Abstract
Two monofunctional NiFeS carbon monoxide (CO) dehydrogenases, designated CODH I and CODH II, were purified to homogeneity from the anaerobic CO-utilizing eubacterium Carboxydothermus hydrogenoformans. Both enzymes differ in their subunit molecular masses, N-terminal sequences, peptide maps, and immunological reactivities. Immunogold labeling of ultrathin sections revealed both CODHs in association with the inner aspect of the cytoplasmic membrane. Both enzymes catalyze the reaction CO + H2O → CO2 + 2 e− + 2 H+. Oxidized viologen dyes are effective electron acceptors. The specific enzyme activities were 15,756 (CODH I) and 13,828 (CODH II) μmol of CO oxidized min−1 mg−1 of protein (methyl viologen, pH 8.0, 70°C). The two enzymes oxidize CO very efficiently, as indicated by kcat/Km values at 70°C of 1.3 · 109 M−1 CO s−1 (CODH I) and 1.7 · 109 M−1 CO s−1 (CODH II). The apparent Km values at pH 8.0 and 70°C are 30 and 18 μM CO for CODH I and CODH II, respectively. Acetyl coenzyme A synthase activity is not associated with the enzymes. CODH I (125 kDa, 62.5-kDa subunit) and CODH II (129 kDa, 64.5-kDa subunit) are homodimers containing 1.3 to 1.4 and 1.7 atoms of Ni, 20 to 22 and 20 to 24 atoms of Fe, and 22 and 19 atoms of acid-labile sulfur, respectively. Electron paramagnetic resonance (EPR) spectroscopy revealed signals indicative of [4Fe-4S] clusters. Ni was EPR silent under any conditions tested. It is proposed that CODH I is involved in energy generation and that CODH II serves in anabolic functions.
Bacteria which utilize CO as a growth substrate include the aerobic carboxidotrophs and the anaerobic acetogens, sulfate-reducers, methanogens, and phototrophs (16, 34, 42, 44, 47). Carboxydothermus hydrogenoformans is a strictly anaerobic, thermophilic, gram-positive eubacterium which was isolated from a volcanic hot spring (55). Phylogenetically, C. hydrogenoformans falls into the group of the low-G+C subphylum of the gram-positive bacteria and shows highest 16S rRNA gene sequence homology to Thermoterrabacterium (52). C. hydrogenoformans utilizes CO under chemolithoautotrophic conditions (55). The bacterium couples the oxidation of CO to CO2 (E0′ = −0.52 V) to the reduction of protons to H2 (E0′ = −0.41 V) in the energy-conserving reaction CO + H2O → CO2 + H2, ΔG0′ = −20 kJ mol−1 (54, 55). The metabolism of C. hydrogenoformans is strictly fermentative because the bacterium is obligately anaerobic and utilizes protons as the characteristic intracellular electron acceptor. On the basis of the formation of H2 as the ultimate fermentation product by C. hydrogenoformans, we propose the terms “hydrogenogenic,” “hydrogenogens,” and “hydrogenogenesis” to refer to the type of metabolism, the physiological group, and the process of H2 formation, respectively. Although C. hydrogenoformans is not a phototroph, it is in many ways similar to the phototrophic bacteria Rhodospirillum rubrum and Rhodocyclus gelatinosus, which utilize CO anaerobically in the dark (34, 58, 59). C. hydrogenoformans is also able to ferment pyruvate to acetate and H2 (56).
Although carbon monoxide dehydrogenases (CODHs) formally catalyze the same reaction (CO + H2O → CO2 + 2 e− + 2 H+), different types of enzymes serving different metabolic functions operate in the various groups of CO-oxidizing bacteria (15, 16, 42, 43, 44, 47). Aerobic CODHs are MoFeS flavoproteins containing [2Fe-2S] clusters, while anaerobic CODHs are NiFeS proteins containing [4Fe-4S] clusters. The high-resolution crystal structures of the MoFeS CODHs from Oligotropha carboxidovorans (12, 26, 43) or Hydrogenophaga pseudoflava (27, 43) show a dimer of two heterotrimers in an (LMS)2 subunit structure. Each heterotrimer is composed of a molybdoprotein (L subunit), a flavoprotein (M subunit), and an iron-sulfur protein (S subunit). The molybdoprotein carries the active site, which contains a 1:1 molar complex of molybdopterin cytosine dinucleotide and a molybdenum atom. The iron-sulfur protein contains the type I and type II [2Fe-2S] centers. The flavoprotein contains the flavin adenine dinucleotide (FAD) cofactor and shows a new flavin-binding type (25, 26). The NiFeS CODHs are either monofunctional or bifunctional (15, 16, 47). The latter are associated and operate in a complex with acetyl coenzyme A (acetyl-CoA) synthase (ACS). The monofunctional CODH from the phototrophic bacterium R. rubrum (6, 7) is inducible in the dark under anaerobic conditions in the presence of CO, shows a micromolar Km for CO (6, 7, 34), and contains a proposed nickel-iron-sulfur cluster (cluster C) (30, 32, 53) and a conventional [4Fe-4S] cluster (cluster B) (32, 47). Radiolabeling studies suggested a catalytically essential nonsubstrate CO ligand (COL) to the Fe atom in the putative [Fe-Ni] center of cluster C (31). Acetogens and methanogens employ the bifunctional CODH-ACS (15, 16, 47). The enzymes are tetramers of two different subunits or pentamers of five different subunits. The subunits harboring the CODH activity contain cluster C and cluster B (16, 47), which is similar to the situation in R. rubrum CODH.
Significant CO:oxidized benzyl viologen (BV) oxidoreductase activity was identified in cytoplasmic fractions of strains of C. hydrogenoformans (46). The CODH structural genes cooF and cooS from C. hydrogenoformans have been sequenced (23). cooS was identified directly downstream of cooF. The genes showed the highest similarity to the cooF genes from the archeon Archaeglobus fulgidus and the cooS gene from the bacterium R. rubrum, respectively.
In this investigation we have identified, purified, and characterized two distinct homodimeric NiFeS CODHs from C. hydrogenoformans. Immunoelectron microscopy showed both enzymes attached to the inner aspect of the cytoplasmic membrane. The N-terminal sequences of CODH II and a 49-kDa chymotryptic peptide match the sequence of CooS. This is the first study describing Ni CODHs from an anaerobic CO-oxidizing hydrogenogenic bacterium.
MATERIALS AND METHODS
Organism and cultivation.
C. hydrogenoformans Z-2901 (DSM 6008) (55) was grown under strictly anaerobic conditions in 50-liter fermentors (Biostat U; Braun Biotech, Melsungen, Germany) at 60°C and pH 6.8 in the following medium (units are milligrams per liter): 1,500 NH4Cl; 200 MgSO4 · 7H2O; 20 CaCl2 · 2H2O; 300 KH2PO4; 300 K2HPO4; 500 NaHCO3; 500 Na-thioglycolate; 500 yeast extract; 1 resazurin; 2 NiCl2 · 6H2O; 10 ammonium ferric(III) citrate; 1 FeSO4 · 7H2O; 1 ZnSO4 · 7H2O; 5 MnSO4 · H2O; 0.1 H3BO3; 1 CoCl2 · 6H2O; 0.1 CuSo4 · 5H2O; 0.1 Na2MoO4 · 2H2O; 0.2 Na2SeO3 · 5H2O; 0.1 KAl(SO4)2 · 12H2O; 1 ml of vitamin solution (61). The fermentors were continuously supplied with 0.5 liters of CO min−1 and stirred at 500 rpm. Bacteria were harvested by centrifugation under N2 and kept frozen at −20°C under N2 until use.
Anaerobic procedures.
All procedures for the preparation of cell extracts, cell fractions, and enzyme purifications were carried out anaerobically under a flow of N2 or in an anoxic glove box chamber (model 1024 anaerobic system; Forma Scientific, Marrietta, Ohio) under an atmosphere of pure N2. All buffers were repeatedly degassed by evacuation, flushed with N2, supplied with 2 mM Na-dithionite, and maintained under a slight overpressure of N2.
Preparation and purification of CODH.
For enzyme purifications, C. hydrogenoformans was harvested at an approximate optical density at 436 nm of 2. About 100 g of bacterial cell mass was suspended in 200 ml of 50 mM Tris-HCl (pH 8.0) containing 2 mM Na-dithionite (buffer A), 0.1 mg of lysozyme per ml, 0.05 mg of DNase I per ml, and 0.2 mM phenylmethylsulfonyl fluoride and incubated for 30 min at 37°C with gentle stirring. The protoplasts thus obtained were subjected to osmotic lysis followed by low-spin centrifugation. The resulting cell-free extracts were subjected to ultracentrifugation for 2 h at 120,000 × g, yielding cytoplasmic and membrane fractions. Intact protoplasts were prepared from bacteria suspended in the above buffer and supplied with 0.6 M sucrose.
Cytoplasmic fractions (250 ml) were subjected to anion exchange chromatography (Macro-Prep High Q; Bio-Rad) on columns (dimensions, 17 by 5 cm) equilibrated with buffer A. Elution was with 700 ml of buffer A followed by 2,800 ml of a linear gradient of 0 to 1 M NaCl in buffer A. Fractions with CODH activity were pooled, supplemented with 1.3 M ammonium sulfate, and gently stirred for 30 min, and the precipitated protein was removed by low-spin centrifugation. The supernatant was loaded onto a hydrophobic interaction chromatography column (Source 15 ISO; Pharmacia; dimensions, 20 by 5 cm), equilibrated with 1.2 M ammonium sulfate in buffer A, and eluted with 800 ml of equilibration buffer followed by 2,400 ml of a decreasing linear gradient of 1.2 to 0 M ammonium sulfate in buffer A. Two separate peaks showing CODH activity appeared which were designated CODH I and CODH II and purified separately. CODH I was subjected to hydrophobic interaction chromatography on butyl-Sepharose 4 Fast Flow (Pharmacia). Columns (dimensions, 10 by 5 cm) were equilibrated with 0.7 M ammonium sulfate in buffer A, and proteins were desorbed with 200 ml of equilibration buffer followed by 800 ml of a decreasing linear gradient of 0.7 to 0 M ammonium sulfate in buffer A. CODH I or CODH II were desalted by gel filtration on Sephadex G-25 in buffer A, subjected separately to anion exchange chromatography on Source 30 Q (Pharmacia; column dimensions, 12 by 2.6 cm), equilibrated with buffer A, and eluted with 130 ml of buffer A followed by 640 ml of a linear gradient of 0 to 0.6 M NaCl in buffer A. CODH I or CODH II were then subjected to gel filtration (Sephacryl S-200; Pharmacia; column dimensions, 60 by 2.6 cm) employing buffer A. Preparations of purified CODH I and CODH II were frozen in liquid N2 and kept at −20°C under N2 until use.
Enzyme assays.
CODH activity was assayed at 70°C, which is the optimal growth temperature for C. hydrogenoformans (55), by following the CO-dependent reduction of oxidized methyl viologen (MV) in a spectrophotometer employing an ɛ578 of 9.7 mM−1 cm−1 (6). For the assays, 1-ml volumes containing 20 mM MV and 2 mM dithioerythritol (DTE) in buffer B (50 mM HEPES-NaOH [pH 8.0]) were flushed with CO in screw-cap cuvettes sealed with a rubber septum. Reactions were initiated by injecting enzyme with a syringe. H2 oxidation activity was assayed under the same conditions except that CO was replaced by H2. One unit of CO or H2 oxidation activity is defined as the reduction of 2 μmol of MV min−1, which is equivalent to 1 μmol of CO or H2 oxidized min−1.
H2 evolution activity was assayed in a gas chromatograph by headspace analysis of the H2 produced with reduced MV or CO as the source of reducing equivalents for the reduction of protons. Assays were carried out at 70°C in 40-ml serum-stoppered vials which were kept shaken at 120 rpm. Assays with reduced MV were composed of 10 ml of buffer B, 2 mM MV, and 60 mM Na-dithionite under a gas atmosphere of N2. Assays with CO were composed of 10 ml of 50 mM buffer B and 2 mM DTE under a gas atmosphere of pure CO. The gas chromatograph (model CP 9000; Chrompack, Middelburg, The Netherlands) was equipped with a thermal conductivity detector (model 903 A), a Hayeser Q column (2.5 m), and a molecular sieve 13× column (1.8 m). The temperatures (in degrees Celsius) were 40 (oven), 80 (injection port), and 200 (detector), respectively. The carrier gas was N2 at a flow rate of 30 ml min−1. One unit of H2 evolution activity is defined as 1 μmol of H2 produced min−1.
The [1-14C]acetyl-CoA–CO exchange activity in cell-free extracts or of purified CODHs was assayed at 30, 50, and 70°C following published procedures (13, 49). ACS activity was examined by following acetate formation from 5-methyltetrahydrofolate, CO, and CoA (21). Extracts of Clostridium thermoaceticum (DSM 2955) grown on glucose, peptone, and yeast extract (39) served as a positive control. Acetate was analyzed with a gas chromatograph, employing the following conditions: gas chromatograph (model 430; Packard Instrument Company, Downers Grove, Ill.); flame ionization detector (model 901); column (Porapak Q, 50/80 mesh, 2.5 m); 175°C (oven), 200°C (injection port and detector). The carrier gas was N2 (30 ml min−1). Samples of 0.5 ml were acidified with 10 μl of concentrated aqueous HCl, precipated protein was removed by low-spin centrifugation, and 2 μl of the supernatant was injected into the column.
Electron acceptor specificity.
Electron acceptor specificity of CO oxidation catalyzed by CODH was tested spectrophotometrically under the conditions of the CODH activity assay with the exception that DTE was omitted from the reaction mixture. The following compounds (100 μM concentrations of each) were examined (extinction coefficient units are per millimolar per centimeter) (11): BV, ɛ560 = 8.7; NAD+ or NADP+, ɛ340 = 6.2; FAD, ɛ450 = 11.3; flavin mononucleotide (FMN), ɛ450 = 12.2; 1-phenyl-2-(4-iodophenyl)-3-(4-nitrophenyl)-2H-tetrazolium chloride (INT)–20 μM 1-methoxy-phenazine methosulfate (MPMS), ɛ496 = 18.0 (35); ubiquinone Q10, ɛ276 = 14.7; methylene blue, ɛ615 = 37.1; phenazine methosulfate (PMS), ɛ387 = 25.0; 2,6-dichlorophenol-indophenol (DCPIP), ɛ600 = 16.1; horse heart cytochrome c, ɛ550 = 29.5.
Kinetic measurements.
The Km for CO was determined by varying the CO concentration in the reaction mixture under MV saturation (20 mM). The different CO concentrations were established by adding appropriate amounts of CO-saturated reaction mixture composed of buffer B, 20 mM MV, and 2 mM DTE to assays containing the same reaction mixture saturated with N2. At 70°C and 1 atm pressure, the CO concentration in CO-saturated reaction mixtures was taken to be 645 μM (38). The actual starting CO concentration was calculated from the final concentration of reduced MV, considering that 1 μmol of CO reduces 2 μmol of MV. The Km for MV was determined in a CO-saturated reaction mixture composed of 50 mM buffer B, 2 mM DTE, and different concentrations of MV.
Peptide mapping by limited proteolysis.
Purified CODH in 50 mM Tris-HCl (pH 8.0) containing 0.5% (wt/vol) sodium dodecyl sulfate (SDS) was denatured by boiling for 2 min. Assay mixtures were diluted to 0.1% SDS by employing 100 mM acetate buffer (pH 4.0) (pepsin digestion) or 100 mM Tris-HCl (pH 8.0) (α-chymotrypsin digestion). Limited proteolysis was carried out at 23°C (pepsin) or 37°C (α-chymotrypsin) at protein-to-protease ratios (by mass) of 4:1 for pepsin and 80:1 for α-chymotrypsin. Proteolytic fragments were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 12 or 15% (wt/vol) running gels.
Immunological methods.
Polyclonal immunoglobulin G (IgG) antibodies directed against CODH I (Sequence Laboratories GmbH, Göttingen, Germany) or CODH II (Eurogentec Bel S.A., Herstal, Belgium) were obtained from immunized rabbits. The antibodies were purified from polyclonal rabbit antisera by affinity chromatography on protein A-Sepharose CL-4B (Pharmacia) according to the instructions of the manufacturer. For Western blotting, alkaline phosphatase-labeled goat anti-rabbit IgGs (Sigma, Deisenhofen, Germany) were employed.
Ouchterlony double immunodiffusion was performed on glass slides using slabs solidified with 0.75% (wt/vol) agarose in 60 mM Na2HPO-KH2PO4 buffer (pH 8.0) containing 0.02% (wt/vol) sodium azide (45). Precipitin bands were allowed to develop for 24 to 72 h at 30°C in a humid atmosphere and then stained with Coomassie brilliant blue G-250.
For immunoelectron microscopy, bacteria were subjected to postembedding immunogold labeling, as described previously (2, 50). After prefixation in the growth medium (1.5% [vol/vol] glutaraldehyde; 30 min at 4°C) the bacteria were washed in 50 mM phosphate buffer (pH 7.4), embedded in 1% (wt/vol) agar in the same buffer, dehydrated in ethanol, fixed with 1.5% (wt/vol) glutaraldehyde (1 h at 4°C), and embedded in Lowicryl K4M resin (Polysciences Inc., Warrington, Pa.) by using the progressive-lowering-of-temperature embedding technique (1, 2).
Ultrathin sections, cut with diamond knives, were mounted on Formvar carbon-coated copper grids and labeled as described previously (2). The sections were treated for 30 min with 0.1 M lysine in phosphate-buffered saline (PBS; 50 mM phosphate buffer containing 0.9% [wt/vol] NaCl, pH 6.9), then with 1% (wt/vol) milk powder solution in PBS, washed in PBS, and incubated for 2 h on drops containing primary antibodies (IgG antibodies directed against CODH I or CODH II) diluted in 2% (wt/vol) bovine serum albumin in PBS. The sections were washed in PBS, incubated for 1 h on drops containing secondary antibodies (gold-labeled goat anti-rabbit IgG; Amersham Pharmacia Biotech, Freiburg, Germany) diluted in 2% (wt/vol) bovine serum albumin in PBS, washed in PBS again, and rinsed with distilled water (three times on drops for a total of 5 min). The sections were stained with 2% (wt/vol) uranyl acetate (up to 5 min) and examined in a Zeiss EM 109 or Zeiss CEM 902A transmission electron microscope (Zeiss, Oberkochen, Germany) at an acceleration voltage of 80 kV. The specificity of labeling was demonstrated with preimmune sera.
The occurrence of CODH I and CODH II at the cytoplasmic membrane, or in the cytoplasm, was examined by counting the gold particles on thin sections. Gold particles occurring within a range of 30 nm from either side of the cytoplasmic membrane were taken as indicative of membrane-associated CODH, and those outside that range were interpreted as cytoplasmic CODH (51). Corrections were made for unspecific background labeling. The software XL-Docu (analySIS 3.0; Soft Imaging System, Münster, Germany) was used for quantitative analysis.
Miscellaneous methods.
Protein estimation employed conventional methods (5, 10). Analytical PAGE was carried out according to Laemmli (37). For SDS-PAGE, 7.5% (wt/vol) stacking gels and 12 or 15% (wt/vol) running gels were used. Native PAGE was performed under anaerobic conditions employing 5% (wt/vol) stacking gels and 7.5% (wt/vol) running gels. Proteins were stained with Coomassie brilliant blue G-250. Protein transfer from acrylamide gels to polyvinylidene difluoride membranes and N-terminal amino acid sequencing were carried out as described previously (24). Isoelectric focusing was carried out on IsoGel agarose plates (FMC Bioproducts, Rockland, Maine) according to the manufacturer's instructions.
The molecular masses of CODH I or CODH II were determined by gel filtration on Superdex 200 (HiLoad 16/60; Pharmacia; column dimensions, 60 by 1.6 cm) equilibrated with buffer A. Ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), and ovalbumin (43 kDa) were employed for standardization.
Metal contents were estimated by inductively coupled plasma mass spectroscopy (model VG Plasmaquad PQ2 turbo plus; Fisons Instruments/VG elemental, Wiesbaden, Germany) and by neutron activation analysis. Iron (17) and acid-labile sulfur (19) were estimated colorimetrically.
X-band electron paramagnetic resonance (EPR) spectra were recorded on a Bruker EMX spectrometer (Karlsruhe, Germany) operated with a helium cryostat under the experimental conditions described previously (28).
Chemicals.
All chemicals employed were obtained from the usual commercial sources. Gases were purchased from Riessner-Gase (Lichtenfels, Germany).
RESULTS
CO oxidation in subcellular fractions.
Under the conditions detailed in Materials and Methods, C. hydrogenoformans grew with a generation time of 4.9 h and a yield of 1.2 mg of protein per ml. During growth, the specific CO oxidation activity in cell-free extracts increased continuously from 20 to 1,800 U mg of protein−1. About 71% of the total CO oxidation activity in extracts appeared to be soluble in the cytoplasmic fraction (Table 1). Less than 1% of the total CO oxidation activity showed up outside protoplasts. CODH is apparently only loosely associated with the cytoplasmic membrane, since washing of the membranes with buffer mobilized about 50% of the membrane-associated CODH fraction. The H2-evolving hydrogenase (74.7 U mg of membrane protein−1) of C. hydrogenoformans is entirely membrane bound and showed only a little H2 oxidation activity (0.9 U mg of membrane protein−1).
TABLE 1.
Purification of CODH I and CODH II from CO-grown C. hydrogenoformans
| Purification state | CODH fraction | Total protein (mg) | Total activitya (kU) | Specific activitya (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Cell-free extract (step 1) | I + II | 13,086 | 15,301 | 1,169 | 100 | 1 |
| Cytoplasmic fraction (step 2) | I + II | 6,260 | 10,814 | 1,728 | 71 | 1.5 |
| Macro-Prep High Q (step 3) | I + II | 2,448 | 11,956 | 4,884 | 78 | 4.2 |
| Source 15 ISO (step 4) | I | 643 | 6,253 | 9,725 | 41 | 8.3 |
| Butyl-Sepharose 4 Fast Flow (step 5) | I | 427 | 5,228 | 12,243 | 34 | 10.5 |
| Source 30 Q (step 6) | I | 366 | 4,601 | 13,692 | 30 | 11.7 |
| Sephacryl S-200 (step 7) | I | 279 | 4,399 | 15,756 | 29 | 13.5 |
| Source 15 ISO (step 8) | II | 249 | 2,963 | 11,890 | 19 | 10.2 |
| Source 30 Q (step 9) | II | 190 | 1,883 | 9,919 | 12 | 8.5 |
| Sephacryl S-200 (step 10) | II | 142 | 1,964 | 13,828 | 13 | 11.8 |
One unit is defined as 1 μmol of CO oxidized min−1 with 20 mM oxidized MV as electron acceptor at 70°C and pH 8.0.
Identification of two distinct CODHs.
We managed to establish a protocol for the purification of two CODHs from C. hydrogenoformans (Table 1). Upon anion exchange chromatography (step 3) CODH activity eluted as a single peak at 0.25 M NaCl. Upon hydrophobic interaction chromatography (steps 4 and 8) two distinct peaks of CODH activity appeared, one at 0.6 M ammonium sulfate (designated CODH I) and a second one at 0.35 M ammonium sulfate (designated CODH II). CODH I and CODH II were purified separately (Table 1). Upon hydrophobic interaction chromatography (step 5) CODH I eluted at 0.05 M ammonium sulfate. Upon anion exchange chromatography (steps 6 and 9) CODH I and CODH II eluted at 0.17 and 0.14 M NaCl, respectively. CODH I was purified 14-fold with a specific activity of 15,756 U mg of protein−1 and a yield of 29%. CODH II was purified 12-fold with a specific activity of 13,828 U mg of protein−1 and a yield of 13%. Both enzymes had the same isoelectric point (pI) of 5.5.
The two CODHs have been purified to homogeneity, as shown by the single bands obtained after native and SDS-PAGE (Fig. 1). They are homodimers with subunit masses of 62.5 kDa (CODH I) and 64.5 kDa (CODH II) (Fig. 1B), which can be deduced by considering the following. Gel filtration of CODH I revealed a Stokes radius of 4.370 nm, corresponding to a molecular mass of 118,768 Da. The corresponding values of CODH II were 4.328 nm and 116,869 Da. CODH I and CODH II showed slightly different mobilities upon native PAGE, corresponding to molecular masses of 155 and 142 kDa, respectively (Fig. 1A). The lowered mobility of CODH I might reflect an increased structural flexibility or a different Stokes radius of the native protein (Fig. 1A).
FIG. 1.
Analysis of CODH I and CODH II by PAGE. CODH I and CODH II were subjected to native PAGE (A), SDS-PAGE (B), peptide mapping (C), and Western blot analysis (D). (A) Native PAGE. Lane 1, molecular mass markers; lane 2, 10 μg of CODH I; lane 3, 10 μg of CODH II. (B) SDS-PAGE. Lane 4, 5 μg of CODH I; lane 5, 5 μg of CODH II; lane 6, molecular mass markers. (C) Peptide mapping of 10 μg of CODH I or CODH II by limited proteolysis with pepsin or α-chymotrypsin for 30 min. Lane 7, pepsin digest of CODH I; lane 8, pepsin digest of CODH II; lane 9, α-chymotrypsin digest of CODH I; lane 10, α-chymotrypsin digest of CODH II (the number 49 refers to the 49-kDa peptide which has been sequenced). (D) Western blot analysis with purified polyclonal IgG antibodies directed against CODH I or CODH II (10 μg of each). Lane 11, CODH I and antibodies directed against CODH I; lane 12, CODH II and antibodies directed against CODH I; lane 13, CODH I and antibodies directed against CODH II; lane 14, CODH II and antibodies directed against CODH II.
The subunit amino-terminal sequences of CODH I (SNWKNSVDDAVDYLLPIAKKAG) and CODH II (AKQNLXKTDRAVQQMLDKAK) as determined by Edman degradation characterize the two enzymes as distinct proteins. The sequences indicate that the methionyl residue at each N terminus was excised. The proteolytic peptide patterns of the CODHs indicate different primary structures (Fig. 1C). The N terminus of a 49-kDa chymotryptic peptide of CODH II (Fig. 1C) was VTTVLPSSRV. Polyclonal IgG antibodies raised against CODH I were specific for that enzyme in Western blots and did not react with CODH II (Fig. 1D), and IgG antibodies raised against CODH II were specific for CODH II and did not react with CODH I (Fig. 1D). Ouchterlony double immunodiffusion employing CODH I, CODH II, and IgG antibodies directed against CODH I showed that the enzymes share no common antigenic determinants (data not shown).
The presence of Ni, Fe, and acid-labile sulfur in both CODHs is apparent from analysis by inductively coupled plasma mass spectroscopy (values in moles of element per mole of enzyme [mean ± standard deviation]: Ni, 1.41 ± 0.01 for CODH I and 1.65 ± 0.03 for CODH II; Fe, 19.83 ± 0.22 for CODH I and 19.25 ± 0.27 for CODH II); neutron activation (Ni, 1.28 ± 0.03 for CODH I and 1.67 ± 0.11 for CODH II; Fe, 22.37 ± 0.27 for CODH I and 24.19 ± 0.98 for CODH II); and colorimetric determination (Fe, 20.7 ± 0.5 for CODH I and 19.6 ± 0.3 for CODH II; acid-labile sulfur, 21.9 ± 1.9 for CODH I and 18.8 ± 1.2 for CODH II). Both enzymes contained less than 0.1 mol of Co, Cr, Cu, Mo, V, or Zn per mol.
Catalytic properties.
The two CODHs catalyze the CO-dependent reduction of various oxidized electron acceptors and show similar electron acceptor specificities (CO oxidation activities at 70°C and pH 8.0, in units per milligram) for CODH I (BV, 5,200 [set at 100%]; MV, 1,400 [27%]; methylene blue, 1,248 [24%]; PMS, 572 [11%]; FAD, 364 [7%]; FMN, 104 [2%]; DCPIP, 52 [1%]) and for CODH II (BV, 3,100 [set at 100%]; MV, 1,000 [32%]; PMS, 341 [11%]; methylene blue, 155 [5%]; FAD, 31 [1%]; FMN, 31 [1%]; DCPIP, 31 [1%]). NADP+, NAD+, INT-MPMS, ubiquinone Q10, and horse heart cytochrome c were not reduced by the two enzymes. At saturating electron acceptor concentrations (20 mM), the following specific CO oxidation activities (in units per milligram) were obtained for CODH I: 29,400 (BV) and 15,800 (MV). For CODH II the activities were 27,000 (BV) and 13,800 (MV).
Both CODHs catalyze the formation of a total of two electrons from CO and water (CO + H2O → CO2 + 2H+ + 2e−). Examination of the amounts of MV reduced in the presence of limiting CO concentrations indicated molar ratios of 2.15 (±0.21):1 (CODH I) and 2.08 (±0.08):1 (CODH II), which are consistent with the functioning of MV as a one-electron acceptor.
CO oxidation followed first-order kinetics. The apparent Km was 4 mM MV for both CODHs, 30 μM CO for CODH I, and 18 μM CO for CODH II (assayed at 70°C and pH 8.0). The apparent Vmax, kcat, and kcat/Km of CODH I were 18,900 μmol min−1 mg−1, 39,000 s−1, and 1.3 · 109 M−1 CO s−1, respectively, and of CODH II were 14,200 μmol min−1 mg−1, 31,000 s−1, and 1.7 · 109 M−1 CO s−1, respectively.
CODH I (CO oxidation activity of 14,400 U mg−1) could oxidize H2 with MV as an electron acceptor (0.81 U mg−1). The reverse reaction, or the formation of H2 from CO, was not catalyzed. CODH II (CO oxidation activity of 10,900 U mg−1) could neither oxidize nor produce H2 from reduced MV, but it could produce H2 from CO (0.14 U mg−1).
Neither CODH catalyzed the exchange of 14C from the carbonyl group of [1-14C]acetyl-CoA with 12C from 12CO. The amount of radioactivity in the aqueous phase containing [1-14C]acetyl-CoA under an atmosphere of pure CO remained constant in the presence of CODH I or CODH II (0.5 mg of CODH ml−1) when assayed for up to 2 h at 30, 50, and 70°C. The same results were obtained when purified CODH was replaced by cell-free extracts, irrespective of whether the extracts were prepared in the presence or absence of dithionite. Cell-free extracts of C. hydrogenoformans (CODH activity of 1,438 U mg−1) were assayed for ACS activity by examination of acetate production from 5-methyltetrahydrofolate, CO, and CoA as detailed in Materials and Methods. Acetate was below the detection limit (0.05 nmol min−1 mg of protein−1) after 2, 4, and 6 h of incubation, reflecting the absence of ACS activity in C. hydrogenoformans. Control experiments with cell-free extracts of C. thermoaceticum (CODH activity of 13.1 U mg−1) showed an ACS activity of 2.8 nmol of acetyl-CoA synthesized min−1 mg of protein−1. An EPR signal with a gav of 2.06 has been attributed to the reduced Ni-X-[4Fe-4S] cluster A, which is the site of acetyl-CoA synthesis in the bifunctional CODH-ACS from acetogens and methanogens (16, 47). In accordance with the absence of ACS activity in both CODHs from C. hydrogenoformans, we were not able to demonstrate this EPR signal in CO-reduced CODH I or CODH II at temperatures between 10 and 130 K. Ribulose-1,5-bisphosphate carboxylase activity, assayed according to published procedures (9), could also not be demonstrated in C. hydrogenoformans.
Both CODHs showed an unusual dependence of CO oxidation activity on temperature, with maxima at 95°C (CODH I, 37.949 kU mg−1) and 105°C (CODH II, 40.666 kU mg−1) and half-lives at 90°C under N2 of 7 h (CODH I) and 2 h (CODH II). The half-lives at 100°C were 7 min for CODH I and 10 min for CODH II. At 114°C, CODH II retained 90% of its maximum activity seen at 105°C, and CODH I retained 28% of its maximum activity seen at 95°C. The pH optima at 70°C were 8.0 (CODH I) and 8.3 (CODH II). The two enzymes were inactivated upon exposure to air at 23°C with half-lives of 20 min (CODH I) and 60 min (CODH II).
Spectral characterization.
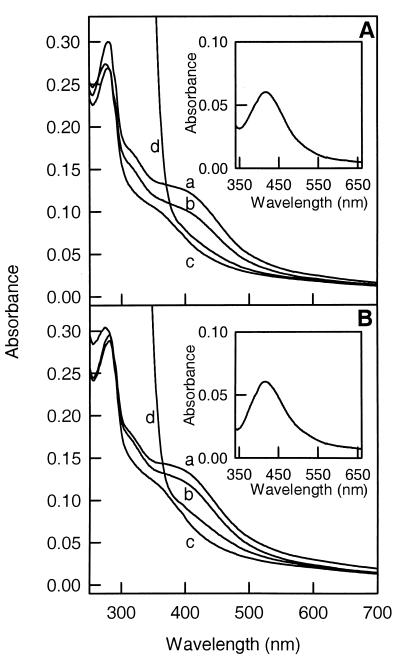
The UV-visible absorption spectra of purified CODH I (Fig. 2A) and CODH II (Fig. 2B) were similar under the various conditions examined. Treatment of either enzyme with CO or dithionite resulted in bleaching of the FeS-like shoulder extending from 350 to 550 nm and centered around 419 nm, referring to a reduced type of spectrum (Fig. 2A and B, traces c and d). The shoulder reappeared when the enzymes treated with CO (Fig. 2A and B, trace c) were exposed to air (Fig. 2A and B, trace b). When dithionite was removed from the as-isolated enzymes kept under N2, the spectra shown in Fig. 2A and B (trace a) were obtained. The spectra are similar to those of the air-oxidized enzymes (Fig. 2A and B, trace b) and therefore are believed to represent an oxidized state. Corresponding oxidized-minus-reduced-difference spectra are shown in the insets. The following extinction coefficients (ɛ, per millimolar per centimeter) were calculated for the two CODHs in the CO-reduced (Fig. 2A and B, trace c) or oxidized (Fig. 2A and B, trace a) state: CODH I, ɛ280 = 191.9 (oxidized) or 172.8 (reduced), ɛ419 = 74.0 (oxidized) or 35.5 (reduced); CODH II, ɛ280 = 200.4 (oxidized) or 203.7 (reduced), ɛ419 = 83.8 (oxidized) or 42.8 (reduced). Assuming an extinction coefficient of about 4 per mM per Fe in a 4Fe cluster, the calculated approximate number of 4Fe clusters is 4.6 in CODH I and 5.2 in CODH II.
FIG. 2.
UV-visible absorption spectra of CODH I (A) and CODH II (B). The enzymes (0.2 mg ml−1) were in 50 mM Tris-HCl (pH 8.0). Conditions for each curve: a, under N2 as isolated, dithionite was removed by gel filtration; b, oxidized with air; c, reduced with pure CO; d, reduced with 2 mM dithionite under N2. Insets: difference spectra of condition a minus condition c.
Between 10 and 75 K, the two CODHs in their as-isolated state revealed only weak paramagnetic signals (Fig. 3A and B, traces a and e). The spectra did not change upon exposure of the enzymes to air, with the exception of a slight increase of the signal at g = 4.29 (Fig. 3A and B, traces b and f). Reduction of the as-isolated enzymes (Fig. 3A and B, traces a and e) with CO (Fig. 3A and B, traces c and g) or dithionite (Fig. 3A and B, traces d and h) exhibited complex EPR signals. The reduced CODH signals originated from two different paramagnetic components (Fig. 3A and B, traces c, d, g and h). One paramagnetic component shows a rhombic signal with g values (gz, gy, gx) of 2.04, 1.94, and 1.90 (gav = 1.94), which is the same for both CODHs. The other paramagnetic component shows a different rhombic signal with g values (gz, gy, gx) of 1.97, 1.87, and 1.77 (gav = 1.86) for CODH I, and g values (gz, gy, gx) of 1.99, 1.80, and 1.74 (gav = 1.84) for CODH II. The two paramagnetic components can be differentiated by their temperature dependence. The rhombic signal at gav = 1.94 (CODH I and II) appeared between 10 and 40 K with maximum intensity at 10 to 25 K, whereas the signals at gav = 1.86 (CODH I) or 1.84 (CODH II) were apparent at 10 K and absent at 25 K. The g values and temperature dependence of the reduced signal at gav = 1.94 (CODH I and II) suggests that it originates from an S = 1/2 [4Fe-4S]1+ cluster (32). The reduced signals at gav = 1.86 (CODH I) or 1.84 (CODH II) can be assigned to a second but faster relaxing S = 1/2 [4Fe-4S]1+ cluster (32). Integration of the spectra and comparison with a spin standard of copper EDTA give a spin concentration for CO-reduced CODH I of 2.74 mol of spin/mol of CODH I for the gav = 1.94 EPR signal and 2.18 mol of spin/mol of CODH I for the gav = 1.86 EPR signal. The spin concentration for CO-reduced CODH II was 2.87 mol of spin/mol of CODH II for the gav = 1.94 EPR signal and 2.17 mol of spin/mol of CODH II for the gav = 1.84 EPR signal. The air-oxidized CODHs at 10 K (Fig. 3A and B, trace b) and 25 K (Fig. 3A and B, trace f) exhibited an axial-type EPR signal near gav of 2.01, which was not detectable above 75 K. We suggest that this signal emerges from an oxidized [3Fe-4S]1+ cluster produced by the oxidative damage of one of the [4Fe-4S] clusters in analogy to previous reports (3, 18, 40). Simultaneously with the axial signal, a small signal at g = 4.29 appeared which presumably originated from Fe3+ liberated through damage of a [4Fe-4S] cluster (3). No EPR signal attributable to Ni(III) or Ni(I) has been observed in either enzyme in their as-isolated, air-oxidized, or CO- or dithionite-reduced states between 10 and 130 K.
FIG. 3.
EPR spectra of CODH I (A) and CODH II (B). CODH I (3.8 mg ml−1) or CODH II (3.1 mg ml−1) were in 50 mM Tris-HCl (pH 8.0). Traces: (a and e) as-isolated freshly prepared enzyme was frozen under N2; (b and f) air-oxidized as-isolated enzyme was kept under air for 12 h at 4°C; (c and g) CO-reduced as-isolated enzyme was kept under pure CO for 60 min at 50°C; (d and h) dithionite-reduced as-isolated enzyme was treated with 4 mM dithionite under pure N2. The relevant g values are indicated in the spectra. General conditions: 10 mW microwave power; 10 G modulation amplitude; 9.47 GHz microwave frequency; temperatures (in degrees K) as indicated.
Intracellular location of the two CODHs.
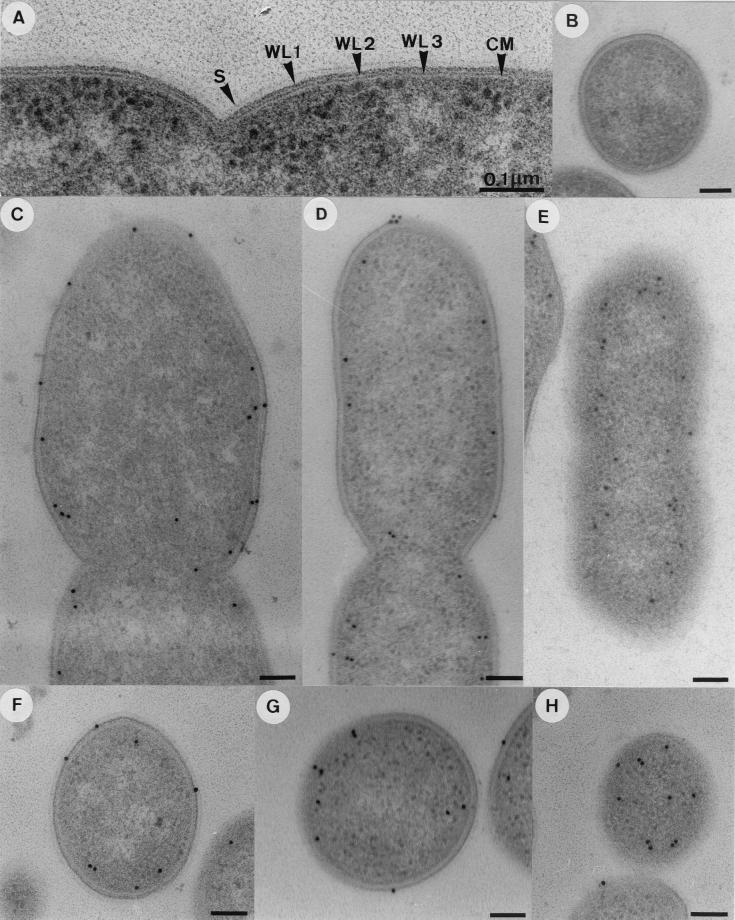
Osmotic lysis, which is a gentle method of cell breakage, revealed 71% of the total CODH activity was in the cytoplasmic fraction and 29% was in the membrane fraction of C. hydrogenoformans (Table 1). Western blotting indicated that all fractions contained CODH I as well as CODH II. It was, therefore, of interest to examine the intracellular location of the two CODHs in intact cells of C. hydrogenoformans. For this purpose, immunogold labeling of ultrathin sections of CO-grown bacteria was employed (Fig. 4). Low-temperature embedding in Lowicryl K4M resin resulted in good preservation of the bacterial ultrastructure (Fig. 4 A). The distribution of the gold label was the same for the specific IgG antibodies directed against CODH I or CODH II and was independent of the growth phase (Table 2; Fig. 4C to H). Longitudinal and cross-sections showed the gold label nearly exclusively close to the cytoplasmic membrane (Fig. 4C, D, F, and G; Table 2). Tangential sections, which leave a major part of the inner aspect of the cytoplasmic membrane exposed, revealed significant labeling (Fig. 4E and H), which is also indicative of a location of the two enzymes at the membrane. The preference of the gold label for the cytoplasmic side of the membrane (Fig. 4C, D, F, and G) suggests a position of both CODHs at the inner side of the membrane. The specificity of labeling is apparent from control sections treated with preimmune sera showing virtually no gold particles (Fig. 4B).
FIG. 4.
Localization of CODH I and CODH II on immunogold-labeled ultrathin sections of CO-grown C. hydrogenoformans. (A) Ultrastructure of the cell envelope after low-temperature embedding. CM, cytoplasmic membrane; S, surface layer; WL1, first wall layer; WL2, second wall layer; WL3, third wall layer. (B) Control labeling was with preimmune serum and gold-labeled secondary IgG antibodies. IgG antibodies directed against CODH I or CODH II were absent. (C and F) Longitudinal and cross-sections after labeling with IgG antibodies directed against CODH I and the gold-labeled secondary IgG antibodies. (D and G) Treated as for panels C and F, except that IgG antibodies directed against CODH II were used. (E) Tangential section parallel to the long cell axis; all other conditions were as for panels C and F. (H) Tangential section through a cell pole; all other conditions were as for panels C and F. Bar, 0.1 μm.
TABLE 2.
Distribution of CODH I and CODH II on ultrathin sections of CO-grown C. hydrogenoformansa
| Source (phase) of bacterial cells | CODH I in:
|
CODH II in:
|
||
|---|---|---|---|---|
| Cytoplasmic membrane, total count/cellb (%c) | Cytoplasm, total count/cellb (%c) | Cytoplasmic membrane, total count/cellb (%c) | Cytoplasm, total count/cellb (%c) | |
| Middle of exponential growth | 6.8 ± 1.2 (99.3 ± 2.9) | 0.1 ± 0.3 (0.7 ± 2.9) | 4.1 ± 1.3 (93.4 ± 9.4) | 0.4 ± 0.5 (6.6 ± 9.4) |
| End of exponential growth | 8.5 ± 1.9 (98.3 ± 3.6) | 0.2 ± 0.4 (1.7 ± 3.6) | 6.1 ± 1.4 (92.4 ± 7.6) | 0.5 ± 0.5 (7.6 ± 7.6) |
| Stationary growth | 9.0 ± 1.7 (98.8 ± 3.3) | 0.1 ± 0.3 (1.2 ± 3.3) | 5.0 ± 1.9 (94.2 ± 8.3) | 0.4 ± 0.5 (5.8 ± 8.3) |
Gold particles found in attachment to the cytoplasmic membrane of C. hydrogenoformans or present in the cytoplasm after postembedding labeling with the immunogold method were counted; for details see Materials and Methods.
Numbers refer to mean values ± standard deviations of gold particles on equally sized cross-sections of about 30 cells on different grids.
Percentage of gold particles in the indicated cell fraction relative to the total count of gold particles per cell, which was set 100%.
Reconstitution of the CO-dependent production of H2 by cytoplasmic membranes.
As already mentioned, cytoplasmic membrane fractions contained less than 30% of the total CO-oxidizing activity present in cell-free extracts (Table 1) along with the entire H2-evolving hydrogenase activity. The experiments reported in Table 3 examined the conditions of the CO-driven formation of H2 by cytoplasmic membranes of C. hydrogenoformans. H2 evolution increased considerably when both CODH I and protein B were present. Under the same conditions, CODH II or protein B alone had no effect. Apparently, protein B functions as a specific electron acceptor of CODH I, transferring the electrons to the hydrogenase. Although oxidized flavins are able to accept electrons from both CODHs, they could not functionally replace protein B. Protein B refers to a greenish-brown-colored protein fraction which eluted at 0.67 M NaCl from the Macro-Prep High Q anion exchanger in step 3 of the CODH purification scheme (Table 1). The fraction presumably contains one or several ferredoxins, according to the negative charge, and a broad shoulder centered around 390 nm and extending from 360 to 500 nm in the oxidized UV-visible absorption spectrum. We have not been able to identify a fraction from the Macro-Prep High Q anion exchanger that would couple the electron transfer from CODH II to the hydrogenase.
TABLE 3.
Effect of CODH I and CODH II on the CO-dependent formation of H2 by cytoplasmic membranes of CO-grown C. hydrogenoformans
| Assay conditionsa | H2 evolutionb (U) |
|---|---|
| Cytoplasmic membranes plus CODH I plus protein B | 30.1 |
| Cytoplasmic membranes plus CODH I | 8.1 |
| Cytoplasmic membranes plus CODH II plus protein B | 8.8 |
| Cytoplasmic membranes plus CODH II | 8.8 |
| Cytoplasmic membranes | 8.1 |
| CODH I | 0 |
| CODH II | 0.7 |
| B protein | 0 |
| CODH I plus B protein | 0 |
| CODH II plus B protein | 0.8 |
Assays contained 5 mg of CODH I or CODH II, 5 mg of protein B, and 1 mg of cytoplasmic membranes as indicated. Cytoplasmic membranes were prepared as detailed in Materials and Methods and washed in buffer A by ultracentrifugation.
H2 formation was measured under an atmosphere of 100% CO.
CO-dependent production of NADPH.
Other than the purified CODHs, cytoplasmic fractions of C. hydrogenoformans catalyzed the CO-dependent reduction of NADP+ (2 mM) with a rate of 0.31 μmol of NADP+ reduced min−1 mg−1. NADPH formation increased 1.5-fold upon addition of CODH II (0.5 mg of CODH II per mg of cytoplasmic protein). CODH I or NAD+ had no effect.
DISCUSSION
C. hydrogenoformans contains two distinct CODHs.
CODH I and CODH II of C. hydrogenoformans are distinct proteins according to the results which we have obtained. CODH I (subunit mass, 62.5 kDa; holoenzyme mass, 125 kDa) and CODH II (subunit mass, 64.5 kDa; holoenzyme mass, 129 kDa) differ slightly in their subunit and holoenzyme masses (Fig. 1A and B) and are immunologically unrelated to each other (Fig. 1D). Their N-terminal sequences and proteolytic peptide patterns (Fig. 1C) indicate different primary structures. The enzymes show different temperature optima and catalytic properties. CODH I and CODH II from C. hydrogenoformans, as well as the CODH from R. rubrum, are very similar in sharing high CO-oxidizing activity, high affinity for CO, and comparable holoenzyme and subunit masses (6, 7).
Searches in the National Center for Biotechnology Information, SwissProt, Protein Information Resource, and The Institute for Genomic Research (TIGR) databases revealed that the N terminus of the CODH II subunit (Fig. 1B), as well as the N terminus of the 49-kDa chymotryptic peptide (Fig. 1C), is located on a DNA stretch from C. hydrogenoformans. This piece of DNA shows highest homology to cooS, which is the CODH subunit of R. rubrum (23). The experimentally determined subunit mass of CODH II (64.5 kDa; Fig. 1B) differs by less than 5% from the mass (67.3 kDa) which can be calculated from the C. hydrogenoformans CooS sequence (23). The mass of the 49-kDa chymotryptic fragment (Fig. 1C) of the CODH II subunit (Fig. 1B) compares favorably to the 49.6 kDa calculated from the CooS sequence. These considerations can be taken as evidence for the relatedness of the CODH II subunit with CooS. The amino acid sequences of C. hydrogenoformans CooS, the CODH from R. rubrum (55% identity), and the CO-oxidizing β-subunit of CODH-ACS from C. thermoaceticum (48% identity) are closely related (23).
Searches in the TIGR database revealed that the N terminus of the CODH I subunit (Fig. 1B) matches the amino acid sequence of an open reading frame identified in the C. hydrogenoformans genome. The deduced protein shows 74% homology to CooS (CODH II). The experimentally determined subunit mass of CODH I of 62.5 kDa (Fig. 1B) differs by only 8% from the mass (67.5 kDa) deduced from the sequence of this protein. These considerations identify the predicted C. hydrogenoformans protein as the CODH I subunit.
Both CODHs are membrane-associated homodimeric NiFeS proteins.
Intact CO-grown cells of C. hydrogenoformans contain more than 92% of the total CO dehydrogenase population in association with the inner aspect of the cytoplasmic membrane (Fig. 4 and Table 2). The intracellular location of both CODHs was independent of the growth phase. Upon cell disintegration, a high proportion (∼ 70%) of both CODHs became solubilized (Table 1), reflecting rather weak noncovalent interactions of the enzymes with the membrane.
Both CODHs of C. hydrogenoformans are homodimers with an α2 subunit structure (Fig. 1A and B) which is similar to the CODH from R. rubrum but different from acetogenic or methanogenic CODHs-ACSs, which are α2β2 tetramers or αβγδɛ pentamers (15, 16, 47). The two C. hydrogenoformans CODHs contain Ni, Fe, and acid-labile sulfur. The mean Ni content values obtained with different methods (CODH I, 1.28 to 1.41 atoms of Ni/mol of dimer; CODH II, 1.65 to 1.67 atoms of Ni/mol of dimer) suggest the presence of 1 atom of Ni per mol of subunit of catalytically fully competent CODH and indicates an ∼67% (CODH I) or ∼83% (CODH II) occupancy of the Ni site. In R. rubrum, the occupancy of the CODH Ni site of ∼65% (6) results from the biosynthesis of a mixture of an enzyme containing the full complement of 2 atoms of Ni/ mol of dimer and a Ni-deficient apo-CODH (8, 31). The absence of EPR signals attributable to Ni(III) or Ni(I) in both C. hydrogenoformans CODHs under all conditions examined is interpreted as a divalent Ni (Fig. 3).
The Fe and acid-labile sulfur content, along with the EPR spectra (Fig. 3) of CODH I and CODH II, suggests the presence of at least two different redox-active [4Fe-4S] clusters per subunit. The rhombic signal at gav = 1.94 (Fig. 3A and B, traces c and g) is assumed to originate from a [4Fe-4S]1+ cluster similar to the reduced electron-transferring B cluster of the CODHs from R. rubrum or C. thermoaceticum (32). The second rhombic signal at gav = 1.86 of CODH I (Fig. 3A, trace c) and gav = 1.84 of CODH II (Fig. 3B, trace c) is believed to come from a faster-relaxing [4Fe-4S]1+ cluster similar to the fully reduced CO-oxidizing Ni-X-[4Fe-4S] cluster (C cluster) of the CODHs from R. rubrum and C. thermoaceticum (32). The cluster C of R. rubrum CODH is suggested to be composed of [4Fe-4S]c and [FeNi] clusters (30, 31, 53). The presumed B and C clusters in CODH I and CODH II would account for 8 atoms of Fe and 8 atoms of labile sulfur per mol of subunit. Therefore, the extra 3 to 4 atoms of Fe and 1.5 to 3 atoms of labile sulfur per mol of CODH subunit remain to be explained. They could be part of a single [2Fe-2S] cluster in each subunit or part of a single [4Fe-4S] cluster per dimer. The presence or a third [4Fe-4S] cluster bridging the two subunits would agree with the visible spectra of both CODHs (Fig. 2A and B), which show no [2Fe-2S] features. The additional Fe could also be part of a binuclear [NiFe] cluster (22, 31, 60).
Presumed functions of the two CODHs in the metabolism of C. hydrogenoformans.
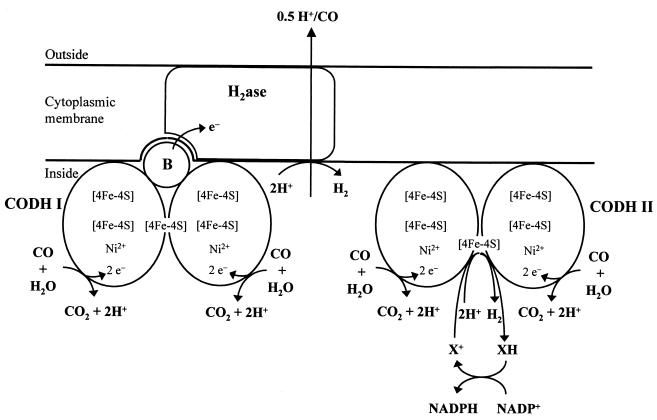
According to the scheme presented in Fig. 5, CODH I is involved in the generation of energy, and CODH II is involved in the assimilation of carbon. CODH I generates the electrons from CO which are subsequently channelled via the ferredoxin-like protein B to a hydrogenase, which is the site where intracellular protons are reduced to H2. Ferredoxins couple the electron transport from CODH to hydrogenase in phototrophs (14), acetogens (48), and methanogens (57). The CO-driven proton respiration in C. hydrogenoformans is coupled to the translocation of H+ across the cytoplasmic membrane. This is indicated by experiments where the application of CO pulses to resting cells led to a transient acidification of the bacterial environment from which H+/CO ratios of 0.5 could be extrapolated under conditions where the membrane potential was dissipated by thiocyanate (KSCN). In R. rubrum (4, 20) or methanogenic Archaea (4, 29, 36, 41), the complex I-related H2-evolving hydrogenases were proposed to be the site of proton translocation. We are, therefore, assuming the same function for the membrane-bound hydrogenase of C. hydrogenoformans, which shows sequence similarities of 80, 95, 72, and 67% (TIGR database) to the large (CooH), small (CooL), proposed H+-translocating (CooK), and TYKY (CooX) subunits of the corresponding [Ni-Fe] hydrogenase of R. rubrum (20).
FIG. 5.
Hypothetical scheme showing the function of the two CODHs in C. hydrogenoformans. CODH I is involved in energy generation and CODH II serves anabolic functions. For details refer to the text. The scheme is not intended to give correct stoichiometries. Abbreviations: B, ferredoxin-like protein B; H2ase, membrane-bound [NiFe] hydrogenase.
The anabolic function of CODH II involves the generation of NADPH and part of the CO2 for carbon assimilation. CODH II is able to reduce NADP+ if a cytoplasmic coupling factor X+/XH is present (Fig. 5). The coupling factor X could be a ferredoxin:NADP+ oxidoreductase, in analogy to the CO-dependent reduction of NADP+ in acetogenic bacteria (33, 48). The ability of CODH II to transfer electrons to protons instead to NADP+ might help to regulate the proper ratio of reducing equivalents and CO2 in carbon assimilation.
This study did not intend to resolve the path of the autotrophic fixation of CO or CO2 in C. hydrogenoformans. However, we can conclude from the absence of ribulose-1,5-bisphosphate carboxylase activity in C. hydrogenoformans and the absence of a corresponding sequence on the genome that the Calvin-Benson-Bassham cycle is not operative. Although the genomic sequence of the bacterium (TIGR database) contains an open reading frame which is 86% homologous to the ACS of C. thermoaceticum, we have not been able to demonstrate ACS activity in C. hydrogenoformans. Alternative paths for carbon fixation in C. hydrogenoformans which must be considered in future research are the reductive citric acid cycle, the 3-hydroxypropionate cycle, the hydration of CO to formate, or the reduction of CO2 to formate through the action of formate dehydrogenase. Indeed, cytoplasmic fractions of C. hydrogenoformans contain formate:oxidized MV oxidoreductase activity (0.7 μmol mg−1).
ACKNOWLEDGMENTS
We thank Dorothea Alber (Hahn-Meitner-Institut, Berlin, Germany) for neutron activation analysis, Botho Bowien (Universität Göttingen, Göttingen, Germany) for analysis of ribulose-1,5-bisphosphate carboxylase activity, Harold Drake (BITÖK, Universität Bayreuth, Bayreuth, Germany) for providing us with the culture of C. thermoaceticum, Rita Grotjahn (Universität Bayreuth) for expert technical assistence, and Anna Wolf (Universität Bayreuth) for reading the manuscript. O.M. is grateful to Reiner Hedderich, Jongyun Heo, Paul W. Ludden, Stephen W. Ragsdale, and Christopher R. Staples for discussion.
V.S. acknowledges a stipend from the Alexander von Humboldt Foundation (Bonn, Germany). This work was financially supported by the Fonds der Chemischen Industrie (Frankfurt am Main, Germany).
ADDENDUM IN PROOF
A crystal structure of reduced CODH II has been solved at 1.6-Å resolution (H. Dobbek, V. Svetlitchnyi, L. Gremer, R. Huber, and O. Meyer, Science, in press). The structure represents the prototype for Ni-containing CODHs from anaerobic bacteria and archaea. It contains five metal clusters, of which clusters B, B′, and a subunit-bridging, surface-exposed cluster D are cubane-type [4Fe-4S] clusters. The active-site cluster C and C′ are navel, asymmetric [Ni-4Fe-5S] clusters. Their integral Ni ion, which is the likely site of CO oxidation, is coordinated by four sulfur ligands with square planar geometry.
REFERENCES
- 1.Acetarin J-D, Carlemalm E, Villiger W. Development of new Lowicryl resins for embedding biological specimens at even lower temperature. J Microsc. 1986;143:81–88. doi: 10.1111/j.1365-2818.1986.tb02766.x. [DOI] [PubMed] [Google Scholar]
- 2.Acker G. Immunoelectron microscopy of surface antigens (polysaccharides) of gram-negative bacteria using pre- and post-embedding techniques. In: Mayer F, editor. Electron microscopy in microbiology. Methods in microbiology. Vol. 20. London, England: Academic Press Ltd.; 1988. pp. 147–174. [Google Scholar]
- 3.Albracht S P. The use of electron-paramagnetic-resonance spectroscopy to establish the properties of nickel and the iron-sulfur cluster in hydrogenase from Chromatium vinosum. Biochem Soc Trans. 1985;13:582–585. doi: 10.1042/bst0130582. [DOI] [PubMed] [Google Scholar]
- 4.Albracht S P, Hedderich R. Learning from hydrogenases: location of a proton pump and of a second FMN in bovine NADH-uniquinone oxidoreductase (Complex I) FEBS Lett. 2000;485:1–6. doi: 10.1016/s0014-5793(00)02172-4. [DOI] [PubMed] [Google Scholar]
- 5.Beisenherz G, Bolze H J, Bücher T, Czok R, Garbade K H, Meyer-Arendt E, Pfleiderer G. Diphosphofructose-Aldolase, Phosphoglyceraldehyd-Dehydrogenase, Milchsäure-Dehydrogenase, Glycerophosphat-Dehydrogenase and Pyruvat-Kinase aus Kaninchenmuskulatur in einem Arbeitsgang. Z Naturforsch B. 1953;8:555–577. [Google Scholar]
- 6.Bonam D, Ludden P W. Purification and characterization of carbon monoxide dehydrogenase, a nickel, zinc, iron-sulfur protein, from Rhodospirillum rubrum. J Biol Chem. 1987;262:2980–2987. [PubMed] [Google Scholar]
- 7.Bonam D, Murrell S A, Ludden P W. Carbon monoxide dehydrogenase from Rhodospirillum rubrum. J Bacteriol. 1984;159:693–699. doi: 10.1128/jb.159.2.693-699.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonam D, McKenna M C, Stephens P J, Ludden P W. Nickel-deficient carbon monoxide dehydrogenase from Rhodospirillum rubrum: in vivo and in vitro activation by exogenous nickel. Proc Natl Acad Sci USA. 1988;85:31–35. doi: 10.1073/pnas.85.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowien B, Meyer F, Codd G A, Schlegel H G. Purification, some properties and quaternary structure of the d-ribulose-1,5-diphosphate carboxylase of Alcaligenes eutrophus. Arch Microbiol. 1976;110:157–166. doi: 10.1007/BF00690223. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Dawson R M C, Elliot D C, Elliot W H, Jones K M. Data for biochemical research. New York, N.Y: Oxford University Press Inc; 1986. [Google Scholar]
- 12.Dobbek H, Gremer L, Meyer O, Huber R. Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcystein. Proc Natl Acad Sci USA. 1999;96:8884–8889. doi: 10.1073/pnas.96.16.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake H L, Hu S-I, Wood H G. Purification of five components from Clostridium thermoaceticum which catalyze synthesis of acetate from pyruvate and methyltetrahydrofolate. Properties of phosphotransacetylase. J Biol Chem. 1981;256:11137–11144. [PubMed] [Google Scholar]
- 14.Ensign S A, Ludden P W. Characterization of the CO oxidation/H2 evolution system of Rhodospirillum rubrum: role of a 22-kDa iron-sulfur protein in mediating electron transfer between carbon monoxide dehydrogenase and hydrogenase. J Biol Chem. 1991;266:18395–18403. [PubMed] [Google Scholar]
- 15.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer R K. Active sites of transition-metal enzymes with a focus on nickel. Curr Opin Struct Biol. 1998;8:749–758. doi: 10.1016/s0959-440x(98)80095-x. [DOI] [PubMed] [Google Scholar]
- 16.Ferry J G. CO dehydrogenase. Annu Rev Microbiol. 1995;49:305–333. doi: 10.1146/annurev.mi.49.100195.001513. [DOI] [PubMed] [Google Scholar]
- 17.Fish W W. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 18.Flint D H, Allen R M. Iron-sulfur proteins with nonredox functions. Chem Rev. 1996;96:2315–2334. doi: 10.1021/cr950041r. [DOI] [PubMed] [Google Scholar]
- 19.Fogo J K, Popowsky M. Spectrophotometric determination of hydrogen sulfide. Anal Chem. 1949;21:732–734. [Google Scholar]
- 20.Fox J D, Kerby R L, Roberts G P, Ludden P W. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J Bacteriol. 1996;178:1515–1524. doi: 10.1128/jb.178.6.1515-1524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fröstl J M, Seifritz C, Drake H L. Effect of nitrate on the autotrophic metabolism of the acetogens Clostridium thermoautotrophicum and Clostridium thermoaceticum. J Bacteriol. 1996;178:4597–4603. doi: 10.1128/jb.178.15.4597-4603.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcin E, Vernede X, Hatchikian E C, Volbeda A, Frey M, Fontecilla-Camps J C. The crystal structure of a reduced [NiFeS] hydrogenase provides an image of the activated catalytic center. Structure. 1999;7:557–566. doi: 10.1016/s0969-2126(99)80072-0. [DOI] [PubMed] [Google Scholar]
- 23.González J M, Robb F T. Genetic analysis of Carboxydothermus hydrogenoformans carbon monoxide dehydrogenase genes cooF and cooS. FEMS Microbiol Lett. 2000;191:243–247. doi: 10.1111/j.1574-6968.2000.tb09346.x. [DOI] [PubMed] [Google Scholar]
- 24.Gremer L, Meyer O. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur J Biochem. 1996;238:862–866. doi: 10.1111/j.1432-1033.1996.0862w.x. [DOI] [PubMed] [Google Scholar]
- 25.Gremer L, Kellner S, Meyer O. A new type of flavin adenin dinucleotide-binding resolved in the molybdo iron-sulfur-flavoprotein carbon monoxide dehydrogenase from Oligotropha carboxidovorans. In: Ghisla S, Kroneck P, Macheroux P, Sund H, editors. Flavins and flavoproteins 1999. Berlin, Germany: Rudolf Weber, Agency for Scientific Publications; 1999. pp. 759–766. [Google Scholar]
- 26.Gremer L, Kellner S, Dobbek H, Huber R, Meyer O. Binding of flavin adenine dinucleotide to molybdenum-containing carbon monoxide dehydrogenase from Oligotropha carboxidovorans. J Biol Chem. 2000;275:1864–1872. doi: 10.1074/jbc.275.3.1864. [DOI] [PubMed] [Google Scholar]
- 27.Hänzelmann P, Dobbek H, Gremer L, Huber R, Meyer O. The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase. J Mol Biol. 2000;301:1221–1235. doi: 10.1006/jmbi.2000.4023. [DOI] [PubMed] [Google Scholar]
- 28.Hänzelmann P, Hofmann B, Meisen S, Meyer O. The redox centers in the molybdo iron-sulfur flavoprotein CO dehydrogenase from the thermophilic carboxidotrophic bacterium Pseudomonas thermocarboxydovorans. FEMS Microbiol Lett. 1999;176:139–145. [Google Scholar]
- 29.Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter K O. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev. 1999;22:353–381. [Google Scholar]
- 30.Heo J, Staples C R, Telser J, Ludden P W. Rhodospirillum rubrum CO-dehydrogenase. Part 2. Spectroscopic investigation and assignment of spin-spin coupling signals. J Am Chem Soc. 1999;121:11045–11057. [Google Scholar]
- 31.Heo J, Staples C R, Halbleib C M, Ludden P W. Evidence for a ligand CO that is required for catalytic activity of CO dehydrogenase from Rhodospirillum rubrum. Biochemistry. 2000;39:7956–7963. doi: 10.1021/bi992958g. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z, Spangler N J, Anderson M E, Xia J, Ludden P W, Lindahl P A, Münck E. Nature of the C-cluster in Ni-containing carbon monoxide dehydrogenases. J Am Chem Soc. 1996;118:830–845. [Google Scholar]
- 33.Hugenholtz J, Ljungdahl L G. Electron transport and electrochemical proton gradient in membrane vesicles of Clostridium thermoautotrophicum. J Bacteriol. 1989;171:2873–2875. doi: 10.1128/jb.171.5.2873-2875.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerby R L, Ludden P W, Roberts G P. Carbon monoxide-dependent growth of Rhodospirillum rubrum. J Bacteriol. 1995;177:2241–2244. doi: 10.1128/jb.177.8.2241-2244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraut M, Hugendieck I, Herwig S, Meyer O. Homology and distribution of CO dehydrogenase structural genes in carboxydotrophic bacteria. Arch Microbiol. 1989;152:335–341. doi: 10.1007/BF00425170. [DOI] [PubMed] [Google Scholar]
- 36.Künckel A, Verholt J A, Thauer R K, Hedderich R. An Escherichia coli hydrogenase 3-type hydrogenase in methanogenic archaea. Eur J Biochem. 1998;252:467–476. doi: 10.1046/j.1432-1327.1998.2520467.x. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Lax E. D'Ans-Lax: Taschenbuch für Chemiker und Physiker. Vol. 1. Berlin, Germany: Springer Verlag; 1967. pp. I–1025. [Google Scholar]
- 39.Lundie L L, Drake H L. Development of a minimally defined medium for the acetogen Clostridium thermoaceticum. J Bacteriol. 1984;158:700–703. doi: 10.1128/jb.159.2.700-703.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menon A L, Hendrix H, Hutchins A, Verhagen M F J, Adams M W W. The δ-subunit of pyruvate ferredoxin oxidoreductase from Pyrococcus furiosus is a redox-active, iron-sulfur protein: evidence for an ancestral relationship with 8Fe-type ferredoxin. Biochemistry. 1998;37:12838–12846. doi: 10.1021/bi980979p. [DOI] [PubMed] [Google Scholar]
- 41.Meuer J, Bartoschek S, Koch J, Künckel A, Hedderich R. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur J Biochem. 1999;265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 42.Meyer O, Frunzke K, Mörsdorf G. Biochemistry of the aerobic utilization of carbon monoxide. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, England: Intercept Ltd.; 1993. pp. 433–459. [Google Scholar]
- 43.Meyer O, Gremer L, Ferner R, Ferner M, Dobbek H, Gnida M, Meyer-Klaucke W, Huber R. The role of Se, Mo, and Fe in the structure and function of carbon monoxide dehydrogenase. Biol Chem. 2000;381:865–876. doi: 10.1515/BC.2000.108. [DOI] [PubMed] [Google Scholar]
- 44.Mörsdorf G, Frunzke K, Gadkari D, Meyer O. Microbial growth on carbon monoxide. Biodegradation. 1992;3:61–82. [Google Scholar]
- 45.Ouchterlony Ö. Diffusion-in-gel methods for immunological analysis. In: Kallos P, Waksman B, editors. Progress in allergy. VI. Basel, Switzerland: Verlag Karger; 1962. pp. 30–154. [DOI] [PubMed] [Google Scholar]
- 46.Pusheva M A, Sokolova T G, Gerhardt M, Svetlichnyi V A. Hydrogenase, formate dehydrogenase and CO dehydrogenase activities of the thermophilic anaerobic carboxydotrophic bacterium Carboxydothermus hydrogenoformans strain Z-2906. Mikrobiologiya. 1992;61:939–944. [Google Scholar]
- 47.Ragsdale S W, Kumar M. Nickel-containing carbon monoxide dehydrogenase/acetyl-CoA synthase. Chem Rev. 1996;96:2515–2539. doi: 10.1021/cr950058+. [DOI] [PubMed] [Google Scholar]
- 48.Ragsdale S W, Ljungdahl L G, DerVartanian V. Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J Bacteriol. 1983;155:1224–1237. doi: 10.1128/jb.155.3.1224-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raybuck S A, Bastian N R, Orme-Johnson W H, Walsh C T. Kinetic characterization of the carbon monoxide-acetyl-CoA (carbonyl group) exchange activity of the acetyl-CoA synthesizing CO dehydrogenase from Clostridium thermoaceticum. Biochemistry. 1988;27:7698–7702. doi: 10.1021/bi00420a019. [DOI] [PubMed] [Google Scholar]
- 50.Rohde M, Mayer F, Meyer O. Immunocytochemical localization of carbon monoxide oxidase in Pseudomonas carboxydovorans. The enzyme is attached to the inner aspect of the cytoplasmic membrane. J Biol Chem. 1984;259:14788–14792. [PubMed] [Google Scholar]
- 51.Rohde M, Gerberding H, Mund T, Kohring G-W. Immunoelectron microscopic localization of bacterial enzymes: pre- and post-embedding labelling techniques on resin-embedded samples. In: Mayer F, editor. Electron microscopy in microbiology. Methods in microbiology. Vol. 20. London, England: Academic Press Ltd.; 1988. pp. 175–210. [Google Scholar]
- 52.Sokolova T G, Gonzáles J M, Kostrikina N A, Chernyh N A, Tourova T P, Kato C, Bonch-Osmolovskaya E A, Robb F T. Carboxydobrachium pacificum gen. nov., sp. nov., a new anaerobic, thermophilic, CO-utilizing bacterium from Okinawa Trough. Int J Syst Evol Microbiol. 2001;51:141–149. doi: 10.1099/00207713-51-1-141. [DOI] [PubMed] [Google Scholar]
- 53.Staples C R, Heo J, Spangler N J, Kerby R L, Roberts G P, Ludden P W. Rhodospirillum rubrum CO-dehydrogenase. Part I. Spectroscopic studies of CODH variant C531A indicate the presence of a binuclear [FeNi] cluster. J Am Chem Soc. 1999;121:11034–11044. [Google Scholar]
- 54.Svetlichny V A, Sokolova T G, Gerhardt M, Kostrikina N A, Zavarzin G A. Anaerobic extremely thermophilic carboxydotrophic bacteria in hydrotherms of Kuril Islands. Microb Ecol. 1991;21:1–10. doi: 10.1007/BF02539140. [DOI] [PubMed] [Google Scholar]
- 55.Svetlichny V A, Sokolova T G, Gerhardt M, Ringpfeil M, Kostrikina N A, Zavarzin G A. Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir island. Syst Appl Microbiol. 1991;14:254–260. [Google Scholar]
- 56.Svetlichny V A, Sokolova T G, Kostrikina N A, Lysenko A M. A new thermophilic anaerobic carboxydotrophic bacterium Carboxydothermus restrictus sp. nov. Mikrobiologiya. 1994;63:294–297. [Google Scholar]
- 57.Terlesky K C, Ferry J G. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J Biol Chem. 1988;263:4075–4079. [PubMed] [Google Scholar]
- 58.Uffen R L. Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate. Proc Natl Acad Sci USA. 1976;73:3298–3302. doi: 10.1073/pnas.73.9.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uffen R L. Metabolism of carbon monoxide by Rhodopseudomonas gelatinosa: cell growth and properties of the oxidation system. J Bacteriol. 1983;155:956–965. doi: 10.1128/jb.155.3.956-965.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volbeda A, Charon M-H, Piras C, Hatchikian E C, Frey M, Fontecilla-Camps J C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 61.Wolin E A, Wolin M J, Wolfe R S. Formation of methane by bacterial extracts. J Biol Chem. 1963;238:2282–2286. [PubMed] [Google Scholar]