Abstract
The emergence of the SARS‐CoV‐2 Omicron variant (B.1.1.529) has created great global distress. This variant of concern shows multiple sublineages, importantly B.1.1.529.1 (BA.1), BA.1 + R346K (BA.1.1), and B.1.1.529.2 (BA.2), each with unique properties. However, little is known about this new variant, specifically its sub‐variants. A narrative review was conducted to summarise the latest findings on transmissibility, clinical manifestations, diagnosis, and efficacy of current vaccines and treatments. Omicron has shown two times higher transmission rates than Delta and above ten times more infectious than other variants over a similar period. With more than 30 mutations in the spike protein's receptor‐binding domain, there is reduced detection by conventional RT‐PCR and rapid antigen tests. Moreover, the two‐dose vaccine effectiveness against Delta and Omicron variants was found to be approximately 21%, suggesting an urgent need for a booster dose to prevent the possibility of breakthrough infections. However, the current vaccines remain highly efficacious against severe disease, hospitalisation, and mortality. Japanese preliminary lab data elucidated that the Omicron sublineage BA.2 shows a higher illness severity than BA.1. To date, the clinical management of Omicron remains unchanged, except for monoclonal antibodies. Thus far, only Bebtelovimab could sufficiently treat all three sub‐variants of Omicron. Further studies are warranted to understand the complexity of Omicron and its sub‐variants. Such research is necessary to improve the management and prevention of Omicron infection.
Keywords: B.1.1.529, BA.1, BA.2, Omicron, SARS‐CoV‐2, subvariant
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- COVID‐19
coronavirus disease 2019
- LMICs
low‐middle‐income countries
- RBD
receptor‐binding domain
- S‐protein
spike protein
- SPR
surface plasmon resonance
- VE
vaccine effectiveness
- VOC
variant of concern
- WHO
World Health Organization
1. INTRODUCTION
A new wave of uncertainty came among researchers and physicians as they navigated with the many knowns of the Omicron variant such as presentation, transmission and genetic variation, all while trying to still discover the many unknowns such as subvariant differences. The morbidity and mortality of COVID‐19 have changed the world despite the advanced research working towards its management and prevention. However, the SARS‐CoV‐2 virus continued to evolve into multiple variants, including the most recent variant of concern (VOC), Omicron (B.1.1.529). This variant was initially reported to the World Health Organization (WHO) by South Africa on 24 November 2021, after a proactive and thorough investigation of a sudden surge of cases, which led to the identification of Omicron through genome sequencing. 1 , 2 , 3 WHO classified Omicron as a VOC on 26 November 2021, along with Alpha, Beta, Gamma, and Delta variants. 3 , 4
Omicron has the highest number of mutations compared to other variants discovered to date, with over 30 mutations located within the spike protein (S‐protein), a critical component in determining the infectivity and antigenicity of the virus. 5 In addition, 15 of these mutations are located in the receptor‐binding domain (RBD) of S‐protein, which are believed to lead to breakthrough infections or reinfections. 5 Surveillance shows that certain sub‐lineages identified of the Omicron variant have shown increased prevalence. 6 As a result, there is a significant concern of Omicron having higher transmission and hospitalisation rates, significantly reduced vaccine effectiveness (VE), development of more severe disease, and increased mortality compared with other variants.
2. DISCUSSION
This review aims to gain in‐depth insight into multiple aspects of the Omicron variant for appropriate management and public health preventive measures in response to this unanticipated crisis.
2.1. Epidemiology
Since the initial sudden surge of cases in South Africa, Omicron was detected in 27 countries within a week, which quickly expanded to 40 countries by December 6th, further increasing to 65 countries in 2 weeks, and a staggering 110 countries as of 23 December 2021. 7 , 8 , 9 , 10 , 11 As of 8 January 2022, there are a total of 552,191 confirmed cases of Omicron in 150 countries, with 115 confirmed deaths (Figure 1). 12 Since that date, we no longer tracked the number of global cases and only a small portion of confirmed COVID‐19 cases have undergone genomic sequencing to confirm the variant; the actual case number is expected to be substantially higher than reported.
FIGURE 1.
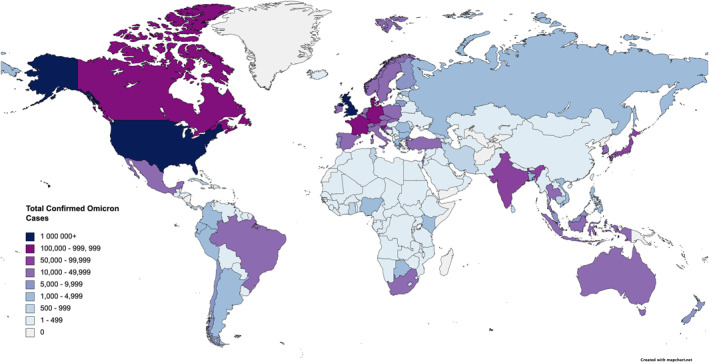
The geographical representation of the omicron case distribution as of 8 January 2022
2.2. Transmissibility
Approximately 50 mutations had been detected in Omicron, with over 30 mutational hotspots in the S‐protein, particularly in the RBD. 5 , 13 , 14 , 15 , 16 Viral sequencing showed that only 35% of S‐protein mutations were found in prior VOCs. 17 The S‐protein is essential to the virus as it binds to angiotensin‐converting enzyme 2 (ACE2) receptors, allowing it to enter host cells. 15 Surface plasmon resonance (SPR) was used to measure mutational affinity and kinetics of the RBD/ACE2 complex in one study, along with Cryo‐electron microscopy in another study to show that S477N, Q498R and N501Y mutations cause S‐proteins to bind to ACE2 receptors with stronger binding affinity, facilitating viral transmissions. 18 , 19 Numerous new modifications proximal to the furin cleavage site also enhances the transmissibility. 20
Overlapping mutations between Omicron and other variants, including T478K, E484A, K417N, K440N, and S446K, are associated with increased neutralising antibody resistance and immune avoidance. 21 This brings concern as Omicron's mutations increase as both antibodies evade and escape death, unlike ancestral variants. 17 , 22 The three predominant sublineages, BA.1, BA.1.1 and BA.2, share 21 mutations. BA.1.1 has an additional R346K mutation with 13 spike mutations, whereas BA.2 was found with 8 spike mutations. Figure 2 depicts the mutations in the Omicron variant, the difference among Omicron subgroups, and common shared mutations with the ancestor variants. 23 , 24 Amongst the primary Omicron sub‐variants, BA.1 is the dominant form overall, followed by BA.1.1 and BA.2. But the BA.2 strain kept rising and became most prevalent in several countries. 6 , 25 Figure 3. 6 , 26
FIGURE 2.
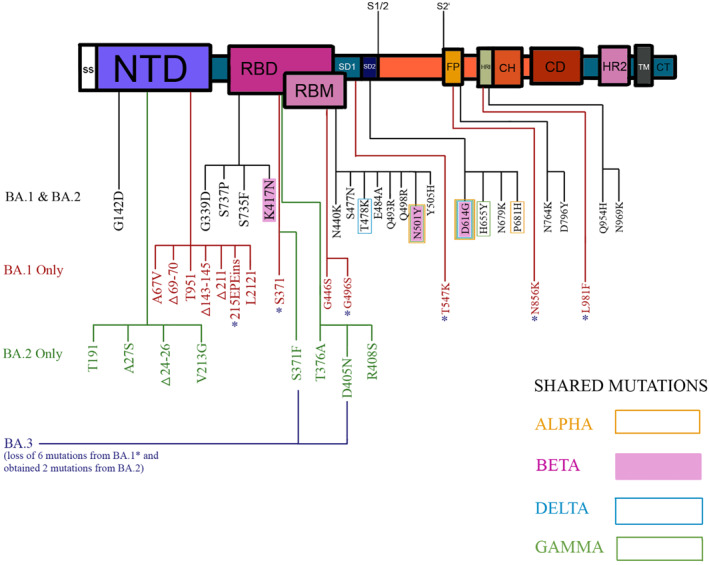
The mutations of the omicron variant highlighting the differences among omicron subgroups and shared mutations with ancestral variants
FIGURE 3.
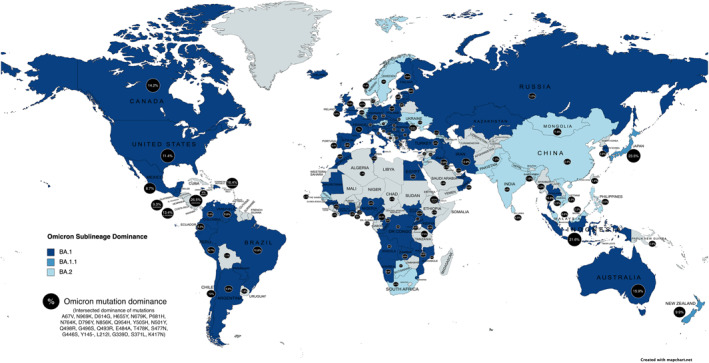
The geographical representation of the omicron sublineage dominance with percentage of mutation dominance across the globe
A study demonstrated that Omicron to be around two timesas infectious as the Delta variant and maybe over ten times more contagious than the other variants. 27 Danish data further suggested that Omicron can replicate 3.19 times (95% confidence interval (95% CI: 2.82–3.61) more than the Delta variant, which is consistent with another study in South Africa showing 4.2 times (95% CI: 2.1–9.1) greater replication. 28 , 29 This reproduction rate was further shown to be 4.0 times in the United Kingdom and 2.5 times in India. 30 Omicron was found to have an estimate of 100.3% higher transmission rate (95% CI: 74.8%–140.0%) than ancestral variants and an estimate of 36.5% higher transmission rate (95% CI: 20.9%–60.1%) than the Delta variant. 31 Still, the substantial increase in the transmission rate of Omicron can also be attributed to a number of other factors such as preferential infection in the upper airways making it easier for infection to pass along, potential to cause immune escape and the effect of lowering of health protocols. 32
The doubling time of Omicron was within 1.5 days after a surge point of 10 cases per 100,000 population, whereas both Delta and Beta were 1.9 days. 20 The overall doubling time for Omicron was 1.5–3.0 days which may explain the exponential case growth in many countries (Figure 4). 33 Interestingly, a recent preprint showed evidence suggesting that BA.2 was 1.4 times higher than BA.1 concerning the effective reproduction number. 25
FIGURE 4.
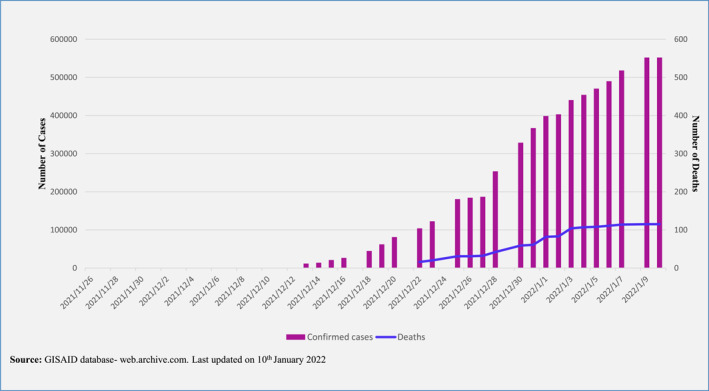
A bar graph depicting the number of confirmed omicron cases with a trend line showing the number of deaths due to omicron
All in all, the unique mutations are seen in Omicron magnify concern of infectivity, treatment efficiency, vaccine breakthrough, and risk of reinfection. 21 , 34 , 35 , 36 This high rate of transmission also urges faster treatment and management to reduce exposure and further population morbidity. 30
2.3. Diagnosis of omicron
To date, there is no exact diagnostic measure to detect Omicron comparable to the efficient RT‐PCR test used for the previous strains. Sensitivity and specificity data on diagnosing Omicron cases with RT‐PCR is predominantly lacking.
The diagnosis of Omicron with a reliable test is a major concern globally, as many of the currently used RT‐PCR, including those based on mutation targets, may produce some level of false‐negative results (>1.4%) when it comes to Omicron detection. 37 , 38 Since modifications in the viral genome may alter the implementation of an assay, for accurate interpretation, clinical manifestations, patient history, and epidemiological data should also be tackled with a negative result. 39 Although limited evidence suggests that the Omicron variant can be detected using RT‐PCR and rapid antigen assays without altering the overall diagnostic accuracy of these assays, the comparative sensitivity data are yet to be available to validate this accuracy. 33 , 38 , 40 The partial detection failure (one of the three target genes) of some RT‐PCR tests may be utilised to detect possible Omicron. 41 Targeting the S gene, only two of the eight assays showed S‐gene dropout with this variant. 42 Despite this, S gene target failure (SGTF) or ‘S gene dropout’ producing a false‐negative result may be used as a proxy marker for screening Omicron. 14 However, a minority of publicly shared sequences (a new Omicron ‘offshoot’) lack this deletion and other VOCs that harbor this deletion. It demands sequence‐based confirmation making it potentially tougher to trace. 33 Assays designed to detect common proteins whose genes were mutated in Omicron may fail to detect actual positive cases. For instance, the spike gene 69–70 deletion existed in ∼1% of the circulating variants questioning its universal application in some populations. 43
Several assay systems are being developed to detect Omicron variants. One method designed to detect Omicron is OmMet, validated in silico through a variant‐specific set of primers for RT‐PCR. 44 It is yet to be verified by laboratories using clinical samples and improved as needed. A newly developed RT‐PCR assay using a new set of primers (targeting mutations in the nsp6 (Orf1a), spike, and nucleocapsid genes) was also found to detect Omicron accurately. 43 Although the ten antigen kits showed similar analytical sensitivity for both Delta (6.5 log10 copies/ml (Ct 25.4)) and Omicron (6.39 log10 copies/ml (Ct 25.8)) variants, all kits failed to detect these variants at the lowest dilutions (5.23 log10 copies/ml, Ct 28.8 and 5.33 log10 copies/ml, Ct 28.8, respectively). 37
2.4. Clinical signs and symptoms or omicron
Early reports from South Africa implied that the clinical presentation of Omicron does not differ from other variants, with no reports of any unusual symptoms as the common presentation is shown in Figure 5. 20 , 45 , 46 A study conducted in Canada has shown that of all confirmed and suspected Omicron cases, 9.6% of patients were asymptomatic, only 10% of cases reported shortness of breath. 47 , 49 Similar symptoms were seen in an outbreak in Norway, and several reported cases in South Korea both showed cough and nasal congestion being predominantly reported symptoms and no severe outcomes. 45 , 48
FIGURE 5.
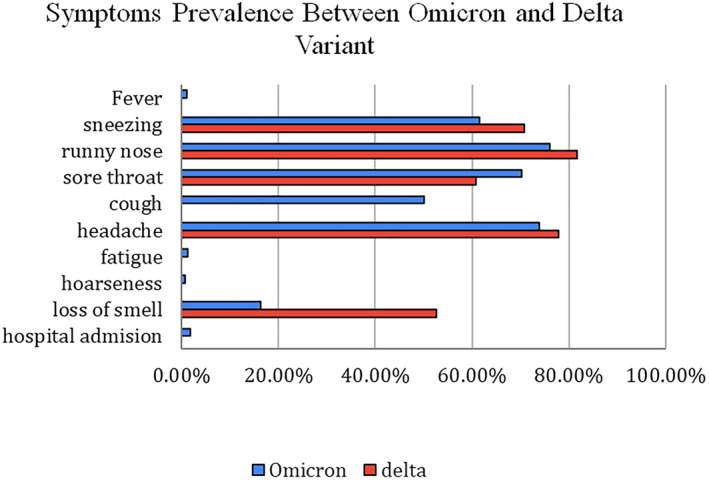
A graph comparing the percentage prevalence of symptoms between the omicron and delta variant
South African reports suggested a milder disease course as Omicron cases surge in the community. The risk of hospitalisation and requirement of advanced care is decreased compared to previous waves when adjusted for vaccination statuses of infected people. However, it must also be noted that this surge (14 November 2021–16 December 2021) predominantly infected a younger age group (30–39 year age group) as compared to prior waves (4 May 2021–13 November 2021) in which the mean age was 49.8 years, which may explain the improved disease prognosis compared to other variants, which can be a confounding factor. 49 , 50 It should be noted that these studies may not accurately depict the findings and may lead to underestimation of severity in younger patients, unknown vaccination rates and unknown history of COVID‐19 infection. Confounding factors such as the age and overall health of the patient populations and improved healthcare preparedness must be considered when interpreting the rate of severe disease, hospitalisation, and death. Notably, according to a preprint, infection experiments using a hamster model from Japanese researchers recently indicated that BA.2 causes more severe disease with more pulmonary damage and body weight loss than the original Omicron strain, BA.1. 25
2.5. Effectiveness of current vaccines on protection against infection and severe disease
To date, several studies have revealed that Omicron reduced the effectiveness of vaccines and neutralised antibodies to an alarming extent which could enhance the risk of breakthrough infections but, fortunately, was restored after obtaining a booster shot (Table 1). 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 According to a recently released preprint, neutralising antibodies in vaccinated and previously infected individuals with the Omicron variant also increase against both Omicron and Delta, 14.4‐fold and 4.4‐fold, respectively, which may decrease the re‐infection with Delta. 69 A recent study demonstrated that receiving the third dose of mRNA COVID‐19 vaccine was more protective with fewer cases of symptomatic infection contrasted with unvaccinated and with primary course alone. However, the adjusted odds ratio (comparison of 3 Doses vs. 2 Doses) for Omicron was significantly higher than for Delta, with 0.34 (95% CI, 0.32–0.36) and 0.16 (95% CI, 0.14–0.17), respectively. This finding supported evidence that booster doses are less effective against Omicron than previous variants, including Delta. 70 These findings had a non‐significant difference among all three Omicron sublineages, including BA.1, BA.1.1, and BA.2. 6
TABLE 1.
Antibody neutralisation level and vaccine efficacy against SARS‐CoV‐2 Omicron variant
| Authors | Country of first author | Neutralisation assay method | Sample size | Primary vaccine | Primary virus neutralisation | Booster dose interval | Booster vaccine | Booster virus neutralisation | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Time post‐primary dose | Antibody neutralisation or vaccine efficacy (%) | Time post‐booster dose | Antibody neutralisation or vaccine efficacy (%) | |||||||
| Ai et al (Dec 2021) | China | Pseudotyped virus | 37 | BBIBP‐CorV (Sinopharm) | 2 weeks | 11.16‐fold reduction | 4–8 months | BBIBP‐CorV (Sinopharm) | 2 weeks | 5.06‐fold increase |
| 4 weeks | 4.95‐fold increase | |||||||||
| ZF2001 (Zifivax) | 2 weeks | 9.95‐fold increase | ||||||||
| 4 weeks | 11.28‐fold increase | |||||||||
| Lu et al (Dec 2021) | China | Live virus | 50 | BNT162b2 (Pfizer/BioNTech) | <1 month | 39.9‐fold reduction | NA | NA | NA | NA |
| CoronaVac (Sinovac) | <1 month | 4.3‐fold reduction | ||||||||
| Yu et al (Dec 2021) a | China | Pseudotyped virus | 292 | BBIBP‐CorV (Sinopharm) | 28 days | 20.1‐fold reduction | 8–9 months | BBIBP‐CorV (Sinopharm) | 28 days | 3.3‐fold increase |
| Muik et al (Dec 2021) a | Germany | Pseudotyped virus | 51 | BNT162b2 (Pfizer/BioNTech) | 21 days | 22.8‐fold reduction | >6 months | BNT162b2 (Pfizer/BioNTech) | 1 month | 23.4‐fold increase |
| Nemet et al (Dec 2021) | Israel | Live virus | 20 | BNT162b2 (Pfizer/BioNTech) | 166 days | 14.9‐fold reduction | NA | BNT162b2 (Pfizer/BioNTech) | 25 days | 96.9‐fold increase |
| Cele et al (Dec 2021) a | South Africa | Live virus | 19 | BNT162b2 (Pfizer/BioNTech) | 10–63 days | 22‐Fold reduction | NA | NA | NA | NA |
| 10–63 days | 73% (vaccinated and previously infected participants) | |||||||||
| 10–63 days | 35% (vaccinated only participants) | |||||||||
| Cameroni et al (Dec 2021) a | Switzerland | Pseudotyped virus | 170 | BNT162b2 (Pfizer/BioNTech) | 14–28 days | 44‐Fold reduction | NA | NA | NA | NA |
| mRNA‐1273 (Moderna) | 14–28 days | 33‐Fold reduction | ||||||||
| ChAdOx1 (AstraZeneca) | 14–28 days | 36‐Fold reduction | ||||||||
| Ad26.COV2.S (Johnson and Johnson) | 14–28 days | No protective effect | ||||||||
| Sinovac | 14–28 days | No protective effect | ||||||||
| Sputnik | 14–28 days | No protective effect | ||||||||
| Andrews et al (Dec 2021) a | UK | Live virus | 581 | BNT162b2 (Pfizer/BioNTech) | 2–9 weeks | 88% | NA | BNT162b2 (Pfizer/BioNTech) | 2 weeks | 75.5% |
| 10–14 weeks | 48·5% | |||||||||
| 15 weeks | 34%–37% | |||||||||
| ChAdOx1 (Vaxzevria, AstraZeneca) | 15 weeks | No protective effect | BNT162b2 (Pfizer/BioNTech) | 2 weeks | 71.4% | |||||
| Dejnirattisai et al (Dec 2021) | UK | Live virus | 43 | ChAdOx1 (AstraZeneca) | 28 days | 13.3 fold reduction | NA | NA | NA | NA |
| BNT162b2 (Pfizer/BioNTech) | 28 days | 29.8 fold reduction | ||||||||
| Mallory et al (Dec 2021) a | UK | hACE2 receptor binding test | 257 | NVX‐CoV2373 (Novavax) | 14 days | 8.2‐fold reduction | 6 months | NVX‐CoV2373 (Novavax) | 28 days | 14.8‐fold increase |
| Doria‐Rose et al (Dec 2021) a | USA | Pseudotyped virus | 7 | mRNA‐1273 (Moderna) | 2 weeks | 8.9‐fold reduction | NA | mRNA‐1273 (Moderna) | 2 weeks | 12.6‐fold increase |
| Edaraet al (Dec 2021) a | USA | Live virus | 138 | BNT162b2 (Pfizer/BioNTech) or mRNA‐1273 (Moderna) | N/A | 30‐Fold reduction | 8 months | BNT162b2 (Pfizer/BioNTech) or mRNA‐1273 (Moderna), Homologous (mostly) | 1–4 weeks | 14‐Fold reduction |
| 6 months | No protective effect | |||||||||
| Garcia‐Beltran et al (Jan 2022) | USA | Pseudotyped virus | 239 | mRNA‐1273 (Moderna) | <3 months | 43‐Fold reduction | NA | mRNA‐1273 (Moderna) | <3 months | 19‐Fold increase |
| BNT162b (Pfizer/BioNTech) | <3 months | 122‐fold reduction | BNT162 (Pfizer/BioNTech) | <3 months | 27‐Fold increase | |||||
| Ad26.COV2.S (Johnson and Johnson) | N/A | NA | mRNA‐1273 (Moderna) | <3 months | 4‐Fold increase | |||||
| Gardner and Kilpatrick (Dec 2021) a | USA | NA | NA | BNT162b2 (Pfizer/BioNTech) | Shortly after vaccination | 84.9% | NA | BNT162b2 (Pfizer/BioNTech) | Shortly after a booster | 91.7% |
| 6 months | 63.1% | |||||||||
| mRNA‐1273 (Moderna) | 6 month | >63.1% | mRNA‐1273 (Moderna) | Shortly after a booster | <91.7% | |||||
| Lusvarghi et al (Dec 2021) a | USA | Pseudotyped virus | 39 | BNT162b2 (Pfizer/BioNTech) | 19–41 days | 25.5‐fold reduction | NA | BNT162b2 (Pfizer/BioNTech) | 26–60 days | 31.8‐fold increase |
| Schmidt et al (Dec 2021) a | USA | Pseudotyped virus | NA | BNT162b2 (Pfizer/BioNTech) or mRNA‐1273 (Moderna) | 1.3 months | 127‐fold reduction | >6 months | BNT162b2 (Pfizer/BioNTech) | 1 month | 38‐Fold increase |
| 5 months | 27‐Fold reduction | |||||||||
| Ad26.COV2.S (Johnson and Johnson) | NA | No protective effect | ||||||||
| Syed et al (Jan 2022) a | USA | SARS‐CoV‐2 virus‐like particles | 38 | BNT162b2 (Pfizer/BioNTech), mRNA‐1273 (Moderna) or Ad26.COV2.S (Johnson and Johnson) | NA | 15‐Fold reduction | NA | BNT162b2 (Pfizer/BioNTech) | NA | Significant increase |
| Zeng et al (Dec 2021) a | USA | Pseudotyped virus | 48 | BNT162b2 (Pfizer/BioNTech) or mRNA‐1273 (Moderna) | 3–4 weeks | 22.9‐fold reduction | NA | BNT162b2 (Pfizer/BioNTech) or mRNA‐1273 (Moderna) | 1–11 weeks | 3.3‐fold reduction |
Abbreviations: NA, not available; USA, United States of America; UK, United Kingdom.
Preprints.
The results released as a preprint from a research group in the U.S. have suggested that Omicron reduced VE by four to ten times; specifically, VE for people who had recently been vaccinated the second dose of Pfizer or Moderna vaccine was about 83% against the symptomatic disease of Delta, but only around 21% against Omicron. 54 Similarly, a study on cancer patients exhibited a considerable increase in protection of neutralising antibodies against this new variant after a booster dose in comparison with individuals who obtained only a two‐dose course. 71 A published study from pseudovirus data that includes variants sharing specific spike mutations, including Omicron, contributes additional proof that the neutralising antibody magnitude overall extends with repeated immunising exposures from prior infection accompanied by vaccination or an extra dose of vaccine. 72
In terms of the impact of vaccines on protection against the outcomes caused by Omicron, preliminary data to date has revealed that the existing vaccines remain efficacious against severe illness, hospitalisation, and mortality. 20 , 22 , 66 According to Pfizer, the fully 2‐dose Pfizer vaccination can provide more than 80% safeguard against severe illness and mortality for 6 months; the booster dose aids in increasing protection by 10%; exceptionally, this may still be effective with Omicron infections. 73 Even though the significant reduction of the neutralisation level with Omicron has been demonstrated, the residual level is still higher than the minimal level, which may be sufficient for protection against severe disease. The latest study released as a preprint has also indicated that vaccine efficacy against severe disease is about 77% for the vaccinated group. 66
Studies have suggested T cells might not prevent infections, but they would be potential for defense against severe outcomes which might be less dependent on antibodies. 20 , 22 , 74 Furthermore, other than neutralising antibody levels, T‐cell immunity is not likely to dramatically decrease with Omicron infection. 66 An additional insight contributing to the vaccine‐induced protecting immunological mechanisms is the maintenance of Omicron Spike‐specific FcγR2a and Fcγ3a binding antibodies across all three vaccines, including the Moderna mRNA‐1273, Pfizer/BioNTech BNT162b2, and Sinovac CoronaVac. 75 For these reasons, vaccine efficacy is likely preserved to prevent severity and mortality followed by infections of this novel variant, albeit the significant loss of antibody neutralisation.
2.6. Effectiveness of current treatments
Not a novelty in medicine, especially in the ICU; however, sparking interest in COVID‐19 is the concept of a cytokine storm. To find the optimal treatment, understanding the pathogenesis is essential. A ‘cytokine storm’ is a clinical syndrome induced by a tremendous number of cytokines released by highly pathogenic viruses. 76 SARS‐CoVssRNAs displayed potent immunostimulatory properties, causing the release of pro‐inflammatory cytokines TNF‐α, IL‐6, and IL‐12. Furthermore, one cytokine can promote other cytokines to boost the pro‐inflammatory response, which the cytokine storm reaction is subsequently significant in acute lung injury (ALI). This cytokine‐release reaction (primarily associated with IL6) is a hypersensitive reaction (HSR), which leads to the activation of direct and indirect complement cascades, generation of anaphylatoxins such as C3a and C5a and release of inflammatory mediators such as histamine, leukotrienes and prostaglandins. In addition, the stimulation of the coagulation system, both direct and indirect, is another feature of cytokine storm reaction. Therefore, a new potential therapeutic approach of COVID‐19, immunomodulators, acts on the mechanism of preventing the cytokine storm targeting anti‐IL6R monoclonal antibodies (mAb) and other molecules associated with the IL‐6/IL‐6R axis. 77
World Health Organization has reported that therapies that target the cytokine storm responses, including corticosteroids, interleukin 6 receptor blockers, and prophylaxis with anticoagulation, are still effective to severe or critical Omicron infected patients. 78 However, Omicron possesses many mutations in the RBD of the S‐protein, which is also the target site of most monoclonal antibody treatments. 21 The difference in the structure of spike glycoprotein may contribute to why certain drugs are not effective against the Omicron variant. 13 Research suggests that certain monoclonal antibodies may be less potent or even non‐effective in treating Omicron, while some remain susceptible to its treatment (Table 2). 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 89
TABLE 2.
Effectiveness of antiviral and monoclonal antibody therapies for treating Omicron variant
| Type of therapy | Source | Antibody ID (mAbs) | Name of medication | Date of first EUA/Approval issuance | Authorised use | Effectiveness against omicron variant | Neutralising activity of mAbs against omicron |
|---|---|---|---|---|---|---|---|
| Antivirals | Pfizer | NA | PAXLOVID (Ritonavir‐boosted nirmatrelvir) | Authorised under FDA EUA on 22 December 2021. | EUA for the treatment of patients with mild to moderate COVID‐19 in high‐risk individuals aged ≥12 years and weighing ≥40 kg. | Remain effective | NA |
| Antivirals | Gilead | NA | Remdesivir | Approved by FDA on 21 January 2022 | Treatment of COVID‐19 in individuals aged ≥12 years and weighing ≥40 kg. | Remain effective | NA |
| Antivirals | Merck | NA | Molnupivarir | Authorised under FDA EUA on 23 December 2021. | EUA for the treatment of mild to moderate COVID‐19 in high‐risk individuals aged ≥18 years. | Remain effective | NA |
| Monoclonal antibodies | Vir | S309 | Sotrovimab | Authorised under FDA EUA on 26 May 2021. | EUA for the treatment of mild to moderate COVID‐19 in individuals aged ≥12 years and weighing ≥40 kg. | Remain effective | <2 to 2.7‐fold reduction |
| Monoclonal antibodies | Regeneron | REGN10933/REGN10987 | REGEN‐COV (Casirivimab + imdevimab) | Authorised under FDA EUA on 21 November 2020. a | EUA for post‐exposure prophylaxis of COVID‐19 or the treatment of mild to moderate COVID‐19 in individuals aged ≥12 years and weighing ≥40 kg. | Ineffective | Non‐neutralising at the highest concentration tested (10,000 ng ml−1) |
| Monoclonal antibodies | Eli Lilly | LY‐COV555/LV‐COV016 | Bamlanivimab + etesevimab | Authorised under FDA EUA on 09 February 2021. a | EUA for post‐exposure prophylaxis of COVID‐19 or the treatment of mild to moderate COVID‐19 in individuals aged ≥12 years and weighing ≥40 kg. | Ineffective | Non‐neutralising at the highest concentration tested (10,000 ng ml−1) |
| Monoclonal antibodies | Celltrion | CT‐P59 | Regdanvimab | Not yet approved by FDA | The extended use in elderly patients aged 50 years and over, or with at least one underlying medical condition with mild symptoms of COVID‐19 | Ineffective | Non‐neutralising at the highest concentration tested (10,000 ng ml−1) |
| Fully approved by Korean Ministry of food and drug safety (MFDS) | Adults with moderate symptoms of COVID‐19. | ||||||
| Monoclonal antibodies | Astra Zeneca | COV2‐2130/COV2‐2196 | Evushield (Tixagevimab + cilgavimab) | Authorised under FDA EUA on 08 December 2021. | EUA for pre‐exposure prophylaxis for prevention of COVID‐19 in individuals aged ≥12 years and weighing ≥40 kg. | Less effective | 12‐Fold reduction |
| Monoclonal antibodies | Eli Lilly | LY‐COV1404 | Bebtelovimab b | Authorised under FDA EUA on 02 February 2022. | EUA for the treatment of mild to moderate COVID‐19 in individuals aged ≥12 years and weighing ≥40 kg. | Remain effective | Retained full neutralisation potency |
Abbreviation: EUA, Emergency Use Authorisation.
Due to the high frequency of the Omicron variant, REGEN‐COV may not be administered for treatment or post‐exposure prevention of COVID‐19 under the Emergency Use Authoriation. And this treatment is not currently authorised for use anywhere in the U.S.
Remain effective to all three sublineages of Omicron (BA.1, BA.1.1, BA.2).
Omicron evolution continues to create its sublineages, including B.1.1.529.1 (BA.1), BA.1 + R346K (BA.1.1), and B.1.1.529.2 (BA.2). Their structure mutation has changed these sublineages, making them distinct to therapeutic interventions, specifically the monoclonal antibodies. A recent study indicated that only bebtelovimab, recently authorised by FDA, could sufficiently cover all three sub‐lineages of Omicron. The inhibitory activity of sotrovimab against BA.2 is a 27‐fold reduction, while its activity remains effective against BA.1 and BA.1.1. 6 Therefore, to control this ever‐evolving virus, it is urgent to prioritise studies that focus on novel protective variant‐specific monoclonal antibody therapy and its neutralising ability.
2.7. A deeper look at the known unknowns
Omicron might have an origin in animals. Cell‐based studies still as preprints found that, different from previous variants, the spike protein of Omicron may be able to bind to the ACE2 proteins of a few kinds of animals, including turkeys, chickens, and mice. 90 , 91 Another preprint also revealed that Omicron binds tightly to rat ACE2 by the presence of N501Y–Q498R combination of mutations. 92 Furthermore, the kinds of single‐nucleotide substitution seen in Omicron genome resembled the scopes typically observed with virus evolution in a mouse. 93 Besides, a few plausible hypotheses for Omicron's fast‐paced evolution are low healthcare facilities and a high immunocompromised population who cannot easily get rid of the virus. Omicron has quickly dominated others when sequenced in South Africa and many other countries following that, as seen in (Figure 6). 33 , 94 , 95 , 96 It is interesting to note that even though the RBD's affinity for ACE2 aids in transmission, it is not the main reason behind higher transmissibility. 97
FIGURE 6.
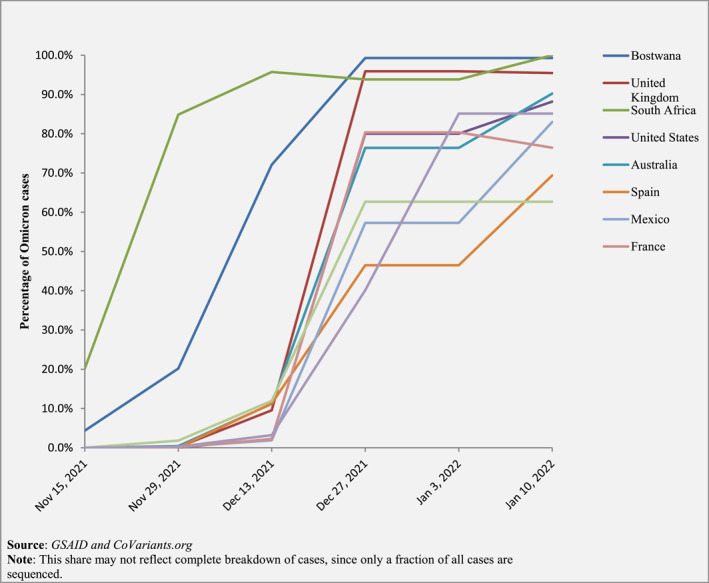
A line graph demonstrating the dominance of omicron across the countries with the most cases as of 10 January 2021
Omicron was initially assessed to spread three to six folds higher than Delta within the same period. 98 A study has revealed that Omicron replicates 70‐fold faster than the Delta and other variants in the human bronchus. However, it multiplies ten times less in lung tissues than in the Delta. This may explain why this novel variant is more contagious but less severe clinically than the original variants, including the Delta variant. 32 Preliminary reports suggest Omicron may present milder and even silent symptoms in patients, with studies showing only 10% of patients reporting shortness of breath. 47 Such symptoms result in diagnosis concern; however, definitive data lacks the prevalence severity of outcomes compared to its counterpart variants. One of the most urgent needs to combat the crisis is to develop highly sensitive and specific new assays (regardless of RT‐PCR or rapid antigens tests) and other new laboratory techniques.
Omicron showed great concern among the pediatric age group with a 5‐time increased hospitalisation rate among children between 0 and 4 years and the highest rate being among children under 6 months of age. 99 However, it was later assessed that the risk of hospitalisation on an individual basis was actually significantly lower than the Delta variant. This surge of cases may be due to lack of vaccination and immunity against the virus among children, yet the real reason is yet to be understood by researchers. However, clinicians worry about the possibility of developing the still misunderstood condition of long COVID and other rare yet serious long‐term consequences such as multisystem inflammatory system. 100 The same remains a mystery among all ages, with increased concern for the elderly or immunocompromised, which is why physicians are urging all to get vaccinated and practice other health precautions.
The emphasis of vaccinations was made as scientists believe T‐cell immunity, rather than antibody neutralisation, is more likely responsible for the severe outcomes of Omicron infections. 20 , 74 Additionally, being the primary site to target several antibodies, the mutated S‐protein in Omicron may result in breakthrough infections, emphasising an urgent need for a widespread booster dose. Further information and studies are also required to evaluate the protective period following a booster dose for a more extended duration and the safety of third‐dose boosters, especially with different types of vaccines. Besides, vaccine companies continue conducting trials for developing Omicron‐specific vaccines as well as boosters. Pfizer and Moderna are currently developing an Omicron vaccine, while Johnson and Johnson and Sputnik V are ongoing booster studies. Notably, preliminary data elucidated that the Omicron subvariant, BA2, had higher transmissibility, higher level of illness severity, and higher resistance to sotrovimab‐an effective monoclonal antibody against ancestral variants ‐ when compared to the original B.1 virus. 6 , 25 These findings may pose BA.2 as the most concerning variant for global health in the foreseeable future, and more strain‐specific studies are essential to be done.
The COVID‐19 vaccine still remains a controversial topic around the world. Public health specialists continue to encourage vaccination to the public as one of the most main measures to stay protected from COVID‐19. There are currently at least 8 fully approved vaccines and over 120 others that are in human clinical trial phases. These vaccines were each created using different biomedical approaches. Both Pfizer and Moderna vaccines employed a synthetic mRNA by encoding for the SARS‐CoV‐2 S‐protein sequence. The Oxford‐AstraZeneca, Gamaleya (Sputnik V), Janssen, and CanSino all developed recombinant vaccines which were based on a DNA sequence encoding for the S‐protein. The Sinopharm, Sinovac, and Bharat Biotech vaccines were produced through the inactivation od the SARS‐CoV‐2 grown in Vero cells. Finally, the Norovax is a recombinant protein subunit vaccine. 101 Despite the development and approval of so many vaccines, the acceptance of vaccination by a community highly influences the success of vaccination programs. Currently, half the global population still remains unvaccinated with a vaccine coverage of less than 20% in low‐middle‐income countries (LMICs). 101
Challenges in vaccination acceptance included various perceptions. A study in the UK and Turkey showed that factors associated with acceptance included anxiety, risk perception, government satisfaction and the belief that the natural origin of the virus correlated to acceptance of vaccination. 102 Another Italian study identified age, gender and socioeconomic status to be associated with vaccine acceptance. 103 An acceptance rate of 71.5% was seen among 19 countries with factors such as the impact on their country and actions taken by the government to heavily influence vaccine acceptance. 104 As one of the top global threats, it was found that vaccine hesitancy was mostly seen among Asian and African ethnicity, Muslim and Buddhists religion, low socioeconomic groups and among young females. 105 Vaccine hesitancy results as a direct consequence of lack of awareness and conflicting beliefs in regard to objective, effectiveness and adverse effects of the vaccine. A survey study indicated that not most people achieved an antibody level to protect against SARS‐CoV2 after receiving the Sinovac COVID‐19 vaccine. It may be more effective if boosted with a heterologous vaccine. 106 Additionally, willingness‐to‐pay (WTP) for vaccination among ten low‐middle‐income countries (LMICs) is also seen to contribute to vaccine hesitancy which is why it remains a priority to have the availability of free vaccination services in LMICs for control of the pandemic, especially among low socioeconomic populations. 107
The discovery of Omicron led to a shocking response by many countries to shed light on the lack of global solidarity, promoting urgent reactions from governments, especially in views of vaccination supply.
National governments accelerate vaccine strategy and boost campaigns, increasing the vaccine gap between rich and developing countries. This imbalance is strongly believed to further develop the rapid spread of the Omicron variant. Notably, severe sickness requiring hospitalization is seen primarily in unvaccinated populations. The WHO statement, ‘none of us is safe until all of us are safe’, emphasizes that global delivery of doses is crucial and requires careful consideration to avoid hazards in the adoption of booster doses. 108 Pfizer research estimates the fully two‐dose vaccination can provide more than 80% safeguard against severe illness and mortality for 6 months, and the booster dose aids in increasing protection by 10%. 73 Therefore, delivering boosters to countries lacking vaccine supply can help obtain >80% protection against serious outcomes.
3. LIMITATIONS
Due to the nature of urgency on the topic, this narrative review was mainly limited to PubMed, Scopus, WHO, FDA, CDC, and ECDC databases to identify potentially eligible studies. The review included all papers reporting the original data related to the Omicron variant. Studies not available in English, not primary research, non‐official news, statements, and abstract‐only papers were excluded. Several recent papers included in this review were preprints, posing a risk to the data quality. Although this review is up to date, the information on Omicron is rapidly evolving with newer findings. There still lacks clarity in various aspects of Omicron, its sub‐variants and how they have been studied to date. Estimates of severity still need to be better understood of various elements such as age and vaccine status. Due to a lack of sequencing, screening and tracking of Omicron, there is an unclear understanding of its epidemiology.
4. CONCLUSION
The newer mutations of the Omicron variant have led to a stronger viral binding affinity, increased transmissibility, and reduced detection by conventional RT‐PCR and rapid antigen tests. Crucial treatments for COVID‐19 remain unchanged except for monoclonal antibody drugs due to their target on the RBD of the SARS‐CoV‐2 S‐protein, which is highly mutated in the Omicron variant. Only bebtelovimab could adequately shield all three sub‐lineages of Omicron. Clinical presentation remains unclear in Omicron infection, especially with the more severe symptoms when compared to its counterpart variants. Most symptoms are found to be milder to date, with no reports of unusual symptoms. Preliminary data elucidated that the Omicron subvariant, BA2, had a higher level of illness severity when compared to the original B.1 virus.
The significant reduction of neutralising antibody concentration leads to a gradual decline in vaccine efficacy and a higher likelihood of immune escape, suggesting the urgent need for a booster dose. Regardless, current vaccines are still expected to reduce severe disease, hospitalisation, and death, further highlighting the importance of promoting vaccination. With this comes the need to work together globally to overcome the socioeconomic bias seen in vaccine availability. Further research and clinical studies are needed to understand all known unknowns of this highly contagious variant to help push towards a vaccine design and booster campaigns equitably to control the COVID‐19 pandemic efficiently.
AUTHOR CONTRIBUTIONS
The idea and study design for this study was first conceived by Nguyen Tien Huy, Trang Thi Bich Le, Tamilarasy Vasanthakumaran, I‐Chun Hung, Mai Ngoc Luu, Hau Nguyen Thi Hien, Zeeshan Ali Khan, Nguyen Thanh An, Van‐Phu Tran, Wei Jun Lee, Jeza Muhamad Abdul Aziz, and Tasnim Ali collected and extracted data. Trang Thi Bich Le, Tamilarasy Vasanthakumaran, I‐Chun Hung, Mai Ngoc Luu, Hau Nguyen Thi Hien, Zeeshan Ali Khan, Nguyen Thanh An, Van‐Phu Tran, Wei Jun Lee, Jeza Muhamad Abdul Aziz, Tasnim Ali, and Shyam Prakash Dumre wrote the first draft. Trang Thi Bich Le, Tamilarasy Vasanthakumaran, I‐Chun Hung, Mai Ngoc Luu, Hau Nguyen Thi Hien, Zeeshan Ali Khan, Nguyen Thanh An, Van‐Phu Tran, Wei Jun Lee, Jeza Muhamad Abdul Aziz, Tasnim Ali, Shyam Prakash Dumre, and Nguyen Tien Huy contributed to the interpretation of findings and commented on subsequent versions of the manuscript with substantial contributions from Trang Thi Bich Le, Tamilarasy Vasanthakumaran, I‐Chun Hung, and Mai Ngoc Luu, Trang Thi Bich Le, Tamilarasy Vasanthakumaran, I‐Chun Hung, Hau Nguyen Thi Hien, and Zeeshan Ali Khan accessed and verified the data. All authors had read and approved the final version and had full access to all the data in the study. All authors had final responsibility for the decision to submit for publication.
CONFLICT OF INTEREST
The authors declare they have no conflict of interest.
Le TTB, Vasanthakumaran T, Thi Hien HN, et al. SARS‐CoV‐2 Omicron and its current known unknowns: a narrative review. Rev Med Virol. 2022;e2398. 10.1002/rmv.2398
Trang Thi Bich Le, Tamilarasy Vasanthakumaran, Hau Nguyen Thi Hien and I‐Chun Hung are equally contributed to the work.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Vaughan A. Omicron emerges. New Sci (1971). 2021;252(3363):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingraham NE, Ingbar DH. The omicron variant of SARS‐CoV‐2: understanding the known and living with unknowns. Clin Transl Med. 2021;11(12):e685. 10.1002/ctm2.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shanmugaraj B, Malla A, Khorattanakulchai N, Phoolcharoen W. SARS‐CoV‐2 omicron variant: could it be another threat? J Med Virol. 2021;94(4):1284‐1288. 10.1002/jmv.27532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espenhain L, Funk T, Overvad M, et al. Epidemiological characterisation of the first 785 SARS‐CoV‐2 Omicron variant cases in Denmark. Eur Surveill Bull Eur sur les Mal Transm Eur Commun Dis Bull. 2021;26(50). 10.2807/1560-7917.es.2021.26.50.2101146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Zhang L, Li Q, et al. The significant immune escape of pseudotyped SARS‐CoV‐2 variant Omicron. Emerg Microb Infect. 2022;11(1):1‐5. 10.1080/22221751.2021.2017757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iketani S, Liu L, Guo Y, et al. Antibody evasion properties of SARS‐CoV‐2 Omicron sublineages. Nature. 2022;604(7906):553‐556. 10.1038/s41586-022-04594-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. European Centre for Disease Prevention and Control . Weekly Epidemiological Update: Omicron Variant of Concern (VOC) – Week 50 (Data as of 19 December 2021) 2021 [cited 2022 January 10th]. https://www.ecdc.europa.eu/en/news‐events/weekly‐epidemiological‐update‐omicron‐variant‐concern‐voc‐week‐50‐data‐19‐december‐2021
- 8. Kazybay B, Ahmad A, Mu C, Mengdesh D, Xie Y. Omicron N501Y mutation among SARS‐CoV‐2 lineages: insilico analysis of potent binding to tyrosine kinase and hypothetical repurposed medicine. Trav Med Infect Dis. 2021;45:102242. 10.1016/j.tmaid.2021.102242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World Health Organization . Weekly Epidemiological Update on COVID‐19–21 December 2021. Edition 71 2021 [cited 2022 January 10th]. https://www.who.int/publications/m/item/weekly‐epidemiological‐update‐on‐covid‐19–‐21‐december‐2021
- 10. Zhao H, Lu L, Peng Z, et al. SARS‐CoV‐2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2‐expressed cells. Emerg Microb Infect. 2021:1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa. J Med Virol. 2021;94(4):1728‐1733. 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 12. Omicron Variant (B.1.1.529) . Data from BNO News/Newsnoded 2022 [cited 2022 January 10th]. https://newsnodes.com/omicron_tracker/?fbclid=IwAR3cZq2HJK12SgypksVmgWgd9DjeGsCBDNk0pZdZFLidU7GyHWk82LPUl1o
- 13. Saxena SK, Kumar S, Ansari S, et al. Characterization of the novel SARS‐CoV‐2 omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol. 2021;94(4):1738‐1744. 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 14. Meo SA, Meo AS, Al‐Jassir FF, et al. Omicron SARS‐CoV‐2 new variant: global prevalence and biological and clinical characteristics. Eur Rev Med Pharmacol Sci. 2021;25(24):8012‐8018. [DOI] [PubMed] [Google Scholar]
- 15. Kannan SR, Spratt AN, Sharma K, et al. Omicron SARS‐CoV‐2 variant: unique features and their impact on pre‐existing antibodies. J Autoimmun. 2022:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu L, Zhou L, Mo M, et al. SARS‐CoV‐2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct Targeted Ther. 2022;7(1):8. 10.1038/s41392-021-00863-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gu H, Krishnan P, Ng DYM, et al. Probable transmission of SARS‐CoV‐2 omicron variant in quarantine hotel. Emerg Infect Dis. 2021;28(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barton MI, MacGowan SA, Kutuzov MA, Dushek O, Barton GJ, van der Merwe PA. Effects of common mutations in the SARS‐CoV‐2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10:e70658. 10.7554/elife.70658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zahradník J, Marciano S, Shemesh M, et al. SARS‐CoV‐2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6(9):1188‐1198. 10.1038/s41564-021-00954-4 [DOI] [PubMed] [Google Scholar]
- 20. Karim SSA, Karim QA. Omicron SARS‐CoV‐2 variant: a new chapter in the COVID‐19 pandemic. Lancet. 2021;398(10317):2126‐2128. 10.1016/s0140-6736(21)02758-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62(2):412‐422. 10.1021/acs.jcim.1c01451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller NL, Clark T, Raman R, et al. Insights on the mutational landscape of the SARS‐CoV‐2 Omicron variant. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 23. Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022;94(5):1808‐1810. 10.1002/jmv.27601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. CoVariants . Shared mutations 2022. updated June 14, 2022. https://covariants.org/shared‐mutations?fbclid=IwAR2D5MNH49lt‐BPdZWJlAxqRh6FRTgEUPFFbj7mLZybjpEVPTKJWJ2ljxMc
- 25. Yamasoba D, Kimura I, Nasser H, et al. Virological characteristics of SARS‐CoV‐2 BA.2 variant. bioRxiv: the preprint server for biology. 2022.
- 26. CoVariants . Overview of variants in countries. updated March 08. 2022. https://covariants.org/per‐country?fbclid=IwAR1Kh5rz8lx5umOc_RBShDI9‐gHf6Q1a6DK4Io4C7‐KInxACGjV7vzYHZgo
- 27. Chen J, Wang R, Gilby NB, et al. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. ArXiv. 2021:arXiv:2112.01318v1. [DOI] [PMC free article] [PubMed]
- 28. Ito K, Piantham C, Nishiura H. Relative instantaneous reproduction number of omicron SARS‐CoV‐2 variant with respect to the delta variant in Denmark. J Med Virol. 2021;94(5):2265‐2268. 10.1002/jmv.27560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishiura H, Ito K, Anzai A, Kobayashi T, Piantham C, Rodriguez‐Morales AJ. Relative reproduction number of SARS‐CoV‐2 omicron (B.1.1.529) compared with Delta variant in South Africa. J Clin Med. 2022;11(1):30. 10.3390/jcm11010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma V, Rai H, Gautam DNS, et al. Emerging evidence on Omicron (B.1.1.529) SARS‐CoV‐2 variant. J Med Virol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang W, Shaman J. SARS‐CoV‐2 transmission dynamics in South Africa and epidemiological characteristics of the Omicron variant. medRxiv: the preprint server for health sciences. 2021.
- 32. HKUMed Finds Omicron SARS‐CoV‐2 Can Infect Faster and Better than Delta in Human Bronchus but with Less Severe Infection in Lung 2021 [cited 2022 January 10th]. https://www.med.hku.hk/en/news/press/20211215‐omicron‐sars‐cov‐2‐infection
- 33. World Health Organization . Enhancing Readiness for Omicron (B.1.1.529): Technical Brief and Priority Actions for Member States 2022 [cited 2022 January 10th]. https://www.who.int/publications/m/item/enhancing‐readiness‐for‐omicron‐(b.1.1.529)‐technical‐brief‐and‐priority‐actions‐for‐member‐states
- 34. Lubin JH, Markosian C, Balamurugan D, et al. Structural models of SARS‐CoV‐2 Omicron variant in complex with ACE2 receptor or antibodies suggest altered binding interfaces. bioRxiv: the preprint server for biology. 2021.
- 35. Papanikolaou V, Chrysovergis A, Ragos V, et al. From delta to Omicron: S1‐RBD/S2 mutation/deletion equilibrium in SARS‐CoV‐2 defined variants. Gene. 2022;814:146134. 10.1016/j.gene.2021.146134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang X, Wu S, Wu B, et al. SARS‐CoV‐2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Targeted Ther. 2021;6(1):430. 10.1038/s41392-021-00852-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deerain J, Druce J, Tran T, et al. Assessment of the analytical sensitivity of ten lateral flow devices against the SARS‐CoV‐2 omicron variant. J Clin Microbiol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hamill V, Noll L, Lu N, et al. Molecular detection of SARS‐CoV‐2 strains and differentiation of Delta variant strains. Transboundary Emerg Infect Dis. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. FDA . SARS‐CoV‐2 Viral Mutations: Impact on COVID‐19 Tests 2021 [cited 2022 January 10th]. https://www.fda.gov/medical‐devices/coronavirus‐covid‐19‐and‐medical‐devices/sars‐cov‐2‐viral‐mutations‐impact‐covid‐19‐tests
- 40. FDA . Coronavirus (COVID‐19) Update: FDA Actively Working to Investigate, Address Potential Impacts of Omicron Variant; Urges Vaccination and Boosters 2021 [cited 2021 January 10th]. https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19‐update‐fda‐actively‐working‐investigate‐address‐potential‐impacts‐omicron
- 41. CDC COVID‐19 Response Team . SARS‐CoV‐2 B.1.1.529 (omicron) variant United States. MMWR Morb Mortal Wkly Rep. 2021;70(50):1731‐1734. 10.15585/mmwr.mm7050e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Metzger C, Lienhard R, Seth‐Smith HMB, et al. PCR performance in the SARS‐CoV‐2 Omicron variant of concern? Swiss Med Wkly. 2021;151:w30120. [DOI] [PubMed] [Google Scholar]
- 43. Erster O, Beth‐Din A, Asraf H, et al. Specific detection of SARS‐COV‐2 B.1.1.529 (omicron) variant by four RT‐qPCR differential assays. medRxiv: the preprint server for health sciences. 2021.
- 44. Petrillo M, Querci M, Corbisier P, et al. Silico design ofSpecific PrimerSets forthe detectionof B.1.1.529 SARS‐CoV‐2 variant of concern (omicron). Zenodo. 2021. [Google Scholar]
- 45. Brandal LT, MacDonald E, Veneti L, et al. Outbreak caused by the SARS‐CoV‐2 omicron variant in Norway. Eur Surveill Bull Eur sur les Mal Transm Eur Commun Dis Bull. 2021;26(50). 10.2807/1560-7917.es.2021.26.50.2101147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Menni C, Valdes AM, Polidori L, et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS‐CoV‐2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399(10335):1618‐1624. 10.1016/s0140-6736(22)00327-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li A, Maier A, Carter M, Guan TH. Omicron and S‐gene target failure cases in the highest COVID‐19 case rate region in Canada. J Med Virol. 2021;94(5):1784‐1786. 10.1002/jmv.27562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee JJ, Choe YJ, Jeong H, et al. Importation and transmission of SARS‐CoV‐2 B.1.1.529 (omicron) variant of concern in Korea. J Kor Med Sci. 2021;36(50):e346. 10.3346/jkms.2021.36.e346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abdullah F, Myers J, Basu D, et al. Decreased severity of disease during the first global omicron variant Covid‐19 outbreak in a large hospital in TSHWANE, South Africa. Int J Infect Dis IJID Off Publ Int Soc Infect Dis. 2021;116:38‐42. 10.1016/j.ijid.2021.12.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID‐19 omicron wave compared with previous waves. JAMA. 2021;327(6):583. 10.1001/jama.2021.24868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doria‐Rose NA, Shen X, Schmidt SD, et al. Booster of mRNA‐1273 vaccine reduces SARS‐CoV‐2 omicron escape from neutralizing antibodies. medRxiv: the preprint server for health sciences. 2021.
- 52. Cameroni E, Saliba C, Bowen JE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 53. Andrews N, Stowe J, Kirsebom F, et al. Effectiveness of COVID‐19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv: the preprint server for health sciences. 2021.
- 54. Gardner BJ, Kilpatrick AM. Estimates of reduced vaccine effectiveness against hospitalization, infection, transmission and symptomatic disease of a new SARS‐CoV‐2 variant, Omicron (B.1.1.529), using neutralizing antibody titers. medRxiv: the preprint server for health sciences. 2021.
- 55. Mallory R, Formica N, Pfeiffer S, et al. Immunogenicity and safety following a homologous booster dose of a SARS‐CoV‐2 recombinant spike protein vaccine (NVX‐CoV2373): a phase 2 randomized placebo‐controlled trial. medRxiv: the preprint server for health sciences. 2021.
- 56. Edara VV, Manning KE, Ellis M, et al. mRNA‐1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS‐CoV‐2 Omicron variant. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 57. Garcia‐Beltran WF, St Denis KJ, Hoelzemer A, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell. 2022;185(3):457‐466.e4. 10.1016/j.cell.2021.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zeng C, Evans JP, Qu P, et al. Neutralization and stability of SARS‐CoV‐2 omicron variant. bioRxiv: the preprint server for biology. 2021.
- 59. Muik A, Lui BG, Wallisch A‐K, et al. Neutralization of SARS‐CoV‐2 Omicron pseudovirus by BNT162b2 vaccine‐elicited human sera. medRxiv: the preprint server for health sciences. 2021. [DOI] [PMC free article] [PubMed]
- 60. Lu L, Mok BW, Chen LL, et al. Neutralization of SARS‐CoV‐2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis Off Publ Infect Dis Soc Am. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Syed AM, Ciling A, Khalid MM, et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS‐CoV‐2 virus‐like particles. medRxiv: the preprint server for health sciences. 2022. [DOI] [PMC free article] [PubMed]
- 62. Ai J, Zhang H, Zhang Y, et al. Omicron variant showed lower neutralizing sensitivity than other SARS‐CoV‐2 variants to immune sera elicited by vaccines after boost. Emerg Microb Infect. 2021:1‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schmidt F, Muecksch F, Weisblum Y, et al. Plasma neutralization properties of the SARS‐CoV‐2 Omicron variant. medRxiv: the preprint server for health sciences. 2021.
- 64. Dejnirattisai W, Shaw RH, Supasa P, et al. Reduced neutralisation of SARS‐CoV‐2 omicron B.1.1.529 variant by post‐immunisation serum. Lancet. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu X, Wei D, Xu W, et al. Reduced sensitivity of SARS‐CoV‐2 Omicron variant to booster‐enhanced neutralization. medRxiv: the preprint server for health sciences. 2021.
- 66. Cele S, Jackson L, Khan K, et al. SARS‐CoV‐2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv: the preprint server for health sciences. 2021.
- 67. Lusvarghi S, Pollett SD, Neerukonda SN, et al. SARS‐CoV‐2 Omicron neutralization by therapeutic antibodies, convalescent sera, and post‐mRNA vaccine booster. bioRxiv: the preprint server for biology. 2021.
- 68. Nemet I, Kliker L, Lustig Y, et al. Third BNT162b2 vaccination neutralization of SARS‐CoV‐2 omicron infection. N. Engl J Med. 2021;386(5):492‐494. 10.1056/nejmc2119358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Khan K, Karim F, Cele S, et al. Omicron infection enhances neutralizing immunity against the Delta variant. medRxiv: the preprint server for health sciences. 2021.
- 70. Accorsi EK, Britton A, Fleming‐Dutra KE, et al. Association between 3 doses of mRNA COVID‐19 vaccine and symptomatic infection caused by the SARS‐CoV‐2 omicron and delta variants. JAMA. 2022;327(7):639‐651. 10.1001/jama.2022.0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zeng C, Evans JP, Chakravarthy K, et al. COVID‐19 mRNA booster vaccines elicit strong protection against SARS‐CoV‐2 Omicron variant in patients with cancer. Cancer Cell. 2021;40(2):117‐119. 10.1016/j.ccell.2021.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Laurie MT, Liu J, Sunshine S, et al. SARS‐CoV‐2 variant exposures elicit antibody responses with differential cross‐neutralization of established and emerging strains including Delta and Omicron. J Infect Dis. 2022;225(11):1909‐1914. 10.1093/infdis/jiab635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Thomas SJ, Moreira ED, Jr. , Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA covid‐19 vaccine through 6 months. N. Engl J Med. 2021;385(19):1761‐1773. 10.1056/nejmoa2110345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Redd AD, Nardin A, Kared H, et al. Minimal cross‐over between mutations associated with Omicron variant of SARS‐CoV‐2 and CD8+ T cell epitopes identified in COVID‐19 convalescent individuals. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 75. Bartsch Y, Tong X, Kang J, et al. Preserved Omicron Spike specific antibody binding and Fc‐recognition across COVID‐19 vaccine platforms. medRxiv: the preprint server for health sciences. 2021.
- 76. Buonaguro FM, Ascierto PA, Morse GD, et al. Covid‐19: time for a paradigm change. Rev Med Virol. 2020;30(5):e2134. 10.1002/rmv.2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. NIH . Characteristics of Immunomodulators 2021 [updated December 16; cited 2022 March 09]. https://www.covid19treatmentguidelines.nih.gov/tables/immunomodulators‐characteristics/
- 78. World Health Organization . Update on Omicron 2021 [cited 2022 January 10th]. https://www.who.int/news/item/28‐11‐2021‐update‐on‐omicron
- 79. NIH . The COVID‐19 Treatment Guidelines Panel's Statement on the Role of Bebtelovimab for the Treatment of High‐Risk, Nonhospitalized Patients with Mild to Moderate COVID‐19 updated March 02; 2022. https://www.covid19treatmentguidelines.nih.gov/therapies/statement‐on‐bebtelovimab/
- 80. Cathcart AL, Havenar‐Daughton C, Lempp FA, et al. The dual function monoclonal antibodies VIR‐7831 and VIR‐7832 demonstrate potent in vitro and in vivo activity against SARS‐CoV‐2. bioRxiv: the preprint server for biology. 2021.
- 81. FDA . Emergency Use Authorization 2022 updated March 02. https://www.fda.gov/emergency%2Dpreparedness%2Dand%2Dresponse/mcm%2Dlegal%2Dregulatory%2Dand%2Dpolicy%2Dframework/emergency%2Duse%2Dauthorization%23coviddrugs
- 82. FDA . Fact Sheet for Health Care Providers Emergency use Authorization (Eua) of Sotrovimab updated February 23; 2022. https://www.fda.gov/media/149534/download
- 83. GILEAD . Gilead Statement on Veklury® (Remdesivir) and the SARS‐CoV‐2 Omicron Variant updated December 1; 2021. https://www.gilead.com/news‐and‐press/company‐statements/gilead‐statement‐on‐veklury‐remdesivir‐and‐the‐sars‐cov‐2‐omicron‐variant
- 84. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS‐CoV‐2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28(3):1‐6. 10.1038/s41591-021-01678-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Westendorf K, Wang L, Žentelis S, et al. LY‐CoV1404 (bebtelovimab) potently neutralizes SARS‐CoV‐2 variants. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 86. Ullrich S, Ekanayake KB, Otting G, et al. Main protease mutants of SARS‐CoV‐2 variants remain susceptible to nirmatrelvir ( PF‐07321332). bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 87. Rosenke K, Okumura A, Lewis MC, et al. Molnupiravir (MK‐4482) is efficacious against Omicron and other SARS‐CoV‐2 variants in the Syrian hamster COVID‐19 model. bioRxiv: the preprint server for biology. 2022.
- 88. Vangeel L, Chiu W, De Jonghe S, et al. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS‐CoV‐2 Omicron and other variants of concern. Antivir Res. 2022;198:105252. 10.1016/j.antiviral.2022.105252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aggarwal A, Stella AO, Walker G, et al. SARS‐CoV‐2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv: the preprint server for health sciences. 2021. [DOI] [PMC free article] [PubMed]
- 90. Peacock TP, Brown JC, Zhou J, et al. The altered entry pathway and antigenic distance of the SARS‐CoV‐2 Omicron variant map to separate domains of spike protein. bioRxiv: the preprint server for biology. 2021.
- 91. Cameroni E, Saliba C, Bowen JE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 92. Bate N, Savva CG, Moody PCE, et al. In vitro evolution predicts emerging CoV‐2 mutations with high affinity for ACE2 and cross‐species binding. bioRxiv: the preprint server for biology. 2021. [DOI] [PMC free article] [PubMed]
- 93. Wei C, Shan K‐J, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS‐CoV‐2 Omicron variant. J Genet Genom. 2021;48(12):1111‐1121. 10.1016/j.jgg.2021.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bai Y, Du Z, Xu M, et al. International risk of SARS‐CoV‐2 Omicron variant importations originating in South Africa. medRxiv: the preprint server for health sciences. 2021. [DOI] [PMC free article] [PubMed]
- 95. Jansen L, Tegomoh B, Lange K, et al. Investigation of a SARS‐CoV‐2 B.1.1.529 (omicron) variant cluster—Nebraska. MMWR Morb Mortal Wkly Rep. 2021;70(5152):1782‐1784. 10.15585/mmwr.mm705152e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Maruki T, Iwamoto N, Kanda K, et al. Two cases of breakthrough SARS‐CoV‐2 infections caused by the Omicron variant (B.1.1.529 lineage) in international travelers to Japan. Clin Infect Dis Off Publ Infect Dis Soc Am. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Fan Y, Li X, Zhang L, Wan S, Zhou F. SARS‐CoV‐2 Omicron variant: recent progress and future perspectives. Signal Transduct Targeted Ther. 2022;7(1):141. 10.1038/s41392-022-00997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Callaway E, Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600(7888):197‐199. 10.1038/d41586-021-03614-z [DOI] [PubMed] [Google Scholar]
- 99. Prevention CfDCa . Hospitalization of Infants and Children Aged 0–4 Years with Laboratory‐Confirmed COVID‐19—COVID‐NET, 14 States, March 2020–February 2022. https://www.cdc.gov/mmwr/volumes/71/wr/mm7111e2.htm [DOI] [PMC free article] [PubMed]
- 100. Radia T, Williams N, Agrawal P, et al. Multi‐system inflammatory syndrome in children & adolescents (MIS‐C): a systematic review of clinical features and presentation. Paediatr Respir Rev. 2021;38:51‐57. 10.1016/j.prrv.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hassan W, Kazmi SK, Tahir MJ, et al. Global Acceptance and Hesitancy of COVID‐19 Vaccination: A Narrative Review. Narra J . 2021;1(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Salali GD, Uysal MS. COVID‐19 vaccine hesitancy is associated with beliefs on the origin of the novel coronavirus in the UK and Turkey. Psychol Med. 2020:1‐3. 10.1017/s0033291720004067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Palamenghi L, Barello S, Boccia S, Graffigna G. Mistrust in biomedical research and vaccine hesitancy: the forefront challenge in the battle against COVID‐19 in Italy. Eur J Epidemiol. 2020;35(8):785‐788. 10.1007/s10654-020-00675-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lazarus JV, Ratzan SC, Palayew A, et al. A global survey of potential acceptance of a COVID‐19 vaccine. Nat Med. 2021;27(2):225‐228. 10.1038/s41591-020-1124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Razai MS, Chaudhry UAR, Doerholt K, Bauld L, Majeed A. Covid‐19 vaccination hesitancy. BMJ. 2021;373:n1138. 10.1136/bmj.n1138 [DOI] [PubMed] [Google Scholar]
- 106. Surawan DP, Sumohadi D, Budhitresna AAG, et al. Titer disparity of anti‐Spike receptor binding domain SARS‐CoV‐2 antibody between vaccinated and naturally infected individuals. Narrative J. 2022;2(1). 10.52225/narra.v2i1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sallam M, Anwar S, Yufika A, et al. Willingness‐to‐pay for COVID‐19 vaccine in ten low‐middle‐income countries in Asia, Africa and South America: a cross‐sectional study. Narrative J. 2022;2(1). 10.52225/narra.v2i1.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Khan NA, Al‐Thani H, El‐Menyar A. The emergence of new SARS‐CoV‐2 variant (Omicron) and increasing calls for COVID‐19 vaccine boosters‐The debate continues. Trav Med Infect Dis. 2021;45:102246. 10.1016/j.tmaid.2021.102246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


