, Belin J M, Gaillardin C, Nicaud J M. Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol. 1999;181:5140–5148. doi: 10.1128/jb.181.17.5140-5148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf K, editor. Nonconventional yeasts in biotechnology. Berlin, Germany: Springer Verlag; 1996. [Google Scholar]
- 37.Zimmer T, Iida T, Schunck W-H, Yoshida Y, Ohta A, Takagi M. Relation between evolutionary distance and enzymatic properties among the members of the CYP52A subfamily of Candida maltosa. Biochem Biophys Res Commun. 1998;251:244–247. doi: 10.1006/bbrc.1998.9450. [DOI] [PubMed] [Google Scholar]