Abstract
The ongoing pandemic of severe acute respiratory coronavirus 2 (SARS‐CoV‐2) is causing a devastating impact on public health worldwide. However, details concerning the profound impact of SARS‐CoV‐2 on host cells remain elusive. Here, we investigated the effects of SARS‐CoV‐2‐encoded viral proteins on the intracellular activity of long interspersed element 1 (L1) retrotransposons using well‐established reporter systems. Several nonstructural or accessory proteins (Nsps) of SARS‐CoV‐2 (i.e., Nsp1, Nsp3, Nsp5, and Nsp14) significantly suppress human L1 mobility, and these viral L1 inhibitors generate a complex network that modulates L1 transposition. Specifically, Nsp1 and Nsp14 inhibit the intracellular accumulation of L1 open reading frame proteins (ORF1p), whereas Nsp3, Nsp5, and Nsp14 repress the reverse transcriptase activity of L1 ORF2p. Given recent findings concerning the roles of L1 in antiviral immune activation and host genome instability, the anti‐L1 activities mediated by SARS‐CoV‐2‐encoded inhibitors suggest that SARS‐CoV‐2 employs different strategies to optimize the host genetic environment.
Keywords: coronavirus, LINE‐1, retrotransposon, SARS‐CoV‐2
1. INTRODUCTION
Human coronavirus infections mostly cause respiratory and enteric diseases. Coronavirus disease (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) continues to widely affect the world. 1 Biologically, coronaviruses are positive‐sense single‐stranded RNA viruses with an envelope. 2 The coronavirus structural proteins include the spike (S), envelope (E), membrane (M), and nucleocapsid (N). The positive‐sense single‐stranded RNA is encapsidated by N proteins, whereas M and E proteins assist in viral incorporation during the particle‐assembly process. The specific binding of the S protein to cellular‐entry receptors, including angiotensin‐converting enzyme 2, initiates coronavirus infection. Genomic RNA contains two large open reading frames (ORF1a and ORF1b) that encode 15–16 nonstructural proteins (Nsps). 2 The coronavirus genome also contains genes that encode accessory proteins that are dispensable for viral replication and growth in vitro. 3
The human genome contains a large proportion of repeated DNA sequences, which can be further characterized according to length as either short (SINEs) or long (LINEs). 4 The human long interspersed element 1 (LINE‐1 or L1), the only autonomously retrotransposition‐competent retrotransposon, belongs to the dominant family of elements driving amplification. 5 L1 is transcribed as a bicistronic mRNA that encodes ORF1p and ORF2p. 4 ORF1p, a 40‐kDa RNA‐binding protein, possesses nucleic acid chaperone activity, whereas ORF2p, a 150‐kDa protein, is an endonuclease that cuts genomic DNA target sites and an endogenous reverse transcriptase (RTase) that generates L1 cDNA (complementary DNA). 5 , 6 ORF1p and ORF2p interact with their own RNAs to form L1 ribonucleoproteins (RNPs) in the cytoplasm. A previous study indicated that ORF1p and ORF2p exhibit highly heterogeneous localization, with ORF1p located either in the cytoplasm or both in the cytoplasm and the nucleus but never exclusively in the nucleus, whereas ORF2p is located in the cytoplasm, only in the nucleus, or in both the cytoplasm and the nucleus. 7 A recent study revealed that multiple‐charged amino acids in the ORF1p C‐terminus are responsible for its subcellular localization by interacting with nuclear binding proteins. 8 L1 RNPs can enter the nucleus, where retrotransposition occurs via a mechanism called target‐site‐primed reverse transcription (TPRT). 9 Nonhomologous end‐joining repair is also involved in L1 retrotransposition. 10 The new L1 copy is then integrated into the genome, although details associated with this process remain unclear. 11
Analyses of the human genome show that L1 occupies 17% of the DNA. 12 Transposable elements (TE) shape the human genome in many different ways, including by driving genome evolution, genomic instability, genomic rearrangements, and genetic innovation. 13 Accumulating evidence indicates that several host‐encoded factors, such as DNA‐repair proteins, are involved in the L1 life cycle. 10 Nicole et al. 14 reported that the DNA damage response protein ataxia telangiectasia mutated, a serine/threonine kinase modulates L1 retrotransposition and is correlated with neurodegenerative disease. However, host cells also maintain restriction factors that suppress L1 transposition activity. 11 For example, the Aicardi–Goutières syndrome‐related sterile α motif and histidine–aspartate domain‐containing protein 1 (SAMHD1), a cellular regulator of LINE‐1 activity, inhibits ORF2p‐mediated L1 reverse transcription. 15 Additionally, the tripartite motif‐containing protein 5α, which is an intracellular restriction factor against retroviral infection, restricts L1 elements. 16
Previous studies have revealed that illegitimate recombination between exogenous RNA viruses and endogenous retrotransposons leads to nonretroviral cDNA Integration. 17 , 18 , 19 Zhang et al. 20 showed that reverse‐transcribed SARS‐CoV‐2 RNA can be integrated into the genome of cultured human cells and expressed in patient‐derived tissues. Additionally, viral–cellular chimeric transcripts have been detected in patient‐derived tissues. 20 In particular, portions of consensus L1 endonuclease‐recognition sequences can flank the integrated viral sequences, which is consistent with the L1‐mediated TPRT and transposition mechanism. 21 However, Smits et al. 22 did not find any evidence of SARS‐CoV‐2 integration into the genome in HEK293T cells infected with SARS‐CoV‐2. Furthermore, exogenous pathogen‐associated molecular patterns epigenetically activated L1 to engage cyclic GMP–AMP synthase, thus enhancing host type‐I interferon (IFN) responses. This suggests that the induction of transposable‐element expression is central to innate sensing. 23 Notably, SARS‐CoV‐2 infection induces low host‐IFN responses and high levels of chemokines. 24 , 25 Yet, the impact of SARS‐CoV‐2 on host L1 remains elusive. Here using well‐established reporter systems and overexpression of viral proteins, we investigated interactions/cross‐talk between SARS‐CoV‐2‐encoded proteins and host L1 to expand our understanding of viral pathogenesis, disease control, and prevention.
2. MATERIALS AND METHODS
2.1. Cells and plasmids
HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 μg/ml penicillin–streptomycin. The human LINE‐1 plasmids 99 PUR L1RP EGFP (enhanced green fluorescent protein) (L1RP EGFP) and 99 PUR JM111 EGFP (JM111), the pc‐L1‐1FH plasmid, and the pEGFP‐N1‐ORF1‐EGFP plasmids were kindly provided by Dr. Haig H. Kazazian, Jr. and John L. Goodier. 26 , 27 , 28 , 29 The JM111 construct containing two missense mutations in the ORF1 region was used as a negative control for L1RP EGFP. 27 The pc‐L1‐1FH plasmid expressed full‐length LINE‐1, and LINE‐1 ORF1p was tagged with FLAG and HA. 28 pEGFP‐N1‐ORF1‐EGFP construct contained L1RP ORF1 sequence tagged with GFP. 29 The pYX014 and pYX017 plasmids were kindly provided by Dr. Wenfeng An. 30 , 31 The SARS‐CoV‐2‐encoded protein expressing vectors were kindly provided by Prof. Pei‐Hui Wang. 32
2.2. L1 retrotransposition with EGFP reporter assay
HEK293T cells were seeded in 24‐well plates and transfected with 1 μg of L1RP EGFP or JM111 constructs. Cells were then puromycin‐selected (3 μg/ml) 48 h posttransfection. The percentage of GFP(+) cells was measured using a BD FACSCalibur Flow Cytometer after 48 h of puromycin selection. The plasmid JM111 construct was used as a negative control. A total of 10 000 single‐cell events were gated using flow cytometry. The percentage of GFP(+) cells was analyzed using the CellQuest Pro (v.5.2).
2.3. L1 retrotransposition with a Luciferase reporter system
HEK293T cells seeded in 24‐well plates were transfected with the pYX014 or pYX017 plasmids. The cells were then puromycin selected 24 h posttransfection. After 3 days of puromycin selection, dual‐luciferase assays were performed according to the manufacturer's instructions (Promega). Firefly and Renilla luciferase were measured using Promega GloMax® from a single sample.
2.4. Cell viability assay
HEK293T cells were pretransfected. The cells were seeded in 96‐well plates 1 day posttransfection. Then the cells were cultured for another 3 days before assessing their viability. Absorbance was detected at a wavelength of 490 nm using a BioTek ELISA reader (BioTek Instruments, Inc.) on adding 3‐(4,5‐dimethylthiazol‐2‐yl)−5‐(3‐carboxymethoxyphenyl)−2‐(4‐sulphophenyl)−2H‐tetrazolium, inner salt (Promega) to each well.
2.5. Immunoblotting
Cell samples were harvested and lysed in RIPA buffer (1 M Tris pH 7.8, 1 M NaCl, 1% NP‐40, 0.5 M EDTA). The cell lysate was separated on 12% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis gels and then transferred to nitrocellulose membranes using a semidry apparatus (Bio‐Rad). Anti‐HA was purchased from Thermo Fisher Scientific. The anti‐glyceraldehyde‐3‐phosphate dehydrogenase antibody was purchased from GenScript Biotech Corp.
2.6. L1 element amplification protocol (LEAP) assay and quantitative real‐time polymerase chain reaction (qRT‐PCR)
The L1 construct pc‐L1‐1FH has been described. 28 HEK293T cells were transfected with the pc‐L1‐1FH plasmid. L1 RNPs were separated by sucrose cushion of 8.5% + 17% gradient centrifugation at 4°C, 178 000g for 2 h as previously described. 33 During the LEAP assay, 2 μl of the L1 RNP sample was added to each cDNA extension reaction solution (500 mM KCl, 50 mM MgCl2, 500 mM Tris‐HCl (pH 7.5), 1 M dithiothreitol, RNasin (40 U/μl), 0.05% (v/v) Tween 20, and dNTP) using the LEAP primer: 5′‐GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN‐3′. L1 mRNA was reversed‐transcribed to L1 cDNA by L1 ORF2p in the RNPs at 37°C for 1 h. To detect the level of the L1 mRNA, L1 RNA was extracted from the L1 RNP and reverse‐transcribed with the same primer using MuLV RT (GoScript Reverse Transcription System; Promega). The synthesized cDNA from both LEAP assay and MuLV RT was then analyzed by qRT‐PCR using the following primers: linker PCR primer, 5′‐GCGAGCACAGAATTAATACGACT‐3′; L1 3′‐end primer, 5′‐ GGGTTCGAAATCGATAAGCTTGGATCCAGAC‐3′, with a standard three‐step method (95°C for 15 s, 60°C for 1 min, and 72°C 2–4 kb/min) as previously described. 33 The method was used for calculations.
2.7. Statistical analysis
This study's statistical data were analyzed using GraphPad Prism software (version 8.0; GraphPad Software Inc.). Data are described as the M ± SD from three replicates of each experiment. Unpaired Student's t test was performed on the data between the two groups. p < 0.01 was considered significant in groups.
3. RESULTS
3.1. SARS‐CoV‐2‐encoded proteins regulate L1‐retrotransposition activity
We screened the effects of individual SARS‐CoV‐2‐encoded proteins on L1 mobility using a well‐established EGFP reporter system. 26 The L1RP EGFP construct contained both the L1RP cassette and the EGFP reporter cassette with its cytomegalovirus (CMV) promoter (Figure 1A). The EGFP reporter cassette was interrupted by introns, and the EGFP marker could only be detected after successful intron removal from the retroelement and L1 integration (Figure 1B). We found that several viral proteins restricted L1‐transposition activity at different levels. In particular, Nsp1, Nsp3, Nsp5, and Nsp14 showed higher inhibitory activity against L1 (Figures 1C and 2A), whereas other viral proteins resulted in modestly enhanced L1 activity. Notably, overexpression of all tested proteins did not significantly influence cell proliferation (Figure 2B) or CMV promoter‐driven GFP expression (Figure 2C) in HEK293T cells. These results suggested that the viral proteins encoded by SARS‐CoV‐2 are specific inhibitors of human transposon integration.
Figure 1.
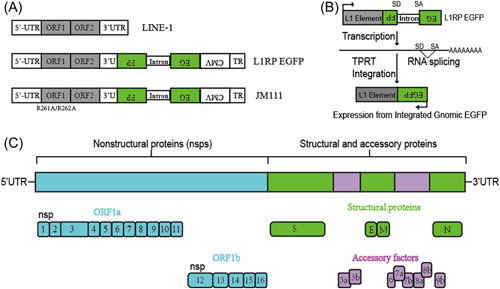
Schematics of the L1RP EGFP and JM111 constructs. (A) The L1RP EGFP construct included both the L1 5′‐UTR and the CMV promoter. The JM111 construct included two point mutations (R261A/R262A) in ORF1, rendering it transposition‐incompetent. (B) Description of the retrotransposition assay. EGFP can only be identified when the intron is removed after RNA splicing and when L1 is successfully transposed into the genome. (C) Genome organization of SARS‐CoV‐2. CMV, cytomegalovirus; EGFP, enhanced green fluorescent protein; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; ORF, open reading frame; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; UTR, untranslated region.
Figure 2.
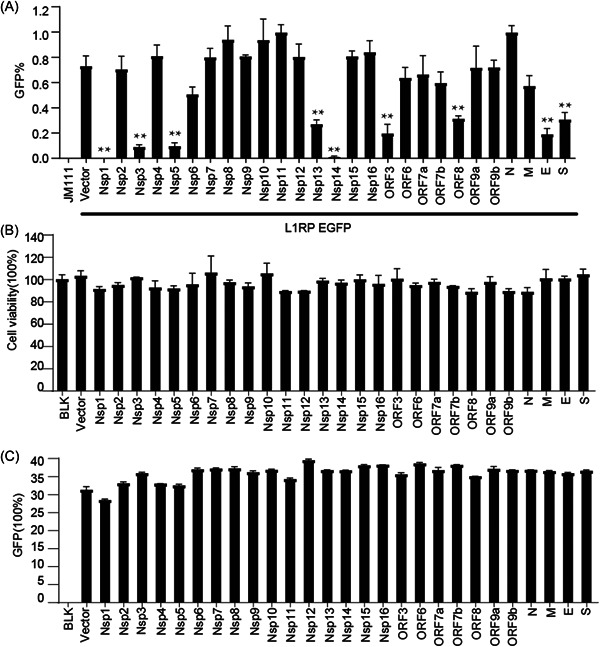
SARS‐CoV‐2‐encoded proteins decrease L1‐retrotransposition frequency. (A) Empty vectors or SARS‐CoV‐2 protein‐expressing plasmids (50 ng) were cotransfected into HEK293T cells along with L1RP EGFP plasmids (1 µg), and the number of EGFP‐positive cells was determined at 4 days posttransfection by flow cytometry. JM111 was used as a negative control for flow cytometric gating. The bar represents retrotransposition efficiency. Experiments were performed in triplicate, and each error bar indicates the standard deviation of three replicates for one experiment. **p < 0.01, Student's t test. (B) HEK293T cells pretransfected with empty vectors or SARS‐CoV‐2 protein‐expressing constructs were seeded on 96‐well plates and cultured for 4 days, followed by MTS staining and measurement of absorbance. The bar represents cell viability. The control‐treated sample was set to 100%. (C) pcDNA3.1‐EGFP plasmids and empty vectors/SARS‐CoV‐2 protein‐expressing plasmids were cotransfected into HEK293T cells, and the number of EGFP‐positive cells was determined at 4 days posttransfection using flow cytometry. The bar represents the percentage of GFP‐positive cells. EGFP, enhanced green fluorescent protein; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; MTS, 3‐(4,5‐dimethylthiazol‐2‐yl)−5‐(3‐carboxymethoxyphenyl)−2‐(4‐sulphophenyl)−2H‐tetrazolium; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.2. SARS‐CoV‐2 Nsp1 modulates L1 activity by inhibiting L1 expression
We then investigated the potential mechanisms of L1 inhibition by the viral protein Nsp1 by using luciferase‐based L1 reporter constructs (pYX014 and pYX017) 30 , 31 to confirm the inhibitory effect of Nsp1 on L1 activity. The pYX014 and pYX017 plasmids contain both firefly luciferase, which serves as an indicator of L1 retrotransposition, and Renilla luciferase, which was used for normalization. The pYX014 plasmid includes full‐length L1RP, whereas the pYX017 plasmid contains a stronger promoter by replacing the L1 5′‐UTR with CAG. 30 , 31 Similar to the EGFP reporter results (Figure 3A), luciferase reporter results showed that L1 mobility was decreased by Nsp1 (Figure 3B). Additionally, immunoblot assay and living‐cell images indicated a twofold downregulation of ORF1p protein levels by Nsp1 as compared with those in the control (Figure 3C,D). We then performed a LEAP 34 assay to assess the reverse‐transcription activity of ORF2p in the L1 RNP complex (Figure 3E). We found that Nsp1 did not decrease L1 ORF2p RTase activity (Figure 3F). These findings suggested that SARS‐CoV‐2 Nsp1 suppresses L1 mobility by targeting L1 expression.
Figure 3.
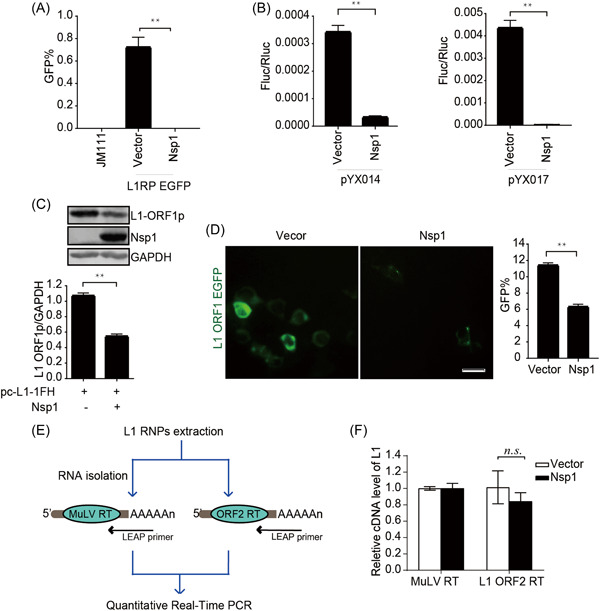
SARS‐CoV‐2 Nsp1 modulates L1 mobility by inhibiting L1 expression. (A) L1RP EGFP plasmids along with empty vectors/SARS‐CoV‐2 Nsp1‐expressing constructs were transfected into HEK293T cells, followed by flow cytometry performed at 4 days posttransfection. JM111 was used as a negative control. The bar graph depicts the percentage of GFP‐positive cells. **p < 0.01. (B) Empty vectors or SARS‐CoV‐2 Nsp1‐expressing constructs were cotransfected along with pYX014/pYX017 into HEK293T cells, followed by dual‐luciferase assays at 4 days posttransfection. The bar graph of the Firefly:Renilla luciferase ratio represents the retrotransposition frequency. **p < 0.01. (C) Empty vectors/SARS‐CoV‐2 Nsp1‐expressing plasmids along with pc‐L1‐1FH were cotransfected into HEK293T cells, followed by western blot analysis at 48‐h posttransfection. **p < 0.01. (D) HEK293T cells were transfected with pEGFP‐N1‐ORF1‐EGFP and empty vectors/SARS‐CoV‐2 Nsp1‐expressing plasmids, followed by living‐cell imaging at 48‐h posttransfection (scale bar, 20 mm). The bar graph represents the percentage of GFP‐positive cells according to flow cytometry results. **p < 0.01. (E) Description of the LEAP assay. HEK293T cells were transfected with pc‐L1‐1FH, and L1 RNP complexes were ultracentrifuged, L1 mRNA was extracted, and L1 cDNA was reverse‐transcribed by MuLV and ORF2p, respectively, followed by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). (F) LEAP assay, followed by quantitation of the products by qRT‐PCR. The bar graph of relative cDNA levels of L1 represents the reverse‐transcription efficiency of ORF2p or MuLV. The relative cDNA level of MuLV reverse transcription was set to 1.0. cDNA, complementary DNA; EGFP, enhanced green fluorescent protein; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; LEAP, L1 element amplification protocol; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; mRNA, messenger RNA; Nsp, nonstructural protein; n.s., not significant; ORF, open reading frame; RNP, ribonucleoprotein; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.3. SARS‐CoV‐2 Nsp3 restricts the reverse‐transcription activity of L1 ORF2p
We confirmed the decreased L1 activity induced by Nsp3 using EGFP‐ and luciferase‐based reporter assays (Figure 4A,B). The results showed that Nsp3 did not decrease the expression of L1 ORF1p according to repeated immunoblot data (Figure 4C), and living‐cell images indicated that Nsp3 overexpression did not dampen L1 ORF1p aggregation in the cytoplasm (Figure 4D). However, the presence of Nsp3 significantly restrained the RTase activity of L1 ORF2p in L1 RNPs (Figure 4E). These results showed that SARS‐CoV‐2 Nsp3 restricts L1 retrotransposition by targeting the reverse‐transcription step.
Figure 4.
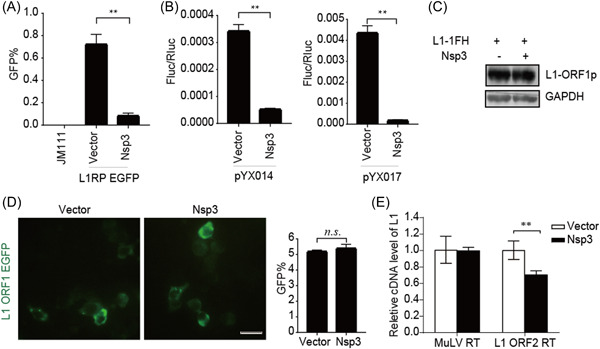
SARS‐CoV‐2 Nsp3 restricts L1 mobility in an ORF2p‐dependent manner. (A) L1RP EGFP plasmids and empty vectors/SARS‐CoV‐2 Nsp3‐expressing constructs were transfected into HEK293T cells, with JM111 used as a negative control, and flow cytometry was performed at 4 days posttransfection. The bar graph depicts the percentage of GFP‐positive cells. **p < 0.01. (B) Empty vector or SARS‐CoV‐2 Nsp3‐expressing constructs were cotransfected along with the pYX014/pYX017 reporter constructs into HEK293T cells, followed by dual‐luciferase assays performed at 4 days posttransfection. The bar graph represents the retrotransposition frequency. **p < 0.01. (C) HEK293T cells were cotransfected with empty vectors/SARS‐CoV‐2 Nsp3 and pc‐L1‐1FH, after which cells were collected at 48‐h posttransfection, and anti‐HA antibodies were used in an immunoblot assay. (D) HEK293T cells were transfected with pEGFP‐N1‐ORF1‐EGFP and empty vectors/SARS‐CoV‐2 Nsp3‐expressing constructs, followed by living‐cell imaging performed at 48‐h posttransfection (scale bar, 20 mm). The bar graph depicts the percentage of GFP‐positive cells according to flow cytometry. (E) LEAP assays, followed by analysis of MuLV RTase and LEAP products by qRT‐PCR. The bar graph of the relative cDNA level of L1 represents reverse‐transcription efficiency of the L1 ORF2p or MuLV. The relative cDNA level of MuLV reverse transcription was set to 1.0. **p < 0.01. cDNA, complementary DNA; EGFP, enhanced green fluorescent protein; LEAP, L1 element amplification protocol; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; Nsp, nonstructural protein; ORF, open reading frame; n.s., not significant; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.4. SARS‐CoV‐2 Nsp5 restricts the reverse‐transcription activity of L1 ORF2p
Both EGFP‐ and luciferase‐based assays further revealed that Nsp5 robustly impaired L1 mobility (Figure 5A,B), although the presence of Nsp5 did not reduce L1 ORF1p expression (Figure 5C) or impact ORF1p subcellular localization (Figure 5D). However, Nsp5 specifically decreased the RTase activity of L1 ORF2p according to LEAP assay results (Figure 5E). These findings suggested that L1 inhibition by Nsp5 involves a reverse‐transcription step during L1 transposition.
Figure 5.
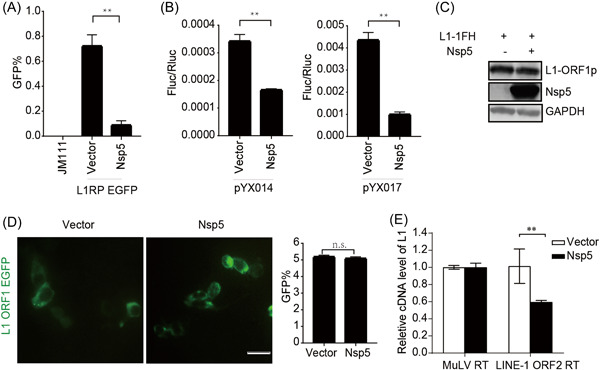
SARS‐CoV‐2 Nsp5 restricts L1 mobility in an ORF2p‐dependent manner. (A) L1RP EGFP plasmids and empty vectors/SARS‐CoV‐2 Nsp5‐expressing constructs were transfected into HEK293T cells, followed by flow cytometry at 4 days posttransfection. JM111 was used as a negative gating control. The bar graph indicates the percentage of GFP‐positive cells. **p < 0.01. (B) Empty vectors or SARS‐CoV‐2 Nsp5‐expressing constructs were cotransfected with the pYX014/pYX017 reporter into HEK293T cells, followed by dual‐luciferase assays performed at 4 days posttransfection. The bar graph of the Firefly:Renilla luciferase ratio depicts the retrotransposition frequency. **p < 0.01. (C) Empty vectors or SARS‐CoV‐2 Nsp5‐expressing plasmids were cotransfected with pc‐L1‐1FH into cells, followed by immunoblot analysis at 48 h posttransfection. (D) HEK293T cells were transfected with pEGFP‐N1‐ORF1‐EGFP and empty vectors/SARS‐CoV‐2 Nsp5‐expressing plasmids, followed by living‐cell imaging performed at 48 h posttransfection (scale bar, 20 mm). The bar graph represents the percentage of GFP‐positive cells according to flow cytometry. (E) LEAP assays, followed by analysis of MuLV RTase and LEAP products by qRT‐PCR. The bar graph of the relative cDNA level of L1 represents reverse‐transcription efficiency of the L1 ORF2p or MuLV. The relative cDNA level of MuLV reverse transcription was set to 1.0. **p < 0.01. cDNA, complementary DNA; EGFP, enhanced green fluorescent protein; LEAP, L1 element amplification protocol; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; Nsp, nonstructural protein; n.s., not significant; ORF, open reading frame; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.5. SARS‐CoV‐2 Nsp14 suppresses L1 retrotransposition
We then identified SARS‐CoV‐2 Nsp14 as a viral suppressor of L1 retrotransposition activity using both reporter assays (Figure 6A,B). Nsp14 overexpression caused about a two‐fold decrease in L1 ORF1p levels (Figure 6C), which was confirmed by living‐cell images (Figure 6D). Moreover, Nsp14 restricted the reverse‐transcription activity of L1 ORF2p in L1 RNPs (Figure 6E). The data suggested that SARS‐CoV‐2 Nsp14 impairs both L1 expression and L1 RNP RTase activity.
Figure 6.
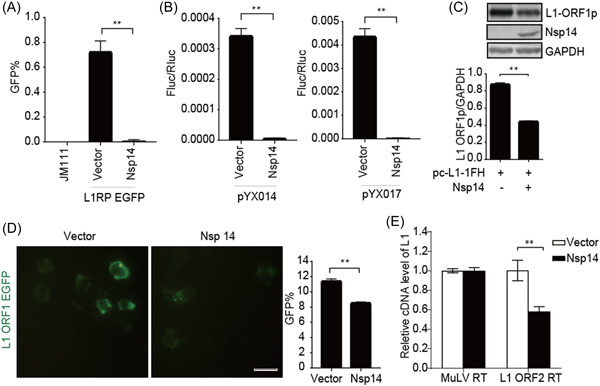
SARS‐CoV‐2 Nsp14 inhibits L1 mobility by inhibiting L1 expression. (A) L1RP EGFP plasmids and empty vectors/SARS‐CoV‐2 Nsp14‐expressing constructs were transfected into HEK293T cells, with JM111 used as a negative control, followed by flow cytometry at 4 days posttransfection. The bar graph depicts the percentage of GFP‐positive cells. **p < 0.01. (B) Empty vectors or SARS‐CoV‐2 Nsp14‐expressing constructs were cotransfected with the pYX014/pYX017 reporter into HEK293T cells, followed by dual‐luciferase assays at 4 days posttransfection. The Firefly:Renilla luciferase ratio depicts the retrotransposition frequency. **p < 0.01. (C) Empty vectors or SARS‐CoV‐2 Nsp14‐expressing plasmids were cotransfected with pc‐L1‐1FH into cells, followed by immunoblot assays at 48‐h posttransfection. **p < 0.01. (D) HEK293T cells were transfected with pEGFP‐N1‐ORF1‐EGFP and empty vectors/SARS‐CoV‐2 Nsp14‐expressing plasmids, followed by living‐cell imaging performed at 48‐h posttransfection (scale bar, 20 mm). The percentage of GFP‐positive cells is presented in the bar graph according to flow cytometry results. **p < 0.01. (E) LEAP assays, followed by analysis of MuLV RTase and LEAP products by qRT‐PCR. The bar graph of the relative cDNA level of L1 represents reverse‐transcription efficiency of the L1 ORF2p or MuLV. The relative cDNA level of MuLV reverse transcription was set to 1.0. **p < 0.01. cDNA, complementary DNA; EGFP, enhanced green fluorescent protein; LEAP, L1 element amplification protocol; L1RP, a full‐length L1 element that has inserted into the retinitis pigmentosa‐2 (RP) gene; Nsp, nonstructural protein; ORF, open reading frame; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
4. DISCUSSION
L1 is an oncogenic retroelement that is silenced early in development via tightly controlled epigenetic mechanisms. 13 Owing to the critical roles of L1 in genetic variability and immune activation, its transposition activity is strictly modulated during virus infection. 23 In this study, we revealed that the SARS‐CoV‐2‐encoded inhibitors Nsp1, Nsp3, Nsp5, and Nsp14 limit L1 activity via various L1‐specific components (Figure 7). Our results add one more piece of evidence to support the arms race relationship between virus and host retroelements. 35
Figure 7.
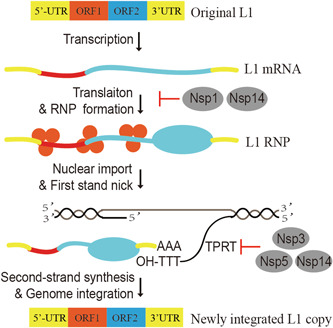
Diagram describing the mechanism of SARS‐CoV‐2‐mediated suppression of L1 transposition by Nsp1, Nsp3, Nsp5, and Nsp14. Mobilization of the full‐length L1 elements involves the 5′‐UTR, two ORFs (ORF1 and ORF2), and the 3′‐UTR according to the following steps. L1 is transcribed and translated into ORF1p and ORF2p, and the functional proteins interact with their own mRNA to form the RNP complex. L1 ORF1p is an RNA‐binding protein that assists L1 RNP translocation from the cytoplasm to the nucleus, whereas ORF2p performs both endonuclease (EN) and RTase activities. Upon L1 RNP nuclear translocation, ORF2p exerts its EN activity to nick a DNA strand at the target site to generate the 3′ hydroxyl group. This is followed by ORF2p‐mediated TPRT involving reverse transcription of L1 RNA using the 3′ hydroxyl group as a primer, resulting in L1 insertion into the genome. mRNA, messenger RNA; Nsp, nonstructural protein; ORF, open reading frame; RNP, ribonucleoprotein; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; UTR, untranslated region.
Nsp1 of SARS‐CoV‐2 exhibits a biological function that suppresses host gene expression via ribosome association, resulting in the shutdown of mRNA translation and blockage of the host immune response. 36 SARS‐CoV‐2 circumvents host translational blockage to produce its own proteins through a comprehensive mechanism involving interactions between Nsp1–C‐terminal domain–40S and Nsp1–N‐terminal domain–5′‐UTR. 37 Nsp14 contains 3′‐to‐5′‐exoribonuclease and guanine‐N7‐methyltransferase domains. 38 An in vitro reconstitution experiment of the SARS‐CoV mRNA cap revealed that viral mRNA cap methylation requires Nsp10, Nsp14, and Nsp16, where Nsp14 acts as an essential initiator of the obligate sequence of methylation events. 39 Thus, the virus hijacks the translational machinery of cells that enables them to survive through the functions of viral proteins, including Nsp1 and Nsp14. 36 In this study, the overexpression of Nsp1 or Nsp14 decreased L1 ORF1p levels, suggesting that SARS‐CoV‐2 Nsp1 and Nsp14 may inhibit L1 mobility through translational blockage of L1 mRNA to complete viral replication. Interestingly, Nsp14 also decreased the RTase activity of L1 RNP compared with Nsp1. However, the detailed mechanisms of L1 RTase inhibition by Nsp14 remain to be characterized.
Similar to other RNA viruses, SARS‐CoV‐2 encodes polyproteins containing Nsps that are then processed by viral proteases. 40 SARS‐CoV‐2 Nsp3 and Nsp5, both viral proteases, are responsible for cleaving viral polyproteins. 41 , 42 Nsp3, the largest multidomain protein encoded by SARS‐CoV‐2 RNA, contains a papain‐like protease, 43 and Nsp5 contains a chymotrypsin‐like cysteine protease, 3CLpro. 3 Nsp3 and Nsp5 together with other SARS‐CoV‐2‐encoded nonstructural proteins facilitate virus replication by assembling the replication and transcription complex. 44 The virus also induces reorganization of host cell membranes into double‐membrane vesicles or convoluted membranes to organize viral replication. 45 Furthermore, both Nsp3 and Nsp5 are detected on the convoluted membranes, indicating membrane‐associated replication. 45 Nsp3 and Nsp5 did not affect the expression or subcellular localization of L1 ORF1p but decreased the reverse‐transcription activity of L1 ORF2p based on the results of the LEAP assay in the present study. Thus, we speculate that Nsp3 and Nsp5 disrupt the function of L1 ORF2p to maintain an optimal microenvironment for viral RNA synthesis. However, because we could not obtain an effective antibody against L1 ORF2p, the potential interaction between the viral proteins and L1 ORF2p should be investigated in future studies. The relationship between viral proteins (Nsp3 and Nsp5) and L1 ORF2p activity remains to be clarified.
Recent studies have revealed that upregulation of transposable element expression is common in virus‐infected cells, which may be triggered by cell stress induced by virus infection and by enhancing global DNA demethylation. 21 , 46 In contrast, we found that SARS‐CoV‐2‐encoded proteins potently suppress L1 retrotransposition when they are co‐expressed in cells. This discrepancy may be due to technical differences and different experimental strategies. Previous studies analyzing the transcriptome of virus‐infected cells were mainly based on next‐generation sequencing data sets; they showed only the general status of SARS‐CoV‐2 infection's influence on TE transposition. Our studies used in vitro assays to confirm the L1 inhibition activity of a single viral protein encoded by SARS‐CoV‐2, without virus infection. In combination, these results suggest that there was viral resistance to the extraordinary activation of TE. Strict transposition, although it may be not completely successful, permitted virus replication and transmission, even to acceptable levels. Marston et al. 47 reported that SARS‐CoV‐2 infection in COVID‐19 patients led to upregulation or downregulation of TE transcription in different tissue samples, which implies a complex relationship between SARS‐CoV‐2 and transposons that requires further investigation.
L1 transposition is an important cause of host genome instability, which can influence the intracellular gene expression of viral cofactors and the host microenvironment for virus replication. Moreover, accumulating evidence implies that L1 transposons are novel host innate immune activators, and nucleic acid derived from L1 transposition can be sensed by the cGAS‐STING or RLR pathway to stimulate host antiviral defenses. 48 Hence, we speculate that the elevated expression of retrotransposons upon virus infection is a host defense response, whereas SARS‐CoV‐2 encodes diverse proteins to control L1 mobility to evade over‐loaded immune activation.
Human cells have developed a number of regulators that coexist with transposons. Especially, many antiviral factors, including apolipoprotein B mRNA‐editing enzyme catalytic polypeptide‐like proteins, zinc finger antiviral proteins, three‐prime exonuclease 1, and SAMHD1, exert potent inhibitory effects on L1 retroelements. 11 To successfully infect host cells, viruses utilize diverse strategies to counteract host antiviral factors. However, the elimination of these antiviral factors upon virus infection might contribute to increased L1 mobility. Accumulating studies have reported that various viruses, such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), and enteroviruses, encode viral L1 inhibitors, generating a complex network to control L1 transposition. 30 , 49 , 50 HIV Vpr decreases the transposition rate of L1 in a cell cycle‐dependent manner. Additionally, Vpr can bind L1 ORF2p, subsequently inhibiting its RTase activity. 30 A recent study also showed that HCV infection results in decreased L1 activity via the redistribution of L1 ORF1p to HCV‐assembly sites in lipid droplets. 49 Further, we recently found that infection with different enteroviruses leads to the suppression of L1 mobility owing to the synergistic effects of diverse viral accessory proteins. 50 Herein, we demonstrated that the newly emerging virus SARS‐CoV‐2 has gained the ability to interfere with L1 activity, which also implies that other coronaviruses have potential L1‐inhibitory features. Future studies should focus on the roles of L1 inhibition during coronavirus infection.
The arms race between different viruses and host transposons supports the non‐negligible suppression of L1 activity during viral transmission. It is important to characterize the altered infectivity of RNA viruses in cells that have integrated viral genetic sequences. 17 , 20 , 35 Whether this cryptic insertion of viral cDNA is a viral strategy for persistent infection and transmission or an introduction of CRISPR‐like immunological “memory” to provide host cells with a molecular fingerprint of the viruses that infect them remains open to discussion and needs to be addressed urgently.
AUTHOR CONTRIBUTIONS
Wei Wei conceived and designed the experiments; Yan Li, Jiaxin Yang and Siyu Shen participated in multiple experiments; Yan Li, Jiaxin Yang, Siyu Shen, Wei Wang, Nian Liu, Haoran Guo, and Wei Wei analyzed the data; Yan Li and Wei Wei wrote the manuscript with help from all authors.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We thank Yuanyuan Li and Guoqiang Zhou for their technical assistance. We thank Dr. Kazazian HH Jr, Dr. John L Goodier, Dr. Pei‐Hui Wang, and Dr. Wenfeng An for their critical reagents. This study was supported by the National Natural Science Foundation of China (32222005 and 82172246), the National Major Project for Infectious Disease Control and Prevention (2018ZX10731‐101‐001‐016), the Department of Science and Technology of Jilin Province (No. 20190304033YY and 20180101127JC), the Open Project of Key Laboratory of Organ Regeneration and Transplantation, Ministry of Education, the Program for JLU Science and Technology Innovative Research Team (2017TD‐08), and Fundamental Research Funds for the Central Universities.
Li Y, Yang J, Shen S, et al. SARS‐CoV‐2‐encoded inhibitors of human LINE‐1 retrotransposition. J Med Virol 2022;1‐11. 10.1002/jmv.28135
Yan Li and Jiaxin Yang contributed equally to this study.
Contributor Information
Haoran Guo, Email: sheng315640@163.com.
Wei Wei, Email: wwei6@jlu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article. All relevant data are within the manuscript and its Supporting Information files.
REFERENCES
- 1. Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS‐CoV‐2. JAMA. 2020;323(22):2249‐2251. [DOI] [PubMed] [Google Scholar]
- 2. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2021;19(3):155‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295(37):12910‐12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scott A, Schmeckpeper BJ, Abdelrazik M, et al. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1(2):113‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belancio VP, Hedges DJ, Deininger P. Mammalian non‐LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18(3):343‐358. [DOI] [PubMed] [Google Scholar]
- 6. Kopera HC, Flasch DA, Nakamura M, Miyoshi T, Doucet AJ, Moran JV. LEAP: L1 element amplification protocol. Methods Mol Biol. 2016;1400:339‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mita P, Wudzinska A, Sun W, et al. LINE‐1 protein localization and functional dynamics during the cell cycle. eLife. 2018;7:e30058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Freeman BT, Sokolowski M, Roy‐Engel AM, Smither ME, Belancio VP. Identification of charged amino acids required for nuclear localization of human L1 ORF1 protein. Mob DNA. 2019;10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macia A, Widmann TJ, Heras SR, et al. Engineered LINE‐1 retrotransposition in nondividing human neurons. Genome Res. 2017;27(3):335‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suzuki J, Yamaguchi K, Kajikawa M, et al. Genetic evidence that the non‐homologous end‐joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5(4):e1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goodier JL. Restricting retrotransposons: a review. Mob DNA. 2016;7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brook Brouha JS, Badge RM, Lutz‐Prigge S, Farley AH, Moran JV, Kazazian HH, Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100(9):5280‐5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coufal N, Garcia‐PerezPeng JL, Peng GE, et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element‐1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci. 2011;108(51):20382‐20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao K, Du J, Han X, et al. Modulation of LINE‐1 and Alu/SVA retrotransposition by Aicardi‐Goutieres syndrome‐related SAMHD1. Cell Rep. 2013;4(6):1108‐1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volkmann B, Wittmann S, Lagisquet J, et al. Human TRIM5α senses and restricts LINE‐1 elements. Proc Natl Acad Sci USA. 2020;117(30):17965‐17976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geuking MB, Weber J, Dewannieux M, et al. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 2009;323(5912):393‐396. [DOI] [PubMed] [Google Scholar]
- 18. Klenerman P, Hengartner H, Zinkernagel RM. A non‐retroviral RNA virus persists in DNA form. Nature. 1997;390(6657):298‐301. [DOI] [PubMed] [Google Scholar]
- 19. Shimizu A, Nakatani Y, Nakamura T, et al. Characterisation of cytoplasmic DNA complementary to non‐retroviral RNA viruses in human cells. Sci Rep. 2014;4(1):5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L, Inmaculada Barrasa M, Hughes SH, Young RA, Jaenisch R, Richards A. Reverse‐transcribed SARS‐CoV‐2 RNA can integrate into the genome of cultured human cells and can be expressed in patient‐derived tissues. Proc Natl Acad Sci U S A. 2021;118(21):e2105968118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yin Y, Liu XZ, He X, Zhou LQ. Exogenous coronavirus interacts with endogenous retrotransposon in human cells. Front Cell Infect Microbiol. 2021;11:609160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smits N, Rasmussen J, Bodea GO, et al. No evidence of human genome integration of SARS‐CoV‐2 found by long‐read DNA sequencing. Cell Rep. 2021;36(7):109530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rookhuizen DC, Bonte P‐E, Ye M. Induction of transposable element expression is central to innate sensing. bioRXiv preprint. 2021. 10.1101/2021.09.10.457789 [DOI] [Google Scholar]
- 24. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu H, Chan JF‐W, Wang Y, et al. Comparative replication and immune activation profiles of SARS‐CoV‐2 and SARS‐CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID‐19. Clin Infect Dis. 2020;71(6):1400‐1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ostertag EM, Luning Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28(6):1418‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moran JV, Holmes SE, Naas TP, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917‐927. [DOI] [PubMed] [Google Scholar]
- 28. Goodier JL, Cheung LE, Kazazian HH, Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8(10):e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goodier JL, Zhang L, Vetter MR, Kazazian HH, Jr. LINE‐1 ORF1 protein localizes in stress granules with other RNA‐binding proteins, including components of RNA interference RNA‐induced silencing complex. Mol Cell Biol. 2007;27(18):6469‐6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawano K, Doucet AJ, Ueno M, et al. HIV‐1 Vpr and p21 restrict LINE‐1 mobility. Nucleic Acids Res. 2018;46(16):8454‐8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie Y, Rosser JM, Thompson TL, Boeke JD, An W. Characterization of L1 retrotransposition with high‐throughput dual‐luciferase assays. Nucleic Acids Res. 2011;39(3):e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J, Cruz‐cosme R, Zhuang M‐W, et al. A systemic and molecular study of subcellular localization of SARS‐CoV‐2 proteins. Signal Transduct Target Ther. 2020;5(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang W, Xu J, Yuan W, et al. APOBEC3DE inhibits LINE‐1 retrotransposition by interacting with ORF1p and influencing LINE reverse transcriptase activity. PLoS One. 2016;11(7):e0157220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kulpa DA, Moran JV. Cis‐preferential LINE‐1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13(7):655‐660. [DOI] [PubMed] [Google Scholar]
- 35. Horie M, Honda T, Suzuki Y, et al. Endogenous non‐retroviral RNA virus elements in mammalian genomes. Nature. 2010;463(7277):84‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thoms M, Buschauer R, Ameismeier M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS‐CoV‐2. Science. 2020;369(6508):1249‐1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi M, Wang L, Fontana P, et al. SARS‐CoV‐2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 2020. 10.1101/2020.09.18.302901 [DOI] [Google Scholar]
- 38. Ogando NS, Zevenhoven‐Dobbe JC, van der Meer Yvonne, Bredenbeek PJ, Posthuma CC, Snijder EJ. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS‐CoV and SARS‐CoV‐2. J Virol. 2021;94(23):e01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouvet M, Debarnot C, Imbert I, et al. In vitro reconstitution of SARS‐Coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshimoto FK. A biochemical perspective of the nonstructural proteins (NSPs) and the spike protein of SARS CoV‐2. Protein J. 2021;40(3):260‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moustaqil M, Ollivier E, Chiu H‐P, et al. SARS‐CoV‐2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infect. 2021;10(1):178‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muramatsu T, Kim YT, Nishii W, Terada T, Shirouzu M, Yokoyama S. Autoprocessing mechanism of severe acute respiratory syndrome coronavirus 3C‐like protease (SARS‐CoV 3CLpro) from its polyproteins. FEBS J. 2013;280(9):2002‐2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gallo A, Tsika AC, Fourkiotis NK, et al. (1)H,(13)C and (15)N chemical shift assignments of the SUD domains of SARS‐CoV‐2 non‐structural protein 3c: “the N‐terminal domain‐SUD‐N”. Biomol NMR Assignments. 2021;15(1):85‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan L, Zhang Y, Ge J, et al. Architecture of a SARS‐CoV‐2 mini replication and transcription complex. Nat Commun. 2020;11(1):5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knoops K, Kikkert M, van den Worm SHE. SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Macchietto MG, Langlois RA, Shen SS. Virus‐induced transposable element expression up‐regulation in human and mouse host cells. Life Sci Alliance. 2020;3(2):e201900536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marston JL, Greenig M, Singh M, et al. SARS‐CoV‐2 infection mediates differential expression of human endogenous retroviruses and long interspersed nuclear elements. JCI Insight. 2021;6(24):e147170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao K, Du J, Peng Y, et al. LINE1 contributes to autoimmunity through both RIG‐I‐ and MDA5‐mediated RNA sensing pathways. J Autoimmun. 2018;90:105‐115. [DOI] [PubMed] [Google Scholar]
- 49. Schobel A, Nguyen‐Dinh V, Schumann GG, Herker E. Hepatitis C virus infection restricts human LINE‐1 retrotransposition in hepatoma cells. PLoS Pathog. 2021;17(4):e1009496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y, Shen S, Guo H, et al. Enterovirus infection restricts long interspersed element 1 retrotransposition. Front Microbiol. 2021;12:706241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article. All relevant data are within the manuscript and its Supporting Information files.


