Abstract
The global emergence of coronavirus disease 2019 (COVID‐19) has caused substantial human casualties. Clinical manifestations of this disease vary from asymptomatic to lethal, and the symptomatic form can be associated with cytokine storm and hyperinflammation. In face of the urgent demand for effective drugs to treat COVID‐19, we have searched for candidate compounds using in silico approach followed by experimental validation. Here we identified celastrol, a pentacyclic triterpene isolated from Tripterygium wilfordii Hook F, as one of the best compounds out of 39 drug candidates. Celastrol reverted the gene expression signature from severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐infected cells and irreversibly inhibited the recombinant forms of the viral and human cysteine proteases involved in virus invasion, such as Mpro (main protease), PLpro (papain‐like protease), and recombinant human cathepsin L. Celastrol suppressed SARS‐CoV‐2 replication in human and monkey cell lines and decreased interleukin‐6 (IL‐6) secretion in the SARS‐CoV‐2‐infected human cell line. Celastrol acted in a concentration‐dependent manner, with undetectable signs of cytotoxicity, and inhibited in vitro replication of the parental and SARS‐CoV‐2 variant. Therefore, celastrol is a promising lead compound to develop new drug candidates to face COVID‐19 due to its ability to suppress SARS‐CoV‐2 replication and IL‐6 production in infected cells.
Keywords: celastrol, COVID‐19, cysteine protease inhibition, molecular docking, SARS‐CoV‐2
1. INTRODUCTION
COVID‐19 (coronavirus disease 2019), caused by the β‐coronavirus SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus 2), was first reported to the World Health Organization in January 2020 after a local pneumonia outbreak of unknown etiology in Wuhan, China (World Health Organization, 2020; N. Zhu et al., 2020). SARS‐CoV‐2 has been rapidly and effectively transmitted from human to human and became a worldwide pandemic that affected more than 446 million people in March 2022 (Dong et al., 2020; Q. Li et al., 2020).
The devastating effects of COVID‐19 on global public health and the economy have demanded urgent efforts to discover potential drug and vaccine candidates to prevent and treat this disease. Since the pandemic's beginning, many immunization strategies have been studied to develop vaccines for COVID‐19. Although there are many vaccine candidates under development (Polack et al., 2020; Voysey et al., 2021; Wouters et al., 2021), there are many challenges to achieve an efficient global immunization, such as production limitations, efficacy levels, restrictions on use, dosing procedures, storage requirements, price, the emergence of SARS‐CoV‐2 lineages, and promotion of durable immunological memory (Baric, 2020; Faria et al., 2021; Fontanet et al., 2021; Teijaro & Farber, 2021; Thomson et al., 2021; Wouters et al., 2021; W. Wang et al., 2020).
Despite the essentiality of effective vaccines, the quick discovery of drugs to prevent and treat SARS‐CoV‐2 infection is a critical demand to face COVID‐19 (Atzrodt et al., 2020). The drug reuse strategy accelerates discovering candidate compounds with known activities that reduce the SARS‐CoV‐2 viral load or promote a better clinical evolution of COVID‐19 (Galindez et al., 2021; Guy et al., 2020). One aspect of the immunopathology of this disease that stands out is non‐hemostatic inflammation associated with cytokine storm involving several mediators, including interleukin‐6 (IL‐6), which is a severity biomarker of this illness (Hadjadj et al., 2020; Han et al., 2020; Tay et al., 2020).
In this sense, bioinformatics and computational biology are powerful and multidimensional in silico tools for drug discovery and repurposing of compounds approved or under clinical trial (Lotfi Shahreza et al., 2018; Pawar, 2020; Shameer et al., 2015; Zhou et al., 2020). Some of the currently employed strategies to find drugs applicable in COVID‐19 have focused on host or virus targets, such as the receptor‐binding domain present in spike glycoprotein and angiotensin‐converting enzyme II (ACE2), which mediate virus−host cell interaction (Gordon et al., 2020; Hanson et al., 2020); enzymes from host or virus, such as hCatL (Zhao et al., 2021), TMPRSS2 (Hoffmann et al., 2020), main protease (Mpro) (Jin et al., 2020), papain‐like protease (PLpro) (Shin et al., 2020), and RNA‐dependent RNA polymerase (RdRp) (Elfiky, 2020); and reversion of the host gene expression signature of SARS‐CoV‐2‐infected cells (Belyaeva et al., 2021; Galindez et al., 2021; Riva et al., 2020). This study used in silico predictions to search for compound candidates that could concomitantly reverse the SARS‐CoV‐2 gene expression induced in host cells, including IL‐6, and target enzymes essential to the SARS‐CoV‐2 life cycle, followed by biological validation.
2. MODELS AND METHODS
2.1. Identification of signatures from SARS‐CoV‐2 in vitro infection model and search for compounds with reversed viral infection signature
The gene expression signature from the SARS‐CoV‐2‐infected primary human lung epithelium cell line (NHBE) was obtained from Blanco‐Melo and coauthors (Blanco‐Melo et al., 2020), by filtering differentially expressed genes (DEGs) after differential expression analysis from independent biological triplicates of SARS‐CoV‐2 (USA‐WA1/2020 strain)‐infected and mock‐treated cells. The signature was constructed based on DEGs selected considering the absolute value of fold change in log2 scale higher than 1 (|log2(FC)| > 1), and significance accepted at adjusted p value (p adj) smaller than .05 (p adj < .05), the p adj means the correction of nominal p values determined by differential expression analysis employing the Benjamini and Hochberg (BH) method (Benjamini & Hochberg, 1995) to minimize the false discovery ratio in the selection of DEGs. The signature was submitted to overrepresentation analysis using Reactome pathways (Jassal et al., 2020) and cluster Profiler R package (T. Wu et al., 2021) to find enriched biological pathways due to viral infection, within BH adjusted p < .05.
We used the obtained signature containing upregulated and downregulated genes to search for compounds with reversed gene expression signatures compared to viral infection. For this purpose, we explored the LINCS L1000 data set (Subramanian et al., 2017) using the web‐based search engine application called L1000CDS2 (Duan et al., 2016) (https://amp.pharm.mssm.edu/L1000CDS2/). We used the upregulated and downregulated genes as the entry point into L1000CDS2, generating a list of 50 best‐ranked signatures to reverse the input signature. These were ranked based on decreasing order of search score (Q score), defined as the overlap between input and the signature DEGs divided by the effective input, which is the intersection length between the input DEGs and the LINCS L1000 genes. In this data set, some compounds presented multiple signatures defined at different concentrations. We also created a new score for compounds based on biological aspects (B score). First, a weighting factor (WF) for each DEG observed in enriched biological pathways perturbated by SARS‐CoV‐2 infection was defined by multiplying the DEG occurrence in all enriched pathways by |log2(FC)|. Next, the B score for each drug was defined as the sum of WFs for the DEGs identified in the drug's signature normalized by the sum of all WFs. Thus, for each compound, the values range from 0 to 1, where 0 indicated no reversion and 1 indicated total reversion of enriched biological pathways. We obtained the graphical representation of drug signature data and scores with pheatmap (Kolde, 2019) and ggplot2 packages (Wickham, 2016) in R software (R Core Team, 2020).
2.2. Docking of selected compounds on SARS‐CoV‐2 protein targets
The compounds capable of reversing the SARS‐CoV‐2 infection signature (selected in Section 2.1) were submitted to molecular docking on the catalytic site of M.pro Here we used ensemble docking to increase the sampling and avoid individual conformational bias. The multiple Mpro structures were obtained from Protein Data Bank(Berman et al., 2000) (Supporting Information: Table S1). We removed the heteroatoms from M,pro then the resulting structures were repaired, and the energy was minimized with FoldX (Schymkowitz et al., 2005), and we used AutoDockTools (Morris et al., 2009) to prepare input structures for docking analysis and grid box definitions (Supporting Information: Table S1). The docking analysis we carried out using Autodock Vina (Trott & Olson, 2010) with an exhaustiveness parameter equal to 20. We plotted graphical energy results with the ggplot2 R package and the molecular model structures we obtained with Pymol (Schrodinger, 2015) and Discovery Studio Visualizer® (version‐2020) (Biovia, 2020).
2.3. The rationale for selecting a predictable candidate lead drug for experimental validation
The selection of candidate compounds in silico for their biological validation considered their abilities to reverse the genetic signature of SARS‐CoV‐2 infection and binding affinities to molecular viral targets. Briefly, 10 best‐repurposed compounds were selected based on Q score values. A compound with high predicted median binding affinity energy for MPro—and thereby with high potential to inhibit the functions of this molecular target—was selected using molecular docking data.
2.4. Effects of celastrol on SARS‐CoV‐2 recombinant M,pro PL,pro papain, and human cathepsin L (hCatL)
Recombinant Mpro and PLpro were expressed from E. coli as previously described (Freire et al., 2021; L. Zhang et al., 2020). hCatL and papain we acquired from Sigma. Fluorescence resonance energy transfer (FRET) peptides corresponded to cleavage at the nonstructural proteins (nsp) sites were synthesized using published protocols (Carmona et al., 2006; Korkmaz et al., 2008) and Z‐FR‐MCA we obtained from Sigma. All buffer salts were reagent‐grade and purchased from Thermo Fisher Scientific or Sigma‐Aldrich. The hydrolytic activities of recombinant Mpro and PLpro we assayed with FRET peptides whose sequences are those of native viral polyprotein processing sites. For Mpro the substrate was Abz‐SAVLQSGFRK(Dnp)‐NH2 [Abz = ortho‐aminobenzoic acid, fluorescent group; and K(Dnp) = N‐ε−2,4‐dinitrophenyl‐l‐lysine, the fluorescence quencher group], and the peptide SAVLQSGFR, which corresponds to the sequence between nsp4 and nsp5 with cleavage at Q‐S peptide bound. For PL,pro the substrate was Abz TLKGGAPIK‐Q‐EDDnp [Q‐EDDnp = N‐(2,4‐dinitrophenyl)‐ethylenediamine, attached to glutamine], and the peptide TLKGGAPIK, which corresponds to the sequence between nsp2 and nsp3 with cleavage at G‐A peptide bound. Details about the uses of these FRET peptides were described in the literature (Carmona et al., 2006; Korkmaz et al., 2008; Okamoto et al., 2010). The assay conditions were: (a) for Mpro and PLpro: buffer 50 mM Tris 1 mM EDTA pH 7.5, enzyme concentration 54 nM for M,pro 2.5 µM for PLpro; (b) for hCatL: buffer 100 mM sodium acetate 1 mM EDTA pH 5.0, enzyme concentration 9.0 nM, and Z‐FR‐MCA as substrate; (c) for papain: buffer 100 mM sodium phosphate, 0.1 mM EDTA pH 6.0, enzyme concentration 87.5 nM, and Z‐FR‐MCA as substrate. Before the assays, we activated hCatL and papain at 25°C in its appropriate buffer with 5 mM DTT for 10 min, followed by gel filtration to remove excess DTT and all assays were performed in degassed buffer solutions.
All assays were performed in a 96‐well black plate (Greiner). The enzymes, celastrol, previously diluted in DMSO, and concentration adjusted with appropriate assay buffer to give a total reaction volume of 200 μl. After 10 min of preincubation at 37°C, we started the reaction by adding the respective substrate. For Mpro and papain, we also assayed longer preincubation times in order to verify the inhibition mechanism and evaluated the stability of the complex celastrol‐Mpro and celastrol‐papain using the same procedure reported for the inhibition of Mpro by dalcetrapib (Niesor et al., 2021). We followed the hydrolysis progress using a SpectraMax M2e (Molecular Devices) with 320 nm excitation and 420 nm emission for FRET peptides and 360 nm excitation and 480 nm emission for MCA substrates.
2.5. Preparation of viral stocks
We used two SARS‐CoV‐2 lineages: one obtained from clinical isolates (SARS‐CoV‐2 Brazil/SPBR‐02/2020 strain) from RT‐PCR‐confirmed COVID‐19 patients; and the SARS‐CoV‐2 gamma variant (MAN_87209) detected in Manaus, State of Amazonas, Brazil, associated with high transmissibility and immune evasion (Faria et al., 2021; Sabino et al., 2021). Here, we named these lineages SARS‐CoV and SARS‐CoV‐2 gamma variant, respectively, and propagated them in monkey Vero CCL‐81 cells (kidney) under strict biosafety level 3 conditions. Briefly, for initial viral passages, Vero CCL‐81 cells were cultured in Dulbecco minimal essential medium (DMEM) supplemented with 10% heat‐inactivated fetal bovine serum (FBS) and antibiotic/antimycotic mix (10,000 U/ml penicillin and 10,000 μg/ml streptomycin). After viral inoculum (1:100 ratio) to the cells, the cultures were incubated (48 h, 37°C, 5% CO2 humidified atmosphere) in DMEM without FBS but supplemented with antibiotic/antimycotic mix and trypsin‐protease inhibitor, L‐1‐tosylamide‐2‐phenylethylchloromethyl ketone (TPCK) host cell treatment (1 μg/μl) to optimize virus adsorption to the cells (Banerjee et al., 2020). After confirming the cytopathic effects of the viral preparation using an inverted Olympus ix51 microscope, infected Vero CCL‐81 cells were detached by scraping, harvested, and centrifuged (10,000×g, 10 min, room temperature). The resulting supernatants we stored at −80°C until use. Finally, virus titration was performed on Vero CCL81 cells using standard limiting dilution to determine the 50% tissue culture infectious dose (TCID50) of viral stock (Harcourt et al., 2020; Reed & Muench, 1938).
2.6. In vitro SARS‐CoV‐2 infection
The SARS‐CoV‐2 infection we assessed in vitro in four cell lines: Vero CCL‐81, human Calu‐3 (lung), and human Caco‐2 (colon). Cells were seeded into 24‐well plates (80,000 cells/well) to ensure 90% of confluence on the day of inoculation/infection. The three cell lines were infected with SARS‐CoV‐2 and treated with celastrol. Cells were infected with SARS‐CoV‐2 at a multiplicity of infection 1.0 in 500 µl of infection media composed of DMEM without FBS, 1% antibiotic/antimycotic mix, and 1 μg/μl trypsin‐TPCK. After 2 h of incubation, the supernatant containing SARS‐CoV‐2 was removed and replaced by celastrol (125, 250, 500, and 1000 nM) or vehicle (0.05% DMSO) diluted in a fresh medium, followed by 48 h of incubation at 37°C and under 5% CO2. Photomicrographs were taken using the Olympus ix51 inverted microscope and analyzed using the QCapture Pro 6.0 software under ×200 magnification (QImaging) (Kang et al., 2020), to examine whether celastrol interfered with SARS‐CoV‐2 cytopathic effects in Vero CCL‐81 cells. The supernatants were collected for RNA extraction, and viral load was quantified using a standard curve. All the infections were conducted in technical triplicate. In this work, the SARS‐CoV‐2 gamma variant was used only to infect Calu‐3 cells, using the same procedure for the SARS‐CoV‐2 lineage described above.
2.7. Cell viability
We determined the cytotoxicity of celastrol (Sigma‐Aldrich) to Vero CCL‐81, Calu‐3, and Caco‐2 using the Alamar Blue Cell Viability protocol (Thermo Fisher Scientific), according to the manufacturer's instructions. Briefly, the cells were seeded into a 96‐well plate to grow as monolayers and treated with celastrol (250 or 1000 nM) in DMSO (0.05%) or DMSO (5%, v/v; cell death reference) for 24 h. Alamar Blue reagent (10%, v/v) was added to the cells, which were later incubated on a plate at 37°C for 4 h. Median fluorescence intensity was measured using the SpectraMax i‐3 (Molecular Devices) microplate reader, with excitation and emission wavelengths set at 530 and 590 nm, respectively. The mean value from the control (untreated cells) was set as 100%, and the viability of cells from each treatment condition was calculated relative to this value in triplicate.
2.8. RT‐PCR
SARS‐CoV‐2 and SARS‐CoV‐2 gamma variant genomes were quantified by RT‐PCR using the same primer‐probe sets for N2 and RNAse‐P housekeeping gene, following USA‐CDC protocols (Table 1) (Lu et al., 2020). We determined the viral genome load from in vitro infection assays testing N2 and RNAse‐P gene by one‐step real‐time RT‐PCR using total nucleic acids extracted with Trizol® (Invitrogen) from 250 µl of culture supernatants. All RT‐PCR tests were carried out using the Step‐One Plus real‐time PCR thermocycler (Applied Biosystems). Briefly, we used 100 ng of RNA for genome amplification, mixed with specific primers (20 µM), probe (5 µM), and TaqPath 1‐Step qRT‐PCR Master Mix (Applied Biosystems) in the following conditions: 25°C for 2 min, 50°C for 15 min, 95°C for 2 min, followed by 45 cycles of 95°C for 3 s and 55°C for 30 s.
Table 1.
Primers/probe sequences for detection of SARS‐CoV‐2 and SARS‐CoV‐2 gamma variant genomes and housekeeping gene
| Primer | Sequence | |
|---|---|---|
| Forward | 5′‐TTA CAA ACA TTG GCC GCA AA‐3′ | |
| N2 | Reverse | 5’‐GCG CGA CAT TCC GAA GAA‐3′ |
| Probe | 5’‐FAM‐ACA ATT TGC CCC CAG CGC TTC AG‐BHQ1‐3′ | |
| Forward | 5′‐AGA TTT GGA CCT GCG AGC G‐3′ | |
| RNAse‐P | Reverse | 5′‐GAG CGG CTG TCT CCA CAA GT‐3′ |
| Probe | 5′‐FAM – TTC TGA CCT GAA GGC TCT GCG CG – BHQ‐1‐3′ |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
A plasmid standard curve, previously plotted, provided the SARS‐CoV‐2 or SARS‐CoV‐2 gamma variant viral load. A 944 bp amplicon was inserted into a TA cloning vector (PTZ57R/T CloneJetTM Cloning Kit Thermo Fisher®), starting from residue 14 of the N gene, including all three sets of primers/probes designed by the CDC protocol (N1, N2, and N3). We quantify the amount of virus produced, with tenfold serial dilution of the plasmid in the range from 106 to 1 plasmid copy. The coefficient of determination (R 2) for the plasmid standard curve was 0.999, with efficiency above 91% reached using any set of primers/probes (Martins et al., 2021).
2.9. IL‐6 quantification
IL‐6 levels were quantified in cell culture supernatants from SARS‐CoV‐2‐infected Caco‐2 and Calu‐3 cells using the Human DuoSet ELISA assay kit (R&D Systems) according to the manufacturer's instructions. The IL‐6 detection limit was 9.38−1200 pg. ml−1.
2.10. Statistical analysis
For obtention of IC50 of celastrol in enzymatic inhibition assay, we plotted and modeled each sample's concentration‐response data by a four‐parameter logistic fit. Raw plate reads for each titration point were first normalized relative to a control containing no inhibitor (100% activity, full activity). We performed data normalization, visualization, and curve fitting using Grafit 5.0 software (Erithacus Software). Raw data for viral load from RT‐PCR were transformed into log scale, normalized, and analyzed by nonlinear regression using dose−response curve fitting and the equation log (inhibitor) versus normalized response with variable slope. All the experimental data are expressed as mean ± standard error of the mean, and they were plotted and analyzed using GraphPad Prism 7 software (Inc., 2016). Statistical significance was analyzed by one‐way analysis of variance followed by Tukey's posttest for multiple comparisons. Differences were considered significant at p < .05. The p values were labeled as *p < .05, **p < .01, ***p < .001, and ****p < .0001.
3. RESULTS
3.1. Identification of potential candidates for drug repurposing to treat COVID‐19
We determined the genetic signature of SARS‐CoV‐2‐infected NHBE cells based on transcriptome data reported in the literature (Blanco‐Melo et al., 2020). Then, we used |log2(FC)| > 1 and p adj < .05 as selection criteria, and identified 64 upregulated and 8 downregulated DEGs in infected NHBE cells (Supporting Information: Table S2). The overrepresentation analysis of selected DEGs in Reactome pathways annotations indicated the existence of 22 significantly enriched biological pathways (Figure 1a, Supporting Information: Table S3). The top enriched pathways were related to interleukin, chemokine, and interferon signaling, according to literature data (Blanco‐Melo et al., 2020), and were closely related to biological pathways involved in SARS‐CoV‐2 human infection (Del Valle et al., 2020; Desai et al., 2020; Fajgenbaum & June, 2020; Nienhold et al., 2020).
Figure 1.
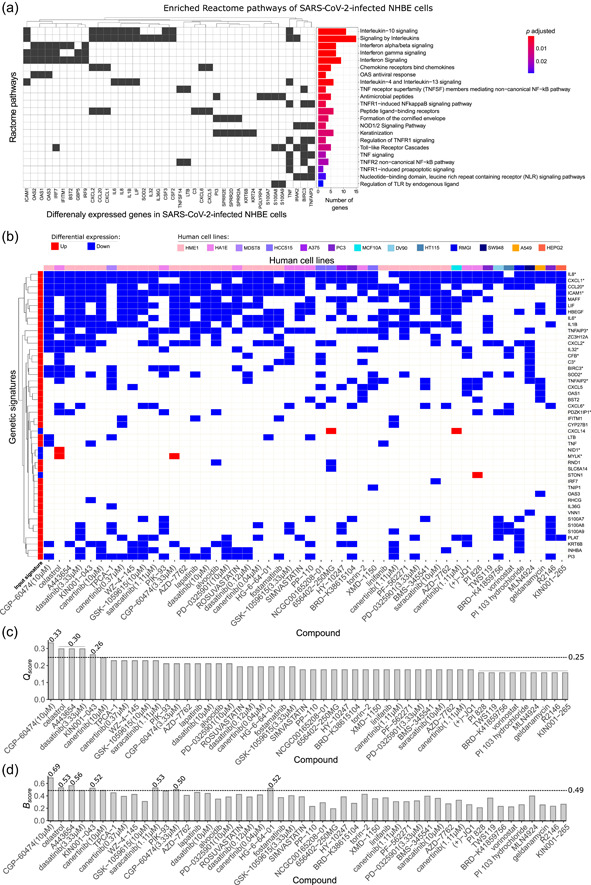
Assessment of gene signature in SARS‐CoV‐2‐infected human bronchial epithelial cells (NHBE) for drug discovery to treat COVID‐19. (a) Enriched biological pathways associated with COVID‐19 from Reactome annotations (p adj < .05) from DEGs identified in NHBE‐infected cells transcriptome data. The genes that enriched each pathway (left) are indicated together with statistical results and pathway description (right). (b) Heatmap of genes from the 50 best‐ranked compound signatures that reversed the genetic signature of SARS‐CoV‐2‐infected NHBE cells in decreasing order of Q score. The signature map annotations are related to up‐ and downregulated genes, and cell lines are indicated in different colors. (c) Compounds in decreasing order of Q score following the output of L1000CDS.2 A dashed line indicates the mean Q score (0.26) threshold. Equal Q score values are displayed over the bars. (d) B score for each compound, considering the enrichment analysis and reverse signature. The dashed line corresponds to the mean B score (0.49). *Genetic signature that justified the biological validation of celastrol. COVID‐19, coronavirus disease 2019; DEGs, differentially expressed genes; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Considering that the genetic signature described here was composed of DEGs associated with COVID‐19 pathophysiology, we searched for candidate compounds that could reverse the genetic signature of NHBE‐infected cells using the L1000CDS2 protocol to define the Q score. The top 50 better‐scored compound signatures derived from 39 compounds were able to modify the expression pattern of 46 DEGs (42 upregulated and 4 downregulated) from the original input of 72 DEGs (Figure 1b; Supporting Information: Table S4). Based on the Q score values from these 50 signatures, a medium value score was defined as 0.26 (Figure 1c). CGP‐60474, a cyclin‐dependent kinase inhibitor (Stanetty et al., 2005), exhibited the best‐ranked Q score (0.33): it downregulated 19 DEGs that were upregulated by SARS‐CoV‐2 infection. The second best‐ranked Q score (0.30) was associated with three compounds that modified the expression levels of 17 DEGs induced by SARS‐CoV‐2: (i) celastrol, a pentacyclic triterpenoid derived from Tripterygium wilfordii Hook F with anti‐inflammatory and anticancer properties (Kannaiyan et al., 2011); (ii) A443654, a potent inhibitor of all members of the protein kinase B (Akt/PKB) family (Luo et al., 2005); and (iii) dasatinib, a small‐molecule inhibitor of multiple tyrosine kinases (Kantarjian et al., 2006) (Figure 1c). In the set of best‐ranked drugs by Q score, CGP‐60474, celastrol, and A443654 also presented B score values higher than the mean score of 0.49 (Figure 1d), suggesting that they are potential reversers of biological pathways related to SARS‐CoV‐2 infection.
3.2. Selection of compounds that bind to viral molecular target Mpro among those that reverse SARS‐CoV‐2‐induced genetic signature
We have searched for candidate molecules with two potential actions: to revert the genetic signature of SARS‐CoV‐2 infection and interfere with critical events of viral infection. For this purpose, 39 compounds previously identified as reversers of the viral genetic signature were submitted to molecular docking on the catalytic site of Mpro (Figure 2a). We determined the pose with the highest binding affinity energy (ΔG bind in kcal. mol−1) for each ensemble's structures of all selected compounds (Figure 2b). The median ΔG bind values for binding to Mpro ranged from −8.7 kcal. mol−1 for the leucine‐rich repeat kinase inhibitor XMD‐1150 (J. Wang et al., 2018) to −5.7 kcal. mol−1 for the histone deacetylase inhibitor vorinostat (McGuire & Lee, 2010). The ensemble dockings led to energetic variations but not to wide dispersion of ΔG bind values, demonstrating that certain molecules maintained their high binding affinity to different conformations of the studied targets. Following the criteria for selecting a candidate to lead the drug for experimental validation as rationalized in methods, we chose the compound with high predicted median binding affinity energy for MPro among the first 10 compounds with high Q score values. Celastrol exhibited the most attractive ΔG bind median energy value for Mpro (−8.3 kcal. mol−1) out of the first 10 best‐ranked molecules based on Q score. The chemical pattern of celastrol‐target interactions was composed of conventional hydrogen bonds, carbon‐hydrogen bonds, Pi interactions, and van der Waals interactions that favored a more attractive binding affinity of celastrol to Mpro with ΔG bind equal to −8.7 kcal. mol−1 (Figure 2c,d). The celastrol binding to Mpro catalytic site was mediated by a sigma Pi interaction between the methyl group of the E‐ring and His41 and a hydrogen bond of the OH group of the A‐ring with Thr25. The mean atomic distance between the B‐ring C6 and the Cys145 sulfur atom—a critical amino acid of the Mpro catalytic site—was 0.63 nm (range: 0.43−1.33 nm) (Figure 2d), suggesting an appropriate geometry for the formation of a covalent bond. In addition, celastrol exhibited one of the highest B score values (0.53) among the 50 signatures analyzed, indicating that this compound had a great potential to revert the genetic signature of SARS‐CoV‐2 infection (Figure 1d). Some molecules with high affinity to the Mpro such as XMD‐1150 were poorly ranked in the reverse signature strategy but they could be relevant due to their predicted binding abilities. The ΔG bind, Q score, and B score values of celastrol supported its selection for biological validation.
Figure 2.
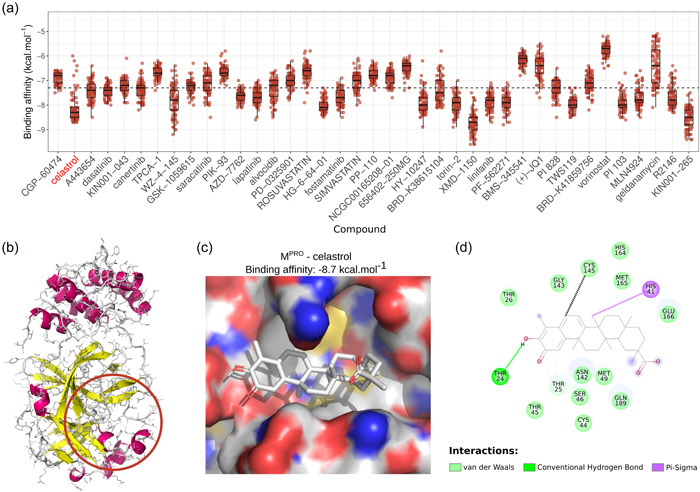
Docking analysis of molecular interactions between Mpro and molecules capable of reversing SARS‐CoV‐2 genetic signature. (a) Boxplots with the affinity binding energies (kcal. mol−1) were obtained from docking analysis between several structural conformations of 39 molecules and each viral site of 83 Mpro structures. The reverser compounds were sorted based on decreasing order of Q score that indicated their potential to revert the genetic signature of SARS‐CoV‐2 infection. Dotted lines indicate median affinity binding energy (−7.3 kcal. mol−1) considering all investigated drugs. (b) Representative structure of Mpro where atoms are represented as lines and secondary structures as a cartoon with helices highlighted in magenta and sheets in yellow. Colored line circles in brown indicate the binding site region used for docking. (c) 3D and (d) 2D representations detailing configuration and chemical interactions between the best‐ranked pose for celastrol in M.pro 2D target‐drug interaction was constructed using the Discovery Studio® software (version‐2020). The distance between the B‐ring C6 and the sulfur atom of the Cys145 residue, which may be related to a possible Michael adduct formation for the best energy poses in each Mpro structure, ranged from 0.43 to 1.33 nm, with an average value of 0.63 nm (black dashed line). M,promain protease; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.3. Celastrol inhibits the activity of papain, hCatL, and the SARS‐CoV‐2 recombinant cysteine proteases Mpro and PLpro
As expected, celastrol inhibited the enzymatic activity of papain, a reference cysteine protease (Mamboya & Amri, 2012). We examined whether this pentacyclic triterpene affected the hydrolytic activity of recombinant M,pro as part of the experimental validation of our in silico strategy, and the activity of PLpro and hCatL, which are other cysteine proteases involved in SARS‐CoV‐2 infection (Dominic et al., 2022; Shin et al., 2020; Zhao et al., 2021). The four cysteine proteases were treated with celastrol for 10 min, and the IC50 values are reported in Table 2. The mean IC50 values ranged from 6.6 to 9.6 μM, as determined using FRET assays. This validated our in silico selection once celastrol inhibited Mpro (IC50 = 6.6 ± 0.6 μM) at same order of model cysteine protease papain (IC50 = 7.0 ± 0.7 μM). In a subsequent time‐dependent inhibition experiment, more extended preincubation periods with celastrol reduced the IC50 values for Mpro and papain (Figure 3a,b), indicating that it was an irreversible inhibitor, as previously reported for other enzymes (Yan et al., 2002). Next, we analyzed the stability of the celastrol‐Mpro and celastrol‐papain complexes using the same procedure reported for the inhibition of Mpro by dalcetrapib (Niesor et al., 2021), where inhibition of protease activity was maintained even after high dilution followed by ultrafiltration. The steps of this procedure and the persistent inhibition of Mpro and papain activity by celastrol after dilution/filtration cycles are reported in Figure 4. Additionally, molecular docking of celastrol resulted in poses with favorable binding energies over catalytic sites of papain (−7.3 kcal. mol−1), PLpro (−7.4 kcal. mol−1), and hCatL (−8.1 kcal. mol−1) (Supporting Information: Figure S1).
Table 2.
Inhibition of the activity of the cysteine proteases M,pro papain, PL,pro and hCatL by celastrol
| Enzyme | IC50 (μM) |
|---|---|
| Mpro | 6.6 ± 0.6 |
| Papain | 7.0 ± 0.7 |
| PLpro | 8.9 ± 0.8 |
| hCatL | 9.6 ± 0.9 |
Abbreviations: hCatL, human cathepsin L; M,pro promain protease; PL,pro papain‐like protease.
Figure 3.
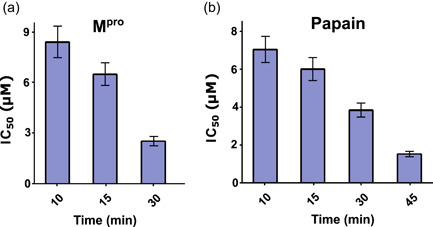
Time‐dependent inhibition of Mpro (a) and papain (b) activity by celastrol, expressed as IC50 value. M,pro main protease.
Figure 4.
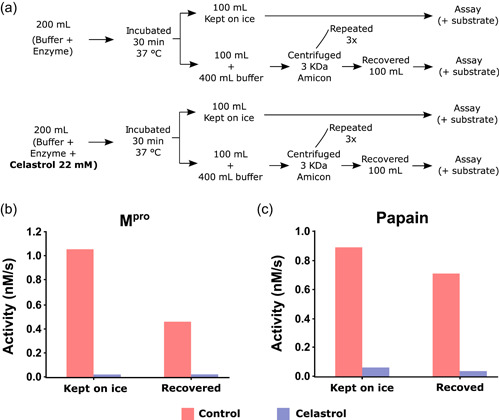
Persistent inhibition of Mpro and papain activity by celastrol. (a) Assay procedure to determine the irreversibility of Mpro and papain inhibition by celastrol. The enzymes were incubated for 30 min with buffer (control) or 22 mM celastrol, as indicated in the Methods section. Part of the incubation mixture was kept on ice and the other part was submitted to three ultrafiltration cycles to remove free celastrol and concentrate the enzymes. The Mpro and papain activity was determined in the concentrate. (b) Mpro and (c) papain activity before and after repeated dilution/filtration cycles. M,pro main protease.
3.4. Celastrol inhibits virus propagation and IL‐6 secretion by infected cells without cytopathic effect in vitro
3.4.1. Celastrol reduces SARS‐CoV‐2 and SARS‐CoV‐2 gamma variant viral load on infected cells
In silico analysis indicated that celastrol is a potential lead compound candidate for the development of drugs to treat COVID‐19 due to its ability to reverse the genetic signature of SARS‐CoV‐2 infection and interact with high affinity with critical molecular viral targets. This in silico prediction was validated using in vitro SARS‐CoV‐2 cell propagation inhibition assays. Celastrol significantly reduced viral load in a monkey cell line Vero CCL‐81 and two human cell lines (Caco‐2 and Calu‐3) (Figure 5a−d). Interestingly, the anti‐SARS‐CoV‐2 effect of celastrol was concentration‐dependent and it drastically inhibited viral replication in Vero CCL‐81 and Calu‐3 cells when tested at 1000 nM. Celastrol at higher concentrations strongly reduced SARS‐CoV‐2 gamma variant replication in Calu‐3 cells (Figure 5). The EC50 values of celastrol for each cell line ranged from 221 to 1000 nM (Supporting Information: Figure S2 and Table S5).
Figure 5.
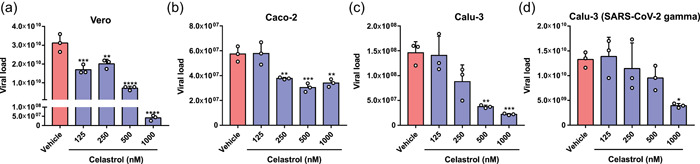
Celastrol suppresses SARS‐CoV‐2 in vitro propagation in nonhuman and human cell lines. RT‐PCR quantification of SARS‐CoV‐2 RNA in supernatants from the infected cell lines (a) Vero CCL‐81, (b) Caco‐2, and (c) Calu‐3, and (d) Calu‐3 infected with SARS‐CoV‐2 gamma variant treated with celastrol at concentrations of 125, 250, 500, and 1000 nM. DMSO solution (0.05%; vehicle) was used as a negative control. Cells were infected using MOI = 1.0 for 2 h and then treated with celastrol for 48 h. The detection levels of SARS‐CoV‐2 RNA were performed in the supernatants of cultures and expressed in viral load using a standard curve described in Section 2. Statistical differences between celastrol treatments and the negative control were analyzed by ANOVA followed by Tukey's posttest. The significance levels were indicated as *p < .05, **p <.01, ***p < .001, and ****p < .0001. ANOVA, analysis of variance; MOI, multiplicity of infection; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
To examine whether cytotoxicity mediated the antiviral effect of celastrol, we determined the cell viability of the three cell lines treated with celastrol at two concentrations, 250 and 1000 nM, which promoted its minimal and maximum antiviral action in human cell lines, respectively. Celastrol‐treated cells and the negative control had similar viability levels, of nearly 100% (Figure 6a−c). Treatment with 5% DMSO (positive control) decreased cell viability by more than 80%. Celastrol also significantly reduced SARS‐CoV‐2 cytopathic effect in a concentration‐dependent manner (Supporting Information: Figure S3).
Figure 6.
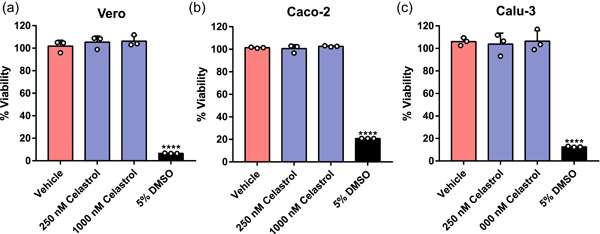
Celastrol does not affect cell viability. Alamar Blue assay was used to measure the potential cytotoxic effect of celastrol. The monkey cell line (a) Vero CCL‐81 and the human cell lines (b) Caco‐2 and (c) Calu‐3 were treated with celastrol at 250 and 1000 nM, for 48 h. DMSO solutions at 0.05% and 5.0% were used as the negative and positive controls of cell death, respectively. Statistical analysis was performed using ANOVA followed by Tukey's posttest to compare treatment with celastrol and 5.0% DMSO. ****p < .0001. ANOVA, analysis of variance.
3.4.2. Celastrol lowers IL‐6 production in SARS‐CoV‐2‐infected human cell lines
SARS‐CoV‐2 infection of human cell lines increased gene expression of inflammatory mediators, such as IL‐6, as demonstrated by in silico analysis, and celastrol reversed this effect (Figure 1). To test this prediction, we determined IL‐6 levels in the supernatant of SARS‐CoV‐2‐infected cells treated or not with celastrol. This compound at 500 and 1000 nM significantly decreased IL‐6 production by Caco‐2‐infected cells, and at 1000 nM it decreased IL‐6 production by Calu‐3‐infected cells (Figure 7a,b). Curiously, Caco‐2‐infected cells produced around 10‐fold lower IL‐6 levels than Calu‐3‐infected cells. This suggested that this phenomenon may be associated with the lower viral load in culture supernatants of Caco‐2 cells compared with samples from Calu‐3 cells (Figure 5b,c), which was also observed in hospitalized patients' samples (Brasen et al., 2021).
Figure 7.
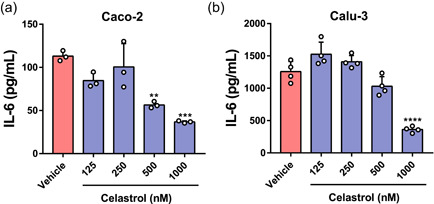
Celastrol inhibits IL‐6 production in SARS‐CoV‐2‐infected human cell lines. ELISA quantification of IL‐6 in the culture supernatants from the human cell lines (a) Caco‐2 and (b) Calu‐3, infected with SARS‐CoV‐2 (MOI = 1.0) for 2 h. Then, after the washing step, infected cells were treated with celastrol for 48 h. DMSO solution at 0.05% (vehicle) was used as the negative control. Statistical differences between celastrol treatment and control were analyzed by ANOVA followed by Tukey's posttest. The significance levels were indicated as **p < .01, ***p < .001, and ****p < .0001. The results are expressed as mean ± standard error of the mean (SEM) of IL‐6 (pg/ml). They are representative of two independent experiments with Caco‐2 cells with three replicates and one experiment with Calu‐3 cells with four replicates. ANOVA, analysis of variance; IL‐6, interleukin‐6; MOI, multiplicity of infection; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
4. DISCUSSION
Here, we identified 39 potential compound candidates to combat SARS‐CoV‐2 infection based on in silico prediction of their abilities to revert the genetic signature of SARS‐CoV‐2‐infected cells and bind with high affinity to viral targets. One of the in silico best‐ranked candidates was celastrol, which could inhibit the virion particles release and IL‐6 secretion by SARS‐CoV‐2‐infected cells. This combination of computational and experimental analyzes corroborated the literature predictions about the potential use of celastrol to face COVID‐19 based on its anti‐inflammatory and antiviral properties (Habtemariam et al., 2020; Xian et al., 2020). Other in silico best‐ranked candidates with antitumor, kinase‐inhibition, and anti‐inflammatory properties have been proposed to treat COVID‐19, such as CGP‐60474, A‐443654, alvocidib, canertinib, dasatinib, vorinostat, and geldanamycin (Belyaeva et al., 2021; Gil et al., 2020; He & Lana, 2020; Taguchi & Turki, 2020).
The perturbed gene expression during SARS‐CoV‐2 infection in vitro modulates biological processes of clinical relevance, such as cytokine‐mediated gene expression, type I interferon pathways, and antiviral responses as detected in our current analysis and literature (Blanco‐Melo et al., 2020). As expected, genes of proinflammatory cytokines (IL‐1β, IL‐6, IL‐8, TNF‐α, and others), chemokines (CXCL1 and CCL20), and some NF‐κB pathway components were upregulated, and are related to the severity and progression of COVID‐19 (Chang et al., 2004; Fajgenbaum & June, 2020; Lin et al., 2020; Rauzi et al., 2016; Tse et al., 2004).
Celastrol was the second best‐ranked compound able to reverse the genetic signature in SARS‐CoV‐2‐infected cells, in agreement with its reported anti‐inflammatory properties (Cascão et al., 2017). This triterpene also dampens HIV‐1 Tat‐induced inflammatory responses, inhibits the production of proinflammatory chemokines, such as CXCL10, IL‐8, and MCP‐1 (Youn et al., 2014), and suppresses replication of Dengue virus serotype 1−4 by promoting IFN‐α expression and stimulating downstream antiviral responses (J.‐S. Yu et al., 2017). Celastrol suppresses the production of cytokines during cytokine storm and chemokines related to worse disease prognosis, such as IL‐8, IL‐6, CXCL1, and CCL20 (Demedts et al., 2007; Liang et al., 2016; Tisoncik et al., 2012), lowering the anti‐inflammatory levels markers (H. Wang et al., 2020), and stimulates type I interferon production and expression of interferon‐stimulated genes against Dengue virus infection (J.‐S. Yu et al., 2017).
Considering that the direct interaction between celastrol and viral targets can also mediate its antiviral activity (Caruso, Singh, et al., 2020; Gupta et al., 2021), we demonstrated its potential ability to bind to essential targets to the viral life cycle. Interestingly, Chen and collaborators (Chen et al., 2022) experimentally demonstrated that celastrol binds to the S1 domain of SARS‐CoV‐2 spike protein (K d = 1.712 µM) and inhibits the viral infection rate of ACE2 overexpressing 293T cell infected with lentivirus particles pseudotyped with SARS‐CoV‐2 spike. A previous study has described the celastrol interaction with the catalytic site of the SARS‐CoV Mpro (Ryu et al., 2010), and here, we demonstrated the irreversibility of celastrol inhibition. Docking simulations on the catalytic site of Mpro revealed that the celastrol binding energy was higher than that of most compounds tested here. Our models suggested that the high binding affinity of celastrol to Mpro involved the sigma Pi interaction between the E‐ring O2 and His41, associated with the hydrogen bond between Thr25 and the hydroxyl group of A‐ring C3, in agreement with literature reports (Caruso, Singh, et al., 2020; Gupta et al., 2021). In addition, our atomic distance analysis suggested that B‐ring C6 interacted with Cys145—a critical amino acid of Mpro catalytic site—by forming Michael's adduct; this inhibition mechanism of celastrol was also proposed in other molecular targets (D. Zhang et al., 2018; Sreeramulu et al., 2009). We also demonstrated that celastrol inhibited two other cysteine proteases PLPro and hCatL, which displayed crucial roles in SARS‐CoV‐2 infection and COVID‐19 aggravation (Shin et al., 2020; Zhao et al., 2021). PLPro is an essential SARS‐CoV‐2 enzyme for processing viral polyproteins and evasion of host antiviral immune response (Shin et al., 2020) and a critical antiviral drug target (Fu et al., 2021). Otherwise, hCatL acts in proteolysis of protein antigens produced by pathogen endocytosis, and its circulating level was positively associated with COVID‐19 severity and an important target for drug development (Zhao et al., 2021). Interestingly, celastrol was able to promote inhibition of host and virus enzymes, a promising antivirus drug for treating COVID‐19, including hCatL and MPro (Dominic et al., 2022; Hashimoto et al., 2021; Ma et al., 2022). The anti‐inflammatory activity of celastrol is associated with downregulation of the NF‐kB pathway mediated by suppression of IKK activation, probably due to the formation of Michael's adduct between its quinone methide and Cys179 from IKK (Caruso, Rossi, et al., 2020; Lee et al., 2006). In silico studies have reported that other quinone derivatives exhibit antiviral properties associated with the ability to form Michael's adduct (Caruso, Rossi, et al., 2020), such as the glucocorticoids methylprednisolone and dexamethasone used to treat severe COVID‐19 patients (Shen et al., 2020; Tomazini et al., 2020).
Next, we demonstrated that celastrol exerted antiviral and anti‐inflammatory effects in vitro, which biologically validated our in silico findings. Celastrol (250−1000 nM) reduced the viral progeny of SARS‐CoV‐2 in Vero CCL‐81 cells and in the pulmonary and intestinal epithelial human cell lines Calu‐3 and Caco‐2, respectively. The antiviral effect of celastrol is selective since it does not inhibit influenza A virus replication (H1N1; PR8) in Madin‐Darby Canine kidney cell line (MDCK) when tested at 150−600 nM (Khalili et al., 2018), but it significantly reduces HIV replication in U937 cells at 150 nM (Narayan et al., 2011), as we found for SARS‐CoV‐2. Celastrol at 1000 nM drastically reduced SARS‐CoV‐2 gamma variant replication in Calu‐3 cells. Interestingly, this lineage accumulates multiple mutations in the spike glycoprotein, including K417T, N501K, and E484K (Faria et al., 2021), similarly to other SARS‐CoV‐2 lineages that have been associated with high transmissibility, resistance to viral neutralization by convalescent sera, and patient's reinfection (Faria et al., 2021; Sabino et al., 2021). These data suggested that the antiviral action of celastrol modulated the replication of SARS‐CoV‐2 lineages with dangerous phenotypes, such as high transmissibility and/or immune escape.
As expected, the reuse of transcriptome data from SARS‐CoV‐2‐infected NHBE cells, a normal human bronchial epithelial cell line, evidenced enriched biological pathways associated with non‐hemostatic inflammation, including upregulation of the IL‐6 gene expression. A recent review paper from our team has reported the relationship between serum IL‐6 levels and different outcomes of COVID‐19 patients (Fraga‐Silva et al., 2021): compared with severe patients, critical and mild patients have higher and lower IL‐6 levels, respectively; and non‐survivors exhibit significantly higher IL‐6 levels than survivors with COVID‐19. In vitro assays using Calu‐3, A549, and NHBE cells have confirmed that SARS‐CoV‐2 infection induces IL‐6 production (Blanco‐Melo et al., 2020). As IL‐6 production is increased in both COVID‐19 patients and in vitro SARS‐CoV‐2 infection models, we examined whether celastrol altered the levels of this cytokine in vitro. Celastrol at 500 and 1000 nM downregulated the infection‐induced IL‐6 production in Caco‐2 and Calu‐3 cells, and such effect was associated with the decreased SARS‐CoV‐2 load. The inhibition of IL‐6 production by celastrol could be associated or not with the reduced viral load observed on infected cells. The modulatory effect of celastrol over IL‐6 release has been described in vitro under sterile inflammatory conditions (Venkatesha et al., 2011). In addition, this triterpene downregulates IL‐6 secretion and gene expression in PC‐3 prostate carcinoma cells, in an NF‐κB‐dependent manner (Chiang et al., 2014), suppresses LPS‐induced IL‐6 production in RAW264.7 macrophages (Kim et al., 2009), and decreases the IL‐6 concentration and mRNA expression but not the virus title and mRNA expression in influenza A‐infected MDCK cells (Khalili et al., 2018). Of note, celastrol reduced lung injury and the release of proinflammatory mediators into the pulmonary airways, including IL‐6 production, in an experimental model of acute respiratory distress syndrome (Majumder et al., 2014).
Our findings demonstrated that celastrol was able to reduce both SARS‐CoV‐2 viral load and IL‐6 production in two human cell lines, suggesting that this compound exerts anti‐SARS‐CoV‐2. Several pharmacological applications of celastrol are associated with COVID‐19 severity, including comorbidities, such as obesity, diabetes, hypertension, and metabolic syndrome (Liu et al., 2015; Nie et al., 2020; Ye et al., 2020; Y. Zhu et al., 2021), and pathological events such as non‐homeostatic production of proinflammatory cytokines, thrombus generation, and formation of neutrophil extracellular traps (Allen et al., 2019; Liu et al., 2019; Y. Yu et al., 2015). Celastrol is currently under clinical trials to treat a variety of diseases, including different types of cancer, neurodegenerative disorders, and inflammatory conditions, such as rheumatoid arthritis, psoriasis, and Crohn's disease (C. Wu et al., 2015; Fang et al., 2017; H.‐Y. Li et al., 2015; J. Li & Hao, 2019; Pinna et al., 2004).
Despite its potential therapeutic effects, there are still some limitations to the use of celastrol, such as its low solubility results in poor bioavailability, in vitro and in vivo toxicity, and adverse effects that remain to be evaluated (J. Wu et al., 2016; T. Zhang et al., 2008). Celastrol was not cytotoxic to all cell lines tested herein, even at the highest concentration (1000 nM), corroborating other reports using Caco‐2 cells (Z. Wang et al., 2018). At 1000 nM, this compound reduced the SARS‐CoV‐2‐cytopathic effect on Vero CCL‐81 cells. A recent study using a mouse model of acute toxicity has demonstrated that celastrol is safe even when orally administered at a high dose (62.5 mg/kg body weight) (Hu et al., 2020). Intraperitoneal administration of celastrol (0.25 mg/kg body weight) increases the gamma irradiated‐mice survival rate by around 70 (H. Wang et al., 2020). To surpass celastrol toxicity, solubility, and pharmacokinetic issues, several pharmaceutical approaches have been proposed, such as nanoencapsulation, liposomes, and sugar‐silica nanoparticles (Niemelä et al., 2015; Sanna et al., 2015; Wolfram et al., 2014). Celastrol has been considered a lead drug for several human illnesses, but its toxicity to humans remains to be determined (Bassanini et al., 2021; Cascão et al., 2017; Habtemariam et al., 2020).
To the best of our knowledge, the present study is the pioneer to use the combination between bioinformatic tools and biological approaches to demonstrate that celastrol inhibits the SARS‐CoV‐2 and SARS‐CoV‐2 gamma variant replication in human cell lines, and downregulates IL‐6 secretion from infected‐human cell lines, reinforcing that celastrol is a potential lead compound to design of new drugs to SARS‐CoV‐2 inhibition and to treat COVID‐19.
AUTHOR CONTRIBUTIONS
Carlos A. Fuzo and Marcelo Dias‐Baruffi designed the study. Carlos A. Fuzo performed in silico analyzes. Ronaldo B. Martins, Juliano P. Souza, Thais M. Lima, Martin K. Amstalden, Thais Canassa De Leo, Marcelo Dias‐Baruffi, and Eurico Arruda designed and executed the experimental SARS‐CoV‐2 infection. Débora Noma Okamoto, Maria Aparecida Juliano, and Luiz Juliano designed and executed the hydrolytic activities of cysteine proteases. Thais F. C. Fraga‐Silva and Vania L. D. Bonato designed and executed the IL‐6 quantification. All authors contributed to writing the manuscript. Marcelo Dias‐Baruffi and Carlos A. Fuzo wrote the final version of the manuscript. Carlos A. Fuzo, Luiz Juliano, Vania L. D. Bonato, Eurico Arruda, and Marcelo Dias‐Baruffi are senior researchers and have made substantial intellectual and material contributions to the work. All authors have approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
We thank Professor Dra. Ester C. Sabino (University of São Paulo‐Brazil) for providing the SARS‐CoV‐2 gamma variant and Associate Professor Dr. Hamilton Cabral (University of São Paulo‐Brazil) for helpful discussions. We are thankful to the Brazilian National Council for Scientific and Technological Development (CNPq) [grant number 312606/2019‐2 to M. D. B.], the Coordination for the Improvement of Higher Educational Personnel (CAPES) [Finance Code 001], and the São Paulo Research Foundation (FAPESP) [grant number 20/05270‐0 to V. L. D. B.].
Fuzo, C. A. , Martins, R. B. , Fraga‐Silva, T. F. C. , Amstalden, M. K. , Canassa De Leo, T. , Souza, J. P. , Lima, T. M. , Faccioli, L. H. , Okamoto, D. N. , Juliano, M. A. , França, S. C. , Juliano, L. , Bonato, V. L. D. , Arruda, E. , & Dias‐Baruffi, M. (2022). Celastrol: A lead compound that inhibits SARS‐CoV‐2 replication, the activity of viral and human cysteine proteases, and virus‐induced IL‐6 secretion. Drug Development Research, 1–18. 10.1002/ddr.21982
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- Allen, S. D. , Liu, Y.‐G. , Kim, T. , Bobbala, S. , Yi, S. , Zhang, X. , Choi, J. , & Scott, E. A. (2019). Celastrol‐loaded PEG‐b‐PPS nanocarriers as an anti‐inflammatory treatment for atherosclerosis. Biomaterials Science, 7(2), 657–668. 10.1039/C8BM01224E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzrodt, C. L. , Maknojia, I. , McCarthy, R. D. P. , Oldfield, T. M. , Po, J. , Ta, K. T. L. , Stepp, H. E. , & Clements, T. P. (2020). A guide to COVID‐19: A global pandemic caused by the novel coronavirus SARS‐CoV‐2. The FEBS Journal, 287(17), 3633–3650. 10.1111/febs.15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A. , Nasir, J. A. , Budylowski, P. , Yip, L. , Aftanas, P. , Christie, N. , Ghalami, A. , Baid, K. , Raphenya, A. R. , Hirota, J. A. , Miller, M. S. , McGeer, A. J. , Ostrowski, M. , Kozak, R. A. , McArthur, A. G. , Mossman, K. , & Mubareka, S . (2020). Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases, 26(9), 2054–2063. 10.3201/eid2609.201495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric, R. S. (2020). Emergence of a highly fit SARS‐CoV‐2 variant. New England Journal of Medicine, 383(27), 2684–2686. 10.1056/NEJMcibr2032888 [DOI] [PubMed] [Google Scholar]
- Bassanini, I. , Parapini, S. , Ferrandi, E. E. , Gabriele, E. , Basilico, N. , Taramelli, D. , & Sparatore, A. (2021). Design, synthesis and in vitro investigation of novel basic celastrol carboxamides as bio‐inspired leishmanicidal agents endowed with inhibitory activity against leishmania Hsp90. Biomolecules, 11(1), 56. 10.3390/biom11010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva, A. , Cammarata, L. , Radhakrishnan, A. , Squires, C. , Yang, K. D. , Shivashankar, G. V. , & Uhler, C. (2021). Causal network models of SARS‐CoV‐2 expression and aging to identify candidates for drug repurposing. Nature Communications, 12(1), 1024. 10.1038/s41467-021-21056-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Berman, H. M. , Westbrook, J. , Feng, Z. , Gilliland, G. , Bhat, T. N. , Weissig, H. , Shindyalov, I. N. , & Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research, 28(1), 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia, D. S. (2020). Discovery studio visualizer.
- Blanco‐Melo, D. , Nilsson‐Payant, B. E. , Liu, W.‐C. , Uhl, S. , Hoagland, D. , Møller, R. , Jordan, T. X. , Oishi, K. , Panis, M. , Sachs, D. , Wang, T. T. , Schwartz, R. E. , Lim, J. K. , Albrecht, R. A. , & tenOever, B. R. (2020). Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell, 181(5), 1036–1045. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasen, C. L. , Christensen, H. , Olsen, D. A. , Kahns, S. , Andersen, R. F. , Madsen, J. B. , Lassen, A. , Kierkegaard, H. , Jensen, A. , Sydenham, T. V. , Madsen, J. S. , Møller, J. K. , & Brandslund, I. (2021). Daily monitoring of viral load measured as SARS‐CoV‐2 antigen and RNA in blood, IL‐6, CRP and complement C3d predicts outcome in patients hospitalized with COVID‐19. Clinical Chemistry and Laboratory Medicine (CCLM), 59(12), 1988–1997. 10.1515/cclm-2021-0694 [DOI] [PubMed] [Google Scholar]
- Carmona, A. K. , Schwager, S. L. , Juliano, M. A. , Juliano, L. , & Sturrock, E. D. (2006). A continuous fluorescence resonance energy transfer angiotensin I‐converting enzyme assay. Nature Protocols, 1(4), 1971–1976. 10.1038/nprot.2006.306 [DOI] [PubMed] [Google Scholar]
- Caruso, F. , Rossi, M. , Pedersen, J. Z. , & Incerpi, S. (2020). Computational studies reveal mechanism by which quinone derivatives can inhibit SARS‐CoV‐2. Study of embelin and two therapeutic compounds of interest, methyl prednisolone and dexamethasone. Journal of Infection and Public Health, 13(12), 1868–1877. 10.1016/j.jiph.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, F. , Singh, M. , Belli, S. , Berinato, M. , & Rossi, M. (2020). Interrelated mechanism by which the methide quinone celastrol, obtained from the roots of Tripterygium wilfordii, inhibits main protease 3CLpro of COVID‐19 and acts as superoxide radical scavenger. International Journal of Molecular Sciences, 21(23), 9266. 10.3390/ijms21239266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascão, R. , Fonseca, J. E. , & Moita, L. F. (2017). Celastrol: A spectrum of treatment opportunities in chronic diseases. Frontiers in Medicine, 4, 69. 10.3389/fmed.2017.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.‐J. , Liu, C. Y.‐Y. , Chiang, B.‐L. , Chao, Y.‐C. , & Chen, C.‐C. (2004). Induction of IL‐8 release in lung cells via activator protein‐1 by recombinant baculovirus displaying severe acute respiratory syndrome‐coronavirus spike proteins: Identification of two functional regions. Journal of Immunology, 173, 7602–7614. 10.4049/jimmunol.173.12.7602 [DOI] [PubMed] [Google Scholar]
- Chen, G.‐Y. , Pan, Y.‐C. , Wu, T.‐Y. , Yao, T.‐Y. , Wang, W.‐J. , Shen, W.‐J. , Ahmed, A. , Chan, S.‐T. , Tang, C.‐H. , Huang, W.‐C. , Hung, M.‐C. , Yang, J.‐C. , & Wu, Y.‐C. (2022). Potential natural products that target the SARS‐CoV‐2 spike protein identified by structure‐based virtual screening, isothermal titration calorimetry and lentivirus particles pseudotyped (Vpp) infection assay. Journal of Traditional and Complementary Medicine, 12(1), 73–89. 10.1016/j.jtcme.2021.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, K.‐C. , Tsui, K.‐H. , Chung, L.‐C. , Yeh, C.‐N. , Chen, W.‐T. , Chang, P.‐L. , & Juang, H.‐H. (2014). Celastrol blocks interleukin‐6 gene expression via downregulation of NF‐κB in prostate carcinoma cells. PLoS One, 9(3), e93151. 10.1371/journal.pone.0093151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle, D. M. , Kim‐Schulze, S. , Huang, H.‐H. , Beckmann, N. D. , Nirenberg, S. , Wang, B. , Lavin, Y. , Swartz, T. H. , Madduri, D. , Stock, A. , Marron, T. U. , Xie, H. , Patel, M. , Tuballes, K. , Van Oekelen, O. , Rahman, A. , Kovatch, P. , Aberg, J. A. , Schadt, E. , … Gnjatic, S. (2020). An inflammatory cytokine signature predicts COVID‐19 severity and survival. Nature Medicine, 26(10), 1636–1643. 10.1038/s41591-020-1051-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demedts, I. K. , Bracke, K. R. , Van Pottelberge, G. , Testelmans, D. , Verleden, G. M. , Vermassen, F. E. , Joos, G. F. , & Brusselle, G. G. (2007). Accumulation of dendritic cells and increased CCL20 levels in the airways of patients with chronic obstructive pulmonary disease. American Journal of Respiratory and Critical Care Medicine, 175(10), 998–1005. 10.1164/rccm.200608-1113OC [DOI] [PubMed] [Google Scholar]
- Desai, N. , Neyaz, A. , Szabolcs, A. , Shih, A. R. , Chen, J. H. , Thapar, V. , Nieman, L. T. , Solovyov, A. , Mehta, A. , Lieb, D. J. , Kulkarni, A. S. , Jaicks, C. , Xu, K. H. , Raabe, M. J. , Pinto, C. J. , Juric, D. , Chebib, I. , Colvin, R. B. , Kim, A. Y. , … Deshpande, V. (2020). Temporal and spatial heterogeneity of host response to SARS‐CoV‐2 pulmonary infection. Nature Communications, 11(1), 6319. 10.1038/s41467-020-20139-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominic, S. M. , Chunlong, M. , Panagiotis, L. , Ang, G. , Alma, T. J. , Xiangzhi, M. , Peter, D. , Xiujun, Z. , Yanmei, H. , Naoya, K. , Brett, H. , Bart, T. , Thomas, M. M. , Antonios, K. , Yan, X. , Yu, C. , & Jun, W. (2022). Structure and inhibition of the SARS‐CoV‐2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Science Advances, 6(50), eabe0751. 10.1126/sciadv.abe0751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, E. , Du, H. , & Gardner, L. (2020). An interactive web‐based dashboard to track COVID‐19 in real time. The Lancet Infectious Diseases, 3099(20), 19–20. 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Q. , Reid, S. P. , Clark, N. R. , Wang, Z. , Fernandez, N. F. , Rouillard, A. D. , Readhead, B. , Tritsch, S. R. , Hodos, R. , Hafner, M. , Niepel, M. , Sorger, P. K. , Dudley, J. T. , Bavari, S. , Panchal, R. G. , & Ma'ayan, A. (2016). L1000CDS2: LINCS L1000 characteristic direction signatures search engine. NPJ Systems Biology and Applications, 2(1), 16015. 10.1038/npjsba.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS‐CoV‐2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592. 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum, D. C. , & June, C. H. (2020). Cytokine storm. New England Journal of Medicine, 383(23), 2255–2273. 10.1056/NEJMra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z. , He, D. , Yu, B. , Liu, F. , Zuo, J. , Li, Y. , Lin, Q. , Zhou, X. , & Wang, Q. (2017). High‐throughput study of the effects of celastrol on activated fibroblast‐like synoviocytes from patients with rheumatoid arthritis. Genes, 8(9), 221. 10.3390/genes8090221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria, N. R. , Mellan, T. A. , Whittaker, C. , Claro, I. M. , Candido, D. S. , Mishra, S. , Crispim, M. A. E. , Sales, F. C. S. , Hawryluk, I. , McCrone, J. T. , Hulswit, R. J. G. , Franco, L. A. M. , Ramundo, M. S. , de Jesus, J. G. , Andrade, P. S. , Coletti, T. M. , Ferreira, G. M. , Silva, C. A. M. , Manuli, E. R. , … Sabino, E. C. (2021). Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science, 372(6544), 815–821. 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanet, A. , Autran, B. , Lina, B. , Kieny, M. P. , Karim, S. S. A. , & Sridhar, D. (2021). SARS‐CoV‐2 variants and ending the COVID‐19 pandemic. The Lancet, 397(10278), 952–954. 10.1016/S0140-6736(21)00370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga‐Silva, T. F. , de, C. , Maruyama, S. R. , Sorgi, C. A. , Russo, E. M. , de, S. , Fernandes, A. P. M. , de Barros Cardoso, C. R. , Faccioli, L. H. , Dias‐Baruffi, M. , & Bonato, V. L. D. (2021). COVID‐19: Integrating the complexity of systemic and pulmonary immunopathology to identify biomarkers for different outcomes. Frontiers in Immunology, 11, 599736. 10.3389/fimmu.2020.599736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire, M. C. L. C. , Noske, G. D. , Bitencourt, N. V. , Sanches, P. R. S. , Santos‐Filho, N. A. , Gawriljuk, V. O. , de Souza, E. P. , Nogueira, V. H. R. , de Godoy, M. O. , Nakamura, A. M. , Fernandes, R. S. , Godoy, A. S. , Juliano, M. A. , Peres, B. M. , Barbosa, C. G. , Moraes, C. B. , Freitas‐Junior, L. H. G. , Cilli, E. M. , Guido, R. V. C. , & Oliva, G. (2021). Non‐toxic dimeric peptides derived from the bothropstoxin‐I are potent SARS‐CoV‐2 and papain‐like protease inhibitors. Molecules, 26(16), 4896. 10.3390/molecules26164896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z. , Huang, B. , Tang, J. , Liu, S. , Liu, M. , Ye, Y. , Liu, Z. , Xiong, Y. , Zhu, W. , Cao, D. , Li, J. , Niu, X. , Zhou, H. , Zhao, Y. J. , Zhang, G. , & Huang, H. (2021). The complex structure of GRL0617 and SARS‐CoV‐2 PLpro reveals a hot spot for antiviral drug discovery. Nature Communications, 12(1), 488. 10.1038/s41467-020-20718-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindez, G. , Matschinske, J. , Rose, T. D. , Sadegh, S. , Salgado‐Albarrán, M. , Späth, J. , Baumbach, J. , & Pauling, J. K. (2021). Lessons from the COVID‐19 pandemic for advancing computational drug repurposing strategies. Nature Computational Science, 1(1), 33–41. 10.1038/s43588-020-00007-6 [DOI] [PubMed] [Google Scholar]
- Gil, C. , Ginex, T. , Maestro, I. , Nozal, V. , Barrado‐Gil, L. , Cuesta‐Geijo, M. Á. , Urquiza, J. , Ramírez, D. , Alonso, C. , Campillo, N. E. , & Martinez, A. (2020). COVID‐19: drug targets and potential treatments. Journal of Medicinal Chemistry, 63(21), 12359–12386. 10.1021/acs.jmedchem.0c00606 [DOI] [PubMed] [Google Scholar]
- Gordon, D. E. , Jang, G. M. , Bouhaddou, M. , Xu, J. , Obernier, K. , White, K. M. , O'Meara, M. J. , Rezelj, V. V. , Guo, J. Z. , Swaney, D. L. , Tummino, T. A. , Hüttenhain, R. , Kaake, R. M. , Richards, A. L. , Tutuncuoglu, B. , Foussard, H. , Batra, J. , Haas, K. , Modak, M. , … Krogan, N. J. (2020). A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature, 583, 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Singh, A. K. , Kushwaha, P. P. , Prajapati, K. S. , Shuaib, M. , Senapati, S. , & Kumar, S. (2021). Identification of potential natural inhibitors of SARS‐CoV2 main protease by molecular docking and simulation studies. Journal of Biomolecular Structure and Dynamics, 39(12), 4334–4345. 10.1080/07391102.2020.1776157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, R. K. , DiPaola, R. S. , Romanelli, F. , & Dutch Rebecca, E. (2020). Rapid repurposing of drugs for COVID‐19. Science, 368(6493), 829–830. 10.1126/science.abb9332 [DOI] [PubMed] [Google Scholar]
- Habtemariam, S. , Nabavi, S. F. , Berindan‐Neagoe, I. , Cismaru, C. A. , Izadi, M. , Sureda, A. , & Nabavi, S. M. (2020). Should we try the antiinflammatory natural product, celastrol, for COVID‐19? Phytotherapy Research, 34(6), 1189–1190. 10.1002/ptr.6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj, J. , Yatim, N. , Barnabei, L. , Corneau, A. , Boussier, J. , Smith, N. , Péré, H. , Charbit, B. , Bondet, V. , Chenevier‐Gobeaux, C. , Breillat, P. , Carlier, N. , Gauzit, R. , Morbieu, C. , Pène, F. , Marin, N. , Roche, N. , Szwebel, T. A. , Merkling, S. H. , … Terrier, B. (2020). Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science, 369(6504), 718–724. 10.1126/science.abc6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H. , Ma, Q. , Li, C. , Liu, R. , Zhao, L. , Wang, W. , Zhang, P. , Liu, X. , Gao, G. , Liu, F. , Jiang, Y. , Cheng, X. , Zhu, C. , & Xia, Y. (2020). Profiling serum cytokines in COVID‐19 patients reveals IL‐6 and IL‐10 are disease severity predictors. Emerging Microbes & Infections, 9(1), 1123–1130. 10.1080/22221751.2020.1770129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, Q. M. , Wilson, K. M. , Shen, M. , Itkin, Z. , Eastman, R. T. , Shinn, P. , & Hall, M. D. (2020). Targeting ACE2–RBD interaction as a platform for COVID‐19 therapeutics: Development and drug‐repurposing screen of an AlphaLISA proximity assay. ACS Pharmacology & Translational Science, 3(6), 1352–1360. 10.1021/acsptsci.0c00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt, J. , Tamin, A. , Lu, X. , Kamili, S. , Sakthivel, S. K. , Murray, J. , Queen, K. , Tao, Y. , Paden, C. R. , Zhang, J. , Li, Y. , Uehara, A. , Wang, H. , Goldsmith, C. , Bullock, H. A. , Wang, L. , Whitaker, B. , Lynch, B. , Gautam, R. , … Thornburg, N. J. (2020). Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerging Infectious Disease Journal, 26(6), 1266–1273. 10.3201/eid2606.200516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, R. , Sakamoto, A. , Deguchi, S. , Yi, R. , Sano, E. , Hotta, A. , Takahashi, K. , Yamanaka, S. , & Takayama, K. (2021). Dual inhibition of TMPRSS2 and cathepsin bprevents SARS‐CoV‐2 infection in iPS cells. Molecular Therapy Nucleic Acids, 26, 1107–1114. 10.1016/j.omtn.2021.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B. , & Lana, G. (2020). Prediction of repurposed drugs for treating lung injury in COVID‐19. Em F1000Research, 9(Número), 609. https://f1000research.com/articles/9-609/v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Kruger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. , Herrler, G. , Wu, N.‐H. , Nitsche, A. , Muller, M. A. , Drosten, C. , & Pohlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W. , Wang, L. , Du, G. , Guan, Q. , Dong, T. , Song, L. , Xia, Y. , & Wang, X. (2020). Effects of microbiota on the treatment of obesity with the natural product celastrol in rats. Diabetes & Metabolism Journal, 44(5), 747–763. 10.4093/dmj.2019.0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal, B. , Matthews, L. , Viteri, G. , Gong, C. , Lorente, P. , Fabregat, A. , Sidiropoulos, K. , Cook, J. , Gillespie, M. , Haw, R. , Loney, F. , May, B. , Milacic, M. , Rothfels, K. , Sevilla, C. , Shamovsky, V. , Shorser, S. , Varusai, T. , Weiser, J. , & D'Eustachio, P. (2020). The reactome pathway knowledgebase. Nucleic Acids Research, 48(D1), D498–D503. 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z. , Du, X. , Xu, Y. , Deng, Y. , Liu, M. , Zhao, Y. , Zhang, B. , Li, X. , Zhang, L. , Peng, C. , Duan, Y. , Yu, J. , Wang, L. , Yang, K. , Liu, F. , Jiang, R. , Yang, X. , You, T. , Liu, X. , … Yang, H. (2020). Structure of Mpro from COVID‐19 virus and discovery of its inhibitors. Nature, 582, 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kang, C. K. , Seong, M.‐W. , Choi, S.‐J. , Kim, T. S. , Choe, P. G. , Song, S. H. , Kim, N.‐J. , Park, W. B. , & Oh, M . (2020). In vitro activity of lopinavir/ritonavir and hydroxychloroquine against severe acute respiratory syndrome coronavirus 2 at concentrations achievable by usual doses. The Korean Journal of Internal Medicine, 35(4), 782–787. 10.3904/kjim.2020.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannaiyan, R. , Shanmugam, M. K. , & Sethi, G. (2011). Molecular targets of celastrol derived from thunder of god vine: Potential role in the treatment of inflammatory disorders and cancer. Cancer Letters, 303(1), 9–20. 10.1016/j.canlet.2010.10.025 [DOI] [PubMed] [Google Scholar]
- Kantarjian, H. , Jabbour, E. , Grimley, J. , & Kirkpatrick, P. (2006). Dasatinib. Nature Reviews Drug Discovery, 5(9), 717–718. 10.1038/nrd2135 [DOI] [PubMed] [Google Scholar]
- Khalili, N. , Karimi, A. , Moradi, M.‐T. , & Shirzad, H. (2018). In vitro immunomodulatory activity of celastrol against influenza A virus infection. Immunopharmacology and Immunotoxicology, 40(3), 250–255. 10.1080/08923973.2018.1440591 [DOI] [PubMed] [Google Scholar]
- Kim, D. H. , Shin, E. K. , Kim, Y. H. , Lee, B. W. , Jun, J.‐G. , Park, J. H. Y. , & Kim, J.‐K. (2009). Suppression of inflammatory responses by celastrol, a quinone methide triterpenoid isolated from Celastrus regelii. European Journal of Clinical Investigation, 39(9), 819–827. 10.1111/j.1365-2362.2009.02186.x [DOI] [PubMed] [Google Scholar]
- Kolde, R. (2019). Implementation of heatmaps that offers more control over dimensions and appearance. https://cran.r-project.org/package=pheatmap
- Korkmaz, B. , Attucci, S. , Juliano, M. A. , Kalupov, T. , Jourdan, M.‐L. , Juliano, L. , & Gauthier, F. (2008). Measuring elastase, proteinase 3 and cathepsin G activities at the surface of human neutrophils with fluorescence resonance energy transfer substrates. Nature Protocols, 3(6), 991–1000. 10.1038/nprot.2008.63 [DOI] [PubMed] [Google Scholar]
- Lee, J.‐H. , Koo, T. H. , Yoon, H. , Jung, H. S. , Jin, H. Z. , Lee, K. , Hong, Y.‐S. , & Lee, J. J. (2006). Inhibition of NF‐κB activation through targeting IκB kinase by celastrol, a quinone methide triterpenoid. Biochemical Pharmacology, 72(10), 1311–1321. 10.1016/j.bcp.2006.08.014 [DOI] [PubMed] [Google Scholar]
- Li, H.‐Y. , Zhang, J. , Sun, L.‐L. , Li, B.‐H. , Gao, H.‐L. , Xie, T. , Zhang, N. , & Ye, Z.‐M. (2015). Celastrol induces apoptosis and autophagy via the ROS/JNK signaling pathway in human osteosarcoma cells: An in vitro and in vivo study. Cell Death & Disease, 6, e1604. 10.1038/cddis.2014.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , & Hao, J. (2019). Treatment of neurodegenerative diseases with bioactive components of Tripterygium wilfordii . The American Journal of Chinese Medicine, 47(4), 769–785. 10.1142/S0192415X1950040X [DOI] [PubMed] [Google Scholar]
- Li, Q. , Guan, X. , Wu, P. , Wang, X. , Zhou, L. , Tong, Y. , Ren, R. , Leung, K. , Lau, E. , Wong, J. Y. , Xing, X. , Xiang, N. , Wu, Y. , Li, C. , Chen, Q. , Li, D. , Liu, T. , Zhao, J. , Liu, M. , … Feng, Z. (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. New England Journal of Medicine, 382(13), 1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, M. , Jiang, Z. , Huang, Q. , Liu, L. , Xue, Y. , Zhu, X. , Yu, Y. , Wan, W. , Yang, H. , & Zou, H. (2016). Clinical association of chemokine (C‐X‐C motif) ligand 1 (CXCL1) with interstitial pneumonia with autoimmune features (IPAF). Scientific Reports, 6(1), 38949. 10.1038/srep38949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L. , Lu, L. , Cao, W. , & Li, T. (2020). Hypothesis for potential pathogenesis of SARS‐CoV‐2 infection‐a review of immune changes in patients with viral pneumonia. Emerging Microbes & Infections, 9(1), 727–732. 10.1080/22221751.2020.1746199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Lee, J. , Salazar Hernandez, M. A. , Mazitschek, R. , & Ozcan, U. (2015). Treatment of obesity with celastrol. Cell, 161(5), 999–1011. 10.1016/j.cell.2015.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Liu, J. , Wang, H. , & Bai, M. (2019). Protective effect of celastrol for burn‐induced acute lung injury in rats. International Journal of Clinical and Experimental Pathology, 12(2), 576–583. [PMC free article] [PubMed] [Google Scholar]
- Lotfi Shahreza, M. , Ghadiri, N. , Mousavi, S. R. , Varshosaz, J. , & Green, J. R. (2018). A review of network‐based approaches to drug repositioning. Briefings in Bioinformatics, 19(5), 878–892. 10.1093/bib/bbx017 [DOI] [PubMed] [Google Scholar]
- Lu, X. , Wang, L. , Sakthivel, S. K. , Whitaker, B. , Murray, J. , Kamili, S. , Lynch, B. , Malapati, L. , Burke, S. A. , & Harcourt, J. (2020). US CDC real‐time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases, 26(8), 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Shoemaker, A. R. , Liu, X. , Woods, K. W. , Thomas, S. A. , de Jong, R. , Han, E. K. , Li, T. , Stoll, V. S. , Powlas, J. A. , Oleksijew, A. , Mitten, M. J. , Shi, Y. , Guan, R. , McGonigal, T. P. , Klinghofer, V. , Johnson, E. F. , Leverson, J. D. , Bouska, J. J. , … Giranda, V. L. (2005). Potent and selective inhibitors of Akt kinases slow the progress of tumors in vivo. Molecular Cancer Therapeutics, 4(6), 977–986. 10.1158/1535-7163.MCT-05-0005 [DOI] [PubMed] [Google Scholar]
- Ma, X.R. , Alugubelli, Y. R. , Ma, Y. , Vatansever, E. C. , Scott, D. A. , Qiao, Y. , Yu, G. , Xu, S. , & Liu, W. R. (2022). MPI8 is potent against SARS‐CoV‐2 by inhibiting dually and selectively the SARS‐CoV‐2 main protease and the host cathepsin L.** ChemMedChem, 17(1), e202100456. 10.1002/cmdc.202100456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder, S. , Zappulla, F. , & Silbart, L. K. (2014). Mycoplasma gallisepticum lipid associated membrane proteins up‐regulate inflammatory genes in chicken tracheal epithelial cells via TLR‐2 ligation through an NF‐κB dependent pathway. PLoS One, 9(11), e112796. 10.1371/journal.pone.0112796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamboya, F. , & Amri, E. (2012). Papain, a plant enzyme of biological importance: A review. American Journal of Biochemistry and Biotechnology, 8(2), 99–104. 10.3844/ajbbsp.2012.99.104 [DOI] [Google Scholar]
- Martins, R. B. , Castro, I. A. , Pontelli, M. , Souza, J. P. , Lima, T. M. , Melo, S. R. , Siqueira, J. P. Z. , Caetano, M. H. , Arruda, E. , & de Almeida, M. T. G. (2021). SARS‐CoV‐2 inactivation by ozonated water: A preliminary alternative for environmental disinfection. Ozone: Science & Engineering, 43(2), 108–111. 10.1080/01919512.2020.1842998 [DOI] [Google Scholar]
- McGuire, C. , & Lee, J. (2010). Brief review of vorinostat. Clinical Medicine Insights: Therapeutics, 2(CMT), S1102 10.4137/CMT.S1102 [DOI] [Google Scholar]
- Morris, G. M. , Huey, R. , Lindstrom, W. , Sanner, M. F. , Belew, R. K. , Goodsell, D. S. , & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan, V. , Ravindra, K. C. , Chiaro, C. , Cary, D. , Aggarwal, B. B. , Henderson, A. J. , & Prabhu, K. S. (2011). Celastrol inhibits tat‐mediated human immunodeficiency virus (HIV) transcription and replication. Journal of Molecular Biology, 410(5), 972–983. 10.1016/j.jmb.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä, E. , Desai, D. , Nkizinkiko, Y. , Eriksson, J. E. , & Rosenholm, J. M. (2015). Sugar‐decorated mesoporous silica nanoparticles as delivery vehicles for the poorly soluble drug celastrol enables targeted induction of apoptosis in cancer cells. European Journal of Pharmaceutics and Biopharmaceutics, 96, 11–21. 10.1016/j.ejpb.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Nienhold, R. , Ciani, Y. , Koelzer, V. H. , Tzankov, A. , Haslbauer, J. D. , Menter, T. , Schwab, N. , Henkel, M. , Frank, A. , Zsikla, V. , Willi, N. , Kempf, W. , Hoyler, T. , Barbareschi, M. , Moch, H. , Tolnay, M. , Cathomas, G. , Demichelis, F. , Junt, T. , & Mertz, K. D. (2020). Two distinct immunopathological profiles in autopsy lungs of COVID‐19. Nature Communications, 11(1), 5086. 10.1038/s41467-020-18854-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesor, E. J. , Boivin, G. , Rhéaume, E. , Shi, R. , Lavoie, V. , Goyette, N. , Picard, M.‐E. , Perez, A. , Laghrissi‐Thode, F. , & Tardif, J.‐C. (2021). Inhibition of the 3CL protease and SARS‐CoV‐2 replication by dalcetrapib. ACS Omega, 6(25), 6, 16584–16591. 10.1021/acsomega.1c01797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, Y. , Fu, C. , Zhang, H. , Zhang, M. , Xie, H. , Tong, X. , Li, Y. , Hou, Z. , Fan, X. , & Yan, M. (2020). Celastrol slows the progression of early diabetic nephropathy in rats via the PI3K/AKT pathway. BMC Complementary Medicine and Therapies, 20(1), 321. 10.1186/s12906-020-03050-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, D. N. , Oliveira, L. C. G. , Kondo, M. Y. , Cezari, M. H. S. , Szeltner, Z. , Juhász, T. , Juliano, M. A. , Polgár, L. , Juliano, L. , & Gouvea, I. E. (2010). Increase of SARS‐CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition. Biological Chemistry, 391(12), 1461–1468. 10.1515/bc.2010.145 [DOI] [PubMed] [Google Scholar]
- Pawar, A. Y. (2020). Combating devastating COVID‐19 by drug repurposing. International Journal of Antimicrobial Agents, 56, 105984. 10.1016/j.ijantimicag.2020.105984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna, G. F. , Fiorucci, M. , Reimund, J.‐M. , Taquet, N. , Arondel, Y. , & Muller, C. D. (2004). Celastrol inhibits proinflammatory cytokine secretion in Crohn's disease biopsies. Biochemical and Biophysical Research Communications, 322(3), 778–786. 10.1016/j.bbrc.2004.07.186 [DOI] [PubMed] [Google Scholar]
- Polack, F. P. , Thomas, S. J. , Kitchin, N. , Absalon, J. , Gurtman, A. , Lockhart, S. , Perez, J. L. , Pérez Marc, G. , Moreira, E. D. , Zerbini, C. , Bailey, R. , Swanson, K. A. , Roychoudhury, S. , Koury, K. , Li, P. , Kalina, W. V. , Cooper, D. , Frenck RW, J. r , Hammitt, L. L. , … Clinical Trial, G. (2020). Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. New England Journal of Medicine, 383(27), 2603–2615. 10.1056/NEJMoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzi, F. , Kirkby, N. S. , Edin, M. L. , Whiteford, J. , Zeldin, D. C. , Mitchell, J. A. , & Warner, T. D. (2016). Aspirin inhibits the production of proangiogenic 15(S)‐HETE by platelet cyclooxygenase‐1. The FASEB Journal, 30(12), 4256–4266. 10.1096/fj.201600530R [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.r-project.org/
- Reed, L. J. , & Muench, H. (1938). A simple method of estimating fifty per cent endpoints. American Journal of Epidemiology, 27(3), 493–497. 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]
- Riva, L. , Yuan, S. , Yin, X. , Martin‐Sancho, L. , Matsunaga, N. , Burgstaller‐Muehlbacher, S. , Pache, L. , De Jesus, P. P. , Hull, M. V. , Chang, M. , Chan, J. F. , Cao, J. , Poon, V. K. , Herbert, K. , Nguyen, T. ‐T. , Pu, Y. , Nguyen, C. , Rubanov, A. , … Chanda, S. K. (2020). A large‐scale drug repositioning survey for SARS‐CoV‐2 antivirals. bioRxiv . 10.1101/2020.04.16.044016 [DOI]
- Ryu, Y. B. , Park, S.‐J. , Kim, Y. M. , Lee, J.‐Y. , Seo, W. D. , Chang, J. S. , Park, K. H. , Rho, M.‐C. , & Lee, W. S. (2010). SARS‐CoV 3CLpro inhibitory effects of quinone‐methide triterpenes from Tripterygium regelii . Bioorganic & Medicinal Chemistry Letters, 20(6), 1873–1876. 10.1016/j.bmcl.2010.01.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino, E. C. , Buss, L. F. , Carvalho, M. , Prete CA, Jr. , Crispim, M. , Fraiji, N. A. , Pereira, R. , Parag, K. V. , da Silva Peixoto, P. , Kraemer, M. , Oikawa, M. K. , Salomon, T. , Cucunuba, Z. M. , Castro, M. C. , de Souza Santos, A. A. , Nascimento, V. H. , Pereira, H. S. , Ferguson, N. M. , … Faria, N. R. (2021). Resurgence of COVID‐19 in Manaus, Brazil, despite high seroprevalence. The Lancet, 397(10273), 452–455. 10.1016/S0140-6736(21)00183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna, V. , Chamcheu, J. C. , Pala, N. , Mukhtar, H. , Sechi, M. , & Siddiqui, I. A. (2015). Nanoencapsulation of natural triterpenoid celastrol for prostate cancer treatment. International Journal of Nanomedicine, 10, 6835–6846. 10.2147/IJN.S93752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, L. L. C. (2015). The JyMOL molecular graphics development component, Version 1.8.
- Schymkowitz, J. , Borg, J. , Stricher, F. , Nys, R. , Rousseau, F. , & Serrano, L. (2005). The FoldX web server: An online force field. Nucleic Acids Research, 33(Suppl_2), W382–W388. 10.1093/nar/gki387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer, K. , Readhead, B. , & Dudley, J. T. (2015). Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Current Topics in Medicinal Chemistry, 15(1), 5–20. 10.2174/1568026615666150112103510 [DOI] [PubMed] [Google Scholar]
- Shen, B. , Yi, X. , Sun, Y. , Bi, X. , Du, J. , Zhang, C. , Quan, S. , Zhang, F. , Sun, R. , Qian, L. , Ge, W. , Liu, W. , Liang, S. , Chen, H. , Zhang, Y. , Li, J. , Xu, J. , He, Z. , Chen, B. , … Guo, T. (2020). Proteomic and metabolomic characterization of COVID‐19 patient sera. Cell, 182(1), 59–72.e15. 10.1016/j.cell.2020.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, D. , Mukherjee, R. , Grewe, D. , Bojkova, D. , Baek, K. , Bhattacharya, A. , Schulz, L. , Widera, M. , Mehdipour, A. R. , Tascher, G. , Geurink, P. P. , Wilhelm, A. , van der Heden van Noort, G. J. , Ovaa, H. , Müller, S. , Knobeloch, K. ‐P. , Rajalingam, K. , Schulman, B. A. , … Dikic, I. (2020). Papain‐like protease regulates SARS‐CoV‐2 viral spread and innate immunity. Nature, 587(7835), 657–662. 10.1038/s41586-020-2601-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreeramulu, S. , Gande, S. L. , Göbel, M. , & Schwalbe, H. (2009). Molecular mechanism of inhibition of the human protein complex Hsp90–Cdc37, a kinome chaperone–cochaperone, by triterpene celastrol. Angewandte Chemie International Edition, 48(32), 5853–5855. 10.1002/anie.200900929 [DOI] [PubMed] [Google Scholar]
- Stanetty, P. , Hattinger, G. , Schnürch, M. , & Mihovilovic, M. D. (2005). Novel and efficient access to phenylamino‐pyrimidine type protein kinase C inhibitors utilizing a negishi cross‐coupling strategy. The Journal of Organic Chemistry, 70(13), 5215–5220. 10.1021/jo0505223 [DOI] [PubMed] [Google Scholar]
- Subramanian, A. , Narayan, R. , Corsello, S. M. , Peck, D. D. , Natoli, T. E. , Lu, X. , Gould, J. , Davis, J. F. , Tubelli, A. A. , Asiedu, J. K. , Lahr, D. L. , Hirschman, J. E. , Liu, Z. , Donahue, M. , Julian, B. , Khan, M. , Wadden, D. , Smith, I. C. , Lam, D. , … Golub, T. R. (2017). A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell, 171(6), 1437–1452. 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, Y. , & Turki, T. (2020). A new advanced in silico drug discovery method for novel coronavirus (SARS‐CoV‐2) with tensor decomposition‐based unsupervised feature extraction. PLoS One, 15(9), e0238907. 10.1371/journal.pone.0238907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, M. Z. , Poh, C. M. , Rénia, L. , MacAry, P. A. , & Ng, L. F. P. (2020). The trinity of COVID‐19: Immunity, inflammation and intervention. Nature Reviews Immunology, 20(6), 363–374. 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro, J. R. , & Farber, D. L. (2021). COVID‐19 vaccines: Modes of immune activation and future challenges. Nature Reviews Immunology, 21(4), 195–197. 10.1038/s41577-021-00526-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, E. C. , Rosen, L. E. , Shepherd, J. G. , Spreafico, R. , da Silva Filipe, A. , Wojcechowskyj, J. A. , Davis, C. , Piccoli, L. , Pascall, D. J. , Dillen, J. , Lytras, S. , Czudnochowski, N. , Shah, R. , Meury, M. , Jesudason, N. , De Marco, A. , Li, K. , Bassi, J. , O'Toole, A. , … Snell, G. (2021). Circulating SARS‐CoV‐2 spike N439K variants maintain fitness while evading antibody‐mediated immunity. Cell, 184(5), 1171–1187.e20. 10.1016/j.cell.2021.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik, J. R. , Korth, M. J. , Simmons, C. P. , Farrar, J. , Martin, T. R. , & Katze, M. G. (2012). Into the eye of the cytokine storm. Microbiology and Molecular Biology Reviews, 76(1), 16 LP–16 32. 10.1128/MMBR.05015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazini, B. M. , Maia, I. S. , Cavalcanti, A. B. , Berwanger, O. , Rosa, R. G. , Veiga, V. C. , Avezum, A. , Lopes, R. D. , Bueno, F. R. , Silva, M. V. A. O. , Baldassare, F. P. , Costa, E. L. V. , Moura, R. A. B. , Honorato, M. O. , Costa, A. N. , Damiani, L. P. , Lisboa, T. , Kawano‐Dourado, L. , Zampieri, F. G. , … Azevedo, L. C. P. , COALITION COVID‐19 Brazil III Investigators . (2020). Effect of dexamethasone on days alive and ventilator‐free in patients with moderate or severe acute respiratory distress syndrome and COVID‐19: The CoDEX randomized clinical trial. Journal of the American Medical Association, 324(13), 1307–1316. 10.1001/jama.2020.17021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O. , & Olson, A. J. (2010). AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse, G. M.‐K. , To, K.‐F. , Chan, P. K.‐S. , Lo, A. W. I. , Ng, K.‐C. , Wu, A. , Lee, N. , Wong, H.‐C. , Mak, S.‐M. , Chan, K.‐F. , Hui, D. S. C. , Sung, J. J.‐Y. , & Ng, H.‐K. (2004). Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS). Journal of Clinical Pathology, 57(3), 260–265. 10.1136/jcp.2003.013276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesha, S. H. , Yu, H. , Rajaiah, R. , Tong, L. , & Moudgil, K. D. (2011). Celastrus‐derived celastrol suppresses autoimmune arthritis by modulating antigen‐induced cellular and humoral effector responses. Journal of Biological Chemistry, 286(17), 15138–15146. 10.1074/jbc.M111.226365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey, M. , Clemens, S. A. C. , Madhi, S. A. , Weckx, L. Y. , Folegatti, P. M. , Aley, P. K. , Angus, B. , Baillie, V. L. , Barnabas, S. L. , Bhorat, Q. E. , Bibi, S. , Briner, C. , Cicconi, P. , Collins, A. M. , Colin‐Jones, R. , Cutland, C. L. , Darton, T. C. , Dheda, K. , Duncan, C. J. A. , … Bijker, E. (2021). Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet, 397(10269), 99–111. 10.1016/S0140-6736(20)32661-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Ahn, K. S. , Alharbi, S. A. , Shair, O. H. , Arfuso, F. , Sethi, G. , Chinnathambi, A. , & Tang, F. R. (2020). Celastrol alleviates gamma irradiation‐induced damage by modulating diverse inflammatory mediators. International Journal of Molecular Sciences, 21(3), 1084. 10.3390/ijms21031084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Erazo, T. , Ferguson, F. M. , Buckley, D. L. , Gomez, N. , Muñoz‐Guardiola, P. , Diéguez‐Martínez, N. , Deng, X. , Hao, M. , Massefski, W. , Fedorov, O. , Offei‐Addo, N. K. , Park, P. M. , Dai, L. , DiBona, A. , Becht, K. , Kim, N. D. , McKeown, M. R. , Roberts, J. M. , … Gray, N. S. (2018). Structural and atropisomeric factors governing the selectivity of pyrimido‐benzodiazipinones as inhibitors of kinases and bromodomains. ACS Chemical Biology, 13(9), 2438–2448. 10.1021/acschembio.7b00638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Wu, Q. , Yang, J. , Dong, K. , Chen, X. , Bai, X. , Chen, X. , Chen, Z. , Viboud, C. , Ajelli, M. , & Yu, H. (2020). Global, regional, and national estimates of target population sizes for covid‐19 vaccination: Descriptive study. BMJ, 371, m4704. 10.1136/bmj.m4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Chen, D. , & Wang, Z. (2018). Effects of diclofenac on the pharmacokinetics of celastrol in rats and its transport. Pharmaceutical Biology, 56(1), 269–274. 10.1080/13880209.2018.1459740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. Springer‐Verlag. https://ggplot2.tidyverse.org [Google Scholar]
- Wolfram, J. , Suri, K. , Huang, Y. , Molinaro, R. , Borsoi, C. , Scott, B. , Boom, K. , Paolino, D. , Fresta, M. , Wang, J. , Ferrari, M. , Celia, C. , & Shen, H. (2014). Evaluation of anticancer activity of celastrol liposomes in prostate cancer cells. Journal of Microencapsulation, 31(5), 501–507. 10.3109/02652048.2013.879932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2020). Coronavirus disease (COVID‐2019) situation reports, 1–45. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Wouters, O. J. , Shadlen, K. C. , Salcher‐Konrad, M. , Pollard, A. J. , Larson, H. J. , Teerawattananon, Y. , & Jit, M. (2021). Challenges in ensuring global access to COVID‐19 vaccines: Production, affordability, allocation, and deployment. The Lancet, 397(10278), 1023–1034. 10.1016/S0140-6736(21)00306-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Jin, H.‐Z. , Shu, D. , Li, F. , He, C.‐X. , Qiao, J. , Yu, X.‐L. , Zhang, Y. , He, Y.‐B. , & Liu, T.‐J. (2015). Efficacy and safety of Tripterygium wilfordii hook F versus acitretin in moderate to severe psoriasis vulgaris: A randomized clinical trial. Chinese Medical Journal, 128(4), 443–449. 10.4103/0366-6999.151069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Hong, C. , Pan, H. , Yang, Q. , Mei, Y. , Ping Dou, Q. , & Yang, H. (2016). Medicinal compound celastrol as a potential clinical anticancer drug: Lessons learned from preclinical studies. Clinical Cancer Drugs, 3(1), 63–73. 10.2174/2212697X03666160112001739 [DOI] [Google Scholar]
- Wu, T. , Hu, E. , Xu, S. , Chen, M. , Guo, P. , Dai, Z. , Feng, T. , Zhou, L. , Tang, W. , Zhan, L. , Fu, X. , Liu, S. , Bo, X. , & Yu, G. (2021). Clusterprofiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation, 2(3), 100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian, Y. , Zhang, J. , Bian, Z. , Zhou, H. , Zhang, Z. , Lin, Z. , & Xu, H. (2020). Bioactive natural compounds against human coronaviruses: A review and perspective. Acta Pharmaceutica Sinica. B, 10(7), 1163–1174. 10.1016/j.apsb.2020.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Z. , Rafferty, B. , Caldwell, G. W. , & Masucci, J. A. (2002). Rapidly distinguishing reversible and irreversible CYP450 inhibitors by using fluorometric kinetic analyses. European Journal of Drug Metabolism and Pharmacokinetics, 27(4), 281–287. 10.1007/BF03192339 [DOI] [PubMed] [Google Scholar]
- Ye, S. , Luo, W. , Khan, Z. A. , Wu, G. , Xuan, L. , Shan, P. , Lin, K. , Chen, T. , Wang, J. , Hu, X. , Wang, S. , Huang, W. , & Liang, G. (2020). Celastrol attenuates angiotensin II–induced cardiac remodeling by targeting STAT3. Circulation Research, 126(8), 1007–1023. 10.1161/CIRCRESAHA.119.315861 [DOI] [PubMed] [Google Scholar]
- Youn, G. S. , Kwon, D.‐J. , Ju, S. M. , Rhim, H. , Bae, Y. S. , Choi, S. Y. , & Park, J. (2014). Celastrol ameliorates HIV‐1 Tat‐induced inflammatory responses via NF‐kappaB and AP‐1 inhibition and heme oxygenase‐1 induction in astrocytes. Toxicology and Applied Pharmacology, 280(1), 42–52. 10.1016/j.taap.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Yu, J.‐S. , Tseng, C.‐K. , Lin, C.‐K. , Hsu, Y.‐C. , Wu, Y.‐H. , Hsieh, C.‐L. , & Lee, J.‐C. (2017). Celastrol inhibits dengue virus replication via up‐regulating type I interferon and downstream interferon‐stimulated responses. Antiviral Research, 137, 49–57. 10.1016/j.antiviral.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Koehn, C. D. , Yue, Y. , Li, S. , Thiele, G. M. , Hearth‐Holmes, M. P. , Mikuls, T. R. , O'Dell, J. R. , Klassen, L. W. , Zhang, Z. , & Su, K. (2015). Celastrol inhibits inflammatory stimuli‐induced neutrophil extracellular trap formation. Current Molecular Medicine, 15(4), 401–410. 10.2174/1566524015666150505160743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Chen, Z. , Hu, C. , Yan, S. , Li, Z. , Lian, B. , Xu, Y. , Ding, R. , Zeng, Z. , Zhang, X. , & Su, Y. (2018). Celastrol binds to its target protein via specific noncovalent interactions and reversible covalent bonds. Chemical Communications, 54(91), 12871–12874. 10.1039/C8CC06140H [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Lin, D. , Sun, X. , Curth, U. , Drosten, C. , Sauerhering, L. , Becker, S. , Rox, K. , & Hilgenfeld, R. (2020). Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved alpha‐ketoamide inhibitors. Science, 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Hamza, A. , Cao, X. , Wang, B. , Yu, S. , Zhan, C.‐G. , & Sun, D. (2008). A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Molecular Cancer Therapeutics, 7(1), 162–170. 10.1158/1535-7163.MCT-07-0484 [DOI] [PubMed] [Google Scholar]
- Zhao, M.‐M. , Yang, W.‐L. , Yang, F.‐Y. , Zhang, L. , Huang, W.‐J. , Hou, W. , Fan, C.‐F. , Jin, R.‐H. , Feng, Y.‐M. , Wang, Y.‐C. , & Yang, J.‐K. (2021). Cathepsin L plays a key role in SARS‐CoV‐2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduction and Targeted Therapy, 6(1), 134. 10.1038/s41392-021-00558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Hou, Y. , Shen, J. , Huang, Y. , Martin, W. , & Cheng, F. (2020). Network‐based drug repurposing for novel coronavirus 2019‐nCoV/SARS‐CoV‐2. Cell Discovery, 6(1), 14. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , Zhao, X. , Huang, B. , Shi, W. , Lu, R. , Niu, P. , Zhan, F. , Ma, X. , Wang, D. , Xu, W. , Wu, G. , Gao, G. F. , & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Wan, N. , Shan, X. , Deng, G. , Xu, Q. , Ye, H. , & Sun, Y. (2021). Celastrol targets adenylyl cyclase‐associated protein 1 to reduce macrophages‐mediated inflammation and ameliorates high fat diet‐induced metabolic syndrome in mice. Acta Pharmaceutica Sinica B, 11(5), 1200–1212. 10.1016/j.apsb.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


