Abstract
African trypanosomiasis is a neglected tropical disease caused by Trypanosoma brucei (T. brucei) and spread by the tsetse fly in sub-Saharan Africa. The trypanosome relies on heat shock proteins for survival in the insect vector and mammalian host. Heat shock protein 90 (HSP90) plays a crucial role in the stress response at the cellular level. Inhibition of its interactions with chaperones and co-chaperones is being explored as a potential therapeutic target for numerous diseases. This study provides an in silico overview of HSP90 and its co-chaperones in both T. brucei brucei and T. brucei gambiense in relation to human and other trypanosomal species, including non-parasitic Bodo saltans and the insect infecting Crithidia fasciculata. A structural analysis of T. brucei HSP90 revealed differences in the orientation of the linker and C-terminal domain in comparison to human HSP90. Phylogenetic analysis displayed the T. brucei HSP90 proteins clustering into three distinct groups based on subcellular localizations, namely, cytosol, mitochondria, and endoplasmic reticulum. Syntenic analysis of cytosolic HSP90 genes revealed that T. b. brucei encoded for 10 tandem copies, while T. b. gambiense encoded for three tandem copies; Leishmania major (L. major) had the highest gene copy number with 17 tandem copies. The updated information on HSP90 from recently published proteomics on T. brucei was examined for different life cycle stages and subcellular localizations. The results show a difference between T. b. brucei and T. b. gambiense with T. b. brucei encoding a total of twelve putative HSP90 genes, while T. b. gambiense encodes five HSP90 genes. Eighteen putative co-chaperones were identified with one notable absence being cell division cycle 37 (Cdc37). These results provide an updated framework on approaching HSP90 and its interactions as drug targets in the African trypanosome.
Keywords: African trypanosomiasis, Trypanosoma brucei, HSP90, molecular chaperones, co-chaperone, heat shock proteins, HSP83
1 Introduction
Trypanosoma brucei (T. brucei), is an extracellular blood- and tissue-borne protozoan parasite transmitted by Glossina (tsetse) fly vectors, which causes devastating diseases in humans, wild animals, and domesticated livestock (Brun et al., 2010). Human African trypanosomiasis (HAT, also known as African sleeping sickness) is a potentially fatal tropical disease found in remote rural regions of sub-Saharan Africa and often coincides with insubstantial health care systems (Fèvre et al., 2008). HAT is caused by two subspecies of T. brucei; the chronic form of the disease, which is endemic to Central and Western Africa, is caused by Trypanosoma brucei (T. b.) gambiense, and the acute zoonotic form, which is endemic to Eastern and Southern Africa, is caused by T. b. rhodesiense (Simarro et al., 2010; Büscher et al., 2017). The livestock disease, nagana, is caused by T. b. brucei and has been shown, along with T. congolense and T. vivax, to have a crippling effect on socioeconomic development within sub-Saharan Africa (Alsan, 2015; Morrison et al., 2016). Recently, atypical human trypanosomiasis was reported to have emerged, with animal Trypanosoma species increasingly being detected in humans (Kumar et al., 2022). Despite the decreasing number of HAT cases and the first recently approved oral treatment called fexinidazole, which has now been added to the updated WHO guidelines as the recommended treatment for first and second stages T b. gambiense HAT (Deeks, 2019; WHO, 2019; Lindner et al., 2020). There is still a need for the development of new and more effective drugs due to lack of a vaccine and increasing parasite resistance (Barrett and Croft, 2012). Molecular chaperones have been identified as an attractive target for drug development against protozoan parasites as this protein family plays essential roles in stress-induced stage differentiation and are vital for disease progression and transmission (Requena et al., 2015; Bentley et al., 2019; Zininga and Shonhai, 2019).
The 90-kDa heat shock protein (HSP90) family contains essential, highly conserved, and abundant molecular chaperones (Csermely et al., 1998; Chen et al., 2006; Johnson, 2012) that facilitate the proper folding and maturation of a large but specific group of substrates called client proteins (Jakob et al., 1995; Hartl et al., 2011; Hoter et al., 2018). More than 400 client proteins of human HSP90 have been identified to date (listed at http://www.picard.ch/), with many of them being implicated in protein folding and degradation, signaling pathways, cellular trafficking, cell cycle regulation, and differentiation (Echeverría et al., 2011; Samant et al., 2012; Taipale et al., 2012). In humans, the HSP90 family normally comprises four isoforms that are localized in various cellular compartments. Two HSP90 isoforms (the stress-inducible form HSP90α/HSPC2 and the constitutive form HSP90β/HSPC3) are localized in the cytosol and in the nucleus (Subbarao Sreedhar et al., 2004; Kampinga et al., 2009; Li et al., 2012); GRP94/HSPC4 is present in the endoplasmic reticulum (ER) (Subbarao Sreedhar et al., 2004; Kampinga et al., 2009; Marzec et al., 2012) and TRAP-1/HSPC5 is found in the mitochondrial matrix (Altieri et al., 2012). Some intracellular HSP90 isoforms are exported and functioned in the extracellular environment to regulate the immune response, cell migration, and invasion (Binder, 2014; Hance et al., 2014; Hunter et al., 2014; Baker-Williams et al., 2019).
HSP90 is a flexible dimeric protein with each monomer containing three domains: an N-terminal nucleotide-binding domain (NBD), a middle client protein-binding domain (MD), and a C-terminal domain (CTD) (Whitesell and Lindquist, 2005; Buchner and Li, 2013; Jackson, 2013). HSP90 is dependent on ATP hydrolysis, and a set of accessory proteins termed co-chaperones, which assist in the recruitment of client proteins and the regulation of the HSP90 reaction cycle (Prodromou, 2012; Röhl et al., 2013). The cytosolic HSP90 isoforms contain a conserved C-terminal MEEVD motif, which acts as a docking site for interaction with co-chaperones that possess the tetratricopeptide repeat (TPR-) domain (Blatch and Lässle, 1999; Prodromou, 1999). Other HSP90 co-chaperones interact with the molecular chaperone through its NBD or M domain (Röhl et al., 2013). Fifty co-chaperones have been identified in the mammalian HSP90 chaperone system to date (Dean and Johnson, 2021): 23 TPR co-chaperones have been characterized, 18 cysteine and histidine-rich domain (CHORD) or SGT1 (CS) domain co-chaperones, and eight co-chaperones without these two domains (Garcia-Ranea et al., 2002; Schopf et al., 2017; Dean and Johnson, 2021). However, the composition of the HSP90 chaperone system appears to vary across organisms, indicating that the role of some co-chaperones may be necessary for activating client proteins in a species-dependent manner, be replaceable with other co-chaperones, or be limited to a distinct subgroup of client proteins (Zuehlke and Johnson, 2010). HSP90 is also subject to post-translational modifications, including s-nitrosylation, phosphorylation, and acetylation, which may influence its activity, cellular localization, or its interaction with co-chaperones, nucleotides, or client proteins (Aoyagi and Archer, 2005; Duval et al., 2007; Rao et al., 2008; Yang et al., 2008). Some HSP90 isoforms are essential for viability, and maintenance of client proteins that are dependent on the chaperone (Citri et al., 2004), making it an attractive drug target for diseases including infectious diseases. Several HSP90 inhibitors, which have been well-studied in the laboratory and clinics for antitumor indications (Porter et al., 2010; Trepel et al., 2010), were also shown to arrest the growth of several kinetoplastids in vitro and have activity against Trypanosoma evansi and T. brucei in mice (Graefe et al., 2002; Pallavi et al., 2010; Meyer and Shapiro, 2013; Meyer et al., 2018). Thus, the repurposing of HSP90 inhibitors designed for cancer treatment is one strategy to evaluate new and effective antitrypanosomal agents (Kaiser et al., 2015).
In Trypanosoma and Leishmania, the HSP90 machinery plays a pivotal role in environmental sensing and life cycle control (Ploeg et al., 1985; Wiesgigl and Clos, 2001; Graefe et al., 2002). In silico analyses of the HSP90/HSPC family of intracellular kinetoplastid parasites has been published (Shonhai et al., 2011; Roy et al., 2012; Figueras et al., 2014; Urményi et al., 2014; Requena et al., 2015), and our study provides an updated and comprehensive analysis for the extracellular parasite, T. brucei. T. brucei exhibits a digenetic lifestyle, and therefore must adapt to fluctuating environmental conditions, such as change in temperature, pH, nutrients, and the pressure from the immune system, as it transitions from the gut of the tsetse fly to the body fluids of its mammalian host (Jones et al., 2008; Roy et al., 2012). A distinct molecular trait of trypanosomes is their dependence on polycistronic transcription akin to prokaryotes, their mRNAs are mainly generated by trans-splicing and there is a dependence on post-transcriptional mechanisms for gene regulation (Preußer et al., 2012). However, correlation studies comparing the previously reported RNA-seq data of transcript abundance and proteomic data from the procyclic form (PF) and bloodstream form (BSF) of the parasite shows that the differences observed between the PF and BSF are two-fold greater at the proteomic level when compared to the transcriptomic level (Urbaniak et al., 2012; Butter et al., 2013). Given the complexities of transcription, its incomplete representation of the life cycle stages of the parasite as well as its lack of control, trypanosome research has largely shifted to rely on proteomic data (Goos et al., 2017). Numerous proteomic studies have been conducted on the parasite, which have compared protein expression at the different life cycle stages (Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013), in the mitochondrion (Panigrahi et al., 2009), mitochondrial importome (Peikert et al., 2017), respiratome (Acestor et al., 2011), mitochondrial membranes (outer, intermembrane space, inner, and matrix) (Acestor et al., 2009), nucleus (Goos et al., 2017), nuclear pore (DeGrasse et al., 2008), glycosomes (Colasante et al., 2006; Güther et al., 2014), flagellum (Broadhead et al., 2006; Subota et al., 2014), and the cell surface (Shimogawa et al., 2015).
T. brucei and other trypanosomatids rely on post-translational modifications (PTMs) to increase their proteome diversity and complexity (Backe et al., 2020). Several studies exploring the phosphoproteome and acetylome of trypanosomes (Nett et al., 2009a, 2009b; Urbaniak et al., 2013; Moretti et al., 2018) have found that phosphorylation and acetylation are the most predominant modifications to occur in T. brucei proteins. Both PTMs are well known for impacting HSP90 intracellular localization in humans as well as their ability to bind co-chaperones, nucleotides, clients (Nett et al., 2009a; Backe et al., 2020), and even inhibitors (Zhang et al., 2020). However, the PTMs regulatory dynamic in the organellar TRAP-1 and GRP94 in humans are yet to be elucidated for a global understanding of this critical chaperone activity regulator system.
The aim of this study was to provide a comprehensive depiction of the T. brucei HSP90 chaperone system based on structural, functional, and evolutionary analyses. In silico tools were used to evaluate the domain conservation, predicted subcellular localizations, syntenic, and phylogenetic analysis of the HSP90 chaperone system in T. brucei with respect to both T. b. brucei and T. b. gambiense. The HSP90 chaperone system was also comparatively analyzed in relation to those found in selected trypanosomastid parasites and Homo sapiens (H. sapiens). The proteomic findings on HSP90 and its co-chaperones from the numerous published proteomic data on T. brucei are presented, and we provide updated insights on the adaptability of the parasite from its stage-specific expressed proteins and provide an overall context for identifying new and potential drug targets for HAT.
2 Materials and methods
2.1 Database mining, sequence analyses, and the determination of the trypanosomastid and human orthologues
A BLASTP search using the amino acid sequences of the HSP90 isoforms from T b. brucei obtained from previous in silico study (Folgueira and Requena, 2007), and the human HSP90AA1/HSPC1, HSPC2, HSP90AB1/HSPC3, HSP90B1/GRP94/HSPC4, and TRAP-1/HSPC5 isoforms were used as queries on the TriTrypDB (version 46) database (http://tritrypdb.org/tritrypdb/) (Aslett et al., 2010) and were conducted in order to determine the HSP90/HSPC complement encoded on the T. b. gambiense genome, as well as identify new T. b. brucei HSP90/HSPC protein members. The e-value was set at an intermediately stringent level of e-10 to identify HSP90/HSPC-related sequences for further analysis. In addition, a keyword search was performed to scan the genome of T b. gambiense for HSP90/HSPC genes on the TriTrypDB database using the terms: “HSP90,” “HSP83,” “heat shock protein,” and “molecular chaperone.” The retrieved amino acid sequences from the various keyword searches were then screened using SMART 7 (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/) (Letunic et al., 2012) and PROSITE (http://prosite.expasy.org/) (Sigrist et al., 2010) for domains annotated by the online servers as “HSP90.” Incomplete sequences for each protein from TriTrypDB were omitted for construction of Table 1.
TABLE 1.
HSP90/HSPC proteins from Trypanosoma brucei with putative orthologues in T. cruzi, L. major, C. fasciculata, B. saltans, and H. sapiens.
| H. sapiens | H. sapiens | T. brucei | T. cruzi c | L. major | C. fasciculata | B. saltans | ||
|---|---|---|---|---|---|---|---|---|
| Name a | Gene ID b | Gene ID b | Gene ID b | Gene ID b | Gene ID b | Gene ID b | Localization d | Reference |
| HSP90-alpha/HSPC2 | 3324 | Tb927.10.10890 | CFAC1_280011900 CFAC1_280012000 | BSAL_87515 | Gunasekera et al. (2012); Urbaniak et al. (2012); Subota et al. (2014); Shimogawa et al. (2015); Dean et al. (2017) | |||
| Tb927.10.10900 | ||||||||
| Tb927.10.10910 | TcCLB.507713.30 | LmjF.33.0312 LmjF.33.0314 LmjF.33.0316 LmjF.33.0318 LmjF.33.0320 LmjF.33.0323 LmjF.33.0326 LmjF.33.0330 LmjF.33.0333 LmjF.33.0336 LmjF.33.0340 LmjF.33.0343 | ||||||
| 3326 | Tb927.10.10920 | C4B63_113g25 | CYT | |||||
| HSP90-beta/HSPC3 | Tb927.10.10930 Tb927.10.10940 | C4B63_113g29 | NUC | |||||
| Tb927.10.10950 Tb927.10.10960 Tb927.10.10970 Tb927.10.10980 | C4B63_113g30 C4B63_113g33 C4B63_84g87 C4B63_84g88 C4B63_84g89 | FLAGELLAR | ||||||
| Tbg972.10.13260 Tbg972.10.13270 Tbg972.10.13280 | Tc_MARK_3,581 | LmjF.33.0346 LmjF.33.0350 LmjF.33.0355 LmjF.33.0360 LmjF.33.0365 | CELL SURFACE | |||||
| GRP94/HSPC4 | 7184 | Tb927.3.3580 Tbg972.3.3850 | C4B63_10g439 Tc_MARK_3058 | LmjF.29.0760 | CFAC1_1,00018800 | BSAL_88715 | ER | Gunasekera et al. (2012); Urbaniak et al. (2012); Subota et al. (2014); Shimogawa et al. (2015) |
| NUC | ||||||||
| FLAGELLAR | ||||||||
| CELL SURFACE | ||||||||
| TRAP-1/HSPC5 | 10131 | Tb927.11.2650 Tbg972.11.2900 | TcCLB.504153.310 | LmjF33.2390 | CFAC1_230028300 | BSAL_33145 | Panigrahi et al. (2009); Subota et al. (2014); Dean et al. (2017) | |
| C4B63_2g430 | MITO | |||||||
| Tc_MARK_6238 | FLAGELLAR |
The nomenclature for the HSP90/HSPC, proteins from T b. brucei, and T b. gambiense were derived according to Folgueira and Requena (2007).
The Gene IDs for the members of the T b. brucei (Tb refers to Tbb), T b. gambiense, T. cruzi, C. fasciculata, B. saltans, and L. major HSP90/HSPC protein family were retrieved from the TriTrypDB database (http://tritrypdb.org/tritrypdb/; Aslett et al., 2010). The Gene IDs for the members of the H. sapiens HSP90/HSPC protein family were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/).
The Gene IDs, for the orthologues, identified by reciprocal BLASTP, analysis, of three strains of T. cruzi are listed. T. cruzi CL, Brener Esmeraldo-like (TcCLB), T. cruzi Dm28c 2018 (C4B63), and T. cruzi marinkelli strain B7 (Tc_MARK).
Subcellular localizations for the T. brucei HSP90/HSPC proteins were either acquired from using the TrypTag database (http://tryptag.org/;Dean, Sunter, and Wheeler 2017) and/or predicted using various proteomic datasets and online prediction software listed in the materials and methods.
CYT-Cytosol and MITO- mitochondrion, CYT-Cytosol; MITO- mitochondrion; NUC- nucleus; ER- endoplasmic reticulum; GYLCO-glycosomes; FLAGELLAR- flagellar; CELL SURFACE- cell surface.
For identification of T. brucei orthologues of selected cytosolic HSP90 co-chaperones, the protein sequences of 50 human co-chaperones were used as queries in a BLASTP search on the TriTrypDB database. Reciprocal BLASTP was conducted to determine if the identified putative T. brucei co-chaperone had the closest match to the desired human co-chaperone. The putative amino acid sequences of the co-chaperones from both T. brucei subspecies were used as queries in a BLASTP search on the National Centre for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov), using the default parameters. If the most similar ortholog in the T. brucei subspecies was identical to the HSP90 co-chaperones sequence used as the first query, the sequence of the second query was selected as an ortholog. Reciprocal BLASTP was also conducted for the identification of human and selected trypanosomastid orthologues of the putative HSP90/HSPC and HSP90 co-chaperone proteins from both T. brucei subspecies.
2.2 Structural analysis of TbHSP83
The retrieved amino acid sequences for hHsp90β (NP_001258899.1), TbbHsp83 (Tb927.10.10890), and TbgHsp83 (Tbg972.10.13260) were analyzed using Jalview (Waterhouse et al., 2009). A multiple sequence alignment was conducted using Clustal with defaults via the Jalview web service. 3D prediction structures were retrieved from the AlphaFold database (https://alphafold.ebi.ac.uk/) by querying the database using the UniProt IDs of the respective proteins. The retrieved structures (TbHsp83- AF-Q389P1-F1; hHsp90 - AF-P08238-F1) were visualized and superimposed using PyMOL molecular graphics system, Version 2.0 Schrödinger, LLC. The FATCAT server (https://fatcat.godziklab.org/) was used to analyze the structural differences between the protein homologs (Li et al., 2020). The two structures were determined to be significantly similar with a p-value of 0.00 (raw FATCAT score 1931.78), 685 equivalent positions, and RMSD of 1.71 Å and two twists.
2.3 Phylogenetic and conserved syntenic analysis
The full-length amino acid sequences for the HSP90/HSPC family in the selected trypanosomastid parasites were obtained from the TriTrypDB database (Aslett et al., 2010), and the human protein sequences were obtained from the NCBI website (www.ncbi.nlm.nih.gov). Accession numbers for the HSP90/HSPC amino acid sequences used in this study are provided in Table 1 and Supplementary Table S1. Multiple sequence alignments were performed using the inbuilt ClustalW program (Larkin et al., 2007) with default parameters in MEGA-X (Kumar et al., 2018) and are listed in Supplementary Figure S1. Maximum-likelihood (ML) was utilized to find the best model of evolution and was selected by the Bayesian information criterion (BIC) implemented in MEGA-X. The amino acid-based HSP90/HSPC ML phylogeny was reconstructed using the JTT (Jones–Taylor–Thornton) model matrix (Jones et al., 1992), with gamma distribution shape parameter (G). The ML phylogenetic tree was constructed using MEGA-X (Kumar et al., 2018). The accuracy of the reconstructed tree was assessed using a bootstrap test using 1,000 replicates with a pairwise gap deletion mode. The phylogenetic tree for HSP90s was unrooted.
The putative HSP90 genes in the three T. cruzi strains homologous to HSP83 identified to be partial, and/or truncated genes were included in the syntenic analysis. Syntenic analysis was conducted to evaluate the conservation of the gene arrangement of the cytosolic HSP83 genes in T. brucei and selected trypanosomastid parasites. The conserved syntenic regions surrounding the selected HSP83 genes were searched by examining the conserved co-localization of neighboring genes on a scaffold of the T. brucei subspecies (T. b. brucei and T. b. gambiense) and selected trypanosomastid parasites for this study using genome information from the TriTrypDB database. The identities of unknown neighbor genes of the selected HSP83 genes were conducted using a BLASTP search on the NCBI database.
2.4 Physiochemical properties, protein expression, and the determination of the organelle distribution for the T. brucei HSP90/HSPC complement
The physiochemical properties, molecular weight (Da), and isoelectric point (pI) of each putative protein was determined using the compute pI/Mw tool from ExPASy (https://web.expasy.org/compute_pi/) (Gasteiger et al., 2005). Data on the previously reported phenotypic RNAi knockdown screen (Alsford et al., 2011), for each member of the HSP90/HSPC complement and identified HSP83 co-chaperones, were retrieved from the TrypsNetDB database (http://trypsnetdb.org/QueryPage.aspx) (Gazestani et al., 2017). The organelle distribution for each putative protein was searched using the TrypTag microscopy project’s online server (http://tryptag.org/) (Dean et al., 2017). This project aims at tagging every trypanosome protein with mNeonGreen (mNG) (Shaner et al., 2013) to determine the protein’s localization within the parasite. Proteomic data from the mitochondrion (Panigrahi et al., 2009), mitochondrial importome (Peikert et al., 2017), respiratome (Acestor et al., 2009), mitochondrial membranes (outer, intermembrane space, inner, and matrix) (Acestor et al., 2009), nucleus (Goos et al., 2017), nuclear pore (DeGrasse et al., 2008), glycosomes (Colasante et al., 2006; Güther et al., 2014), flagellum (Broadhead et al., 2006; Subota et al., 2014), and cell surface (Shimogawa et al., 2015) were also used for the prediction of the organelle distribution for the T. brucei HSP90 complements and HSP90/HSPC complements and HSP83 co-chaperones.
2.5 Identification of potential post-translational modification sites for the T. brucei HSP83 proteins
Data mining from a collection of relevant databases on T. brucei PTMs (Nett et al., 2009b; Urbaniak et al., 2013; Moretti et al., 2018; Zhang et al., 2020) for the relevant proteins was retrieved using the previously identified accession numbers. Information on the respective PTMs (modification sites, modification types, and modified residue) were obtained, and the modified residues were mapped onto Supplementary Figure S2 for all HSP90 isoforms from T. brucei subspecies (T. b. brucei and T. b. gambiense) with orthologues from other trypanosomatids and from human, then analyzed for determination of conserved and specific PTMs among the T. brucei HSP90 complements.
3 Results and discussion
3.1 Determination of the T. b. brucei and T. b. gambiense HSP90/HSPC complements
The protozoan parasite T. brucei comprises three subspecies, with the genomes of T. b. gambiense and T b. brucei already sequenced (Jackson et al., 2010; Gibson, 2012). Any information obtained from the genome of the non-human infective T. brucei subspecies, T. b. brucei, can be inferred for the human infective subspecies, T. b. rhodesiense, as the T. b. brucei TREU927 strain displays the full range of known T. brucei phenotypes and possesses similar biological and genetic characteristics (Gibson, 2012). However, the T. b. gambiense genome was sequenced due to the subspecies displaying profoundly different biological and genetic characteristics (Jackson et al., 2010). Genome-wide identification and in silico analyses of the HSP90/HSPC complement in both T. brucei subspecies was conducted to provide an overview of the T. brucei HSP90 family. The ortholog of the cytosolic HSP90 member in T. brucei is termed HSP83 (Mottram et al., 1989), while in this study we refer to the ER ortholog as GRP94 and the mitochondrial ortholog as TRAP-1. However, to underscore whether discussing a protein from T. b. gambiense or T. b. brucei, the abbreviations Tbg and Tbb were used in this study, respectively. The orthologous relationships of the HSP90 family from T. b. brucei and T. b. gambiense to the selected organisms in this study are presented in Table 1, and a comprehensive domain organization of the predicted T. brucei HSP90 proteins is illustrated in Supplementary Figure S3.
Twelve putative HSP90 genes were identified to be encoded on the T. b. brucei genome (Table 1), which is consistent with previous findings (Mottram et al., 1989; Folgueira and Requena, 2007), while T. b. gambiense was identified in this study to only have five putative HSP90 genes. The reduction in the HSP90 gene numbers found in T. b. gambiense could be a consequence of the reduced genome size observed in the human infective subspecies (Dero et al., 1987). The intraspecific genomic variation is largely associated with tandem or segmental duplications observed in T b. brucei (Jackson et al., 2010). This study also identified an unassigned putative HSP90 gene (Tb11. v5.0543) in the animal infective subspecies, T b. brucei, but this sequence could not be assembled into a chromosome and was part of a bin scaffold that was not considered during reannotation efforts. For the putative HSP90 genes identified in this study for T b. brucei, 10 of the 12 putative HSP90 genes identified were found to be homologous to HSP83, whereas in T b. gambiense, three of the five putative HSP90 genes were homologous to HSP83 (Table 1). The remaining two HSP90 genes found in both T b. brucei (Tb927.3.3580 and Tbg972.3.3850) and T b. gambiense (Tb927.11.2650 and Tbg972.11.2900) showed significant identity to the ER and mitochondrial resident paralogues of HSP90, GRP94, and TRAP-1, respectively (Table 1). This indicates that a single-gene copy for GRP94 and TRAP-1 is encoded on the genome in both T. brucei subspecies. Phylogenetic analysis shows that the T. brucei HSP90/HSPC family also comprises three distinct HSP90 groups (HSP83, GRP94, and TRAP-1), which cluster into clades according to protein sequence and subcellular localization (Supplementary Figure S4).
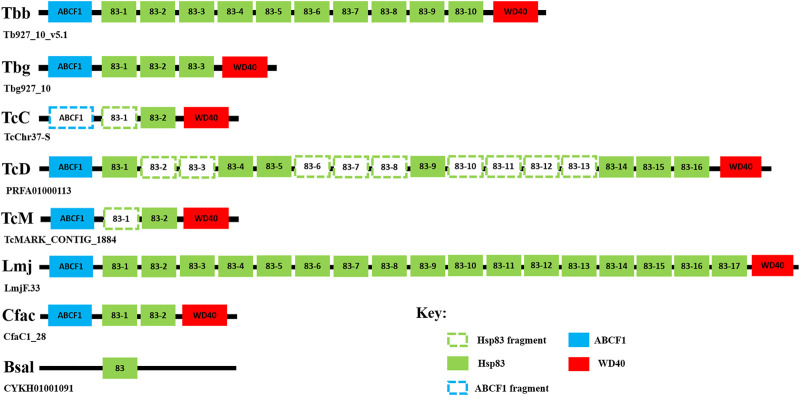
Previous literature reported that 11 HSP90 genes are encoded on the Trypanosoma cruzi (T. cruzi) genome (Shonhai et al., 2011). In this study we included three different T. cruzi strains: CL Brener Esmeraldo-like (TcCLB), Dm28c 2018 (C4B63), and marinkelli strain B7 (Tc_MARK) to determine the HSP90/HSPC complement in the American trypanosome. It was identified in this study that the T. cruzi CL Brener Esmeraldo-like strain has two HSP90 genes, the Dm28c 2018 strain has nine HSP90 genes, and the marinkelli strain B7 has three HSP90 genes (Table 1). However, this study identified that many of the HSP90 genes homologous to HSP83 in the three T. cruzi strains were found to be partial and/or truncated genes. In our syntenic analysis, these partial and/or truncated genes were included as they are probably a result of the methodology utilized to sequence the various T. cruzi strains, and it is very likely that the truncated sequences are full-length in the genome (Figure 1). The obvious discrepancy in numbers of genes among the T. cruzi strains, and its numerous partial and/or truncated HSP90 sequences has been recently reviewed. This review highlights the difficulties in T. cruzi genome analyses (Herreros-Cabello et al., 2020); the first genome sequenced that is still widely accepted as the main reference has close to 50% repetitions in its sequence (El-Sayed et al., 2005a; 2005b) and though newer genomes have been sequenced using short-read sequencing methods as in the case of the T cruzi marinkelli strain B7, these methods end up causing fragmented chromosomes due to their inability to create a complete chromosome from their short reads technique (Franzén et al., 2012; Herreros-Cabello et al., 2020). Other trypanosomatids included in this study were the non-parasitic Bodo saltans (B. saltans) (Deschamps et al., 2011) and the insect infecting Crithidia fasciculata (C. fasciculata) (Wallace, 1966), which were found to have three and four putative HSP90 genes, respectively (Table 1). Both these trypanosomatids were found to possess genes encoding for all three HSP90 isoforms, though C. fasciculata was identified to possess two HSP83 genes (Table 1).
FIGURE 1.
Syntenic analysis of the gene arrangement of the HSP83 genes in T. brucei and selected trypanosomatids. The conserved syntenic regions surrounding the selected HSP83 genes were searched by examining the conserved co-localization of neighboring genes on chromosome 10 on a scaffold of the T. brucei subspecies, T b. brucei (Tbb), and T b. gambiense (Tbg), and selected trypanosomatids: T. cruzi CL Brener Esmeraldo-like (TcC), T. cruzi Dm28c 2018 (TcD) strain, T. cruzi marinkelli strain B7 (TcM), L. major (Lmj), B. saltans (Bsal), and C. fasciculata (Cfac). The genome information used for this study was acquired from the TriTrypDB database (http://tritrypdb.org/tritrypdb/) (Aslett et al., 2010). The identities of unknown neighbor genes of the selected HSP83 genes were conducted using a BLASTP search on the NCBI database. Abbreviations: ABCF1: ATP-binding cassette sub-family F member 1; WD40: WD40-repeat protein.
Early genomic studies suggested that the human genome contained 16 HSP90 genes (five functional and 11 pseudogenes), which have been categorized, according to the proposed standardized guidelines for HSP nomenclature, into four isoforms under the superfamily name HSPC (Chen et al., 2006; Kampinga et al., 2009). In contrast to the trypanosomatids, humans have two isoforms of HSP90 localized in the cytoplasm: the inducible form HSP90α/HSPC2 and the constitutive form HSP90β/HSPC3 (Subbarao Sreedhar et al., 2004). Phylogenetic analysis has suggested that the two cytosolic isoforms arose from gene duplication, and the organelle HSP90s (GRP94/HSPC4 and TRAP-1/HSPC5) developed from a common ancestor (Gupta, 1995; Emelyanov, 2002; Chen et al., 2005).
3.2 HSP83
The ortholog of the cytosolic HSP90 member in trypanosomatids as mentioned previously is commonly referred to as HSP83 and has been found to be an essential and highly abundant protein that is encoded by multiple gene copies organized in a head-to-tail tandem array (Folgueira and Requena, 2007). It has been identified in this study and previous studies (Mottram et al., 1989; Folgueira and Requena, 2007) that T b. brucei has been shown to encode for 10 tandem copies of HSP83 (Figure 1), whereas T b. gambiense genome encodes for three tandem copies of HSP83 (Figure 1). Syntenic analysis revealed that the TbbHSP83 and TbgHSP83 genes are both located on chromosome 10 in a head-to-tail orientation, with the same genomic organization being observed in both T. brucei subspecies (Figure 1). A discrepancy in HSP83 gene copy numbers was observed for the three T. cruzi strains used in this study (Figure 1). Syntenic analysis revealed that the T. cruzi Dm28c 2018 (C4B63) strain has 16 tandem copies of HSP83, though nine were partial sequences (Figure 1), whereas both the CL Brener Esmeraldo-like (TcCLB) and marinkelli strain B7 (Tc_MARK) encode for two HSP83 genes, with one partial gene each (Figure 1). Leishmania major (Lmj) contained the largest HSP90 family with a total of 19 HSP90 genes (Table 1), 17 tandem copies were found to be homologous to HSP83, and these findings agree with previous studies (Folgueira and Requena, 2007; Shonhai et al., 2011; Requena et al., 2015), also correlating with the high abundance of the protein being observed in L. major and several other Leishmania spp. (Brandau et al., 1995). Syntenic regions surrounding the HSP83 genes were found to be virtually conserved across the selected trypanosomatids, with B. saltans being the exception (Figure 1). Thus, the discrepancy in gene copy number of HSP83 in the two T. brucei subspecies and among the trypanosomatids may have arisen from the differences in their life cycles.
Data mining of transcriptomic and proteomic datasets revealed that all identified TbbHSP83 (TbbHSP83 1–10) proteins are constitutively expressed at all life cycle stages of the parasite, as well as expressed at all phases of the cell cycle (Gunasekera et al., 2012; Urbaniak et al., 2012). The protein expression of the TbbHSP83 proteins were all reported to be upregulated at the BSF stage (Urbaniak et al., 2012), despite gene regulation being unchanged in both the bloodstream and procyclic life cycle stages (Gunasekera et al., 2012). All TbbHSP83 proteins were also present in the cell surface proteome (Subota et al., 2014), though only TbbHSP83-10 (Tb927.10.10980) was found to be present in the flagellar proteome (Shimogawa et al., 2015).
The amplification of HSP genes in protozoan parasites has been reported previously (Urményi et al., 2014; Requena et al., 2015; Drini et al., 2016; Bentley et al., 2019) and is considered a means by which the parasites increase chaperone levels to maintain proteostasis under normal and stressful conditions (Wiesgigl and Clos, 2001). The heat shock response is a highly conserved transcriptional program that in most organisms involves increased heat shock gene transcription (de Nadal et al., 2011). However, in trypanosomatids, control of gene expression occurs almost exclusively at the post-transcriptional level, and that HSP synthesis during heat shock depends on regulation of mRNA turnover and translational control (Clayton and Shapira, 2007; Requena, 2011). In T. brucei, post-transcriptional regulation of chaperone mRNAs is facilitated by a zinc finger protein, ZC3H11 (Droll et al., 2013). The mRNA transcript levels of TbbHSP83 in BSF parasites increases >2-fold after heat shock (Ooi et al., 2020) and is stabilized by ZC3H11 to promote the survival of the parasite (Droll et al., 2013). Treatment of T b. brucei BSF parasites with 17-AAG sensitized the parasites to heat shock, as well as caused severe morphological abnormalities and cell cycle disruption (Meyer and Shapiro, 2013). Pharmacological inhibition of HSP83 activity in several Leishmania spp. induced morphological and biochemical promastigote-to-amastigote differentiation (Wiesgigl and Clos, 2001; Bente et al., 2003; Hombach et al., 2013), which mimics environmental triggers such as heat shock and acidic milieu, indicating a pivotal role for HSP83 in trypanosomatid protists in environmental sensing and life cycle control. Interestingly, treatment of T. cruzi bloodstream trypomastigotes with geldanamycin, induced morphological changes in the parasites but not life cycle progression (Graefe et al., 2002). Therefore, HSP90 cellular homeostasis as a key factor for the control of stage differentiation appears to be dependent on the tropism of the parasite and the different regulatory pathways for life cycle control. It would be interesting to investigate if the pharmacological inhibition of HSP83 affects cellular differentiation among the three T. brucei subspecies.
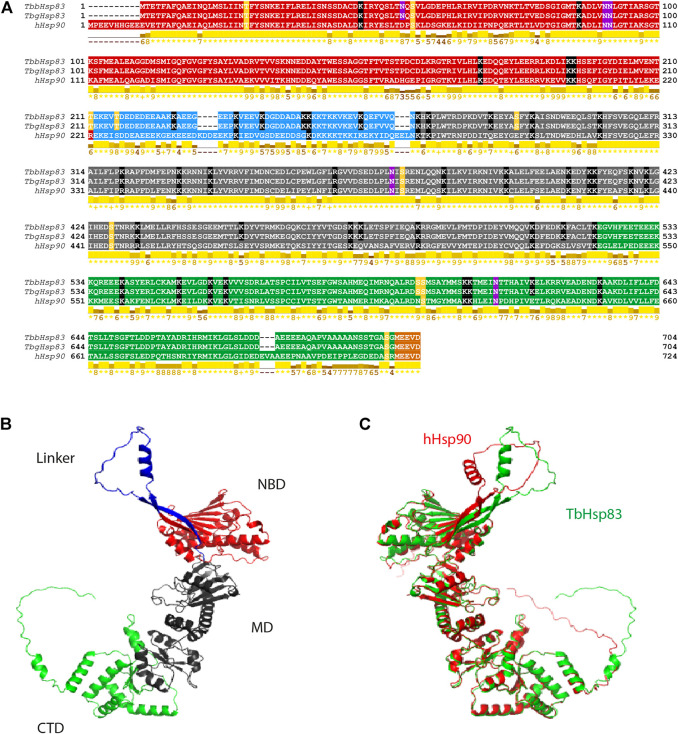
The monophyletic cluster of the cytosolic HSP83 proteins suggests a general conservation of function, structure, and sequence in the trypanosomatids HSP83 homologs (Supplementary Figure S4). In the amino acid sequences of TbbHSP83 and TbgHSP83 there was a single substitution at D461 to E in TbgHSP83 (Figure 2A). In comparison, hHSP90 was 63% identical in sequence to TbbHSP83 (Figure 2). The three HSP90 proteins displayed the characteristic domains (Figure 2A): the ATP-binding N-terminal domain (NTD); the middle domain (MD), which plays a role in ATPase activity and is responsible for interacting with client proteins and co-chaperones; and the C-terminal domain (CTD), which is responsible for HSP90 dimerization and interaction with the TPR domain-containing chaperones via a C-terminal (MEEVD) motif (Hoter et al., 2018). In addition, the NTD and MD are joined together via a charged linker (Jahn et al., 2014). This linker varies in size and is notably shorter in trypanosomatids compared to its human counterpart (Figure 2A) (Silva et al., 2013). Comparison of the hHSP90 to both TbbHSP83 and TbgHSP83 revealed that the amino acid sequence of the NBD was 68% identical, MD 69% identical, and CTD 60% identical (Figure 2). Conversely, the yeast HSP90 proteins (HSP82 and HSC82) were 97% identical in sequence (a difference of 16 amino acid residues) and yet the two proteins exhibit differences in stability, function, and chemical sensitivity (Girstmair et al., 2019). Residues D78 and E32 are conserved in humans and T. brucei HSP90 proteins (Figure 2A). Residue D79 (D78 in T. brucei) was previously described to be located deep in the inner region of the ATP-binding pocket of yeast HSP90 and determined to form a hydrogen bond with ATP and together with E33 (E32 in T. brucei) are important for ATP binding (Panaretou et al., 1998). Mutations of these two residues in yeast HSP90 led to a loss of viability (Panaretou et al., 1998). In comparison to humans, TbHSP83 revealed a 50- to 60-fold higher sensitivity to the HSP90 ATPase inhibitor 17-AAG (Jones et al., 2008). The side chain of residue I171 in TbHSP83 was found to be in contact with L33 and indirectly with I34 (Pizarro et al., 2013), the latter two residues have been implicated in radicicol resistance (Prodromou et al., 2012). Small sequence variations in HSP90 appear to result in large variations in chemical sensitivity between hHSP90 and TbHSP83 (Jones et al., 2008; Prodromou et al., 2012; Pizarro et al., 2013).
FIGURE 2.
Sequence alignment and 3D structural analysis of TbHSP90. (A) Multiple sequence alignment of hHSP90, TbbHSP83, and TbgHSP83. The NTD is highlighted in red, the charged linker domain in blue, the MD in gray, the CTD in green, and the MEEVD motif in orange. The PTMs are highlighted: yellow–phosphorylation, black—acetylation, and pink—N-glycosylation. Conserved residues involved in ATP interaction are highlighted in brown. Conservation based on physico-chemical properties is shown as a numerical index at the bottom of the alignment: “*” denotes score of 11 where amino acid residues are identical; “+” denotes score of 10 and indicates all properties are conserved. (B) Predicted 3D structure of the TbHSP83 monomer and (C) superimposed 3D structures of TbHSP83 (green) and hHSP90 (red).
The overall 3D structures of the TbHSP83 and human HSP90 are similar (Figure 2B). The RMDS between the human and Tb structures is 1.71 Å for a Cα superposition of the full-length proteins. The regions of sequence variation reveal themselves more evidently in the flexibility of the protein (Pizarro et al., 2013; López et al., 2021). The major differences that can be seen in the 3D structures are in the charged linker domain and the C-terminal domain (Figure 2B). The orientation of both regions differs to that of the human HSP90 protein. Furthermore, the C-terminal domain has an extension that contains a short helical structure present in TbHSP83 that is absent in hHSP90 (residues 678–682, EEEEA). The CTD is responsible for dimerization and interaction with co-chaperones, and this may result in the possibility of unique interactors of the TbHSP83 protein. The spatial differences seen in the linker are a direct result of the differences in the length. The hHSP90 has a linker that is seven residues longer than that of TbHSP83 and has a higher overall negative formal charge (Figure 2A). The variation in length of the linker has been noted across different species, the varying length could affect flexibility, and the varying charge could affect transient domain interactions that exist between the NTD and the linker (Jahn et al., 2014; López et al., 2021).
Post-translational modifications, and particularly phosphorylation of tyrosine, serine, and threonine residues, at multiple sites of cytosolic HSP90 is a well-known chaperone activity modulator mechanism in many organisms (Miyata and Yahara, 1992; Mollapour et al., 2011; Mollapour and Neckers, 2012; Hombach-Barrigah et al., 2019), HSP90 steady-state phosphorylation is species-specific relative to the different cellular environments (Mollapour and Neckers, 2012). Two phosphorylation sites, S53 and S286, were found to be unique to T. brucei HSP83 and conserved in all 10 cytosolic HSP83 proteins. The phosphorylation sites T211, T216, and S597 were conserved in all analyzed trypanomastids in this study, while S374 and S698 were found to be conserved in all proteins including humans (Supplementary Figure S2). The same phospho-modified residues were previously described for the cytosolic HSP83 ortholog from L. donovani (Hombach-Barrigah et al., 2019). Silencing mutations of L. donovani HSP90 T211 and T216 reduced parasite growth, while mutation of S594 reduced growth and infectivity (Hombach-Barrigah et al., 2019). The phosphorylation of L. donovani HSP90 at T21 plays a role in the binding of co-chaperones, and mutation of this residue irreversibly inhibited the growth of the promastigote stage (Hombach-Barrigah et al., 2019); however, this residue has not been detected as a T. brucei phospho-site (Cunningham et al., 2008; Mollapour et al., 2011; Hombach-Barrigah et al., 2019). The equivalent site in yeast HSP90 (T22) was found to be essential for dimerization and ATPase activity (Cunningham et al., 2008). Acetylation and ubiquitination sites were also predicted and mapped. The predicted N-glycosylation sites, N90, N372, and N612 were conserved in all cytosolic HSP90s analyzed in this study, while N51 was determined to be specific to T. brucei HSP83 (Supplementary Figure S2). Two ubiquitination sites identified in T. brucei HSP83 as K394 and K560 were found conserved in all analyzed cytosolic HSP90 isoforms in this study (Supplementary Figure S2).
3.3 TRAP-1
The mitochondrial isoform of the HSP90/HSPC family was first identified in association with the mammalian tumor necrosis factor 1 (TNF-1) protein, hence termed TRAP-1 (Song et al., 1995). It was promptly suggested as a member of the 90-kDa molecular chaperone family due to strong homology with other HSP90 members (Song et al., 1995). Since then, TRAP-1/HSPC5 orthologues have been identified in a variety of eukaryotic and prokaryotic organisms and was also recently referred to as HSP84 in T. brucei (Meyer and Shapiro, 2021). RNAi knockdown of HSP84 showed growth defects and led to loss of kinetoplasts in bloodstream form trypanosomes (Meyer and Shapiro, 2021). Our study identified a single entry for a putative TRAP-1 gene annotated in the genomes of both T b. brucei (Tb927.11.2650) and T b. gambiense (Tbg972.11.2900) (Table 1). The selected kinetoplastids in this study were also identified to encode a single copy of TRAP-1 (Table 1), which are consistent with previous studies (Folgueira and Requena, 2007), except for T. cruzi, which was previously stated to encode for two TRAP-1 orthologues (Folgueira and Requena, 2007; Shonhai et al., 2011). Phylogenetic analysis indicates a general conservation in trypanosomatid TRAP-1 proteins (Supplementary Figure S4), though little experimental characterization of these genes has been conducted in kinetoplastids. It is predicted that the cellular role of the trypanosomatid TRAP-1 proteins will be orthologous to human TRAP-1, whose major functions is to maintain mitochondrial integrity, modulate mitochondrial metabolism, and protect against mitochondrial apoptosis (Altieri et al., 2012). Furthermore, human TRAP-1 counteracts protein aggregation inside the mitochondria and supports protein folding (Siegelin et al., 2011), leading to healthy, intact mitochondria.
Mammalian TRAP-1 orthologues are localized predominantly in the mitochondrial matrix, where it exists as at least six different protein variants, resulting from splicing patterns, amino acid additions and/or deletions (Cechetto and Gupta, 2000; Felts et al., 2000). The translation of the main TRAP-1 mRNA generates a precursor protein of 704 amino acids that contains a putative 59-amino acid, N-terminal mitochondrial import sequence, which is removed upon organelle import (Felts et al., 2000; Schleiff and Becker, 2011). It was predicted that both TbbTRAP-1 and TbgTRAP-1 localize in the mitochondria, as the proteins possess a positively charged N-terminal leader sequence (Supplementary Figure S1). Proteomic and localization studies confirmed that TbbTRAP-1 localizes to the mitochondria (Panigrahi et al., 2009; Dean et al., 2017), but interestingly the protein is also present in the flagella of T b. brucei BSF parasites (Subota et al., 2014) (Table 1). The subcellular distribution of TbbTRAP-1 during the parasite’s life cycle could be related to the shape and functional plasticity of the T. brucei single mitochondrion, which undergoes profound alterations to adapt to the different host environments (Osellame et al., 2012). Phenotypic knockdown of TbbTRAP-1 had a detrimental effect on the survival and fitness of the parasite at the procyclic stage of its life cycle and negatively affected parasite differentiation (Alsford et al., 2011). Thus, T. brucei TRAP-1 proteins may be an important modulator of mitochondrial bioenergetics at the procyclic stage, as well as play an integral role in parasite pathogenesis.
The phosphorylation sites, S363 and S453, were conserved in the middle domain of TRAP-1 for all trypanosome proteins, while S439 was conserved in TRAP-1 for all proteins studied including humans (Supplementary Figure S2). Several amino acids were identified as potential targets for post-translational modifications in human TRAP-1, yet the phosphorylation mechanism remains to be revealed (Altieri et al., 2012). Acetylation sites found specific for TbbTRAP-1 include K109, K480, and K601 (Supplementary Figure S2). Most of the PTMs of HSP90 and other inferences stated are yet to be verified experimentally.
3.4 GRP94
The glucose-regulated 94 kDa protein (GRP94) is a HSP90 family member residing in the lumen of the endoplasmic reticulum (ER) (Argon and Simen, 1999), where it is involved in the maturation of membrane-resident and secreted protein clients (Marzec et al., 2012). GRP94 is present as a single gene in all metazoa, although the gene is not found in many unicellular organisms such as bacteria, archaea, yeast, and most fungi (Marzec et al., 2012). This study identified a single putative entry for the GRP94 gene in both T. brucei subspecies and the selected trypanosomatids in this study (Table 1). These findings are consistent with previous findings for T. brucei and L. major (Folgueira and Requena, 2007), though previous reports indicated that T. cruzi CL Brener Esmeraldo-like strain encodes three GRP94 orthologues (Folgueira and Requena, 2007; Shonhai et al., 2011). However, this study identified that only one GRP94 gene in T. cruzi CL Brener Esmeraldo-like strain (TcCLB.506,989.190) was found to encode for a full-length sequence. The genome of this T. cruzi strain needs to be further investigated to determine if these partial sequences of the GRP94 genes (TcCLB.506591.4 and TcCLB.503811.10) are due to sequencing errors.
Both TbbGRP94 and TbgGRP94 genes are present on chromosome III and are shown to encode for proteins considerably longer in amino acid sequence when compared to HSP83 (Supplementary Figure S1), which is characteristic of GRP94 protein members (Stechmann and Cavalier-Smith, 2003; Johnson, 2012). GRP94 proteins share structural similarity with cytosolic HSP90 proteins, though the N-terminus contains an ER signal peptide while the C-terminal MEEVD peptide is replaced with the KDEL motif that is required for retention in the ER (Argon and Simen, 1999). Sequence analysis of TbbGRP94 and TbgGRP94 indicates that the GRP94 protein shares domain architecture with typical GRP94 proteins including the possession of an N-terminal ER signal peptide (Supplementary Figure S1). However, a variation in the C-terminal ER retention motif, KDEL, is observed in all the trypanosomatid orthologues of GRP94; AGDL in Trypanosoma spp., KEEL in B. saltans, and EGDL in C. fasciculata and all Leishmania spp (Supplementary Figure S1). Transcriptomic and proteomic studies revealed that TbbGRP94 is expressed at all life cycles and throughout the phases of the cell cycle (Supplementary Table S2). Proteomic studies confirm the presence of GRP94 in flagella and cell surface (Subota et al., 2014; Shimogawa et al., 2015).
In trypanosomatids, the first recognized and characterized GRP94 gene was in Leishmania infantum (L. infantum). The GRP94 ortholog in L. infantum was shown to localize in the ER and share many of the activities of GRP94s of other eukaryotes (Descoteaux, 2002). Unlike GRP94 in mammalian cells, LinGRP94 is not essential for cell viability, and LinGRP94 mRNA is induced developmentally rather than by canonical GRP94-inducing stresses (Descoteaux, 2002). The protein was highly immunogenic during Leishmania infection (Larreta et al., 2000, 2002), and essential for lipophosphoglycan (LPG) assembly (Descoteaux, 2002), an abundant surface glycolipid of Leishmania promastigotes that is critical to parasite virulence (Yao et al., 2003). Effectively, the critical role of GRP94 in Leishmania appears to be adapted to the synthesis of glycoconjugates and directing the host immune response implicating a pivotal role in parasite virulence (Descoteaux, 2002). Though whether this specialized role is conserved in T. brucei and other trypanosomatids will need to be elucidated. The function and cellular roles of TbGRP94 should be explored, given the immunogenic and antigenic properties shown by the L. infantum GRP94, as this protein could constitute a valuable molecule for diagnostic purposes, and quite possibly a potential candidate for studies of protective immunogenicity. N-glycosylation sites, N137, N370, and N639, were conserved across all species studies (Supplementary Figure S2). GRP94 phosphorylation sites, S63 and S372, were conserved for all species analyzed, while S625 was conserved in T. brucei and T. cruzi (Supplementary Figure S2). K472 and K504 acetylation sites were conserved in all the trypanosome species, while K515, K542, R587, and Q646 were unique to T. brucei (Supplementary Figure S2).
3.5 The T. brucei HSP90 co-chaperone system
In all organisms, HSP90 is a dynamic protein that undergoes a conformational cycle that is directionally determined, in large part by ATP binding/hydrolysis, and a cohort of proteins termed co-chaperones (Panaretou et al., 1998; Prodromou, 1999; Johnson and Brown, 2009). The HSP90 co-chaperone system in intracellular protozoan parasites has been explored in previous studies (Seraphim et al., 2013; Figueras et al., 2014). Thus, using the human and trypanosomatid systems, this study analyzed the composition of the T. brucei HSP83 co-chaperone system. It was determined in this study that T. brucei possesses a similar number of co-chaperones compared to humans (Table 2), with one notable absence being cell division cycle 37 (Cdc37). The absence of a gene encoding for Cdc37 has also been noted in several intracellular protozoan parasites (Chua et al., 2014; Figueras et al., 2014; Tatu and Neckers, 2014; Hombach-Barrigah et al., 2019) and was not evident in 10 out of 19 divergent eukaryotic species examined in a study by Johnson and Brown (2009). Cdc37 is a co-chaperone that has a specialized and indispensable role in the maturation and/or stabilization of a large subset of protein kinases in mammalian cells (Smith and Workman, 2009). The absence of Cdc37 in some species is that clients that are dependent on a specific co-chaperone in one species may not require HSP90 for function in other species, thus the protein kinases in protozoan parasites may have evolved in such a way that the proteins bind a different co-chaperone or are independent of HSP90 for function. Since little is known about why a protein becomes dependent on HSP90 for activity or stability, it poses interesting questions on the mechanism by which the maturation and regulation of protein kinases in protozoan parasite is mediated dependent or independent of HSP83. Exploration of this mechanism may provide a potential avenue for chemotherapeutics since protein kinases are also an attractive drug target in infectious disease, such as African trypanosomiasis. The identified HSP83 co-chaperones in both T. brucei subspecies are listed in Table 2. In addition, the HSP90 co-chaperones were categorized in this study based on the presence of a TPR or CS (CHORD and SGT1) domain.
TABLE 2.
HSP83/HSPC co-chaperones from Trypanosoma brucei with their putative orthologues in T. cruzi, L. major, C. fasciculata, B. saltans, and H. sapiens.
| H. sapiens | H. sapiens | T. brucei | T. cruzi 3 | L. major | C. fasciculata | B. saltans | |||
|---|---|---|---|---|---|---|---|---|---|
| Name | Gene ID1 | Gene ID1 | Gene ID1 | Gene ID1 | Gene ID1 | Gene ID1 | Localization2 | Reference | |
| A: TPR-containing Hsp90 co-chaperones | |||||||||
| STi1/HOP | 10963 | Tb927.5.2940 | Tc_MARK_9009 | LmjF08.1110 | CFAC1_020023900 | BSAL_57725 | CYTO | Gunasekera et al. (2012); Urbaniak et al. (2012); Butter et al. (2013); Shimogawa et al. (2015); Dean et al. (2017) | |
| NUC | |||||||||
| Tbg972.5.4130 | C4B63_59g115 | CELL SURFACE (BSF, PF) | |||||||
| PP5 | 5536 | Tb927.10.13670 Tbg972.10.16800 | TcCLB.507.993.190 | LmjF.18.0150 | CFAC1_140007400 | BSAL_15705 | CYTO (BSF, PF) | Gunasekera et al. (2012); Urbaniak et al. (2012); Butter et al. (2013); Dean et al. (2017) | |
| C4B63_4g368 | |||||||||
| Cyp40 | 5481 | Tb927.9.9780 Tbg972.9.5630 | TcCLB.506,885.400 Tc_MARK_4311 | LmjF.35.4770 | CFAC1_300099000 | BSAL_06490 | CYTO | Oberholzer et al. (2011); Dean et al. (2017) | |
| C4B63_2g294 | FLAGELLAR (BSF) | ||||||||
| DnaJC7 | 7266 | Tb927.10.4900 | TcCLB.504.203.60 | LmjF.36.0500 | CFAC1_250012000 | BSAL_30720 | CYTO | Urbaniak et al. (2012); Butter et al. (2013); Dean et al. (2017) | |
| Tc_MARK_8493 | |||||||||
| Tbg972.10.5950 | C4B63_13g112 | NUC (BSF, PF) | |||||||
| FKBP5 | 2289 | Tb927.10.16100 Tbg972.10.19710 | TcCLB.511.353.10 Tc_MARK_4665 | LmjF.19.1530 | CFAC1_210025000 | BSAL_03610 BSAL_65235 | CYTO | Gunasekera et al. (2012); Urbaniak et al. (2012); Butter et al. (2013); Subota et al. (2014); Shimogawa et al. (2015); Dean et al. (2017) | |
| C4B63_157g28 C4B63_171g30 | FLAGELLAR (BSF, PF) | ||||||||
| SGT | 6449 | Tb927.6.4000 Tbg972.6.3780 | TcCLB.511.737.10 Tc_MARK_2022 | LmjF.30.2740 | CFAC1_260051600 | BSAL_66445 | CYTO | Gunasekera et al. (2012); Urbaniak et al. (2012); Butter et al. (2013); Subota et al. (2014); Shimogawa et al. (2015); Dean et al. (2017) | |
| FLAGELLAR | |||||||||
| C4B63_18g260 | CELL SURFACE (BSF, PF) | ||||||||
| TPR domain protein | 7268 | Tb927.10.11380 | TcCLB.507.709.70 | LmjF.33.0700 | CFAC1_280016100 | CYTO | Urbaniak et al. (2012); Butter et al. (2013); Dean et al. (2017) | ||
| Tbg972.10.13740 | Tc_MARK_3,620 | ||||||||
| C4B63_84g34 | |||||||||
| B: CS-containing Hsp90 co-chaperones | |||||||||
| p23 | 10728 | Tb927.9.10230 Tb927.10.2620 Tbg972.9.5930 Tbg972.10.3260 | TcCLB.509.551.70 TcCLB.506.407.60 | LmjF.35.4470 LmjF.34.0210 | CFAC1_300096200 CFAC1_290030000 | BSAL_38665 | CYTO | Dean et al. (2017) | |
| FLAGELLAR | |||||||||
| C4B63_2g235 C4B63_47g40 | NUC | ||||||||
| CHORD | 26548 | Tb927.1.3170 | TcCLB.507.837.40 | LmjF.20.1620 | CFAC1_170031900 | BSAL_00100 | CYTO (BSF, PF) | Gunasekera et al. (2012); Urbaniak et al. (2012); Butter et al. (2013); Dean et al. (2017) | |
| Tc_MARK_7,282 | |||||||||
| Tbg972.1.1930 | C4B63_93g75 | ||||||||
| Aarsd1 | 80755 | Tb927.9.6650 | TcCLB.504.883.50 | LmjF.15.0690 | CFAC1_240038900 | BSAL_93700 | CYTO | Dean et al. (2017) | |
| Tc_MARK_5844 | |||||||||
| Tbg972.9.3510 | C4B63_136g36 | ||||||||
| PIH1D1 | 55011 | Tb927.9.10490 Tbg972.9.6100 | TcCLB.506.147.150 | LmjF.35.4330 | CFAC1_300094700 | CYTO | Dean et al. (2017) | ||
| Tc_MARK_4354 | |||||||||
| C4B63_80g31 | |||||||||
| PIH1D3 | 139212 | Tb927.3.4410 Tbg972.3.4850 | TcCLB.507.257.160 | LmjF.29.1850 | CFAC1_190042200 | BSAL_07820 | CYTO (BSF, PF) | Urbaniak et al. (2012); Butter et al. (2013); Dean et al. (2017) | |
| Tc_MARK_3,141 | |||||||||
| C4B63_10g277 | BSAL_56185 | ||||||||
| Ubiquitin hydrolase | 10869 | Tb927.9.13430 Tbg972.9.8400 | TcCLB.510.749.40 | LmjF.35.2410 | CFAC1_300073100 | BSAL_27670 | CYTO | Dean et al. (2017) | |
| Tc_MARK_4527 | |||||||||
| C4B63_2g83 | |||||||||
| CS domain/TPR repeat | 161582 | Tb927.11.16050 Tbg972.11.17990 | TcCLB.509.073.90 | LmjF.32.2850 | CFAC1_190042200 | BSAL_62790 | CYTO | Dean et al. (2017) | |
| Tc_MARK_3396 | |||||||||
| C4B63_2g215 | |||||||||
| NADH-cytochrome B5 reductase | 51167 | Tb927.11.3750 Tbg972.11.4290 | TcCLB.511.047.40 | LmjF.13.1060 | CFAC1_220039400 | BSAL_93355 | CYTO | Dean et al. (2017) | |
| Tc_MARK_6965 | |||||||||
| C4B63_46g107 | |||||||||
| LRRC | 23639 | Tb927.3.4770 Tbg972.3.5330 | TcCLB.503.671.30 | LmjF.29.2210 | CFAC1_200030000 | BSAL_68670 | CYTO | Butter et al. (2013); Dean et al. (2017) | |
| Tc_MARK_3,178 | |||||||||
| C4B63_23g85 | |||||||||
| C: Non-TPR, non-CS-containing Hsp90 co-chaperones | |||||||||
| Aha1 | 10598 | Tb927.10.13710 Tbg972.10.16840 | TcCLB.507.993.150 | LmjF.18.0210 | CFAC1_140008400 | BSAL_15670 | CYTO | Gunasekera et al. (2012); Urbaniak et al. (2012); Butter et al. (2013); Dean et al. (2017) | |
| Tc_MARK_4860 | |||||||||
| C4B63_4g357 | NUC (BSF, PF) | ||||||||
| Phosphoducin | 79031 | Tb927.11.4930 | TcCLB.507.669.130 | LmjF.24.0170 | CFAC1_21006700 | BSAL_83285 | CYTO | Dean et al. (2017) | |
| Tc_MARK_6540 | |||||||||
| Tbg972.11.5550 | C4B63_28g45 | ||||||||
The Gene IDs for the T b. brucei (Tb refers to Tbb), T b. gambiense, T. cruzi, C. fasciculata, B. saltans, and L. major HSP83/HSPC co-chaperones were retrieved from the TriTrypDB database (http://tritrypdb.org/tritrypdb/; Aslett et al., 2010). The Gene IDs for the members of the H. sapiens HSP90/HSPC co-chaperones were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/).
The Gene IDs, for the orthologues, identified by reciprocal BLASTP analysis, of three strains of T. cruzi are listed. T. cruzi CL, Brener Esmeraldo-like (TcCLB), T. cruzi Dm28c 2018 (C4B63), and T. cruzi marinkelli strain B7 (Tc_MARK).
Subcellular localizations for the T. brucei HSP83/HSPC co-chaperone proteins were acquired from using the TrypTag database (http://tryptag.org/; Dean et al., 2017) and/or determined using various proteomic datasets listed in the materials and methods.
CYT-Cytosol; MITO- mitochondrion; NUC- nucleus; ER- endoplasmic reticulum; GYLCO-glycosomes; FLAGELLAR- flagellar; CELL SURFACE- cell surface.
3.6 TPR-containing HSP83 co-chaperones
Seven putative TPR-containing co-chaperones were identified in this study.
3.6.1 Stress-inducible protein 1 (STI1)
Stress-inducible protein 1 (STI1), also known as HSP70/HSP90-organizing protein (HOP or STIP1) in mammals, is one of the best studied co-chaperones in the HSP90 reaction cycle (Chang et al., 1997; Johnson et al., 1998) as it acts as an adapter protein, mediating the interaction between HSP70 and HSP90 through its TPR domains (Brinker et al., 2002; Odunuga et al., 2003; Baindur-Hudson et al., 2015). STI1/HOP is a widely conserved HSP90 co-chaperone and has been annotated and characterized across diverse organisms including several kinetoplastid protists. Initially thought to be an indispensable protein, recent discoveries in yeast and some eukaryotes show that direct interaction can take place in vitro between HSP70 and HSP90 in the absence of HOP (Kravats et al., 2018; Bhattacharya et al., 2020). A single STI1/HOP gene was found encoded in both T. brucei subspecies (Table 2). Nine TPR motifs arranged into three TPR domains (TPR1, TPR2A, and TPR2B) in addition to two domains rich in proline and aspartic acid (DP1 and DP2) were predicted (Scheufler et al., 2000; Nelson et al., 2003). Both STI1/HOP orthologues in T. cruzi and L. major were found to immunoprecipitate with HSP83 and HSP70 and co-localize with these chaperones in the cytoplasm and/or around nucleus (Webb et al., 1997; Schmidt et al., 2011). The expression of HOP isoforms was increased in response to different environmental stresses (Webb et al., 1997; Schmidt et al., 2011), with LmjHOP being upregulated when the parasites are exposed under heat stress conditions (Webb et al., 1997), whereas only nutritional stress-induced expression of TcSTI1 in the late growth phase of epimastigotes (Schmidt et al., 2011). The HSP90-STI1 complex in L. major and T. cruzi has been shown to be pivotal to parasite differentiation (Webb et al., 1997; Hombach et al., 2013). Proteomic analysis in T. brucei indicates that TbbSTI1 is part of the cell surface (PF) proteome during the procyclic stage (Shimogawa et al., 2015). Though TbbSti1 is present in both the BSF and PF stages of the parasite, it was more highly expressed in the bloodstream form (Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013). These data suggest that the STI1 ortholog in both T. brucei subspecies should function as an adapter protein for TbHSP83 and TbHSP70s, participating in the foldosome apparatus necessary for maintaining proteostasis, cytoprotection, and modulating parasite differentiation.
3.6.2 Protein phosphatase 5 (PP5)
Protein phosphatase 5 (PP5) is a member of the PPP family of serine/threonine protein phosphatases and it associates with HSP90 in complexes during client protein maturation (Cohen, 1997; Chinkers, 2001; Golden et al., 2008). PP5 is characteristically unique from other PPP family members, in which it possesses an N-terminal TPR domain (Borthwick et al., 2001), which mediates interaction with HSP90 (Chen et al., 1996). This interaction enables PP5 to modify the phosphorylation status of HSP90 client proteins (Golden et al., 2008). The gene for PP5 in T b. brucei (TbbPP5) has been extensively studied. TbbPP5 encodes a ∼52-kDa protein that possesses the canonical N-terminal TPR domain and phosphatase catalytic domain (Anderson et al., 2006). TbbPP5 interacted with TbbHSP90 in vivo and co-localized with the chaperone in the cytosol of PRO parasites (Jones et al., 2008). Both TbbPP5 and TbbHSP90, upon heat shock and geldanamycin treatment, accumulated in the nucleus (Jones et al., 2008), indicating that both TbbPP5 and TbbHSP90 translocate to the nucleus when the parasites are exposed to proteotoxic stresses (Jones et al., 2008). TbbPP5 was detected in both BSF and PF stages of the parasite but upregulated in the procyclic form (Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013). Overexpression of TbbPP5 was found to partially negate the effect of geldanamycin treatment on cell growth, which indicates that the co-chaperone enhances the chaperoning function of TbbHSP90 and promotes the folding and maturation process of important regulatory molecules, which facilitate cell growth.
3.6.3 Peptidyl-prolyl cis–trans-isomerases (PPIases)
The immunophilin superfamily consists of highly conserved proteins with rotamase or peptidyl-prolyl cis–trans-isomerase (PPIase) activity that accelerates protein folding by mediating the isomerization of X-Pro-peptide bonds (Galat, 2003; Pratt et al., 2004). The best characterized PPIases belong to two families, the cyclophilin-type (Cyp) and the FKB-506 drug-binding protein type (FKBP) (Steiner and Haughey, 2010). Data mining of the T. brucei genome identified that Cyp40 and a putative FKB-506 binding like protein (FKBPL) are present in the extracellular parasite proteome (Table 2). Investigation of the domain structure and sequence conservation indicate that both Cyp40 and FKBPL in T. brucei were shown to display the characteristic two-domain structure of a N-terminal PPIase domain and a C-terminal TPR domain (data not shown). Though it must be noted that the C-terminal TPR domain in kinetoplastid Cyp40 underwent substantial evolutionary modification (Yau et al., 2010), thus potentially impacting Cyp40-HSP83 interactions. Future structure/function studies should explore the effect these modifications have on the isomerase and chaperone activities of the protein in comparison to its human counterpart.
Studies conducted on the Cyp40 ortholog in L. donovani have revealed that the protein functions in Leishmania stage-specific morphogenesis, motility, and the development of infectious-stage parasites (Yau et al., 2010, 2014). The study conducted by Yau et al. (2014) also suggested that LdCyp40 and LdFKBP2 functions in regulating Leishmania cytoskeletal dynamics. Given the capacity of Cyp40 and FKBP52 to compete for molecular partners (Ratajczak et al., 2003), LdCyp40 may interact with microtubules to promote tubulin polymerization as a means of counteracting LdFKBP52-mediated depolymerization. RNAi-mediated knockdown of both Cyp40 and FKBPL in T b. brucei parasites demonstrated that these proteins are essential at the BSF stage and are required for parasite differentiation (Alsford et al., 2011; Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013). Proteomic data predicted these proteins to reside in the cytosol and flagellar (Oberholzer et al., 2011; Subota et al., 2014). Together these data indicate that T. brucei Cyp40 and FKBPL may play essential roles in morphogenesis, motility, and the development of infectious-stage parasites.
3.6.4 J-protein 52
The J-protein family is a major subset of co-chaperones for the HSP70 chaperone machinery, and they are broadly classified into four subtypes (I–IV). The J-protein family from T. brucei has been explored previously (Bentley et al., 2019). It was shown in that study that J52 is one of six type III J proteins in T. brucei that possesses the TPR domain (others are J42, J51, J52, J53, J65, and J67) (Bentley et al., 2019). J52 is predicted to reside in the cytosol together with J51 and J42 (Bentley et al., 2019). DnaJC7/Tpr2, the human ortholog of J52 was first identified as a cytosolic protein via a two-hybrid screen for interaction with a GAP-related segment (GRD) of neurofibromin. It was reported to encode seven TPR units and possess a domain of high similarity to the DnaJ family (Murthy et al., 1996). DnaJC7 also regulates the multichaperone system involving HSP70 and HSP90 but in a nucleotide-independent manner with HSP90. DnaJC7 is predominantly thought to be involved in retrograde transport of client proteins from HSP90 to HSP70 (Brychzy et al., 2003; Moffatt et al., 2008). Proteomic analysis showed J52 to be upregulated in the procyclic form of the parasite (Urbaniak et al., 2012; Butter et al., 2013).
3.6.5 Small glutamine-rich TPR-containing protein (SGT)
The small glutamine-rich TPR-containing protein (SGT) is a co-chaperone involved in a specific branch of the global cellular quality control network that determines the fate of secretory and membrane proteins that mislocalize to the cytosol (Leznicki and High, 2012; Wunderley et al., 2014). Human SGT is a modular protein characterized by three characteristic sequence motifs, namely, an N-terminal dimerization domain, central TPR domain, and a glutamine-rich region at the C terminus (Roberts et al., 2015). The SGT orthologues in trypanosomatids are atypical (Table 2), as these proteins all lack the characteristic glutamine-rich region and contain a substituted region with charged amino acid residues (Ommen et al., 2010). Proteomic analysis identified TbbSGT to be upregulated in the procyclic form of the parasite and was found in the flagellar and cell surface proteome (Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013; Subota et al., 2014; Shimogawa et al., 2015). The SGT ortholog in L. donovani is an essential protein for L. donovani promastigote growth and viability (Ommen et al., 2010). LdSGT was shown to form large, stable complexes that included HSP83, HSP70, HIP, HOP, J-proteins, and HSP100 (Ommen et al., 2010), whereas recombinant L. braziliensis SGT was shown to interact with both LbHSP90 and HsHSP70-1A (Coto et al., 2018). Therefore, the orthologous proteins in T b. brucei and T b. gambiense may have developed the same activity and assist in the formation of the T. brucei HSP83 chaperone system. Though future studies should be conducted to elucidate SGT-HSP70/HSP83 interaction in T. brucei.
3.6.6 Tetratricopeptide repeat protein 4 (TTC4)
The co-chaperone TTC4 is the Tetratricopeptide repeat protein 4, a member of the TPR family that has been isolated and characterized in humans and is implicated in the pathogenesis of skin melanomas (Su et al., 1999; Poetsch et al., 2000). An ortholog of TTC4 has been characterized in Drosophila (Pit47) with both proteins shown to be nucleoplasmic; both contain three TPR motifs and are abundant in proliferating tissue (Crevel et al., 2008). A putative ortholog of TTC4 was found in T. brucei and other organisms in this study except in B. saltans. Proteomic analysis identified T. brucei TTC4 to be upregulated in the procyclic form of the parasite (Urbaniak et al., 2012; Butter et al., 2013).
3.7 CS-containing HSP83 co-chaperones
Nine putative CS-containing co-chaperones were identified in this study.
3.7.1 p23
The co-chaperone p23 is a small acidic protein that binds the HSP90 NBD to stabilize the closed conformation of HSP90, inhibiting ATPase activity and preventing client protein release from the complex (Young, 2000; McLaughlin et al., 2006). In addition to its HSP90 co-chaperone function, p23 has its own chaperoning activity in vitro and can suppress the aggregation of denatured proteins (Bose et al., 1996; Freeman et al., 1996). In silico analysis of the genomes of both T. brucei subspecies revealed that the parasite possesses two evolutionarily divergent p23 orthologues, and subsequently these orthologous proteins were named p23a and p23b (Table 2). The possession of two putative p23 proteins was found to be conserved in all the trypanosomatids in this study except B. saltans (Table 2). The Tbp23a and Tbp23b proteins share 28% identity to each other and share 33 and 26% identity, respectively, to human p23. In addition, RNAi knockdown of these proteins showed that each p23 protein is essential to parasite viability at specific stages of the life cycle (Alsford et al., 2011). The orthologues of these proteins have been explored in two Leishmania spp. (Batista et al., 2015). Both proteins in L. braziliensis possessed intrinsic chaperone activity, but they have different client protein specificities; they also inhibit LbrHSP83 ATPase activity to different extents (Batista et al., 2015). Such functional differences might be important in both HSP90 regulation and in their interactions with client proteins during the life stage transformations of kinetoplastid parasites. However, to support these assertions, more functional and in vivo studies of trypanosomatid p23a and p23b proteins are needed.
3.7.2 The cysteine- and histidine-rich domain-containing protein (CHORD)
The cysteine- and histidine-rich domain-containing protein (CHORD) is characterized by six cysteine and three histidine residues as well as a C-terminal CS domain as the characteristic domains of the CHORD-containing proteins (Wu et al., 2005). In humans there are two CHORD domains, CHORD-I was found to be dispensable toward the HSP90 interaction while CHORD-II is essential (Wu et al., 2005). CHORD was identified as an ADP-dependent HSP90 co-chaperone in humans as its interaction was shown to be stimulated by high ADP:ATP ratio in cell culture lysates (Gano and Simon, 2010). Data mining identified a single CHORD gene in T. brucei genome, and all other organisms were studied, and the CHORD protein was found upregulated in procyclic form of T. brucei parasite (Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013).
3.7.3 Alanyl-tRNA synthetase domain-containing 1 name (Aarsd1)
The mammalian Aarsd1 gene is a complex gene with large number of exons. The gene gained its name—alanyl-tRNA synthetase domain-containing 1 name (Aarsd1) from the shared homology of its 3′ exons to the editing domain of tRNA synthetases (Echeverría et al., 2016). As a co-chaperone with 44% identity to p23 in its CS domain, it is primarily expressed in the heart and skeletal issues and competes with p23 for binding to HSP90 (Taipale et al., 2014; Echeverría et al., 2016). Aarsd1 has previously been identified in T. brucei with its involvement in preventing misaminoacylation (Beebe, 2003; Cestari et al., 2013). Data mining revealed a single Aarsd1 gene in all the organisms studied.
3.7.4 Protein interacting with HSP90 domain-containing protein 1 (PIH1D1/PIH1)
Protein interacting with HSP90 domain-containing protein 1 (PIH1D1/PIH1) also called Nop17 (Zhao et al., 2008) is involved in pre-RNA processing (Gonzales et al., 2005) and functions as an adapter protein that aids in recruiting clients (Henri et al., 2018). PIH1s a component of the R2TP (RUVBL1-RUVBL2-RPAP3-PIH1D1) complex, which has been found to be conserved in many species including yeast and humans (Henri et al., 2018; Martino et al., 2018). The human PIH1 contains an N-terminal domain with which it binds phosphorylated substrates and a C-terminal CS domain to bind other substrates of the R2TP complex (Hořejší et al., 2014). Data mining revealed the ortholog of PIH1 in T. cruzi and L. major and is the putative pre-RNA processing protein/Nop17 but the ortholog in T. brucei is alternatively named a component of motile flagella 56 (CMF56). This protein is absent in B. saltans.
3.7.5 PIH1D3
PIH1D3 in humans participates in axonemal dynein assembly in the testis and the respiratory system and mutations in PIH1D3 have been shown to be a prominent cause of primary ciliary dyskinesia (Olcese et al., 2017). The ortholog of PIH1D3 in all organisms studied is the pre-RNA processing protein/NOP17. Proteomic analysis showed the presence of the PIH1D3 protein in both the bloodstream and procyclic forms of the parasite.
3.7.6 NADH cytochrome B5 reductase 4 (Ncb5or)
Ncb5or is a soluble flavohemoprotein with an N-terminal cytochrome b5-like domain and a C-terminal cytochrome b5 reductase domain (Zhu et al., 2004); it is present in a wide range of tissues in humans including some cancerous cell lines and supposedly functions as an oxygen sensor (Zhu et al., 1999). It contains the CS motif similar to p23 with which it mediates protein–protein interactions (Garcia-Ranea et al., 2002). Orthologues of Ncb5or are present in all organisms studied.
3.7.7 Leucine-rich repeat containing protein (LRRC)
Leucine-rich repeat containing proteins in eukaryotes share functional links with the co-chaperone SGT and together they are involved in the HSP90 chaperone machinery complex activation (Stuttmann et al., 2008). An ortholog of the LRRC protein was found in all organisms studied and it was upregulated in the procyclic form of T. brucei parasites (Butter et al., 2013).
3.7.8 Ubiquitin carboxyl-terminal hydrolase (Usp)
Ubiquitin carboxyl-terminal hydrolase 19 (Usp19) in humans has been implicated in various cancers and as a prognostic biomarker in renal cell carcinoma therapy (Shahriyari et al., 2019; Hu et al., 2020). A putative ortholog for ubiquitin carboxyl-terminal hydrolase was found in all organisms studied.
3.7.9 Dyslexia susceptibility 1 candidate gene 1 protein (DYX1C1)
Dyslexia susceptibility 1 candidate gene 1 protein (DYX1C1) in humans has been characterized to possess three TPR domains and is expressed in many tissues including the brain (Taipale et al., 2003). The ortholog in T. brucei is a putative CS domain/TPR repeat protein.
3.8 Non-TPR, non-CS-containing HSP83 co-chaperones
3.8.1 Activator of HSP90 ATPase homolog 1 (Aha1)
Aha1 has been identified as the primary activator of the ATPase activity of HSP90 and it acts independently of the other co-chaperones. Homologs of Aha1 have been identified across species from yeast to mammals; Aha1 binds with both its N- and C-terminal domain to the NBD and MD of HSP90 to facilitate the dimerization of the chaperone (Mayer et al., 2002; Koulov et al., 2010; Retzlaff et al., 2010). Data mining of the T. brucei genome identified that the parasite encodes for a single Aha1 gene (Table 2). The Aha1 ortholog in L. braziliensis (LbrAha1) has been characterized, where it was shown to be a cognate protein that shared several structural and functional properties with the human and yeast orthologues. This suggested similar functional mechanism among these proteins despite the low degree of conservation in the amino acid sequence (Seraphim et al., 2013). Recombinant LbrAha1 stimulated the weak ATPase activity of recombinant LbrHSP83 by around 10-fold, exhibiting a cooperative behavior according to the model that two LbrAha1 molecules can act on one LbHSP83 dimer (Seraphim et al., 2013). Data from proteomic analysis in T. brucei revealed that TbbAha1 is upregulated in the BSF stage of the parasite (Gunasekera et al., 2012; Urbaniak et al., 2012; Butter et al., 2013) as well as being essential to parasite viability at this stage of life cycle (Alsford et al., 2011).
3.8.2 Phosphoducin (Pdc)
Phosphoducins in eukaryotes and other members of the phosphoducin family have been shown to function as chaperones/co-chaperones in the G-protein coupled receptors signal transduction pathways (Savage et al., 2000; Willardson and Howlett, 2007). Data mining revealed an ortholog of phosphoducin was found in all organisms studied.
4 Conclusion
The HSP90 family contains an abundant and essential group of proteins, which are highly conserved and implicated in a myriad of cellular functions. Due to their role in cellular proteostasis, they have been implicated in the pathology of many diseases which warrants their targeting as therapeutics (Samant et al., 2012). In this article, we report an in silico overview of HSP90 and its co-chaperones in both T. b. brucei and T. b. gambiense in relation to human and other trypanosomal species, including non-parasitic Bodo saltans and the insect infecting Crithidia fasciculata. T. b. brucei was found to have 12 putative HSP90 proteins, 10 of which are cytosolic (HSP83). Multiple copies of HSP83 may allow the parasite to reach a high synthesis level of the proteins in an organism that relies on post-transcriptional regulation, and this explains its high levels in the cell even under non-stress conditions (Requena et al., 2015). The expansion of the HSP90 chaperone complement also reiterates its importance in the biology and functioning of these protozoan parasites (Folgueira and Requena, 2007; Shonhai et al., 2011; Urményi et al., 2014). HSP83 was also found in both stages of the parasite but upregulated in the bloodstream form (BSF), this is similar to previous findings of much higher transcripts of HSP83 in bloodstream forms of T. brucei reflecting their temperature induced role of differentiation (Ploeg et al., 1985). The upregulation of HSP83 together with the co-chaperone Sti1 in the BSF may be a further indication of their heat inducibility and involvement in cell defiance, as seen in HSP70 (Urményi et al., 2014).
The protein sequence identities between human and T. brucei HSP90 proteins was the lowest in the linker and C-terminal domains; furthermore, the 3D structure revealed differences in the secondary structure and orientations of both regions. These differences may result in an altered mechanism of interacting with co-chaperones. This study identified 18 co-chaperones in the T. brucei HSP83 chaperone system, which is less than the current number of 50 co-chaperones in the human system, confirming that the HSP90 chaperone machinery is species-specific (Johnson and Brown, 2009; Dean and Johnson, 2021). We predict that additional co-chaperones of T. brucei will be uncovered, some of which will be unique to trypanosomes and possibly T. brucei; and this will provide an interface for targeting chaperone/co-chaperone interactions as potential drug targets. Many of the recently discovered co-chaperones in humans are linked to human diseases including cancer (Dean and Johnson, 2021), and while orthologues have been found in T. brucei their roles remain to be elucidated. Many of these co-chaperones in T. brucei need to be further explored. So far, only the cytosolic HSP90 has been shown to require the function of co-chaperones, the other forms of HSP90 function in the absence of co-chaperones (Richter et al., 2007; Masgras et al., 2017). HSP90 partners with co-chaperones in order to maintain homeostasis; however, these partnerships appear to be dictated by the client protein being chaperoned (Radli and Rüdiger, 2017; Sahasrabudhe et al., 2017). A detailed report for clients in HSP90 is still largely absent (Roy et al., 2012). Previous studies have indicated that inhibitors targeting HSP83 have been shown to cure mice of T. brucei infection, although the toxicity of inhibitors to HSP90 in higher eukaryotes is attributed to a functional loss of client proteins and possible cell cycle arrest (Meyer and Shapiro, 2013). Most of the identified HSP90 client proteins in mammals are kinases (Taipale et al., 2012). Despite the fact that most clients for T. brucei HSP90 have not been identified, over 170 protein kinases (about 30% of the number present in their human host), have been recognized (Parsons et al., 2005; Nett et al., 2009b). In addition to being regulated by co-chaperones, HSP90 is also regulated by various post-translational modifications. Some of these PTM sites have been indicated as potential regulatory sites which affect the binding affinity of inhibitors in PfHSP90 (Pallavi et al., 2010). A number of unique PTM sites were identified in the TbHSP90 proteins and these could be targeted by inhibitors. The T. brucei HSP90, its co-chaperone network, post-translational modifications, and its regulatory mechanisms as well as the subtle structural differences compared to human HSP90 all provide a context for a HSP90-targeted therapy in T. brucei.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization, SB and AB; methodology, SB, MJ, and PM-T; formal analysis, SB, MJ, and PM-T; writing—original draft preparation, SB, MJ, and PM-T; writing—review and editing, SB, MJ, PM-T, AE, and AB; supervision, AE and AB; and funding acquisition, AE and AB. All authors have read and agreed to the published version of the manuscript.
Funding
MJ is a recipient of the 2018 DAAD In-Region Scholarship, PM-T is a recipient of the OWSD Scholarship. ALE is supported by the South African Research Chairs Initiative of the Department of Science and Technology (DST) and the National Research Foundation (NRF) (Grant No 98566), the NRF Competitive Programme for Rated Researchers (Grant No 129262) and Rhodes University (RGG). Open access publication fees were received from Rhodes University (grant no. PP85/2022 and Chair Support Grant - CHAIR03/2020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.947078/full#supplementary-material
References
- Acestor N., Panigrahi A. K., Ogata Y., Anupama A., Stuart K. D. (2009). Protein composition of Trypanosoma brucei mitochondrial membranes. Proteomics 9, 5497–5508. 10.1002/pmic.200900354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acestor N., Zíková A., Dalley R. A., Anupama A., Panigrahi A. K., Stuart K. D. (2011). Trypanosoma brucei mitochondrial respiratome: Composition and organization in procyclic form. Mol. Cell. Proteomics 10, M110006908. 10.1074/mcp.M110.006908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsan M. (2015). The effect of the TseTse fly on african development. Am. Econ. Rev. 105, 382–410. 10.1257/aer.20130604 [DOI] [Google Scholar]
- Alsford S., Turner D. J., Obado S. O., Sanchez-Flores A., Glover L., Berriman M., et al. (2011). High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21, 915–924. 10.1101/gr.115089.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri D. C., Stein G. S., Lian J. B., Languino L. R. (2012). TRAP-1, the mitochondrial Hsp90. Biochim. Biophys. Acta 1823, 767–773. 10.1016/j.bbamcr.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S., Jones C., Saha L., Chaudhuri M. (2006). Functional characterization of the serine/threonine protein phosphatase 5 from Trypanosoma brucei. J. Parasitol. 92, 1152–1161. 10.1645/GE-916R1.1 [DOI] [PubMed] [Google Scholar]
- Aoyagi S., Archer T. K. (2005). Modulating molecular chaperone Hsp90 functions through reversible acetylation. Trends Cell Biol. 15, 565–567. 10.1016/j.tcb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Argon Y., Simen B. B. (1999). GRP94, an ER chaperone with protein and peptide binding properties. Semin. Cell Dev. Biol. 10, 495–505. 10.1006/scdb.1999.0320 [DOI] [PubMed] [Google Scholar]
- Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B. P., Carrington M., et al. (2010). TriTrypDB: A functional genomic resource for the trypanosomatidae. Nucleic Acids Res. 38, D457–D462. 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backe S. J., Sager R. A., Woodford M. R., Makedon A. M., Mollapour M. (2020). Post-translational modifications of Hsp90 and translating the chaperone code. J. Biol. Chem. 295, 11099–11117. 10.1074/jbc.REV120.011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baindur-Hudson S., Edkins A. L., Blatch G. L. (2015). Hsp70/Hsp90 organising protein (hop): Beyond interactions with chaperones and prion proteins. Subcell. Biochem. 78, 69–90. 10.1007/978-3-319-11731-7_3 [DOI] [PubMed] [Google Scholar]
- Baker-Williams A. J., Hashmi F., Budzyński M. A., Woodford M. R., Gleicher S., Himanen S. V., et al. (2019). Co-Chaperones TIMP2 and AHA1 competitively regulate extracellular HSP90:client MMP2 activity and matrix proteolysis. Cell Rep. 28, 1894–1906. e6. 10.1016/j.celrep.2019.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M. P., Croft S. L. (2012). Management of trypanosomiasis and leishmaniasis. Br. Med. Bull. 104, 175–196. 10.1093/bmb/lds031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista F. A. H., Almeida G. S., Seraphim T. V., Silva K. P., Murta S. M. F., Barbosa L. R. S., et al. (2015). Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities. FEBS J. 282, 388–406. 10.1111/febs.13141 [DOI] [PubMed] [Google Scholar]
- Beebe K., Ribas De Pouplana L., Schimmel P. (2003). Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22, 668–675. 10.1093/emboj/cdg065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bente M., Harder S., Wiesgigl M., Heukeshoven J., Gelhaus C., Krause E., et al. (2003). Developmentally induced changes of the proteome in the protozoan parasite Leishmania donovani. PROTEOMICS 3, 1811–1829. 10.1002/pmic.200300462 [DOI] [PubMed] [Google Scholar]
- Bentley S. J., Jamabo M., Boshoff A. (2019). The Hsp70/J-protein machinery of the African trypanosome, Trypanosoma brucei. Cell Stress Chaperones 24, 125–148. 10.1007/s12192-018-0950-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya K., Weidenauer L., Luengo T. M., Pieters E. C., Echeverría P. C., Bernasconi L., et al. (2020). The Hsp70-Hsp90 co-chaperone Hop/Stip1 shifts the proteostatic balance from folding towards degradation. Nat. Commun. 11, 5975. 10.1038/s41467-020-19783-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder R. J. (2014). Functions of heat shock proteins in pathways of the innate and adaptive immune system. J. Immunol. 193, 5765–5771. 10.4049/jimmunol.1401417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G. L., Lässle M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Borthwick E. B., Zeke T., Prescott A. R., Cohen P. T. W. (2001). Nuclear localization of protein phosphatase 5 is dependent on the carboxy-terminal region. FEBS Lett. 491, 279–284. 10.1016/S0014-5793(01)02177-9 [DOI] [PubMed] [Google Scholar]
- Bose S., Weikl T., Bugl H., Buchner J. (1996). Chaperone function of hsp90-associated proteins. Science 274, 1715–1717. 10.1126/science.274.5293.1715 [DOI] [PubMed] [Google Scholar]
- Brandau S., Dresel A., Clos J. (1995). High constitutive levels of heat-shock proteins in human-pathogenic parasites of the genus Leishmania. Biochem. J. 310, 225–232. 10.1042/bj3100225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker A., Scheufler C., von der Mülbe F., Fleckenstein B., Herrmann C., Jung G., et al. (2002). Ligand discrimination by TPR domains: Relevance and selectivity of EEVD-RECOGNITION in Hsp70·Hop·Hsp90 complexes. J. Biol. Chem. 277, 19265–19275. 10.1074/jbc.M109002200 [DOI] [PubMed] [Google Scholar]
- Broadhead R., Dawe H. R., Farr H., Griffiths S., Hart S. R., Portman N., et al. (2006). Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440, 224–227. 10.1038/nature04541 [DOI] [PubMed] [Google Scholar]
- Brun R., Blum J., Chappuis F., Burri C. (2010). Human african trypanosomiasis. Lancet 375, 148–159. 10.1016/S0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- Brychzy A., Rein T., Winklhofer K. F., Hartl F. U., Young J. C., Obermann W. M. J. (2003). Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 22, 3613–3623. 10.1093/emboj/cdg362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner J., Li J. (2013). Structure, function and regulation of the Hsp90 machinery. Biomed. J. 36, 106–117. 10.4103/2319-4170.113230 [DOI] [PubMed] [Google Scholar]
- Büscher P., Cecchi G., Jamonneau V., Priotto G. (2017). Human african trypanosomiasis. Lancet 390, 2397–2409. 10.1016/S0140-6736(17)31510-6 [DOI] [PubMed] [Google Scholar]
- Butter F., Bucerius F., Michel M., Cicova Z., Mann M., Janzen C. J. (2013). Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite’s host adaptation machinery. Mol. Cell. Proteomics 12, 172–179. 10.1074/mcp.M112.019224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cechetto J. D., Gupta R. S. (2000). Immunoelectron microscopy provides evidence that tumor necrosis factor receptor-associated protein 1 (TRAP-1) is a mitochondrial protein which also localizes at specific extramitochondrial sites. Exp. Cell Res. 260, 30–39. 10.1006/excr.2000.4983 [DOI] [PubMed] [Google Scholar]
- Cestari I., Kalidas S., Monnerat S., Anupama A., Phillips M. A., Stuart K. (2013). A multiple aminoacyl-tRNA synthetase complex that enhances tRNA-aminoacylation in african trypanosomes. Mol. Cell. Biol. 33, 4872–4888. 10.1128/MCB.00711-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Nathan D. F., Lindquist S. (1997). In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17, 318–325. 10.1128/MCB.17.1.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Piel W. H., Gui L., Bruford E., Monteiro A. (2005). The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 86, 627–637. 10.1016/j.ygeno.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Chen B., Zhong D., Monteiro A. (2006). Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics 7, 156. 10.1186/1471-2164-7-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-S., Silverstein A. M., Pratt W. B., Chinkers M. (1996). The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J. Biol. Chem. 271, 32315–32320. 10.1074/jbc.271.50.32315 [DOI] [PubMed] [Google Scholar]
- Chinkers M. (2001). Protein phosphatase 5 in signal transduction. Trends Endocrinol. Metab. 12, 28–32. 10.1016/S1043-2760(00)00335-0 [DOI] [PubMed] [Google Scholar]
- Chua C.-S., Low H., Sim T.-S. (2014). Co-chaperones of Hsp90 in Plasmodium falciparum and their concerted roles in cellular regulation. Parasitology 141, 1177–1191. 10.1017/S0031182013002084 [DOI] [PubMed] [Google Scholar]
- Citri A., Kochupurakkal B. S., Yarden Y. (2004). The achilles heel of ErbB-2/HER2: Regulation by the Hsp90 chaperone machine and potential for pharmacological intervention. Cell Cycle 3, 50–59. 10.4161/cc.3.1.607 [DOI] [PubMed] [Google Scholar]
- Clayton C., Shapira M. (2007). Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156, 93–101. 10.1016/j.molbiopara.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Cohen P. T. W. (1997). Novel protein serine/threonine phosphatases: Variety is the spice of life. Trends biochem. Sci. 22, 245–251. 10.1016/S0968-0004(97)01060-8 [DOI] [PubMed] [Google Scholar]
- Colasante C., Ellis M., Ruppert T., Voncken F. (2006). Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 6, 3275–3293. 10.1002/pmic.200500668 [DOI] [PubMed] [Google Scholar]
- Coto A. L. S., Seraphim T. V., Batista F. A. H., Dores-Silva P. R., Barranco A. B. F., Teixeira F. R., et al. (2018). Structural and functional studies of the Leishmania braziliensis SGT co-chaperone indicate that it shares structural features with HIP and can interact with both Hsp90 and Hsp70 with similar affinities. Int. J. Biol. Macromol. 118, 693–706. 10.1016/j.ijbiomac.2018.06.123 [DOI] [PubMed] [Google Scholar]
- Crevel G., Bennett D., Cotterill S. (2008). The human TPR protein TTC4 is a putative Hsp90 Co-chaperone which interacts with CDC6 and shows alterations in transformed cells. PLoS ONE 3, e0001737. 10.1371/journal.pone.0001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P., Schnaider T., So″ti C., Prohászka Z., Nardai G. (1998). The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 79, 129–168. 10.1016/S0163-7258(98)00013-8 [DOI] [PubMed] [Google Scholar]
- Cunningham C. N., Krukenberg K. A., Agard D. A. (2008). Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J. Biol. Chem. 283, 21170–21178. 10.1074/jbc.M800046200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Ammerer G., Posas F. (2011). Controlling gene expression in response to stress. Nat. Rev. Genet. 12, 833–845. 10.1038/nrg3055 [DOI] [PubMed] [Google Scholar]
- Dean M. E., Johnson J. L. (2021). Human Hsp90 cochaperones: Perspectives on tissue-specific expression and identification of cochaperones with similar in vivo functions. Cell Stress Chaperones 26, 3–13. 10.1007/s12192-020-01167-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S., Sunter J. D., Wheeler R. J. (2017). TrypTag.org: A trypanosome genome-wide protein localisation resource. Trends Parasitol. 33, 80–82. 10.1016/j.pt.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks E. D. (2019). Fexinidazole: First global approval. Drugs 79, 215–220. 10.1007/s40265-019-1051-6 [DOI] [PubMed] [Google Scholar]
- DeGrasse J. A., Chait B. T., Field M. C., Rout M. P. (2008). High-yield isolation and subcellular proteomic characterization of nuclear and subnuclear structures from trypanosomes. Methods Mol. Biol. 463, 77–92. 10.1007/978-1-59745-406-3_6 [DOI] [PubMed] [Google Scholar]
- Dero B., Zampetti-Bosseler F., Pays E., Steinert M. (1987). The genome and the antigen gene repertoire of Trypanosoma brucei gambiense are smaller than those of T. b. brucei. Mol. Biochem. Parasitol. 26, 247–256. 10.1016/0166-6851(87)90077-6 [DOI] [PubMed] [Google Scholar]
- Deschamps P., Lara E., Marande W., López-García P., Ekelund F., Moreira D. (2011). Phylogenomic analysis of kinetoplastids supports that trypanosomatids arose from within bodonids. Mol. Biol. Evol. 28, 53–58. 10.1093/molbev/msq289 [DOI] [PubMed] [Google Scholar]
- Descoteaux A., Avila H. A., Zhang K., Turco S. J., Beverley S. M. (2002). LeishmaniaLPG3 encodes a GRP94 homolog required for phosphoglycan synthesis implicated in parasite virulence but not viability. EMBO J. 21, 4458–4469. 10.1093/emboj/cdf447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drini S., Criscuolo A., Lechat P., Imamura H., Skalický T., Rachidi N., et al. (2016). Species- and strain-specific adaptation of the HSP70 super family in pathogenic trypanosomatids. Genome Biol. Evol. 8, 1980–1995. 10.1093/gbe/evw140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droll D., Minia I., Fadda A., Singh A., Stewart M., Queiroz R., et al. (2013). Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 9, e1003286. 10.1371/journal.ppat.1003286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M., Le Bœuf F., Huot J., Gratton J.-P. (2007). Src-mediated phosphorylation of Hsp90 in response to vascular endothelial growth factor (VEGF) is required for VEGF receptor-2 signaling to endothelial NO synthase. Mol. Biol. Cell 18, 4659–4668. 10.1091/mbc.e07-05-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverría P. C., Bernthaler A., Dupuis P., Mayer B., Picard D. (2011). An interaction network predicted from public data as a discovery tool: Application to the Hsp90 molecular chaperone machine. PLOS ONE 6, e26044. 10.1371/journal.pone.0026044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverría P. C., Briand P.-A., Picard D. (2016). A remodeled Hsp90 molecular chaperone ensemble with the novel cochaperone Aarsd1 is required for muscle differentiation. Mol. Cell. Biol. 36, 1310–1321. 10.1128/MCB.01099-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed N. M., Myler P. J., Bartholomeu D. C., Nilsson D., Aggarwal G., Tran A.-N., et al. (2005a). The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415. 10.1126/science.1112631 [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M., Myler P. J., Blandin G., Berriman M., Crabtree J., Aggarwal G., et al. (2005b). Comparative genomics of trypanosomatid parasitic Protozoa. Science 309, 404–409. 10.1126/science.1112181 [DOI] [PubMed] [Google Scholar]
- Emelyanov V. V. (2002). Phylogenetic relationships of organellar Hsp90 homologs reveal fundamental differences to organellar Hsp70 and Hsp60 evolution. Gene 299, 125–133. 10.1016/S0378-1119(02)01021-1 [DOI] [PubMed] [Google Scholar]
- Felts S. J., Owen B. A. L., Nguyen P., Trepel J., Donner D. B., Toft D. O. (2000). The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 275, 3305–3312. 10.1074/jbc.275.5.3305 [DOI] [PubMed] [Google Scholar]
- Fèvre E. M., Wissmann B. v., Welburn S. C., Lutumba P. (2008). The burden of human african trypanosomiasis. PLoS Negl. Trop. Dis. 2, e333. 10.1371/journal.pntd.0000333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras M. J., Echeverria P. C., Angel S. O. (2014). Protozoan HSP90-heterocomplex: Molecular interaction network and biological significance. Curr. Protein Pept. Sci. 15, 245–255. 10.2174/1389203715666140331114233 [DOI] [PubMed] [Google Scholar]
- Folgueira C., Requena J. M. (2007). A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol. Rev. 31, 359–377. 10.1111/j.1574-6976.2007.00069.x [DOI] [PubMed] [Google Scholar]
- Franzén O., Talavera-López C., Ochaya S., Butler C. E., Messenger L. A., Lewis M. D., et al. (2012). Comparative genomic analysis of human infective Trypanosoma cruzi lineages with the bat-restricted subspecies T. cruzi marinkellei. BMC Genomics 13, 531. 10.1186/1471-2164-13-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B. C., Toft D. O., Morimoto R. I. (1996). Molecular chaperone machines: Chaperone activities of the cyclophilin cyp-40 and the steroid aporeceptor-associated protein p23. Science 274, 1718–1720. 10.1126/science.274.5293.1718 [DOI] [PubMed] [Google Scholar]
- Galat A. (2003). Peptidylprolyl cis/trans isomerases (immunophilins): Biological diversity - targets - functions. Curr. Top. Med. Chem. 3, 1315–1347. 10.2174/1568026033451862 [DOI] [PubMed] [Google Scholar]
- Gano J. J., Simon J. A. (2010). A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol. Cell. Proteomics 9, 255–270. 10.1074/mcp.M900261-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea J. A., Mirey G., Camonis J., Valencia A. (2002). p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529, 162–167. 10.1016/S0014-5793(02)03321-5 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The proteomics protocols handbook springer protocols handbooks. Editor Walker J. M. (Totowa, NJ: Humana Press; ), 571–607. 10.1385/1-59259-890-0:571 [DOI] [Google Scholar]
- Gazestani V. H., Yip C. W., Nikpour N., Berghuis N., Salavati R. (2017). TrypsNetDB: An integrated framework for the functional characterization of trypanosomatid proteins. PLoS Negl. Trop. Dis. 11, e0005368. 10.1371/journal.pntd.0005368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. (2012). The origins of the trypanosome genome strains Trypanosoma brucei brucei TREU 927, T. b. gambiense DAL 972, T. vivax Y486 and T. congolense IL3000. Parasit. Vectors 5, 71. 10.1186/1756-3305-5-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girstmair H., Tippel F., Lopez A., Tych K., Stein F., Haberkant P., et al. (2019). The Hsp90 isoforms from S. cerevisiae differ in structure, function and client range. Nat. Commun. 10, 3626. 10.1038/s41467-019-11518-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden T., Swingle M., Honkanen R. E. (2008). The role of serine/threonine protein phosphatase type 5 (PP5) in the regulation of stress-induced signaling networks and cancer. Cancer Metastasis Rev. 27, 169–178. 10.1007/s10555-008-9125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales F. A., Zanchin N. I. T., Luz J. S., Oliveira C. C. (2005). Characterization of Saccharomyces cerevisiae Nop17p, a novel nop58p-interacting protein that is involved in pre-rRNA processing. J. Mol. Biol. 346, 437–455. 10.1016/j.jmb.2004.11.071 [DOI] [PubMed] [Google Scholar]
- Goos C., Dejung M., Janzen C. J., Butter F., Kramer S. (2017). The nuclear proteome of Trypanosoma brucei. PLoS ONE 12, e0181884. 10.1371/journal.pone.0181884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graefe S. E. B., Wiesgigl M., Gaworski I., Macdonald A., Clos J. (2002). Inhibition of HSP90 in trypanosoma cruzi induces a stress response but No stage differentiation. Eukaryot. Cell 1, 936–943. 10.1128/EC.1.6.936-943.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera K., Wüthrich D., Braga-Lagache S., Heller M., Ochsenreiter T. (2012). Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics 13, 556. 10.1186/1471-2164-13-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S. (1995). Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol. Biol. Evol. 12, 1063–1073. 10.1093/oxfordjournals.molbev.a040281 [DOI] [PubMed] [Google Scholar]
- Güther M. L. S., Urbaniak M. D., Tavendale A., Prescott A., Ferguson M. A. J. (2014). High-confidence glycosome proteome for procyclic form Trypanosoma brucei by epitope-tag organelle enrichment and SILAC proteomics. J. Proteome Res. 13, 2796–2806. 10.1021/pr401209w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hance M. W., Nolan K. D., Isaacs J. S. (2014). The double-edged sword: Conserved functions of extracellular hsp90 in wound healing and cancer. Cancers (Basel) 6, 1065–1097. 10.3390/cancers6021065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Bracher A., Hayer-Hartl M. (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Henri J., Chagot M.-E., Bourguet M., Abel Y., Terral G., Maurizy C., et al. (2018). Deep structural analysis of RPAP3 and PIH1D1, two components of the HSP90 Co-chaperone R2TP complex. Structure 26, 1196–1209. e8. 10.1016/j.str.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Herreros-Cabello A., Callejas-Hernández F., Gironès N., Fresno M. (2020). Trypanosoma cruzi genome: Organization, multi-gene families, transcription, and biological implications. Genes. 11, 1196. 10.3390/genes11101196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach A., Ommen G., Chrobak M., Clos J. (2013). The Hsp90-Sti1 interaction is critical for Leishmania donovani proliferation in both life cycle stages. Cell. Microbiol. 15, 585–600. 10.1111/cmi.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach-Barrigah A., Bartsch K., Smirlis D., Rosenqvist H., MacDonald A., Dingli F., et al. (2019). Leishmania donovani 90 kD heat shock protein – impact of phosphosites on parasite fitness, infectivity and casein kinase affinity. Sci. Rep. 9, 5074. 10.1038/s41598-019-41640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hořejší Z., Stach L., Flower T. G., Joshi D., Flynn H., Skehel J. M., et al. (2014). Phosphorylation-dependent PIH1D1 interactions define substrate specificity of the R2TP cochaperone complex. Cell Rep. 7, 19–26. 10.1016/j.celrep.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoter A., El-Sabban M. E., Naim H. Y. (2018). The HSP90 family: Structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 19, 2560. 10.3390/ijms19092560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Su Y., Fei X., Wang X., Zhang G., Su C., et al. (2020). Ubiquitin specific peptidase 19 is a prognostic biomarker and affect the proliferation and migration of clear cell renal cell carcinoma. Oncol. Rep. 43, 1964–1974. 10.3892/or.2020.7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. C., O’Hagan K. L., Kenyon A., Dhanani K. C. H., Prinsloo E., Edkins A. L. (2014). Hsp90 binds directly to fibronectin (FN) and inhibition reduces the extracellular fibronectin matrix in breast cancer cells. PLoS One 9, e86842. 10.1371/journal.pone.0086842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Sanders M., Berry A., McQuillan J., Aslett M. A., Quail M. A., et al. (2010). The genome sequence of trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl. Trop. Dis. 4, e658. 10.1371/journal.pntd.0000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. E. (2013). Hsp90: Structure and function. Top. Curr. Chem. 328, 155–240. 10.1007/128_2012_356 [DOI] [PubMed] [Google Scholar]
- Jahn M., Rehn A., Pelz B., Hellenkamp B., Richter K., Riefa M., et al. (2014). The charged linker of the molecular chaperone Hsp90 modulates domain contacts and biological function. Proc. Natl. Acad. Sci. U. S. A. 111, 17881–17886. SAPP.PDF. 10.1073/pnas.1414073111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U., Meyer I., Bügl H., André S., Bardwell J. C. A., Buchner J. (1995). Structural organization of procaryotic and eucaryotic Hsp90. Influence of divalent cations on structure and function. J. Biol. Chem. 270, 14412–14419. 10.1074/jbc.270.24.14412 [DOI] [PubMed] [Google Scholar]
- Johnson B. D., Schumacher R. J., Ross E. D., Toft D. O. (1998). Hop modulates hsp70/hsp90 interactions in protein folding. J. Biol. Chem. 273, 3679–3686. 10.1074/jbc.273.6.3679 [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Brown C. (2009). Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones 14, 83–94. 10.1007/s12192-008-0058-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L. (2012). Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta 1823, 607–613. 10.1016/j.bbamcr.2011.09.020 [DOI] [PubMed] [Google Scholar]
- Jones C., Anderson S., Singha U. K., Chaudhuri M. (2008). Protein phosphatase 5 is required for Hsp90 function during proteotoxic stresses in Trypanosoma brucei. Parasitol. Res. 102, 835–844. 10.1007/s00436-007-0817-z [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kaiser M., Maes L., Tadoori L. P., Spangenberg T., Ioset J.-R. (2015). Repurposing of the open access malaria box for kinetoplastid diseases identifies novel active scaffolds against trypanosomatids. J. Biomol. Screen. 20, 634–645. 10.1177/1087057115569155 [DOI] [PubMed] [Google Scholar]
- Kampinga H. H., Hageman J., Vos M. J., Kubota H., Tanguay R. M., Bruford E. A., et al. (2009). Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14, 105–111. 10.1007/s12192-008-0068-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulov A. V., LaPointe P., Lu B., Razvi A., Coppinger J., Dong M.-Q., et al. (2010). Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol. Biol. Cell 21, 871–884. 10.1091/mbc.e09-12-1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravats A. N., Hoskins J. R., Reidy M., Johnson J. L., Doyle S. M., Genest O., et al. (2018). Functional and physical interaction between yeast Hsp90 and Hsp70. Proc. Natl. Acad. Sci. U. S. A. 115, E2210–E2219. 10.1073/pnas.1719969115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Gupta S., Bhutia W. D., Vaid R. K., Kumar S. (2022). Atypical human trypanosomosis: Potentially emerging disease with lack of understanding. Zoonoses Public Health 69, 259–276. 10.1111/zph.12945 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Larreta R., Guzman F., Patarroyo M. E., Alonso C., Requena J. M. (2002). Antigenic properties of the Leishmania infantum GRP94 and mapping of linear B-cell epitopes. Immunol. Lett. 80, 199–205. 10.1016/S0165-2478(01)00331-5 [DOI] [PubMed] [Google Scholar]
- Larreta R., Soto M., Alonso C., Requena J. M. (2000). Leishmania infantum: Gene cloning of the GRP94 homologue, its expression as recombinant protein, and analysis of antigenicity. Exp. Parasitol. 96, 108–115. 10.1006/expr.2000.4553 [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., High S. (2012). SGTA antagonizes BAG6-mediated protein triage. Proc. Natl. Acad. Sci. U. S. A. 109, 19214–19219. 10.1073/pnas.1209997109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Soroka J., Buchner J. (2012). The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 1823, 624–635. 10.1016/j.bbamcr.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Li Z., Jaroszewski L., Iyer M., Sedova M., Godzik A. (2020). Fatcat 2.0: Towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 48, W60–W64. 10.1093/NAR/GKAA443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner A. K., Lejon V., Chappuis F., Seixas J., Kazumba L., Barrett M. P., et al. (2020). New WHO guidelines for treatment of gambiense human african trypanosomiasis including fexinidazole: Substantial changes for clinical practice. Lancet. Infect. Dis. 20, e38–e46. 10.1016/S1473-3099(19)30612-7 [DOI] [PubMed] [Google Scholar]
- López A., Elimelech A. R., Klimm K., Sattler M. (2021). The charged linker modulates the conformations and molecular interactions of Hsp90. Chembiochem. 22, 1084–1092. 10.1002/CBIC.202000699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F., Pal M., Muñoz-Hernández H., Rodríguez C. F., Núñez-Ramírez R., Gil-Carton D., et al. (2018). RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex. Nat. Commun. 9, 1501. 10.1038/s41467-018-03942-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M., Eletto D., Argon Y. (2012). GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta 1823, 774–787. 10.1016/j.bbamcr.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masgras I., Sanchez-Martin C., Colombo G., Rasola A. (2017). The chaperone TRAP1 as a modulator of the mitochondrial adaptations in cancer cells. Front. Oncol. 7, 58. 10.3389/fonc.2017.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. P., Nikolay R., Bukau B. (2002). Aha, another regulator for Hsp90 chaperones. Mol. Cell 10, 1255–1256. 10.1016/S1097-2765(02)00793-1 [DOI] [PubMed] [Google Scholar]
- McLaughlin S. H., Sobott F., Yao Z., Zhang W., Nielsen P. R., Grossmann J. G., et al. (2006). The Co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 356, 746–758. 10.1016/j.jmb.2005.11.085 [DOI] [PubMed] [Google Scholar]
- Meyer K. J., Caton E., Shapiro T. A. (2018). Model system identifies kinetic driver of Hsp90 inhibitor activity against african trypanosomes and plasmodium falciparum. Antimicrob. Agents Chemother. 62, e00056. 10.1128/AAC.00056-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. J., Shapiro T. A. (2021). Cytosolic and mitochondrial Hsp90 in cytokinesis, mitochondrial DNA replication, and drug action in trypanosoma brucei. Antimicrob. Agents Chemother. 65, e0063221–21. 10.1128/AAC.00632-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. J., Shapiro T. A. (2013). Potent antitrypanosomal activities of heat shock protein 90 inhibitors in vitro and in vivo . J. Infect. Dis. 208, 489–499. 10.1093/infdis/jit179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y., Yahara I. (1992). The 90-kDa heat shock protein, HSP90, binds and protects casein kinase II from self-aggregation and enhances its kinase activity. J. Biol. Chem. 267, 7042–7047. 10.1016/S0021-9258(19)50533-6 [DOI] [PubMed] [Google Scholar]
- Moffatt N. S. C., Bruinsma E., Uhl C., Obermann W. M. J., Toft D. (2008). Role of the cochaperone Tpr2 in Hsp90 chaperoning. Biochemistry 47, 8203–8213. 10.1021/bi800770g [DOI] [PubMed] [Google Scholar]
- Mollapour M., Neckers L. (2012). Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta 1823, 648–655. 10.1016/j.bbamcr.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M., Tsutsumi S., Truman A. W., Xu W., Vaughan C. K., Beebe K., et al. (2011). Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol. Cell 41, 672–681. 10.1016/j.molcel.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti N. S., Cestari I., Anupama A., Stuart K., Schenkman S. (2018). Comparative proteomic analysis of lysine acetylation in trypanosomes. J. Proteome Res. 17, 374–385. 10.1021/acs.jproteome.7b00603 [DOI] [PubMed] [Google Scholar]
- Morrison L. J., Vezza L., Rowan T., Hope J. C. (2016). Animal african trypanosomiasis: Time to increase focus on clinically relevant parasite and host species. Trends Parasitol. 32, 599–607. 10.1016/j.pt.2016.04.012 [DOI] [PubMed] [Google Scholar]
- Mottram J. C., Murphy W. J., Agabian N. (1989). A transcriptional analysis of the Trypanosoma brucei hsp83 gene cluster. Mol. Biochem. Parasitol. 37, 115–127. 10.1016/0166-6851(89)90108-4 [DOI] [PubMed] [Google Scholar]
- Murthy A. E., Bernards A., Church D., Wasmuth J., Gusella J. F. (1996). Identification and characterization of two novel tetratricopeptide repeat-containing genes. DNA Cell Biol. 15, 727–735. 10.1089/dna.1996.15.727 [DOI] [PubMed] [Google Scholar]
- Nelson G. M., Huffman H., Smith D. F. (2003). Comparison of the carboxy-terminal DP-repeat region in the co-chaperones Hop and Hip. Cell Stress Chaperones 8, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett I. R. E., Davidson L., Lamont D., Ferguson M. A. J. (2009a). Identification and specific localization of tyrosine-phosphorylated proteins in trypanosoma brucei. Eukaryot. Cell 8, 617–626. 10.1128/EC.00366-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett I. R. E., Martin D. M. A., Miranda-Saavedra D., Lamont D., Barber J. D., Mehlert A., et al. (2009b). The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8, 1527–1538. 10.1074/mcp.M800556-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M., Langousis G., Nguyen H. T., Saada E. A., Shimogawa M. M., Jonsson Z. O., et al. (2011). Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious trypanosoma brucei. Mol. Cell. Proteomics 10, M111010538. 10.1074/mcp.M111.010538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odunuga O. O., Hornby J. A., Bies C., Zimmermann R., Pugh D. J., Blatch G. L. (2003). Tetratricopeptide repeat motif-mediated hsc70-mSTI1 interaction: Molecular characterization of the critical contacts for successful binding and specificity. J. Biol. Chem. 278, 6896–6904. 10.1074/jbc.M206867200 [DOI] [PubMed] [Google Scholar]
- Olcese C., Patel M. P., Shoemark A., Kiviluoto S., Legendre M., Williams H. J., et al. (2017). X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat. Commun. 8, 14279. 10.1038/ncomms14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ommen G., Chrobak M., Clos J. (2010). The co-chaperone SGT of Leishmania donovani is essential for the parasite’s viability. Cell Stress Chaperones 15, 443–455. 10.1007/s12192-009-0160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi C. P., Benz C., Urbaniak M. D. (2020). Phosphoproteomic analysis of mammalian infective Trypanosoma brucei subjected to heat shock suggests atypical mechanisms for thermotolerance. J. Proteomics 219, 103735. 10.1016/j.jprot.2020.103735 [DOI] [PubMed] [Google Scholar]
- Osellame L. D., Blacker T. S., Duchen M. R. (2012). Cellular and molecular mechanisms of mitochondrial function. Best. Pract. Res. Clin. Endocrinol. Metab. 26, 711–723. 10.1016/j.beem.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavi R., Roy N., Nageshan R. K., Talukdar P., Pavithra S. R., Reddy R., et al. (2010). Heat shock protein 90 as a drug target against Protozoan infections BIOCHEMICAL characterization of hsp90 from plasmodium falciparum and trypanosoma evansi and evaluation of its inhibitor as a candidate drug. J. Biol. Chem. 285, 37964–37975. 10.1074/jbc.M110.155317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B., Prodromou C., Roe S. M., O’Brien R., Ladbury J. E., Piper P. W., et al. (1998). ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo . EMBO J. 17, 4829–4836. 10.1093/emboj/17.16.4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A. K., Ogata Y., Zíková A., Anupama A., Dalley R. A., Acestor N., et al. (2009). A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450. 10.1002/pmic.200800477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M., Worthey E. A., Ward P. N., Mottram J. C. (2005). Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6, 127. 10.1186/1471-2164-6-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peikert C. D., Mani J., Morgenstern M., Käser S., Knapp B., Wenger C., et al. (2017). Charting organellar importomes by quantitative mass spectrometry. Nat. Commun. 8, 15272. 10.1038/ncomms15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro J. C., Hills T., Senisterra G., Wernimont A. K., Mackenzie C., Norcross N. R., et al. (2013). Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design. PLoS Negl. Trop. Dis. 7, e2492. 10.1371/JOURNAL.PNTD.0002492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploeg L. V. der, Giannini S. H., Cantor C. R. (1985). Heat shock genes: Regulatory role for differentiation in parasitic protozoa. Science 228, 1443–1446. 10.1126/science.4012301 [DOI] [PubMed] [Google Scholar]
- Poetsch M., Dittberner T., Cowell J. K., Woenckhaus C. (2000). TTC4, a novel candidate tumor suppressor gene at 1p31 is often mutated in malignant melanoma of the skin. Oncogene 19, 5817–5820. 10.1038/sj.onc.1203961 [DOI] [PubMed] [Google Scholar]
- Porter J. R., Fritz C. C., Depew K. M. (2010). Discovery and development of Hsp90 inhibitors: A promising pathway for cancer therapy. Curr. Opin. Chem. Biol. 14, 412–420. 10.1016/j.cbpa.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Pratt W. B., Galigniana M. D., Harrell J. M., DeFranco D. B. (2004). Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell. Signal. 16, 857–872. 10.1016/j.cellsig.2004.02.004 [DOI] [PubMed] [Google Scholar]
- Preußer C., Jaé N., Bindereif A. (2012). mRNA splicing in trypanosomes. Int. J. Med. Microbiol. 302, 221–224. 10.1016/j.ijmm.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Prodromou C., SiliGardi G., O'BRien R., Woolfson D. N., Regan L., Panaretou B., et al. (1999). Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18, 754–762. 10.1093/emboj/18.3.754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C. (2012). The ‘active life’ of Hsp90 complexes. Biochim. Biophys. Acta 1823, 614–623. 10.1016/j.bbamcr.2011.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radli M., Rüdiger S. G. D. (2017). Picky hsp90—every game with another mate. Mol. Cell 67, 899–900. 10.1016/j.molcel.2017.09.013 [DOI] [PubMed] [Google Scholar]
- Rao R., Fiskus W., Yang Y., Lee P., Joshi R., Fernandez P., et al. (2008). HDAC6 inhibition enhances 17-AAG–mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood 112, 1886–1893. 10.1182/blood-2008-03-143644 [DOI] [PubMed] [Google Scholar]
- Ratajczak T., Ward B., Minchin R. (2003). Immunophilin chaperones in steroid receptor signalling. Curr. Top. Med. Chem. 3, 1348–1357. 10.2174/1568026033451934 [DOI] [PubMed] [Google Scholar]
- Requena J., M. (2011). Lights and shadows on gene organization and regulation of gene expression in Leishmania. Front. Biosci. 16, 2069–2085. 10.2741/3840 [DOI] [PubMed] [Google Scholar]
- Requena J. M., Montalvo A. M., Fraga J. (2015). Molecular chaperones of Leishmania: Central players in many stress-related and -unrelated physiological processes. Biomed. Res. Int. 2015, e301326. 10.1155/2015/301326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M., Hagn F., Mitschke L., Hessling M., Gugel F., Kessler H., et al. (2010). Asymmetric activation of the Hsp90 dimer by its cochaperone Aha1. Mol. Cell 37, 344–354. 10.1016/j.molcel.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Richter K., Reinstein J., Buchner J. (2007). A grp on the Hsp90 mechanism. Mol. Cell 28, 177–179. 10.1016/j.molcel.2007.10.007 [DOI] [PubMed] [Google Scholar]
- Roberts J. D., Thapaliya A., Martínez-Lumbreras S., Krysztofinska E. M., Isaacson R. L. (2015). Structural and functional insights into small, glutamine-rich, tetratricopeptide repeat protein alpha. Front. Mol. Biosci. 2, 71. 10.3389/fmolb.2015.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A., Rohrberg J., Buchner J. (2013). The chaperone Hsp90: Changing partners for demanding clients. Trends biochem. Sci. 38, 253–262. 10.1016/j.tibs.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Roy N., Nageshan R. K., Ranade S., Tatu U. (2012). Heat shock protein 90 from neglected protozoan parasites. Biochim. Biophys. Acta 1823, 707–711. 10.1016/j.bbamcr.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Sahasrabudhe P., Rohrberg J., Biebl M. M., Rutz D. A., Buchner J. (2017). The plasticity of the Hsp90 Co-chaperone system. Mol. Cell 67, 947–961. e5. 10.1016/j.molcel.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Samant R. S., Clarke P. A., Workman P. (2012). The expanding proteome of the molecular chaperone HSP90. Cell Cycle 11, 1301–1308. 10.4161/cc.19722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage J. R., McLaughlin J. N., Skiba N. P., Hamm H. E., Willardson B. M. (2000). Functional roles of the two domains of phosducin and phosducin-like protein. J. Biol. Chem. 275, 30399–30407. 10.1074/jbc.M005120200 [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., et al. (2000). Structure of TPR domain–peptide complexes: Critical elements in the assembly of the hsp70–hsp90 multichaperone machine. Cell 101, 199–210. 10.1016/S0092-8674(00)80830-2 [DOI] [PubMed] [Google Scholar]
- Schleiff E., Becker T. (2011). Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12, 48–59. 10.1038/nrm3027 [DOI] [PubMed] [Google Scholar]
- Schmidt J. C., Soares M. J., Goldenberg S., Pavoni D. P., Krieger M. A. (2011). Characterization of TcSTI-1, a homologue of stress-induced protein-1, in Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz 106, 70–77. 10.1590/s0074-02762011000100012 [DOI] [PubMed] [Google Scholar]
- Schopf F. H., Biebl M. M., Buchner J. (2017). The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18, 345–360. 10.1038/nrm.2017.20 [DOI] [PubMed] [Google Scholar]
- Seraphim T. V., Alves M. M., Silva I. M., Gomes F. E. R., Silva K. P., Murta S. M. F., et al. (2013). Low resolution structural studies indicate that the activator of Hsp90 ATPase 1 (Aha1) of Leishmania braziliensis has an elongated shape which allows its interaction with both N- and M-domains of Hsp90. PLoS One 8, e66822. 10.1371/journal.pone.0066822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahriyari L., Abdel-Rahman M., Cebulla C. (2019). BAP1 expression is prognostic in breast and uveal melanoma but not colon cancer and is highly positively correlated with RBM15B and USP19. PLoS One 14, e0211507. 10.1371/journal.pone.0211507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., Baird M. A., et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum . Nat. Methods 10, 407–409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa M. M., Saada E. A., Vashisht A. A., Barshop W. D., Wohlschlegel J. A., Hill K. L. (2015). Cell surface proteomics provides insight into stage-specific remodeling of the host-parasite interface in trypanosoma brucei. Mol. Cell. Proteomics 14, 1977–1988. 10.1074/mcp.M114.045146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonhai A., Maier G., Przyborski A. M., Blatch L. (2011). Intracellular Protozoan parasites of humans: The role of molecular chaperones in development and pathogenesis. Protein Pept. Lett. 18, 143–157. 10.2174/092986611794475002 [DOI] [PubMed] [Google Scholar]
- Siegelin M. D., Dohi T., Raskett C. M., Orlowski G. M., Powers C. M., Gilbert C. A., et al. (2011). Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Invest. 121, 1349–1360. 10.1172/JCI44855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J. A., Cerutti L., de Castro E., Langendijk-Genevaux P. S., Bulliard V., Bairoch A., et al. (2010). PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166. 10.1093/nar/gkp885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva K. P., Seraphim T. V., Borges J. C. (2013). Structural and functional studies of Leishmania braziliensis Hsp90. Biochim. Biophys. Acta 1834, 351–361. 10.1016/J.BBAPAP.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Simarro P. P., Cecchi G., Paone M., Franco J. R., Diarra A., Ruiz J. A., et al. (2010). The atlas of human african trypanosomiasis: A contribution to global mapping of neglected tropical diseases. Int. J. Health Geogr. 9, 57. 10.1186/1476-072X-9-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. R., Workman P. (2009). Targeting CDC37: An alternative, kinase-directed strategy for disruption of oncogenic chaperoning. Cell Cycle 8, 362–372. 10.4161/cc.8.3.7531 [DOI] [PubMed] [Google Scholar]
- Song H. Y., Dunbar J. D., Zhang Y. X., Guo D., Donner D. B. (1995). Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J. Biol. Chem. 270, 3574–3581. 10.1074/jbc.270.8.3574 [DOI] [PubMed] [Google Scholar]
- Stechmann A., Cavalier-Smith T. (2003). Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J. Mol. Evol. 57, 408–419. 10.1007/s00239-003-2490-x [DOI] [PubMed] [Google Scholar]
- Steiner J. P., Haughey N. J. (2010). “Immunophilin ligands,” in Encyclopedia of movement disorders. Editors Leo V., Katie K. (Amsterdam, Netherland: Elsevier; ), 66–68. 10.1016/B978-0-12-374105-9.00254-9 [DOI] [Google Scholar]
- Stuttmann J., Parker J. E., Noël L. D. (2008). Staying in the fold: The SGT1/chaperone machinery in maintenance and evolution of leucine-rich repeat proteins. Plant Signal. Behav. 3, 283–285. 10.4161/psb.3.5.5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., Roberts T., Cowell J. K. (1999). TTC4, a novel human gene containing the tetratricopeptide repeat and mapping to the region of chromosome 1p31 that is frequently deleted in sporadic breast cancer. Genomics 55, 157–163. 10.1006/geno.1998.5633 [DOI] [PubMed] [Google Scholar]
- Subbarao Sreedhar A., Kalmár É., Csermely P., Shen Y.-F. (2004). Hsp90 isoforms: Functions, expression and clinical importance. FEBS Lett. 562, 11–15. 10.1016/S0014-5793(04)00229-7 [DOI] [PubMed] [Google Scholar]
- Subota I., Julkowska D., Vincensini L., Reeg N., Buisson J., Blisnick T., et al. (2014). Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localization and dynamics. Mol. Cell. Proteomics 13, 1769–1786. 10.1074/mcp.M113.033357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Kaminen N., Nopola-Hemmi J., Haltia T., Myllyluoma B., Lyytinen H., et al. (2003). A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc. Natl. Acad. Sci. U. S. A. 100, 11553–11558. 10.1073/pnas.1833911100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D., Karras G. I., et al. (2012). Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001. 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Tucker G., Peng J., Krykbaeva I., Lin Z.-Y., Larsen B., et al. (2014). A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158, 434–448. 10.1016/j.cell.2014.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatu U., Neckers L. (2014). Chaperoning parasitism: The importance of molecular chaperones in pathogen virulence. Parasitology 141, 1123–1126. 10.1017/S0031182014000778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepel J., Mollapour M., Giaccone G., Neckers L. (2010). Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537–549. 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak M. D., Guther M. L. S., Ferguson M. A. J. (2012). Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS ONE 7, e36619. 10.1371/journal.pone.0036619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak M. D., Martin D. M. A., Ferguson M. A. J. (2013). Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 12, 2233–2244. 10.1021/pr400086y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urményi T. P., Silva R., Rondinelli E. (2014). The heat shock proteins of Trypanosoma cruzi. Subcell. Biochem. 74, 119–135. 10.1007/978-94-007-7305-9_5 [DOI] [PubMed] [Google Scholar]
- Wallace F. G. (1966). The trypanosomatid parasites of insects and arachnids. Exp. Parasitol. 18, 124–193. 10.1016/0014-4894(66)90015-4 [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. (2009). Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. 10.1093/BIOINFORMATICS/BTP033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J. R., Campos-Neto A., Skeiky Y. A., Reed S. G. (1997). Molecular characterization of the heat-inducible LmSTI1 protein of Leishmania major. Mol. Biochem. Parasitol. 89, 179–193. 10.1016/s0166-6851(97)00115-1 [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S. L. (2005). HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772. 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- WHO (2019). WHO interim guidelines for the treatment of gambiense human African trypanosomiasis. Geneva: World Health Organization. Available at: http://www.ncbi.nlm.nih.gov/books/NBK545514/ (Accessed August 7, 2022). [PubMed] [Google Scholar]
- Wiesgigl M., Clos J. (2001). The heat shock protein 90 of Leishmania donovani. Med. Microbiol. Immunol. 190, 27–31. 10.1007/s004300100074 [DOI] [PubMed] [Google Scholar]
- Willardson B. M., Howlett A. C. (2007). Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell. Signal. 19, 2417–2427. 10.1016/j.cellsig.2007.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Luo S., Jiang H., Li H. (2005). Mammalian CHORD-containing protein 1 is a novel heat shock protein 90-interacting protein. FEBS Lett. 579, 421–426. 10.1016/j.febslet.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Wunderley L., Leznicki P., Payapilly A., High S. (2014). SGTA regulates the cytosolic quality control of hydrophobic substrates. J. Cell Sci. 127, 4728–4739. 10.1242/jcs.155648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Rao R., Shen J., Tang Y., Fiskus W., Nechtman J., et al. (2008). Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 68, 4833–4842. 10.1158/0008-5472.CAN-08-0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Donelson J. E., Wilson M. E. (2003). The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 132, 1–16. 10.1016/S0166-6851(03)00211-1 [DOI] [PubMed] [Google Scholar]
- Yau W.-L., Blisnick T., Taly J.-F., Helmer-Citterich M., Schiene-Fischer C., Leclercq O., et al. (2010). Cyclosporin A treatment of Leishmania donovani reveals stage-specific functions of cyclophilins in parasite proliferation and viability. PLoS Negl. Trop. Dis. 4, e729. 10.1371/journal.pntd.0000729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau W.-L., Pescher P., MacDonald A., Hem S., Zander D., Retzlaff S., et al. (2014). The L eishmania donovani chaperone cyclophilin 40 is essential for intracellular infection independent of its stage-specific phosphorylation status: LdCyP40 null mutant analysis. Mol. Microbiol. 93, 80–97. 10.1111/mmi.12639 [DOI] [PubMed] [Google Scholar]
- Young J. C., Hartl F. U. (2000). Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 19, 5930–5940. 10.1093/emboj/19.21.5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Jiang N., Zhang K., Zheng L., Zhang D., Sang X., et al. (2020). Landscapes of protein posttranslational modifications of african trypanosoma parasites. iScience 23, 101074. 10.1016/j.isci.2020.101074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., et al. (2008). Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578. 10.1083/jcb.200709061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Larade K., Jackson T. A., Xie J., Ladoux A., Acker H., et al. (2004). NCB5OR is a novel soluble NAD(P)H reductase localized in the endoplasmic reticulum. J. Biol. Chem. 279, 30316–30325. 10.1074/jbc.M402664200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Qiu H., Yoon H.-W. P., Huang S., Bunn H. F. (1999). Identification of a cytochrome b -type NAD(P)H oxidoreductase ubiquitously expressed in human cells. Proc. Natl. Acad. Sci. U. S. A. 96, 14742–14747. 10.1073/pnas.96.26.14742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zininga T., Shonhai A. (2019). Small molecule inhibitors targeting the heat shock protein system of human obligate Protozoan parasites. Int. J. Mol. Sci. 20, 5930. 10.3390/ijms20235930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuehlke A., Johnson J. L. (2010). Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers 93, 211–217. 10.1002/bip.21292 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.