Abstract
The spike trimer of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is an effective target for inducing neutralizing antibodies by coronavirus disease 2019 (COVID‐19) vaccines. However, the diversity of spike protein from emerging SASR‐CoV‐2 variants has become the major challenge for development of a universal vaccine. To investigate the immunogenicity of spike proteins from various circulating strains including wild type, Delta, and Omicron variants, we produced various natural spike trimers and designed three vaccination strategies, that is, individual, sequential, and bivalent regimens to assess autologous and heterogenous antibody responses in a mouse model. The results indicated that monovalent vaccine strategy with individual spike trimer could only induce binding and neutralizing antibodies against homologous viruses. However, sequential and bivalent immunization with Delta and Omicron spike trimers could induce significantly broader neutralizing antibody responses against heterogenous SARS‐CoV‐2. Interestingly, the spike trimer from Omicron variant showed superior immunogenicity in inducing antibody response against recently emerging XE variant. Taken together, our data supported the development of novel vaccination strategies or multivalent vaccine against emerging variants.
Keywords: immunogenicity, SARS‐CoV‐2, spike, trimer, variant
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has caused a devastating impact on the global pubic health security. The COVID‐19 vaccine inducing neutralizing antibodies against SARS‐COV‐2 has showed significant effect on the control of the pandemic. However, the emergence of SARS‐CoV‐2 variants of concern (VOCs) bringing new waves of the infection even several approved COVID‐19 vaccines are widely available and used. At present, five SARS‐COV‐2 VOCs have been identified: for example, Alpha (B.1.1.7), 1 Beta (B.1.351), 2 Gamma (P.1), 3 Delta (B.1.617.2), 4 and Omicron (B.1.1.529) 5 were associated with increased transmissibility. 6 Since emerging in November 2021, Omicron variant has evolved into many subtypes and recombinant strains, including early BA.1, BA.2, BA.3, and recently emerged BA.4, BA.5, and BA.2.12.1, as well as XD variant (Delta AY.4 and BA.1 recombinant), XE variant (BA.1 and BA.2 recombinant), and XF variant (Delta and the BA.1 recombinant). 7 XE recombinant variant appears to be roughly 10% more transmissible than its parent variant BA.2, implying that the XE may have the potential for greater range prevalence in the near future. 8 VOCs showed increased transmissibility and might have the potential for increasing disease severity when compared with the wild‐type virus. 9
The diverse spikes of SASR‐CoV‐2 not only affect the replication and infection ability of the virus, but also have an influence on host immune response. Natural spike trimer is the dominated immunogen that induces humoral immune response, thus becomes the main target of neutralizing antibodies and currently approved COVID‐19 vaccines. 10 , 11 , 12 Generally, antigenic drift could occur in the glycoprotein of emerging SARS‐CoV‐2 variants; 13 thus, antibodies induced by parent strain might not afford sufficient cross‐neutralizing effect against these variants. The immune escape of VOCs to the current vaccines based on wild‐type SARS‐CoV‐2 has become the major obstacle to end the pandemic. 14 Therefore, it is necessary to fully explore the difference of immunogenicity of spike proteins from different emerging SARS‐CoV‐2 variants. In addition, vaccines and vaccination strategies inducing potent and broad neutralizing antibody responses against various variants, for example, Delta and Omicron, are urgently needed.
In this study, we applied three immunization strategies including monovalent, sequential, and divalent vaccination to explore the difference of resultant humoral immune responses, aiming to provide theoretical basis for development of efficient COVID‐19 vaccine targeting the viral diversity. Specifically, the binding antibody titers and neutralizing antibody titers of three vaccination strategies, including individual, sequential, and bivalent regimens based on wild type, Delta, and Omicron spike trimer were tested in mouse model. We observed that even though all of the three strategies could induce cross‐binding antibody, only sequential and bivalent immunization with Delta and Omicron spike trimers induced broader neutralizing antibody that even neutralized the newly emerging variants, XE recombinant strain.
2. METHODS AND MATERIALS
2.1. Construction of plasmids and cell lines
SARS‐CoV‐2 surface glycoprotein gene (GenBank: MN_908947) with C‐terminal 19 amino acids deletion was codon‐optimized for Homo sapiens and cloned into eukaryotic expression plasmid pcDNA3.1(+) between HindIII and BamHI sites to generate the spike expression plasmids pcDNA3.1(+)‐OPS. The lentiviral packaging plasmid pNL4‐3 Luc+R‐E‐ carrying an Env‐defective, luciferase‐expressed HIV‐1 genome was generously gifted by Binlian Sun, Jianghan University. Wild‐type SARS‐CoV‐2 pseudovirus was produced by cotransfection of HEK293T cells with pNL4‐3 Luc+R‐E‐ and pcDNA3.1(+)‐OPS. To produce various spike pseudoviruses, pcDNA3.1(+)‐OPS plasmid was subsequently used as a template to generate plasmids encoding various spike mutants, including D614G, emerging Delta, Omicron, and XE variants. The amino acid mutations of each variant in this study were shown in Figure 1A. Codon‐optimized DNA sequences coding various spike were further modified and respectively cloned into the pTT5 vector for expressing corresponding trimeric spike proteins, and the specific regions of T4 fibritin and 6×His tag were fused to the C‐terminal of spike protein. The illustration of various trimeric spike proteins production was indicated in Figure 1A.
Figure 1.
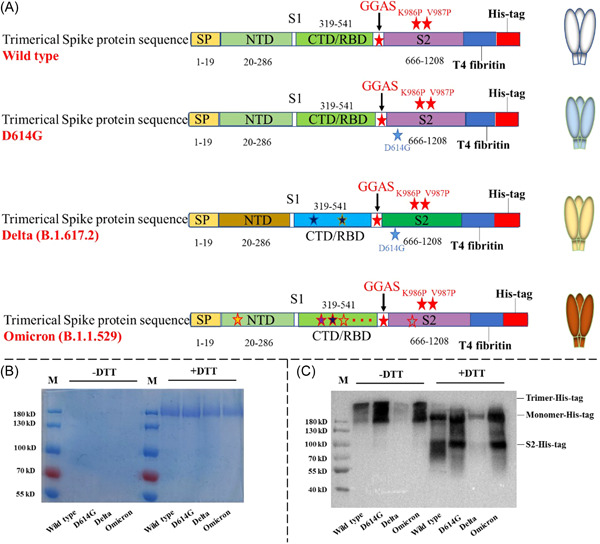
Design, expression and identification of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) spike trimers from wild type, D614G, Delta, and Omicron. (A) Organization of SARS‐CoV‐2 spike protein constructs. (B) Reduced and nonreduced sodium dodecyl‐sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) of SARS‐CoV‐2 spike trimer from wild type, D614G, Delta, and Omicron. (C) Identification of SARS‐CoV‐2 spike trimers from wild type, D614G, Delta, and Omicron with anti‐His tag antibody.
To simulate virus entry assay in vitro, HEK293T cell line exogenously expressing hACE2 (HEK293T/hACE2) was constructed by lentivirus transduction for pseudovirus neutralization assay. HEK293T cells were maintained in high glucose Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovie serum (ExCell), penicillin (100 IU/ml), streptomycin (100 μg/ml) in a 5% CO2 environment at 37°C and passaged every 3 days. HEK293T/hACE2 cells were maintained in above‐mentioned medium with puromycin (2 μg/ml) and passaged every 2 days. HEK293F cells for trimeric spike protein expression were maintained in Chemically Defined Medium (Union‐Biotech Co. Ltd) supplemented with penicillin (100 IU/ml), streptomycin (100 μg/ml) in 37°C shaker at 120 r.p.m. with 5% CO2.
2.2. Preparation and purification of immunogens
pTT5 plasmids encoding various spike trimeric protein, including wild type, D614G, Delta, Omicron, were constructed as described above. To stabilize spike trimeric structure, furin cleavage site RRAR was substituted with GGAS, while proline mutations were introduced in the location of K986 and V987 of spike protein, and T4 trimerization domain 15 , 16 was fused in the C‐terminal of spike protein (Figure 1A). To acquire high‐purity spike trimer, 6×His tag was added in the C‐terminal of entire domain for purification. The plasmids expressing various spike trimers (pTT5‐Spike‐T4‐His) were respectively amplified and purified using TIANGEN HighPure Maxi Plasmid Kit (TIANGEN Biotech Co. Ltd). The method for spike trimer expression in HEK293F cells was based on a published method 17 with moderate modification. Briefly, individual plasmids (pTT5‐Spike‐T4‐His) containing different spike coding sequences were respectively transfected into HEK293F cells at a density of 2 × 106 cells/ml. At 5 days posttransfection, cell culture supernatants were collected and target proteins were purified using HisTrap FF pre‐packed column (GE Healthcare). Recombinant proteins were washed with Buffer A (0.5 M NaCl, 50 mM NaH2PO4) and eluted from the column with Buffer B (0.5 M NaCl, 50 mM NaH2PO4, 400 mM imidazole, pH 7.4) on ÄKTA Pure system. The protein solution was concentrated into Tris‐HCl pH 8.0 by centrifugation with 10 kDa molecular weight cut off membrane centrifugal filter units (Millipore). Recombinant proteins viewed as immunogens were dispensed in aliquots of 0.5 ml each and stored at −80°C for future use.
2.3. Sodium dodecyl‐sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) and western blotting
Recombinant proteins were quantified using an A280 measurement (ND5000, BioTeke Corporation). To confirm the purity of immunogens, samples of each spike protein were analyzed by SDS‐PAGE and Coomassie staining. The monomeric and trimerical spike proteins were confirmed by reduced and nonreduced SDS‐PAGE, respectively. Briefly, the protein samples were first incubated with Nondenatured Gel Protein Sample Loading Buffer (5×, –dithiothreitol [DTT]) and SDS‐PAGE Sample Loading Buffer (5×, +DTT) respectively, then separated using 10% SDS‐PAGE, transferred to a polyvinylidene difluoride membrane, and conducted western blot analysis to verify the expression of spike protein using mouse anti‐His‐tag mAb at a 1:2000 dilution. A horseradish peroxidase (HRP)‐conjugated secondary anti‐Mouse IgG antibody, diluted 1:5000 in tris‐buffered saline (TBS), was used for detection. The antibody‐bound monomer and trimer were visualized by enhanced chemiluminescence in the Tannon 5200 Multi Imaging Systems.
2.4. Immunization
Three vaccination strategies with various spike trimer, including monovalent, sequential and divalent regimens, were designed in this study. Female mice (Balb/c, n = 5 per group) aged 6–8 weeks were immunized intramuscularly at Days 0 and 21 with 7.5 μg (50 μl) of trimer (based on peptidic mass) mixed with equivalent‐volume Quick‐Antibody adjuvant (Beijing Biodragon Immunotechnologies Co., Ltd). The phosphate‐buffered saline (PBS) mixed with same adjuvant was designed as a control group. M1 (wild‐type spike trimer for the first and second doses), M2 (Delta spike trimer for the first and second doses), and M3 (Omicron spike trimer for the first and second doses) groups were classified as monovalent delivery strategy; S1 (wild‐type spike trimer for the first and Delta spike trimer for the second dose), S2 (Delta spike trimer for the first and wild‐type spike trimer for the second dose), S3 (Delta spike trimer for the first and Omicron spike trimer for the second dose), and S4 (Omicron spike trimer for the first and Delta spike trimer for the second dose) groups were classified as sequential delivery strategy; D1 (mixture of wild type and Delta spike trimer for the first and second doses) and D2 (mixture of Delta and Omicron spike trimer for the first and second doses) groups were classified as divalent delivery strategy. Two weeks post the second dose (Day 35), all mice were killed. Sera were collected and split into two aliquots for enzyme‐linked immunosorbent assay (ELISA) and pseudovirus neutralization assay, respectively.
2.5. ELISA assay
ELISA was used to measure the binding antibody titers of the mouse immune sera against various spike trimers. In brief, 96‐well ELISA plates were coated with 2 μg/ml of spike trimer overnight at 4°C. Coated wells were subsequently blocked with 200 μl blocking buffer (TBS containing 5% bovine serum albumin) for 2 h at 37°C. Plates were then washed three times with 200 μl washing buffer (TBS containing 0.1% Tween 20). A series dilution of the immune sera were prepared and incubated with the spike‐coated wells for 2 h at 37°C, while the normal mouse serum was used as a negative control. The plates were washed three times before incubation with 1:10 000 dilution of HRP‐conjugated Goat anti‐Mouse IgG for 1h at 37°C. The plates were washed three times and 100 μl 3,3',5,5'‐tetramethylbenzidine (Beyotime) substrate was added to each well. Reactions were stopped with 2 M HCl after 15 min incubation. Plates were read at 450 nm using a microplate reader (Molecular Devices). ELISA endpoint titers were defined as the highest dilution of serum to give an absorbance >2.1‐fold of the negative control values. Data analysis was conducted using GraphPad Prism 8.0.
2.6. Pseudovirus neutralization assay
To detect the neutralizing antibody titer of the immune sera, neutralization assay with pseudovirus was performed as described in our previous study. 18 Briefly, HEK293T/hACE2 cells were seeded in 96‐well plates at 10 000 cells/well in culture medium 24 h before the assay. The sera from each vaccination group were twofold serially diluted in culture medium with the initial dilution of 1:100 (dilution range of 1:100 to 1:128 000). Fifty microliters of pseudovirus with the values of relative luminescence unit (RLU) at ~1.0 × 105 were incubated with diluted sera at 37°C for 1 h, which were subsequently added to HEK293T/hACE2 cells. After 48 h incubation at 37°C with 5% CO2, culture supernatants were removed and the values of RLU were measured by Britelite plus Reporter Gene Assay System (PerkinElmer). Fifty percent pseudovirus neutralization titer (NTIC50) was determined by fitting nonlinear regression curves in GraphPad Prism 8.
2.7. Statistical analysis
ELISA binding titers were defined as the highest dilution of serum to give an absorbance greater than 2.1‐fold of the negative control values. The NTIC50 was defined as the dilution of serum at which the RLU values were reduced by 50% compared with the pseudovirus control wells calculated by the GraphPad Prism 8.0 (GraphPad Software). The difference of geometric mean titer (GMT) in two independent groups was tested for statistical significance with a Mann–Whitney U test in the GraphPad Prism 8. The difference of GMT in two paired groups was tested for statistical significance with a Wilcoxon matched‐pairs signed‐rank test in the GraphPad Prism 8. Differences were considered statistically significant at *p < 0.05, **p < 0.01, or ***p < 0.001.
3. RESULTS
3.1. Construction, expression and verification of SARS‐CoV‐2 spike trimer as an immunogen
To evaluate the immunogenicity of spike protein from various SARS‐CoV‐2 variants, we prepared the spike trimer based on codon‐optimized sequences from wild type, D614G, Delta, and Omicron SARS‐CoV‐2 in the HEK293F expression platform. To simulate the native trimeric structure of the spike protein of SARS‐CoV‐2, furin cleavage site RRAR was substituted with GGAS amino acid sequence, while proline mutations were introduced in the location of K986 and V987, and T4 trimerization domain was fused in the C‐terminal of extracellular domain of the protein (Figure 1A). Spike proteins purified from the transfection supernatant were analyzed with reduced and non‐reduced SDS‐PAGE, respectively. The results indicated that the size of monomeric spike protein was ~180 kDa in reduced SDS‐PAGE analysis (Figure 1B), which is consistent with the previous report. 19 , 20 Nonreduced SDS‐PAGE analysis just showed the dispersive band rather than clearly single band, which might result from that the molecular weight of trimeric spike was so large that limited spike trimer was stained by Coomassie blue or able to move out of the well. However, two clear bands representing monomeric spike and trimeric spike, at ~180 and ~600 kDa, were showed in western blotting with anti‐His antibody following nonreduced SDS‐PAGE (Figure 1C). Following reduced SDS‐PAGE, two clear bands representing monomeric spike and S2 subunit, at ~180 and ~80kDa, were observed in the western blot analysis (Figure 1C).
3.2. Design of monovalent, sequential, and divalent immunization strategies with spike trimers from wild type, Delta, and Omicron variants
We evaluated the immunogenicity of three different immunization strategies in groups of five mice per group. The mouse specie, vaccines, vaccination strategies, and bleeding schedules are summarized in Figure 2. The combination methods of each group, dosage of immunogens, and more details were listed in Table 1. Specifically, M1, M2, and M3 groups received two identical single immunogens and classified as monovalent immunization strategy; S1, S2, S3, and S4 groups received two vaccinations but with two different single immunogens and classified as sequential immunization strategy; D1 and D2 groups received two vaccinations but with two mixed immunogens and classified as divalent immunization strategy. To detect the antispike trimer‐binding antibody titers and the neutralizing antibody titers against autologous and heterogenous SARS‐CoV‐2, all sera were collected for performing ELISA and pseudovirus neutralization assay.
Figure 2.
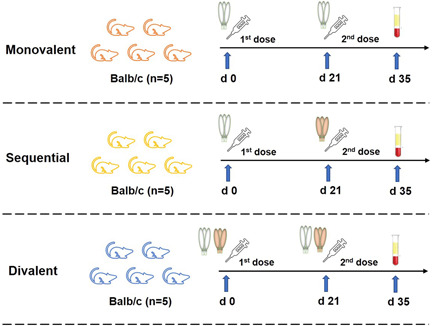
Immunization schedule of monovalent, sequential, and divalent vaccination strategies. Mice (Balb/c) were immunized at Days 0 and 21 with 7.5 μg of trimer (based on peptidic mass) mixed with equivalent volume (50 μl) of Quick‐Antibody adjuvant. The phosphate‐buffered saline (PBS) mixed with same adjuvant was used as a control group. Detailed groups were shown in Table 1. Two weeks post the second dose (Day 35), all mice were killed and sera were collected for test.
Table 1.
Vaccination strategies and dosage of immunogens
| Delivery strategy | Group | First dose | Second dose | μg/dose | n |
|---|---|---|---|---|---|
| Control | C | PBS | PBS | ‐ | 5 |
| Monovalent | M1 | Wild type | Wild type | 7.5 | 5 |
| M2 | Delta | Delta | 7.5 | 5 | |
| M3 | Omicron | Omicron | 7.5 | 5 | |
| Sequential | S1 | Wild‐type | Delta | 7.5 | 5 |
| S2 | Delta | Wild type | 7.5 | 5 | |
| S3 | Delta | Omicron | 7.5 | 5 | |
| S4 | Omicron | Delta | 7.5 | 5 | |
| Divalent | D1 | Wild type/Delta | Wild type/Delta | 3.75 + 3.75 | 5 |
| D2 | Delta/Omicron | Delta/Omicron | 3.75 + 3.75 | 5 |
Abbreviation: PBS, phosphate‐buffered saline.
3.3. Antispike trimer‐binding antibody responses
All three immunization strategies were capable of inducing antispike trimer‐binding antibody responses with titers over 1:10 000. Furthermore, the binding antibody titers induced by Omicron spike trimer (M3) was higher than the other two groups (M1 and M2) in monovalent immunization strategy, which revealed that the immunogenicity of spike trimer of Omicron might be superior to that from wild‐type and Delta variants. Interestingly, the binding antibody titers against three spike trimers in S1 group were all higher than that in S2 group, similar result was also shown between S3 and S4 groups (Figure 3). These results indicated that the development of cross‐antibodies might be associated with the immunization order of different immunogens. In the divalent immunization strategy, the cross‐binding antibody titers induced in D2 group were significantly higher than that in D1 group (Figure 3), which might result from the superior immunogenicity of Omicron spike trimer. Overall, the cross‐binding antibody titers induced by sequential and divalent immunization strategies based on Delta and Omicron spike trimer were significantly higher than that induced by monovalent immunization strategy (Figure 3).
Figure 3.
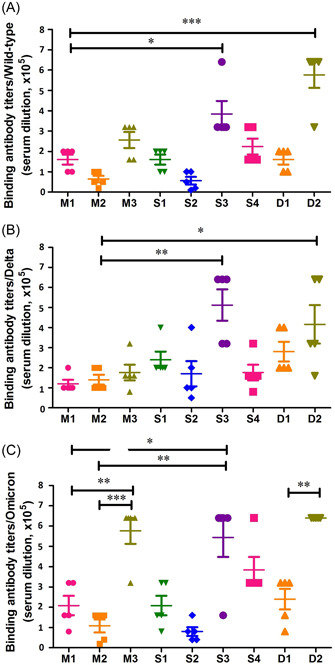
Antispike trimer‐binding antibody titers in monovalent, combination, and sequential vaccination regimens. 96‐well enzyme‐linked immunosorbent assay (ELISA) plates were coated with 2 μg/ml of spike trimer from wild type (A), Delta (B), and Omicron (C) severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants. A series dilution of the mouse immune sera from three groups were prepared and incubated with the spike‐coated wells, respectively. Endpoint titers were defined as the highest dilution of serum to give an OD450 > 2.1‐fold of the negative control value. The difference of binding antibody titer was tested for statistical significance with a unpaired t test in the GraphPad Prism 8. Differences were considered statistically significant at *p < 0.05, **p < 0.01, or ***p < 0.001.
3.4. Neutralizing antibody responses against autologous and heterogenous pseudoviruses
To assess whether sequential and divalent immunization strategies also had superior neutralization against VOCs, we next compared the neutralizing antibody responses against autologous and heterogenous SARS‐CoV‐2 induced by these immunization strategies. A panel of pseudoviruses displaying various spikes derived from D614G, Delta, Omicron, and XE variants, was used to test the neutralizing activities of mice sera from three immunization strategies.
Monovalent immunizations elicited high levels of neutralizing antibody responses against autologous SARS‐CoV‐2 pseudovirus, with most of GMT of NTIC50 over 1000 (Figure 4A–C). However, the NTIC50 GMT against some evolving SARS‐CoV‐2 variants showed pronounced decline, which revealed that immune escape was ubiquitous in monovalent immunizations. For example, immune sera from M1 group exhibited significant reduction (~5‐fold) in neutralizing pseudovirus displaying Omicron (GMT, 240.8) and XE spikes (GMT, 265.7) (Figure 4C,D). Similarly, the neutralization against D614G spike pseudovirus was significantly reduced when compared with that against Omicron spike pseudovirus in the M3 group (Figure 4A,C). In the sequential immunizations, immune sera from S3 group exhibited higher level in neutralizing pseudovirus displaying Delta spike (Figure 4B) but did not show significant difference in neutralizing Omicron variant compared with that in S4 group (Figure 4C). Interestingly, S4 group induced higher level of neutralizing antibody titer against XE spike pseudovirus than any other group (Figure 4D). In the divalent immunization strategy, D1 regimen with wild‐type and Delta spike immunogens could elicit high levels of neutralizing antibodies against SARS‐CoV‐2 pseudoviruses displaying wild type, Delta, and XE spikes, with GMT of NTIC50 over 1000 (Figure 4A,B,D), but with less titer (<1000) against Omicron variant. On the contrary, D2 regimen with Delta and Omicron immunogens elicited high levels of neutralization antibody responses against all of the variants.
Figure 4.
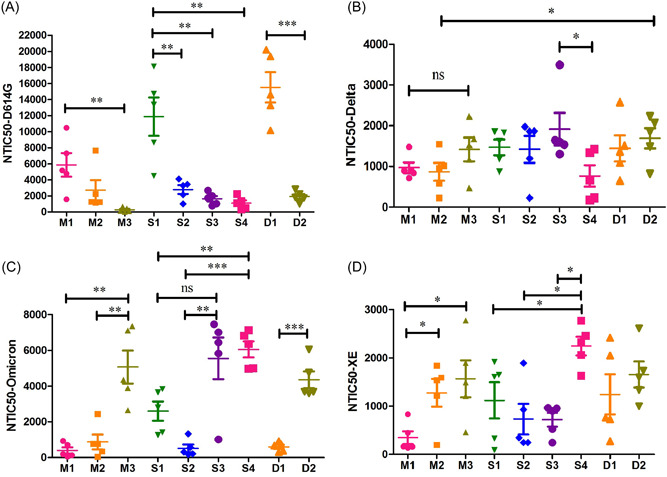
Neutralizing activity against D614G (A), Delta (B), Omicron (C), and XE (D) pseudoviruses induced by monovalent, combination, and sequential vaccination regimens. Fifty percent pseudovirus neutralization titer (NTIC50) representing the relative luminescence unit (RLU) values were reduced by 50% compared with the pseudovirus control well. The difference of geometric mean titer (GMT) was tested for statistical significance with a Wilcoxon matched‐pairs signed‐rank test in the GraphPad Prism 8. Differences were considered statistically significant at *p < 0.05, **p < 0.01, or ***p < 0.001.
We further compared the neutralizing antibody titers among three immunization strategies, each NTIC50 against SARS‐CoV‐2 pseudoviruses displaying wild‐type, Delta, and XE spikes from nine groups was shown in Figure 5A. To investigate the breadth of neutralizing antibody responses of different immunization strategies, we analyzed the relative neutralizing activities of each individual against variant spike pseudoviruses based on the NTIC50 against D614G spike pseudoviruse. The heat map of neutralizing activities exhibited the specific and cross‐reactive neutralizing antibody responses of each group (Figure 5B). Comparative analysis showed that immunization strategies (M3, S3, S4, and D2 groups) containing Omicron spike trimer could elicit more potent and broader neutralizing antibody responses. The result also confirmed that Omicron spike trimer possess superior immunogenicity over any other spikes in the study.
Figure 5.
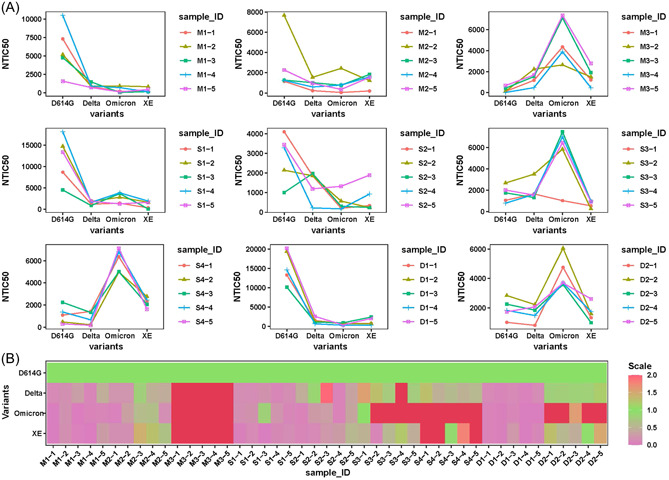
Comparative analysis of neutralizing antibodies titer among the individual, sequential, and combinational regimens. (A) The neutralizing antibodies titers against D614G, Delta, Omicron, and XE pseudoviruses in each group. (B) Heat map of the relative neutralizing activities of mice sera against pseudoviruses bearing the variant spike proteins compared to the D614G pseudovirus (purple denotes reduction; red denotes increase).
4. DISCUSSION
Sequential and cocktail immunization strategies are frequently mentioned in the development of human immunodeficiency virus (HIV) vaccine circumventing the extreme viral diversity. 21 , 22 Generally, these two immunization regimens are expected to induce broader neutralizing antibody responses against multiple HIV strains with highly diverse sequences. Different from HIV, SARS‐CoV‐2 has an exonuclease NSP14, which could correct the nucleotide mismatches occurred in the process of viral genome transcription; 23 thus, the overall mutation frequency is lower than that of HIV. However, the recent emergence of new circulating mutant strains, such as Omicron, has raised public concern about the protection efficiency of SARS‐CoV‐2 vaccine and antibody therapy. On the other hand, the breakthrough infection of COVID‐19 is increasing, 24 , 25 demonstrating the urgent need for alternative vaccine and/or immunization strategies to induce broad neutralizing antibody responses against various emerging variants. In this study, we assessed the immunogenicity of spike trimers derived from various SARS‐CoV‐2 variants by comparing the levels of binding and neutralizing antibodies induced by individual, sequential, or combinational immunizations.
Our data demonstrated that individual immunization of spike trimer could induce high level binding and neutralizing antibody responses against autologous SARS‐CoV‐2. However, monovalent immunization of spike trimer was unsuccessful to induce the generation of neutralizing antibody responses against multiple heterologous variants, named broad neutralizing antibody response. For example, immune sera from mice vaccinated with prototype spike trimer exhibited lower level of neutralization activity against the SARS‐CoV‐2 pseudovirus displaying Omicron spike (Figure 4C). Similarly, sera from Omicron spike trimer vaccination also showed limited neutralization activity against the D614G spike pseudovirus (Figure 4A). Notably, sequential and cocktail immunizations based on Delta and Omicron spike trimers both exhibited superior ability in inducing neutralizing antibody responses against tested emerging variants. Broad neutralizing antibody responses against SARS‐CoV‐2 variants induced by sequential and combinational immunizations might derived from the antibody maturation by presenting new epitopes and the humoral response on more conserved epitopes of spike trimer. In addition, higher level of neutralizing antibody responses against the emerging XE variant were induced in the regimens containing the Omicron spike trimer including M3, S3, S4, D1, and D2 groups. Hence, Omicron spike trimer could be regarded as a superior immunogen to develop novel COVID‐19 vaccine preventing emerging SARS‐CoV‐2 variants.
A potential limitation of this study lies in the lack of authentic virus neutralization assay. Additionally, challenge experiments in animals were not performed, and protective efficacy of different delivery strategies were not evaluated in this study. However, ELISA experiments and pseudovirus neutralization experiments could equally assess the prevention and protection of these immunization strategies against emerging variants. Even so, challenge experiments should be performed to further confirm whether the neutralizing antibody responses is sufficient to protect the challenge from emerging SARS‐CoV‐2 variants. Another extended study about trivalent immunization strategy based on wild type, Delta and Omicron spike trimers could be performed to assess the broad neutralizing antibody responses against SASR‐CoV‐2. In conclusion, our proposed sequential and cocktail immunization strategies had theoretical guidance for the development and inoculation of COVID‐19 vaccines.
AUTHOR CONTRIBUTIONS
Chengchao Ding, Zhirong Liu, and Yong Gao conceived, designed, and supervised the experiments. Chengchao Ding, Shuangshuang Ni, and Xiangyu Zhang wrote the manuscript. Jun He, Yong Sun, and Qingmin Mei performed the neutralization experiments. Jiajia Xie and Lina Huang performed the animal experiment. Shuangshuang Ni prepared and purified the protein. All of authors approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We thank GISAID and associated laboratories and researchers for the shared sequence information of SARS‐CoV‐2. We thank Director Jinjin Ten and Yan Wang of the Anhui Provincial Center for Disease Control and Prevention for providing animal house; we also thank Dr. Tengchuan Jin from University of Science and Technology of China for providing HEK293F cells for this study. This study was supported by the Key Research and Development Project of Anhui Province (202104j07020042; 202104a07020032; 2022e07020005), the Natural Science Foundation of Anhui Province (2208085QH257; 2208085QH258; 2208085MH260), the Postdoctoral Research Foundation of China (2021M693076; 2020M670084ZX), and the Fundamental Research Funds for the Central Universities (WK9110000166; WK9110000167).
Ding C, Ni S, Zhang X, et al. Evaluation of humoral immune responses induced by different SARS‐CoV‐2 spike trimers from wild‐type and emerging variants with individual, sequential, and combinational delivered strategies. J Med Virol. 2022;1‐9. 10.1002/jmv.28081
Chengchao Ding, Shuangshuang Ni, and Xiangyu Zhang contributed equally to this study.
Contributor Information
Chengchao Ding, Email: ding10118026@163.com.
Yong Gao, Email: ygao387@ustc.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Silverman RA, Ceci A, Cohen A, et al. Vaccine effectiveness during outbreak of COVID‐19 alpha (B.1.1.7) variant in men's correctional facility, United States. Emerging Infect Dis. 2022;28(7):1313‐1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moline HL, Keaton A, Rice W, et al. Effectiveness of COVID‐19 mRNA vaccines against infection during an outbreak of SARS‐CoV‐2 beta (B.1.351) variant in a skilled nursing facility–Virginia, March‐April 2021. Clin Infect Dis . 2022:ciac526. Published online June 27, 2022. [DOI] [PMC free article] [PubMed]
- 3. Tagliamonte MS, Mavian C, Zainabadi K, et al. Rapid emergence and spread of severe acute respiratory syndrome coronavirus 2 gamma (P.1) variant in Haiti. Clin Infect Dis. 2022;74(11):2057‐2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan S, Xiao K, Li D, et al. Preclinical immunological evaluation of an intradermal heterologous vaccine against SARS‐CoV‐2 variants. Emerg Microbes Infect. 2022;11(1):212‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ai J, Zhang H, Zhang Y, et al. Omicron variant showed lower neutralizing sensitivity than other SARS‐CoV‐2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. 2022;11(1):337‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi JY, Smith DM. SARS‐CoV‐2 variants of concern. Yonsei Med J. 2021;62(11):961‐968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chakraborty C, Bhattacharya M, Sharma AR, Dhama K. Recombinant SARS‐CoV‐2 variants XD, XE, and XF: the emergence of recombinant variants requires an urgent call for research–correspondence. Int J Surg. 2022;102:106670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Basky G, Vogel L. XE, XD & XF: what to know about the Omicron hybrid variants. Can Med Assoc J. 2022, 194(18):E654‐e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elie B, Roquebert B, Sofonea MT. Variant‐specific SARS‐CoV‐2 within‐host kinetics. J Med Virol. 2022;94(8):3625‐3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao Y, Yisimayi A, Bai Y, Huang W. Humoral immune response to circulating SARS‐CoV‐2 variants elicited by inactivated and RBD‐subunit vaccines. Cell Res. 2021;31(7):732‐741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keech C, Albert G, Cho I, et al. Phase 1‐2 trial of a SARS‐CoV‐2 recombinant spike protein nanoparticle. Vaccine. 2020;383(24):2320‐2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walsh EE, Frenck RW Jr, Falsey AR, et al. Safety and immunogenicity of two RNA‐based Covid‐19 vaccine candidates. N Engl J Med. 2020;383(25):2439‐2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verma S, Patil VM, Gupta MK. Mutation informatics: SARS‐CoV‐2 receptor‐binding domain of the spike protein. Drug Discov Today . 2022:103312. Published online July 3, 2022. [DOI] [PMC free article] [PubMed]
- 14. Xia S, Wang L, Zhu Y. Origin, virological features, immune evasion and intervention of SARS‐CoV‐2 Omicron sublineages. Signal Transduct Target Ther. 2022;7(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stuible M, Gervais C, Lord‐Dufour S, et al. Rapid, high‐yield production of full‐length SARS‐CoV‐2 spike ectodomain by transient gene expression in CHO cells. J Biotech. 2021;326:21‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Ulitzky L, Silberstein E, Taylor DR, Viscidi R. Immunogenicity and protection efficacy of monomeric and trimeric recombinant SARS coronavirus spike protein subunit vaccine candidates. Viral Immunol. 2013;26(2):126‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esposito D, Mehalko J, Drew M, et al. Optimizing high‐yield production of SARS‐CoV‐2 soluble spike trimers for serology assays. Protein Expr Purif. 2020;174:105686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding C, He J, Zhang X, et al. Crucial mutations of spike protein on SARS‐CoV‐2 evolved to variant strains escaping neutralization of convalescent plasmas and RBD‐specific monoclonal antibodies. Front Immunol. 2021;12:693775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS‐CoV‐2. Emerg Microbes Infect. 2020;9(1):680‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bricault CA, Kovacs JM, Nkolola JP, et al. A multivalent clade C HIV‐1 Env trimer cocktail elicits a higher magnitude of neutralizing antibodies than any individual component. J Virol. 2015;89(5):2507‐2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malherbe DC, Doria‐Rose NA, Misher L, et al. Sequential immunization with a subtype B HIV‐1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85(11):5262‐5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14‐nsp10 complex. Proc Natl Acad Sci USA. 2015;112(30):9436‐9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naito T, Yan Y. Real‐world evidence for the effectiveness and breakthrough of BNT162b2 mRNA COVID‐19 vaccine at a medical center in Japan. Hum Vaccin Immunother. 2022;18(1):1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang SL, Ripen AM. COVID‐19 breakthrough infections and humoral immune response among BNT162b2 vaccinated healthcare workers in Malaysia. Emerg Microbes Infect. 2022;11(1):1262‐1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


