Abstract
In streptococci, HPr, a phosphocarrier of the phosphoenolpyruvate:sugar phosphotransferase transport system (PTS), undergoes multiple posttranslational chemical modifications resulting in the formation of HPr(His∼P), HPr(Ser-P), and HPr(Ser-P)(His∼P), whose cellular concentrations vary with growth conditions. Distinct physiological functions are associated with specific forms of HPr. We do not know, however, the cellular thresholds below which these forms become unable to fulfill their functions and to what extent modifications in the cellular concentrations of the different forms of HPr modify cellular physiology. In this study, we present a glimpse of the diversity of Streptococcus salivarius ptsH mutants that can be isolated by positive selection on a solid medium containing 2-deoxyglucose and galactose and identify 13 amino acids that are essential for HPr to properly accomplish its physiological functions. We also report the characterization of two S. salivarius mutants that produced approximately two- and threefoldless HPr and enzyme I (EI) respectively. The data indicated that (i) a reduction in the synthesis of HPr due to a mutation in the Shine-Dalgarno sequence of ptsH reduced ptsI expression; (ii) a threefold reduction in EI and HPr cellular levels did not affect PTS transport capacity; (iii) a twofold reduction in HPr synthesis was sufficient to reduce the rate at which cells metabolized PTS sugars, increase generation times on PTS sugars and to a lesser extent on non-PTS sugars, and impede the exclusion of non-PTS sugars by PTS sugars; (iv) a threefold reduction in HPr synthesis caused a strong derepression of the genes coding for α-galactosidase, β-galactosidase, and galactokinase when the cells were grown at the expense of a PTS sugar but did not affect the synthesis of α-galactosidase when cells were grown at the expense of lactose, a noninducing non-PTS sugar; and (v) no correlation was found between the magnitude of enzyme derepression and the cellular levels of HPr(Ser-P).
Histidine-containing protein, heat-stable protein, and heteromorphous protein are all epithets that have been used to designate HPr, the bacterial phosphocarrier of the phosphoenolpyruvate:sugar phosphotransferase transport system (PTS) (19, 41). Results obtained over the past decade unequivocally indicate that the qualifier “heterofunctional protein” also applies to this small, approximately 9-kDa protein (27, 31, 35).
The PTS is a multienzyme complex that sequentially catalyzes the transport and phosphorylation of sugars (27, 32) and plays a cardinal role in regulatory processes that allow bacteria to metabolize sugars differently depending on environmental conditions (31, 33, 35, 45). The HPr of gram-positive bacteria can be phosphorylated on His15 at the expense of phosphoenolpyruvate by enzyme I (EI) of the PTS and on Ser46 by the ATP-dependent HPr(Ser) kinase-phosphatase (HPrK) (27, 31, 33, 35, 45). HPr(His∼P) not only is involved in sugar transport but also controls transcription of several genes by transferring its phosphate group to histidine residues of antiterminators and transcriptional activators with PTS regulation domains (34). HPr(His∼P) also controls glycerol kinase of Enterococcus faecalis and Enterococcus casseliflavus (4, 5) and the lactose permease of Streptococcus thermophilus, and possibly of Lactobacillus bulgaricus, Pediococcus pentosoceus, and Leuconostoc lactis, by reversible phosphorylation (25, 26, 47). HPr(Ser-P) is not involved in sugar transport. However, this form of HPr controls transcription of catabolic genes in conjunction with a DNA-binding protein called CcpA that recognizes a specific DNA sequence called CRE (catabolite-responsive element) located in the promoter region of target operons. In Bacillus subtilis, the association of CcpA with a number of CRE sequences is promoted by HPr(Ser-P) (6, 8, 13, 17) and results in the activation or inhibition of gene transcription depending on whether the CRE sequence is located upstream or downstream from the promoter sequence (16, 35). HPr(Ser-P) also allosterically controls the activity of sugar permeases in lactococci, lactobacilli, enterococci, and streptococci (7, 48, 50–52).
To properly accomplish their diverse functions, the different forms of HPr must be synthesized at the appropriate concentrations. Previous work has already demonstrated that cellular levels of HPr in streptococci vary two- to threefold with culture conditions and that the relative proportions of the different forms of HPr also change with respect to growth rate (14, 36, 43, 44). However, we do not know to what extent these variations alter the capacity of HPr to fulfill its functions.
To shed more light on the relationship between cellular concentrations of HPr and its physiological functions, we sought to characterize mutants producing lower levels of HPr than that of the wild-type strain. In a previous study conducted with Streptococcus salivarius ATCC 25975, we observed that several types of PTS mutants, including ptsH mutants, could be obtained by positive selection on 2-deoxyglucose (2DG) in the presence of various metabolizable sugars (11). Preliminary data suggested that selection in the presence of galactose favored the isolation of ptsH mutants. We thus decided to verify whether selection in the presence of 2DG and galactose engendered a bias toward the isolation of ptsH mutants and, if so, to use this approach to isolate mutants producing lower levels of HPr. In this paper, we present a glimpse of the diversity of ptsH mutants that can be isolated by plating S. salivarius on a solid medium containing 2DG and galactose and report the characterization of two S. salivarius mutants that synthesize approximately two- and threefold less HPr and EI.
MATERIALS AND METHODS
Strains and growth conditions.
S. salivarius ATCC 25975 was kindly provided by I. R. Hamilton (University of Manitoba). ptsH mutants were isolated by positive selection for resistance to 5 mM 2DG in the presence of 200 mM galactose. Cells were grown at 37°C in a medium containing 10 g of tryptone and 5 g of yeast extract (Difco Laboratories), 2.5 g of NaCl, and 2.5 g of disodium phosphate per liter. Sugars were sterilized by filtration (Millipore filter, 0.22-μm pore size) and added aseptically to the medium to give the appropriate concentrations. When the parental strain was grown in this medium without sugar, the culture reached a maximum optical density at 660 nm (OD660) of approximately 0.1. Generation times were determined by growing the cells at 37°C in the presence of 0.1% (wt/vol) sugar in tubes (16 by 125 mm) containing 10 ml of medium. The tubes were inoculated with 0.1 ml of an overnight culture grown in the presence of 0.1% sugar. Growth was monitored by monitoring the OD660. For growth studies in media containing two sugars, the bacteria were grown in tubes containing 15 ml of medium supplemented with 0.1% glucose or fructose (PTS sugar) and 0.2% lactose or galactose (non-PTS sugar). For some studies, the cells were grown in the presence of 0.2% lactose or galactose, and when the OD660 reached approximately 0.35, glucose or fructose was added to a final concentration of 0.1%. Samples (0.25 to 0.5 ml) were taken at intervals, heated at 100°C for 10 min to stop metabolism, centrifuged to remove cells, and then stored at −20°C for sugar assays.
Identification of ptsH mutants.
Clones were grown in 3 ml of culture medium containing 5 mM 2DG and 0.5% galactose. When the OD660 reached 0.45, the cells were harvested by centrifugation and resuspended in 150 μl of a solution containing 125 mM Tris-HCI (pH 6.8), 10% (vol/vol) glycerol, 10% (wt/vol) Nonidet P-40, and 0.7 μM β-mercaptoethanol. They were then lysed with a Sonifier cell disrupter using the pulse mode (15 pulses) at energy level 5 (model W-350; Branson Sonic Power Co.). During the sonication treatment, the recipient containing the cell suspension was kept on ice. The extract was incubated at 100°C for 5 min, and the proteins were separated by polyacrylamide gel electrophoresis under native conditions with separating gels containing 12.5% acrylamide as described by Robitaille et al. (29). The position of HPr in the gel was determined by Western blotting as described by Robitaille et al. (29) using specific anti-HPr rabbit polyclonal antibodies. Presumptive HPr mutants were selected by comparing their HPr electrophoretic patterns with that of the wild-type strain on the basis of two criteria: the intensity and electrophoretic mobility of the proteins that reacted positively with the anti-HPr antibodies.
HPr and EI determinations.
The cytoplasmic fraction was prepared as described previously (24), and the quantification of the cellular forms of HPr was carried out by crossed immunoelectrophoresis as already reported (43). A standard curve was obtained using purified S. salivarius HPr. Since the classic techniques used to determine protein concentrations gave erroneous results with HPr, we determined its concentration in purified preparations by amino acid analysis following acid hydrolysis. The protein concentration of a purified preparation of HPr measured in this way was two- to sixfold lower than that determined by the methods of Lowry and Bradford. The amount of EI was determined by rocket immunoelectrophoresis using specific rabbit polyclonal antibodies obtained against purified EI as described previously (10), except that Tris-Tricine was replaced by Tris-barbiturate. A standard curve was obtained using purified S. salivarius EI.
Sequencing of ptsH, ptsI, and hprK.
The nucleotide sequences of the ptsH and ptsI genes were determined as described by Gauthier et al. (11). The sequence of the region upstream from the −35 box of the pts promoter was obtained after amplification of a 398-bp DNA fragment by PCR using two oligonucleotides (5′-TATCTTTACAGCTGACTTAG-3′ and 5′-GCTGGACGTGCGTGGAT-3′) that annealed with a region of the idh gene located 252 bp upstream from the −35 region of the pts promoter and with a region in the ptsH gene. The nucleotide sequence of the hprK gene that codes for the HPrK was determined after amplification of a 2,040-bp DNA fragment by PCR using two oligonucleotides (5′-ATGATTGGCCCTGGTGCTA-3′ and 5′-ACCACATGACGGGTACGAAG-3′) that annealed 103 nucleotides upstream and 1,917 nucleotides downstream from the initiation codon of hprK, respectively. The following primers were used to determine the sequences on both strands of the hprK gene and its promoter region: 5′-TATGATTGGCCCTGGTGCTA-3′, 5′-GGTAAGAGTGAAACAGGG-3′, and 5′-CCAAGACGATCAAGACC-3′. The PCR was performed using a DNA Thermal Cycler 480 (Perkin-Elmer) in a total volume of 100 μl containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.2 μM forward and reverse primers, and 200 μM (each) four deoxynucleotide triphosphates. The mixtures were incubated at 94°C for 5 min prior to the addition of 2.5 U of Taq DNA polymerase (Perkin-Elmer). The reactions were carried out for 25 cycles with the following temperature profile: 94°C for 90 50°C for 60 and 72°C for 2 min. All cycles were performed with an autoextension cycle that adds 5 per cycle to the third step of the temperature profile. At the end of the amplification process, the samples were incubated for 10 min at 72°C. The amplicons were purified using a Gene Clean Spin kit (Bio 101). The PCR was carried out with DNA extracted by suspending bacterial colonies into 100 μl of distilled water and placing the samples in boiling water for 1 min (21).
Uptake experiments and glucose consumption by resting cells.
The uptake of [14C]2DG was performed with cells grown in 0.2% glucose and harvested at mid-log phase. Uptake was carried out at 10°C in 50 mM potassium phosphate buffer (pH 7.0) as described previously (46). Glucose consumption by resting cells was carried out as followed. Cells were grown in the presence of 0.2% glucose, and growth was stopped by the addition of chloramphenicol (50 μg ml−1). The cells were harvested by centrifugation, washed twice with 10 mM MgSO4, and resuspended in 100 mM sodium phosphate (pH 7.0) at 20 mg (wet weight) per ml. The cell suspension (10 ml) was maintained at 37°C and gently mixed on a magnetic stirrer. Glucose was added to a final concentration of 0.2%, and the pH was maintained by automatic titration with 0.3 N NaOH.
Enzyme assays.
Cells were grown in 500 ml of medium containing 0.2% sugar. Chloramphenicol (50 μg per ml) was added to stop cell growth in the exponential phase. For measurement of HPrK activities, the cellular extracts were prepared by sonication as described by Brochu and Vadeboncoeur (2). For the other enzymes, the cells were harvested by centrifugation, washed once with 50 mM potassium phosphate (pH 7.0) containing 5 mM β-mercaptoethanol, and then frozen at −40°C. The cells were disrupted by grinding with alumina in the presence of 0.1 mM phenylmethylsulfonyl fluoride and 1 μM pepstatin A as previously described (40). The broken cell suspensions were centrifuged first at 3,000 × g for 5 min at 4°C to remove intact cells and alumina and then at 16,000 × g for 20 min to remove cell debris. The supernatant (cellular extract) was then dialyzed at 4°C for 20 h against 10 mM sodium phosphate (pH 7.0) and used to assay enzyme activities. β-Galactosidase activity was assayed using O-nitrophenyl-β-galactopyranoside (ONPG) as the substrate (15). α-Galactosidase activity was assayed using p-nitrophenyl-α-galactopyranoside as the substrate (22). Galactokinase activity was assayed by measuring the rate of phosphorylation of [14C]galactose at the expense of ATP as described previously (42). HPrK activities were measured with [γ-32P]ATP and purified HPr from S. salivarius as described previously (2). In all cases, enzyme assays were performed under conditions where the rate of reaction was kept constant with the time of incubation and proportional to the enzyme concentration.
Sugar assays.
The glucose concentration was measured using a peroxidase-glucose oxidase assay (Sigma). Lactose was assayed in the presence of glucose or fructose by measuring the concentration of glucose or galactose in samples before and after hydrolysis with β-galactosidase for 1 h at 37°C in 233 mM citrate buffer (pH 6.6) containing 60 mM MgSO4 and 0.05 U of β-galactosidase (Worthington) per μl. Galactose was determined using a peroxidase-galactose oxidase assay (1). Fructose was measured by the resorcinol method (30).
Protein assay.
Protein concentrations were measured using the method of Peterson (23) with bovine serum albumin as the standard.
RESULTS
Isolation of ptsH mutants.
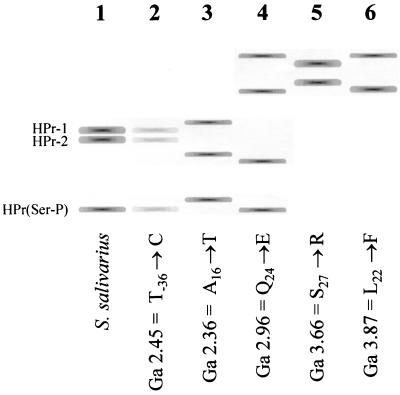
In a previous study, we reported that several types of PTS-negative mutants could be isolated by plating S. salivarius on a medium containing 2DG and a metabolizable sugar (11). We observed that the frequency at which mutants could be isolated was highest with the combination 2DG and galactose and that selection on galactose seemed to favor the isolation of ptsH mutants. To determine whether selection on this combination of sugars actually results in a bias toward the isolation of ptsH mutants, we decided to repeat the experiment on a larger scale by analyzing 546 mutants obtained by plating S. salivarius on a solid medium containing 2DG and galactose. To rapidly detect ptsH mutants, the HPr of the selected clones was analyzed by polyacrylamide gel electrophoresis and Western blotting as described in Materials and Methods. A typical wild-type HPr electrophoretic pattern obtained using this procedure is shown in lane 1 of Fig. 1. We observed that approximately 50% of the clones that we analyzed exhibited several distinctive aberrant HPr electrophoretic mobility patterns. Some examples are shown in Fig. 1. DNA sequencing analysis of these clones confirmed that they were mutated in ptsH. The spectrum of ptsH mutations obtained with this approach is summarized in Table 1. A consensus amino acid sequence for gram-positive bacterial HPrs was deduced from a sequence comparison of 19 HPrs from gram-positive bacteria (Fig. 2). With the exception of the H7→L substitution and the A70→T substitution, we observed that all of the ptsH mutations conferring resistance to 2DG resulted in the substitution of a conserved residue. The mutations occurred mainly in the region of α-helix 1 and in the N-terminal regions of α-helices 2 and 3.
FIG. 1.
Electrophoretic patterns of wild-type HPr and of selected HPr mutants. Cells were lysed by sonication, and the resulting cellular extracts were incubated at 100°C for 5 min. The proteins were then separated by polyacrylamide gel electrophoresis under native conditions. The position of HPr in the gel was determined by Western blotting using anti-HPr-specific rabbit polyclonal antibodies. Wild-type free HPr migrated as a doublet since a portion of the HPr is not processed by the methionine aminopeptidase. This phenomenon results in a population of HPr consisting of two forms; HPr-1 (without Met) and HPr-2 (with Met) (29, 41). Because the cellular extract was heated before electrophoresis, HPr(His∼P) and the doubly phosphorylated product could not be observed. Lane 1, pattern of the wild-type strain; lane 2, example of pattern observed with mutants with mutations in the promoter or the 5′ UTR of the pts operon; lanes 3 to 6, examples of aberrant patterns observed with mutants with point mutations in ptsH. “Ga” indicates strain designations.
TABLE 1.
HPr mutants obtained by selection on 5 mM 2DG and 200 mM galactose
| Change in nucleotide sequence |
Change in the amino acid sequence of HPr |
|---|---|
| T to C at position −36 | No changea |
| A to G at position −10 | |
| A to T at position −10 | |
| G to T at position +1 | |
| C to G at position +21 | |
| C to A at position +21 | |
| A to C at position +28 | |
| A to G at position +45 (ptsH rbsb) | |
| A to G at position +55 (initiation codon ATG for GTG) | |
| C to T at position 72 | His to Leu at position 7 |
| G to A at position 99 | Ala to Thr at position 16 |
| G to T at position 99 | Ala to Ser at position 16 |
| C to A at position 112 | Thr to Asn at position 20 |
| C to T at position 117 | Leu to Phe at position 22 |
| G to A at position 120 | Val to Ile at position 23 |
| C to G at position 123 | Gln to Glu at position 24 |
| C to A at position 134 | Ser to Arg at position 27 |
| TTT to GTA at positions 140–142 | Phe to Val at position 29 |
| TTT to GTT at positions 140–142 | |
| T to C at position 146 | Ser to Leu at position 31 |
| T to C at position 194 | Ile to Thr at position 47 |
| A to G at position 196 | Met to Val at position 48 |
| Insertion of the sequence CTGATG between position 257 and 258 | Insertion of the dipeptide Ala-Asp between position 68 and 69 |
| G to A at position 258 | Asp to Asn at position 69 |
| G to A at position 261 | Ala to Thr at position 70 |
The first nine mutations occurred in the region extending from the ptsHI promoter (−35 box) to the ATG start codon and caused decreased amounts of HPr.
rbs, ribosome binding site.
FIG. 2.
Locations of substituted amino acids in ptsH mutants selected in the presence of 5 mM 2DG and 200 mM galactose. The first row indicates the secondary structures common to HPrs from gram-positive bacteria: S, β-strand; H, α-helix. The second row shows the consensus amino acid sequence of HPrs from gram-positive bacteria deduced from a multiple alignment analysis of the amino acid sequences of 19 HPrs from the following bacteria: Staphylococcus carnosus, Staphylococcus aureus, Staphylococcus epidermidis, Listeria monocytogenes, S. mutans, Streptococcus bovis, Streptococcus pyogenes, S. salivarius, Streptococcus pneumoniae, Streptococcus equi, Lactococcus lactis, Lactobacillus casei, Lactobacillus sake, E. faecalis, B. subtilis, Bacillus megaterium, Bacillus stearothermophilus, Bacillus halodurans, and Clostridium acetobutylicum. Uppercase indicates that the amino acid is conserved in all sequences, whereas lowercase indicates that it is conserved in at least 15 out of 19 sequences. The third row shows the amino acid sequence of S. salivarius HPr. The fourth row indicates the location and nature of the substitution in S. salivarius ptsH mutants isolated in the presence of 2DG and galactose.
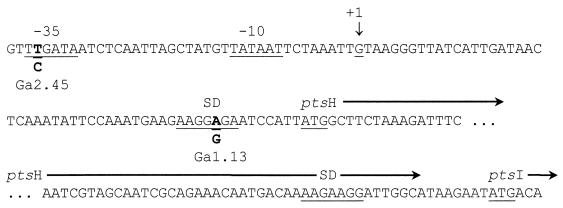
Some mutants that were isolated in the presence of 2DG and galactose exhibited an HPr electrophoretic pattern similar to that of the parental strain but contained much less HPr, suggesting that they bore a mutation that reduced HPr synthesis (Fig. 1, lane 2). In S. salivarius, the genes coding for HPr and EI, designated ptsH and ptsI, respectively, form the pts operon. Transcription of the pts operon is initiated from a single promoter located upstream from ptsH (9). The promoter consists of conserved −35 and −10 boxes separated by 17 nucleotides and followed by a 5′ untranslated region (5′ UTR) of 54 nucleotides comprising the ptsH ribosome binding site (Fig. 3). DNA sequence analysis of mutants possessing lower amounts of HPr revealed that they all possessed mutations in these regions (Table 1). Two mutants were selected for further study: Ga 2.45 had a point mutation in the −35 sequence substituting a T for a C at position −36, while mutant Ga 1.13 had a point mutation in the Shine-Dalgarno sequence of ptsH substituting an A for a G at position +45 (Fig. 3). No other mutations were detected in the pts operons of these mutants, nor in the 252-bp region upstream from the −35 box of the pts promoter.
FIG. 3.
Nucleotide sequence of the 5′ end of the S. salivarius pts operon. The following DNA signal sequences are underlined: the −35 and −10 boxes of the promoter, the transcriptional start point (+1), the ribosome binding sites (SD) of ptsH and ptsI, and the start codons of ptsH and ptsI. Mutations in Ga 2.45 and Ga 1.13 are indicated in boldface.
Cellular levels of HPr and EI.
The intracellular levels of the different forms of HPr were determined in glucose- and fructose-grown cells harvested at mid-log phase (Table 2). As already reported (12, 43), rapidly growing wild-type cells contained mostly HPr(Ser-P) and the doubly phosphorylated product HPr(Ser-P)(His∼P) and very low levels of HPr(His∼P) and unphosphorylated HPr. Total HPr was approximately two fold lower in Ga 1.13 and about threefold lower in Ga 2.45. The mutations not only decreased the total amount of HPr but also changed the levels of the different forms of HPr in the cells. The main changes were a 2- to 4-fold decrease in the level of HPr(Ser-P) and a 1.4- to 6-fold decrease in the level of the doubly phosphorylated product. To determine whether the decrease observed in the levels of HPr(Ser-P) could result from a diminution in the activity of HPrK, we sequenced hprK from both mutants and measured HPrK activities in glucose-grown wild-type and mutant cells. No mutation was detected in hprK and in hprK promoter regions of both mutants. The HPrK activities, determined on two separate cultures and expressed as picomoles HPr(Ser-P) produced per microgram of protein per minute were 2.0 ± 0.2 for the wild-type strain, 2.3 ± 0.7 for mutant Ga 1.13, and 2.0 ± 0.4 for mutant Ga 2.45. These results indicated that the drop in the cellular levels of HPr(Ser-P) in Ga 1.13 and Ga 2.45 could not be attributed to a decrease in the synthesis or the activity of HPrK.
TABLE 2.
Intracellular levels of HPr and EI
| Strain | Growth sugar | Cellular concna
|
|||||
|---|---|---|---|---|---|---|---|
| HPr | HPr(His∼P) | HPr(Ser-P) | HPr(Ser-P)(His∼P) | Total HPr | EI | ||
| Wild type | Glucose | 0.3 ± 0.8 | 3.4 ± 1.3 | 50.8 ± 11.7 | 13.9 ± 10.2 | 85.6 ± 23.9 | 0.20 ± 0.04 |
| Fructose | 0.8 ± 0.9 | 2.4 ± 1.0 | 50.9 ± 14.5 | 55.1 ± 7.3 | 109.0 ± 23.8 | 0.21 ± 0.05 | |
| Ga 1.13 | Glucose | 1.6 ± 0.3 | 3.0 ± 0.3 | 24.5 ± 0.3 | 13.9 ± 0.3 | 43.0 ± 1.1 | 0.10 ± 0.02 |
| Fructose | ND | 3.3 ± 0.1 | 11.9 ± 1.6 | 38.9 ± 3.0 | 54.1 ± 4.7 | 0.13 ± 0.02 | |
| Ga 2.45 | Glucose | 3.5 ± 1.0 | 6.9 ± 1.0 | 11.3 ± 1.0 | 2.3 ± 0.4 | 24.0 ± 3.4 | 0.07 ± 0.01 |
| Fructose | 1.4 ± 0.1 | 2.9 ± 0.6 | 18.7 ± 4.1 | 16.1 ± 1.6 | 39.1 ± 6.4 | 0.07 ± 0.01 | |
Values represent the means ± standard deviations of four determinations performed on cells from two different cultures. Results for HPr are expressed as micrograms per milligram of cytoplasmic protein. Results for EI are expressed as nanomoles of EI subunit per milligram of cytoplasmic protein. ND, not detected.
The mutation in the −35 promoter region (Ga 2.45) caused a three fold decrease in the amount of EI, irrespective of the culture conditions tested, a decrease that paralleled the decline in total HPr. The mutation in the ribosome binding site of ptsH was expected to reduce the levels of HPr but not that of EI. Surprisingly, we observed that the level of EI was reduced about twofold in this mutant.
Generation times.
The generation times of the wild-type and mutant strains were determined for cells growing at the expense of either 0.1% (wt/vol) glucose or fructose, each a PTS sugar, or galactose or lactose, each a non-PTS sugar (Table 3). The wild type grew at virtually the same rate on all the sugars tested. The growth of the mutants on PTS sugars decreased by various amounts depending on the strain and the sugar. The generation times of Ga 1.13 increased by a factor of about 1.3 when cells were grown on glucose and fructose, whereas those of Ga 2.45 increased by a factor of 2 on glucose and by a factor of 1.7 on fructose. The growth of both mutants on the non-PTS sugar galactose was almost the same, with generation times approximately 1.3-fold longer than that of the wild-type strain. The growth of the mutants on lactose was virtually the same, the generation times increasing by less than 1.2 times.
TABLE 3.
Generation times
| Growth sugar (0.1%) | Generation time (min) at 37°C for straina:
|
||
|---|---|---|---|
| Wild type | Mutant Ga 1.13 | Mutant Ga 2.45 | |
| Glucose | 32 ± 1 | 41 ± 1 | 66 ± 1 |
| Fructose | 30 ± 1 | 44 ± 1 | 50 ± 1 |
| Galactose | 35 ± 0 | 42 ± 2 | 44 ± 2 |
| Lactose | 33 ± 5 | 34 ± 2 | 38 ± 2 |
The values are the means of three determinations ± standard deviations.
Uptake of 2DG and rate of glucose consumption by resting cells.
The rate of 2DG transport by the wild type was 1.1 ± 0.1 nmol of 2DG/min/mg (dry weight) of cells, that of Ga 1.13 was 1.4 ± 0.1 nmol of 2DG/min/mg (dry weight) of cells, and that of Ga 2.45 was 1.4 ± 0.1 nmol of 2DG/min/mg (dry weight) of cells. The experiments were conducted in duplicate. The results suggested that a decrease in the amounts of HPr and EI by a factor of at least three did not reduce the rate at which resting cells took up a nonmetabolizable PTS sugar. We also determined the rate of glucose consumption by cells suspended in a phosphate buffer. The values, expressed as micrograms of glucose consumed per minute per milligram (dry weight) of cells, were 52 ± 3 for the wild-type strain, 44 ± 4 for Ga 1.13, and 32 ± 1 for Ga 2.45. The experiments were done in duplicate. These results suggested that a twofold reduction of EI and/or HPr cellular levels was sufficient to reduce the rate at which cells metabolize PTS sugars.
Growth in media containing a PTS sugar and a non-PTS sugar.
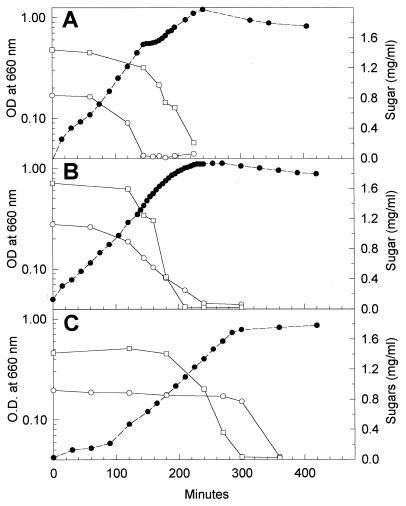
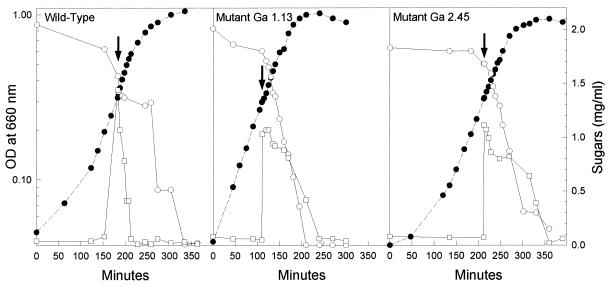
Growth of the parental strain in media containing glucose or fructose and either lactose or galactose is diauxic, the PTS sugars being used before the non-PTS sugars (12, 24). Growing the mutants under such conditions never resulted in diauxic growth. Growth of Ga 1.13 in the presence of a PTS sugar and a non-PTS sugar gave rise to a continuous S-shaped growth curve, and both sugars were used at the same time. This is exemplified by the growth of Ga 1.13 in a mixture of glucose and galactose (Fig. 4B). The growth of Ga 2.45 in mixtures of sugars also resulted in a single, uninterrupted growth phase. However, unlike Ga 1.13, Ga 2.45 always metabolized sugars sequentially, the non-PTS sugar being used before the PTS sugar. The growth of Ga 2.45 in a mixture of glucose and galactose is illustrated in Fig. 4C.
FIG. 4.
Growth patterns of the wild-type strain (A), mutant Ga 1.13 (B), and mutant Ga 2.45 (C) when grown in a medium containing glucose (a PTS sugar) and galactose (a non-PTS sugar). An 0.5-ml aliquot of glucose-grown cells was transferred into 15 ml of fresh medium containing approximately 0.1% (wt/vol) glucose and 0.2% (wt/vol) galactose. The symbols represent the OD660 (●) and the consumption of glucose (○) and galactose (□).
Activities of β-galactosidase, galactokinase, and α-galactosidase.
The incapacity of the mutants to prevent the metabolism of non-PTS sugars in the presence of glucose or fructose prompted us to determine the activities of β-galactosidase, galactokinase, and α-galactosidase, three inducible enzymes involved in the metabolism of the non-PTS sugars lactose, galactose, and melibiose in S. salivarius. The activities of these enzymes were very low in glucose- and fructose-grown wild-type cells (Table 4) (12, 24). However, we reproducibly observed that fructose caused a stronger repression of these enzymes than did glucose, despite the fact that the levels of HPr(Ser-P) and HPr(His∼P) were virtually the same in both fructose- and glucose-grown wild-type cells (Table 2). The enzymes were derepressed in both mutants, but the level of derepression varied according to the enzyme, the growth sugar, and the mutant strain. As a general rule, we observed that (i) the levels of enzyme activities were lower in fructose-grown cells than in glucose-grown cells and (ii), when compared with the parental strain, derepression was stronger in Ga 2.45 than in Ga 1.13.
TABLE 4.
Enzyme activities
| Strain | Enzyme | Sp act after growth on sugara:
|
||||
|---|---|---|---|---|---|---|
| Glucose | Fructose | Lactose | Galactose | Melibiose | ||
| Wild type | Galactokinase | 3 ± 0 | 1 ± 0 | 132 ± 1 | 400 ± 157 | ND |
| β-Galactosidase | 32 ± 1 | 10 ± 1 | 762 ± 6 | ND | ND | |
| α-Galactosidase | 57 ± 12 | 17 ± 6 | 93 ± 8 | ND | 634 ± 96 | |
| Ga 1.13 | Galactokinase | 6 ± 1 | 3 ± 0 | 124 ± 3 | 207 ± 3 | ND |
| β-Galactosidase | 40 ± 2 | 34 ± 1 | 338 ± 6 | ND | ND | |
| α-Galactosidase | 314 ± 50 | 214 ± 23 | 157 ± 11 | ND | 617 ± 45 | |
| Ga 2.45 | Galactokinase | 77 ± 10 | 40 ± 1 | 90 ± 13 | 455 ± 2 | ND |
| β-Galactosidase | 366 ± 8 | 267 ± 8 | 505 ± 12 | ND | ND | |
| α-Galactosidase | 589 ± 24 | 351 ± 26 | 46 ± 11 | ND | 552 ± 80 | |
Specific activities are expressed as nanomoles of product milligram of protein−1 minute−1 ± standard deviations. Values are means of two to three assays. ND, not determined.
Galactokinase and β-galactosidase were slightly derepressed in glucose- and fructose-grown Ga 1.13, but the activities remained very low and far below the activities measured in fully induced wild-type cells (Table 4). These enzymes, however, were strongly derepressed in Ga 2.45, which possessed 10 to 40 times more activity than did the parental strain after growth on PTS sugars.
Unlike the other enzymes, α-galactosidase was significantly derepressed in both mutants after growth on glucose and fructose. Derepression was, however, highest in Ga 2.45, as was the case for β-galactosidase and galactokinase. In glucose-grown Ga 2.45, the gene coding for α-galactosidase was obviously expressed at its maximal rate, as the level of activity was identical to that in cells grown on melibiose, the inducing sugar. Interestingly, the enzyme was only slightly or not at all derepressed in the mutants grown on lactose, a noninducing non-PTS sugar.
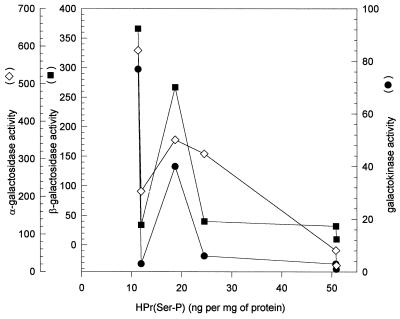
HPr(Ser-P) has been shown to be involved in the control of gene expression in gram-positive bacteria. Using data reported in Tables 2 and 4, we verified whether there was a correlation between the magnitude of galactokinase, β-galactosidase, and α-galactokinase derepression and the cellular levels of HPr(Ser-P) measured in the wild-type and mutant strains after growth on glucose and fructose. As illustrated in Fig. 5, there was no correlation between the cellular amounts of HPr(Ser-P) and enzyme activities. For instance, the levels of HPr(Ser-P) were identical in fructose-grown cells of Ga 1.13 and in glucose-grown cells of Ga 2.45 (Table 2) while the levels of β-galactosidase were 10-fold lower in mutant Ga 1.13 (34 ± 1) than in mutant Ga 2.45 (Table 4).
FIG. 5.
Galactokinase, β-galactosidase, and α-galactosidase activities as a function of cellular levels of HPr(Ser-P). Cellular levels of HPr(Ser-P) are indicated in Table 2, while enzyme activities are indicated in Table 4. The values used are those that have been determined for glucose- and fructose-grown cells.
Inducer exclusion by growing cells.
The inability of the mutants to prevent the metabolism of non-PTS sugars in the presence of PTS sugars may also result from their incapacity to exert inducer exclusion. We have previously shown that, when glucose is added to wild-type S. salivarius cells growing in the presence of lactose, galactose, or melibiose, the metabolism of the non-PTS sugar immediately stops and resumes only when the glucose is depleted (24). In the work presented here, we extended our study by looking at the effect of fructose on the metabolism of lactose and galactose. As illustrated in Fig. 6, the addition of fructose to growing wild-type cells also stopped the metabolism of lactose, which resumed only when the fructose was exhausted. Similar results were obtained with cells growing on galactose (data not shown). In contrast, we observed that the addition of glucose or fructose to growing mutant cells did not prevent the metabolism of lactose or galactose. The effect of fructose on the utilization of lactose is illustrated in Fig. 6 as an example. Similar results were obtained with the other combinations of sugars. All experiments were done in duplicate, and results were reproducible.
FIG. 6.
Effect of fructose on lactose metabolism by growing cells. Cells were grown overnight in the presence of 0.2% lactose. An 0.5-ml aliquot was used to inoculate 15 ml of fresh medium containing 0.2% (wt/vol) lactose. When the culture reached mid-log phase, the medium was supplemented with 0.1% (wt/vol) fructose (indicated by the arrows). The symbols represent the OD660 (●) and the consumption of lactose (○) and that of fructose (□).
DISCUSSION
Thompson and Chassy (37) have already demonstrated that the toxicity of 2DG in lactococci is caused by the establishment of a futile cycle that leads to the dissipation of phosphoenolpyruvate and ATP. Other studies have reported that nonmetabolizable phosphorylated sugars are toxic by interfering with gene expression, permease, or enzyme activities (20, 28, 38, 39). Although the toxicity of 2DG in S. salivarius has never been scrutinized, it is reasonable to assume that these mechanisms contribute to some extent to the toxicity of 2DG in this microorganism, since this sugar analog is transported in S. salivarius by the glucose-mannose PTS and accumulates in the cell as a phosphorylated derivative (40). On the basis of these hypotheses, our results suggested that decreasing the cellular amounts of HPr and EI by a factor of two or three or introducing point mutations in HPr prevented the establishment of a lethal futile energy cycle and probably restricted the intracellular accumulation of 2DG-phosphate to nontoxic levels. These mechanisms, however, could not explain why selection on galactose favored the isolation of ptsH mutants. Further research is required to explain this enigmatic result. Selection of mutants on 2DG and galactose has proved, however, to be useful to identify amino acids that are important for HPr to exert its functions. Indeed, we have identified 13 amino acids that are essential for HPr to properly accomplish its physiological functions in S. salivarius. Interestingly, most of these amino acids are well conserved in gram-positive bacterial HPrs. Ten of these amino acids were found between positions 16 and 48, suggesting that helix 1, the loop between helix 1 and β-strand 2, and helix 2 are critical structural determinants with respect to streptococcal HPr regulatory functions (Fig. 2). These results are consistent with the proposal that HPr interacts with other PTS proteins and proteins under its control via regions located at its surface comprising α-helices 1 and 2 and the loops preceding helix 1 and following helix 2 (17, 49, 53) and with the findings of Jones et al. (17), who showed that residues 14 to 17 and 21 to 27 of B. subtilis HPr are involved in the interaction with the transcriptional regulator CcpA. We also found that residues at positions 69 and 70 (D and A, respectively) were important for normal streptococcal HPr functions, since the replacement of these amino acids by N and T, respectively, conferred resistance to 2DG. These results are consistent with those reported by Koch et al. (18), who showed that residues at these positions in E. coli HPr (D and E) play important roles in controlling conformational aspects of HPr. It is noteworthy that, despite the large number of clones that we analyzed, we did not find any HPr mutants with a mutation replacing Ser46.
A significant proportion of the mutants possessed a mutation in the pts promoter or the 5′ UTR located upstream from the ptsH initiation codon. In all cases, these mutations resulted in a decrease in the cellular amount of HPr, revealing the importance of specific nucleotides in the efficiency of the pts promoter and the essential role of the 5′ UTR in the expression of the pts operon. In mutant Ga 2.45, the second T of the TTG sequence located at the 5′ end of the −35 box of the pts promoter was replaced by a C, causing a threefold decrease in cellular amounts of HPr and EI. This demonstrated the importance of this nucleotide for optimal recognition of the promoter by the major streptococcal RNA polymerase. This mutation reduced the levels of HPr and EI to the same extent, a result that is understandable, as this mutation, by affecting the rate of transcription, will reduce the expression of all the genes making up the operon. The results obtained with Ga 1.13, which underwent an A-to-G transition in the ribosome binding site of ptsH, were, however, unexpected. Indeed, this type of mutation should affect only HPr levels, since this DNA signal sequence should not interfere with the rate of transcription of the operon or with the rate of translation of ptsI. Surprisingly, we found that the amount of EI in this mutant was reduced by a factor of two. This may be explained in terms of a translational coupling between ptsH and ptsI or by the involvement of HPr in the expression of ptsI.
The growth of the wild-type strain in media containing a PTS sugar (glucose or fructose) and a non-PTS sugar (lactose or galactose) is diauxic, the PTS sugar being metabolized before the non-PTS sugar (12, 24). The growth of mutants Ga 1.13 and Ga 2.45 under the same conditions was never diauxic, and both mutants had lost the ability to metabolize PTS sugars preferentially, indicating that a twofold decrease in HPr and EI cellular levels was sufficient to prevent the cells from selectively metabolizing PTS sugars in preference to non-PTS sugars. Diauxic growth is a manifestation of at least two physiological processes, inducer exclusion and inhibition of gene expression. Results reported in this paper indicated that reducing the cellular levels of the general PTS proteins two- to threefold interfered with both processes.
A reduction in the synthesis of EI and HPr resulted in a decline in the levels of the two predominant forms of HPr found in rapidly growing streptococcal cells, HPr(Ser-P) and HPr(Ser-P)(His∼P) (43). This drop could not be attributed to a defect in HPrK, as no mutation was found in ptsK of Ga 1.13 and Ga 2.45, and both mutants possessed the same levels of HPrK activity as that of the wild-type strain. Thus, a two- to threefold reduction in the expression of the pts operon was sufficient to alter the proportion of the different forms of HPr in the cell. The sharpest decrease in the amount of the doubly phosphorylated product was observed in glucose-grown cells of Ga 2.45, which contained less EI than did mutant Ga 1.13. This result suggested that the low level of HPr(Ser-P)(His∼P) observed with Ga 2.45 was the consequence of a combination of two events: a reduction in the concentration of the substrate HPr(Ser-P) and a reduction in the amount of EI, the enzyme that catalyzes the phosphorylation on His15. However, results obtained with fructose-grown cells of the same mutant were not consistent with this explanation, as they contained the same amount of EI and almost the same amount of HPr(Ser-P) as glucose-grown cells but seven times more HPr(Ser-P)(His∼P) (Table 2). The generation times of Ga 2.45 indicated, however, that this mutant grew slower on glucose than on fructose (Table 3). This difference in growth rate would obviously modify the levels of several glycolytic and other types of intermediates in the cells and therefore might influence the activities of the HPrK and EI. It therefore appears that the amount of HPr(Ser-P)(His∼P) is dictated by several determinants, including the amounts of HPr and EI and the nature of the sugar that supports growth, as well as the rate at which the cells divide. As the physiological functions of the doubly phosphorylated product remain to be elucidated for streptococci, it remains unclear whether the physiological defects observed for the mutants were caused, to some extent, by a decrease in the cellular amounts of this form of HPr. However, several lines of evidence have demonstrated that HPr(Ser-P) plays a major role in the exclusion of non-PTS sugars by PTS sugars in gram-positive bacteria (7, 31, 48, 50–52). Our results suggested that a twofold decrease in the levels of HPr(Ser-P) in S. salivarius was sufficient to prevent growing cells from excluding non-PTS sugars when a PTS sugar became available. In gram-positive bacteria, HPr(Ser-P) is also involved in the regulation of gene transcription (6, 8, 13, 17). Results presented in this paper unequivocally indicated that reducing the synthesis of HPr by a factor of three was enough to cause a strong derepression of galactokinase, β-galactosidase, and α-galactosidase, three inducible enzymes in S. salivarius. Our results also suggested that the expression of the genes coding for these enzymes was controlled by HPr in a hierarchical manner. Indeed, a twofold reduction of total HPr caused a significant derepression of α-galactosidase, while the synthesis of galactokinase and β-galactosidase was only slightly affected. On the other hand, a threefold reduction of HPr caused a strong derepression of the three enzymes. However, we did not find a correlation between the magnitude of enzyme derepression and the cellular levels of HPr(Ser-P). Moreover, we observed that α-galactosidase, which was strongly derepressed in both mutants after growth on glucose and fructose, was only slightly or not at all derepressed in lactose-grown cells. These results are consistent with those reported for an S. salivarius mutant in which Ile47 is replaced by Thr (mutant G22.4) (12). The enzymes β-galactosidase and α-galactosidase are derepressed in this mutant, even though it possesses cellular levels of HPr(Ser-P) similar to those found in the wild-type strain. Moreover, derepression of β-galactosidase and α-galactosidase in mutant G22.4 is observed after growth on glucose or fructose, each a PTS sugar, but not after growth on lactose or galactose, each a non-PTS sugar (12). It thus appears that PTS-mediated repression of catabolic genes in streptococci involves a complex regulatory circuit in which HPr plays a dominant role, but not uniquely as HPr(Ser-P), and that genes under the control of the PTS, such as the melibiose genes, might be subjected to different regulatory mechanisms depending on whether the cells grow at the expense of a PTS sugar or a non-PTS sugar.
The mutants took up 2DG at the same rate as that of the parental strain, suggesting that a threefold reduction of HPr and EI cellular levels had no effect on the rate of sugar transport by the PTS. However, the uptake of a nonmetabolizable sugar such as 2DG does not result in ATP synthesis. Under these conditions, cells do not produce HPr(Ser-P), and all the HPr is available for sugar transport. Therefore, the results obtained from the 2DG uptake experiments suggested that decreasing the amount of EI by a factor of at least three would not change the transport capacity of the PTS as long as the amount of HPr available for transport was not limiting. On the other hand, we observed a decline in the rate of glucose consumption in the mutants compared with the parental strain, suggesting that when cells were able to generate HPr(Ser-P), a twofold reduction in HPr and EI synthesis was sufficient to affect the rate at which a PTS sugar was metabolized. This may explain to some extent the fact that the generation times of the mutants on PTS sugars were longer than those of the wild-type strain. This might not, however, be the only factor that reduced the growth of the mutants. The observation that some enzymes were derepressed to different degrees in the mutants (Table 4) suggested that the increase in generation times observed when they were growing on PTS sugars, and possibly also on non-PTS sugars, resulted from a futile expenditure of energy engendered by the synthesis of useless proteins.
We have shown in this paper that a two- to threefold reduction of HPr and EI synthesis is sufficient to modify several aspects of the cell physiology. A reduction of this magnitude could obviously occur when growth is reduced due to adverse chemical or physical conditions. For example, Thevenot et al. (36) have shown that the amount of total HPr is reduced about 2 times and that of HPr(Ser-P) is reduced about 15 times in Streptococcus mutans cells cultured at a dilution rate of 0.1 h−1 (which corresponds to a doubling time of 7 h) in the presence of 10 mM glucose compared with that for cells growing at the same rate in the presence of 100 mM glucose. Considering the fact that oral streptococci live in conditions of starvation and slow growth for long periods (3), it is reasonable to assume that in their natural habitat these bacteria contain reduced amounts of HPr, enabling them to take up PTS and non-PTS sugars nonpreferentially.
ACKNOWLEDGMENTS
This research was supported by the Medical Research Council of Canada operating grants MT 6979 and MOP 36338.
We thank Michel Frenette for useful discussions and comments on the manuscript.
REFERENCES
- 1.Avigad G, Amaval D, Ascensio C, Horecker B L. The D-galactose oxydase of Polyporus circinatus. J Biol Chem. 1962;237:2736–2743. [PubMed] [Google Scholar]
- 2.Brochu D, Vadeboncoeur C. The HPr(Ser) kinase of Streptococcus salivarius: purification, properties, and cloning of the hprK gene. J Bacteriol. 1999;181:709–717. doi: 10.1128/jb.181.3.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burne R A. Oral streptococci … products of their environment. J Dent Res. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 4.Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Claiborne A. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J Biol Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher J, Bauer B, Sauerwald H. Regulation of glycerol metabolism in Enterococcus faecalis by phosphoenolpyruvate-dependent phosphorylation of glycerol kinase catalyzed by enzyme I and HPr of the phosphotransferase system. J Bacteriol. 1993;175:3730–3733. doi: 10.1128/jb.175.12.3730-3733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 7.Dossonnet V, Monedero V, Zagorec M, Galinier A, Pérez-Martinez G, Deutscher J. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J Bacteriol. 2000;182:2582–2590. doi: 10.1128/jb.182.9.2582-2590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 9.Gagnon G, Vadeboncoeur C, Frenette M. Regulation of the ptsH and ptsI gene expression in Streptococcus salivarius ATCC 25975. Mol Microbiol. 1995;16:1111–1121. doi: 10.1111/j.1365-2958.1995.tb02336.x. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier L, Bourassa S, Brochu D, Vadeboncoeur C. Control of sugar utilization in oral streptococci. Properties of phenotypically distinct 2-deoxyglucose-resistant mutants of Streptococcus salivarius. Oral Microbiol Immunol. 1990;5:352–359. doi: 10.1111/j.1399-302x.1990.tb00440.x. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier L, Thomas S, Gagnon G, Frenette M, Trahan L, Vadeboncoeur C. Positive selection for resistance to 2-deoxyglucose gives rise, in streptococcus salivarius, to seven classes of pleiotropic mutants, including ptsH and ptsI missense mutants. Mol Microbiol. 1994;13:1101–1109. doi: 10.1111/j.1365-2958.1994.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier M, Brochu D, Eltis L D, Thomas S, Vadeboncoeur C. Replacement of isoleucine-47 by threonine in the HPr protein of Streptococcus salivarius abrogates the preferential metabolism of glucose and fructose over lactose and melibiose but does not prevent the phosphorylation of HPr on serine-46. Mol Microbiol. 1997;25:695–705. doi: 10.1046/j.1365-2958.1997.4981870.x. [DOI] [PubMed] [Google Scholar]
- 13.Gösseringer R, Küster L, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 14.Gunnewijk M G W, Poolman B. Phosphorylation state of HPr determines the level of expression and the extent of phosphorylation of the lactose transport protein of Streptococcus thermophilus. J Biol Chem. 2000;245:34073–34079. doi: 10.1074/jbc.M003512200. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton I R, Lo G C Y. Co-induction of β-galactosidase and the lactose-P-enolpyruvate phosphotransferase system in Streptococcus salivarius and Streptococcus mutans. J Bacteriol. 1978;136:900–908. doi: 10.1128/jb.136.3.900-908.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 17.Jones B E, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 18.Koch S, Sutrina S L, Wu L F, Reizer J, Schnetz K, Rak B, Saier M H., Jr Identification of a site in the phosphocarrier protein, HPr, which influences its interactions with sugar permeases of the bacterial phosphotransferase system: kinetic analyses employing site-specific mutants. J Bacteriol. 1996;178:1126–1133. doi: 10.1128/jb.178.4.1126-1133.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundig W, Ghosh S, Roseman S. Phosphate bound histidine in a protein as an intermediate in a novel phospho-transferase system. Proc Natl Acad Sci USA. 1964;52:1067–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.London J, Hausman S. A study of the xylitol-mediated transient inhibition of ribitol utilization by Lactobacillus casei. J Bacteriol. 1982;150:657–661. doi: 10.1128/jb.150.2.657-661.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lortie L A, Dubreuil J D, Harel J. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J Clin Microbiol. 1991;29:656–659. doi: 10.1128/jcm.29.3.656-659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCleary B V. α-Galactosidase from luciferine and guar seed. Methods Enzymol. 1988;160:627–632. [Google Scholar]
- 23.Peterson G L. Determination of total protein. Methods Enzymol. 1983;91:95–119. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
- 24.Plamondon P, Brochu D, Thomas S, Fradette J, Gauthier L, Vaillancourt K, Buckley N, Frenette M, Vadeboncoeur C. Phenotypic consequences resulting from a methionine-to-valine substitution at position 48 in the HPr protein of Streptococcus salivarius. J Bacteriol. 1999;181:6914–6921. doi: 10.1128/jb.181.22.6914-6921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poolman B, Knol J, Mollet B, Nieuwenhuis B, Sulter G. Regulation of bacterial sugar-H+ symport by phosphoenolpyruvate-dependent enzyme I/HPr-mediated phosphorylation. Proc Natl Acad Sci USA. 1995;92:778–782. doi: 10.1073/pnas.92.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poolman B, Knol J, van der Does C, Henderson P J F, Liang W J, Leblanc G, Pourcher T, Mus-Veteau I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol Microbiol. 1996;19:911–922. doi: 10.1046/j.1365-2958.1996.397949.x. [DOI] [PubMed] [Google Scholar]
- 27.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiner A M. Xylitol and d-arabitol toxicities due to derepressed fructose, galactitol, and sorbitol phosphotransferases of Escherichia coli. J Bacteriol. 1977;132:166–173. doi: 10.1128/jb.132.1.166-173.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robitaille D, Gauthier L, Vadeboncoeur C. The presence of two forms of the phosphocarrier protein HPr of the phosphoenolpyruvate:sugar phosphotransferase system in streptococci. Biochimie. 1991;73:573–581. doi: 10.1016/0300-9084(91)90025-v. [DOI] [PubMed] [Google Scholar]
- 30.Roe J H. A colorimetric method for the determination of fructose in blood and urine. J Biol Chem. 1934;107:15–22. [Google Scholar]
- 31.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 32.Saier M H, Jr, Reizer J. Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1992;174:1433–1438. doi: 10.1128/jb.174.5.1433-1438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saier M H, Jr, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 34.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 35.Stülke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- 36.Thevenot T, Brochu D, Vadeboncoeur C, Hamilton I R. Regulation of ATP-dependent P-(Ser)-HPr formation in Streptococcus mutans and Streptococcus salivarius. J Bacteriol. 1995;177:2751–2759. doi: 10.1128/jb.177.10.2751-2759.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson J, Chassy B M. Novel phosphoenolpyruvate-dependent futile cycle in Streptococcus lactis: 2-deoxy-d-glucose uncouples energy production from growth. J Bacteriol. 1982;151:1454–1465. doi: 10.1128/jb.151.3.1454-1465.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson J, Chassy B M. Regulation of glycolysis and sugar phosphotransferase activities in Streptococcus lactis: growth in the presence of 2-deoxy-d-glucose. J Bacteriol. 1983;154:819–830. doi: 10.1128/jb.154.2.819-830.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trahan L. Xylitol: a review of its action on mutants streptococci and dental plaque—its clinical significance. Int Dent J. 1995;45:77–92. [PubMed] [Google Scholar]
- 40.Vadeboncoeur C. Structure and properties of the phosphoenolpyruvate:glucose phosphotransferase system of oral streptococci. Can J Microbiol. 1984;30:495–502. doi: 10.1139/m84-073. [DOI] [PubMed] [Google Scholar]
- 41.Vadeboncoeur C. HPr: heteromorphous protein. Res Microbiol. 1995;146:525–530. doi: 10.1016/0923-2508(96)80558-4. [DOI] [PubMed] [Google Scholar]
- 42.Vadeboncoeur C, Bourgeau G, Mayrand D, Trahan L. Control of sugar utilization in the oral bacteria Streptococcus salivarius and Streptococcus sanguis by the phosphoenolpyruvate:glucose phosphotransferase system. Arch Oral Biol. 1983;28:123–131. doi: 10.1016/0003-9969(83)90119-x. [DOI] [PubMed] [Google Scholar]
- 43.Vadeboncoeur C, Brochu D, Reizer J. Quantitative determination of the intracellular concentration of the various forms of HPr, a phosphocarrier protein of the phosphoenolpyruvate:sugar phosphotransferase system in growing cells of oral streptococci. Anal Biochem. 1991;196:24–30. doi: 10.1016/0003-2697(91)90112-7. [DOI] [PubMed] [Google Scholar]
- 44.Vadeboncoeur C, Frenette M, Lortie L A. Regulation of the pts operon in low G+C Gram-positive bacteria. J Mol Microbiol Biotechnol. 2000;2:483–490. [PubMed] [Google Scholar]
- 45.Vadeboncoeur C, Pelletier M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 46.Vadeboncoeur C, Trahan L. Glucose transport in Streptococcus salivarius. Evidence for the presence of a distinct phosphoenolpyruvate:glucose phosphotransferase system which catalyses the phosphorylation of α-methyl glucoside. Can J Microbiol. 1982;28:190–199. doi: 10.1139/m82-025. [DOI] [PubMed] [Google Scholar]
- 47.Vaughan E E, David S, de Vos W M. The lactose transporter in Leuconostoc lactis is a new member of the LacS subfamily of galactoside-pentose-hexuronide translocators. Appl Environ Microbiol. 1996;62:1574–1582. doi: 10.1128/aem.62.5.1574-1582.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viana R, Monedero V, Dossonnet V, Vadeboncoeur C, Pérez-Martinez G, Deutscher J. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol Microbiol. 2000;36:570–584. doi: 10.1046/j.1365-2958.2000.01862.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Sondej M, Garrett D S, Peterkofsky A, Clore M G. A common interface on HPr for interaction with its partner proteins. J Biol Chem. 2000;275:16401–16403. doi: 10.1074/jbc.C000167200. [DOI] [PubMed] [Google Scholar]
- 50.Ye J J, Minarcik J, Saier M H., Jr Inducer expulsion and the occurrence of an HPr(Ser-P)-activated sugar-phosphate phosphatase in Enterococcus faecalis and Streptococcus pyogenes. Microbiology. 1996;142:585–592. doi: 10.1099/13500872-142-3-585. [DOI] [PubMed] [Google Scholar]
- 51.Ye J J, Reizer J, Cui X, Saier M H., Jr ATP-dependent phosphorylation of serine-46 in the phosphocarrier protein HPr regulated lactose/H+ symport in Lactobacillus brevis. Proc Natl Acad Sci USA. 1994;91:3102–3106. doi: 10.1073/pnas.91.8.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye J J, Reizer J, Cui X, Saier M H., Jr Inhibition of the phosphoenolpyruvate:lactose phosphotransferase system and activation of a cytoplasmic sugar-phosphate phosphatase in Lactococcus lactis by ATP-dependent metabolite-activated phosphorylation of serine 46 in the phosphocarrier protein HPr. J Biol Chem. 1994;269:11837–11844. [PubMed] [Google Scholar]
- 53.Zhu P P, Herzberg O, Peterkofsky A. Topography of the interaction of HPr(Ser) kinase with HPr. Biochemistry. 1998;37:11762–11770. doi: 10.1021/bi980455p. [DOI] [PubMed] [Google Scholar]