Abstract
The relative importance of the frxA and rdxA nitroreductase genes of Helicobacter pylori in metronidazole (MTZ) susceptibility and resistance has been controversial. Jeong et al. (J. Bacteriol. 182:5082–5090, 2000) had interpreted that Mtzs H. pylori were of two types: type I, requiring only inactivation of rdxA to became resistant, and type II, requiring inactivation of both rdxA and frxA to become resistant; frxA inactivation by itself was not sufficient to confer resistance. In contrast, Kwon et al. (Antimicrob. Agents Chemother. 44:2133–2142, 2000) had interpreted that resistance resulted from inactivation either of frxA or rdxA. These two interpretations were tested here. Resistance was defined as efficient colony formation by single cells from diluted cultures rather than as growth responses of more dense inocula on MTZ-containing medium. Tests of three of Kwon's Mtzs strains showed that each was type II, requiring inactivation of both rdxA and frxA to become resistant. In additional tests, derivatives of frxA mutant strains recovered from MTZ-containing medium were found to contain new mutations in rdxA, and frxA inactivation slowed MTZ-induced killing of Mtzs strains. Northern blot analyses indicated that frxA mRNA, and perhaps also rdxA mRNA, were more abundant in type II than in type I strains. We conclude that development of MTZ resistance in H. pylori requires inactivation of rdxA alone or of both rdxA and frxA, depending on bacterial genotype, but rarely, if ever, inactivation of frxA alone, and that H. pylori strains differ in regulation of nitroreductase gene expression. We suggest that such regulatory differences may be significant functionally during human infection.
Helicobacter pylori is a genetically diverse bacterial species that chronically infects the stomachs of more than half of all people worldwide. Its long-term carriage is a major cause of chronic gastritis and peptic ulcer disease and is an early risk factor for gastric cancer (for reviews see references 5, 8, 28, and 32). Resistance to metronidazole (MTZ) is common and is important clinically as a primary cause of failure of MTZ-based anti-Helicobacter therapies (for reviews see references 10, 15, and 24). Frequencies of clinical isolates that are MTZ resistant range from only 10% in Japan (25) to 90% or more in India (26), and up to 50% or more of strains in the United States and Western Europe also are resistant (frequency varies among countries) (8, 23). These geographic differences probably reflect frequencies of MTZ use against other, mostly parasitic and anaerobic, infections and thus inadvertent MTZ exposure of resident H. pylori strains. Recent studies have implicated mutations in the chromosomal genes rdxA (HP0954) and frxA (HP0642) in the development of resistance (7, 9, 14, 16, 30, 35). These genes encode related nitroreductases that can convert MTZ from a harmless prodrug to products such as hydroxylamine that are both bactericidal and mutagenic (9, 29).
There has been disagreement about the quantitative contributions of rdxA and frxA to MTZ susceptibility and resistance. On the one hand, Kwon and associates had concluded that inactivation of either gene by itself could make any typical H. pylori strain resistant to MTZ (Mtzr) (21), and that following frxA inactivation, growth on MTZ-containing agar was not associated with mutation of rdxA (20). In contrast, we had concluded that rdxA inactivation is usually or always needed for a Mtzs strain to become Mtzr (16, 17). Two types of Mtzs strains were distinguished, however, based on relative levels of FrxA nitroreductase activity. Most common were strains in which rdxA inactivation by itself was sufficient to cause resistance to moderate levels of MTZ (type I strains). Once rdxA had been inactivated in such type I strains, additional mutations that inactivated frxA (the second nitroreductase gene) resulted in higher levels of resistance, but frxA inactivation by itself had little, if any, effect on MTZ susceptibility if rdxA was still functional. Other strains, designated type II, required inactivation of both frxA and rdxA to become Mtzr (16, 17). In contrast to Kwon et al. (21), we had not found any Mtzs clinical isolate that was rendered Mtzr by frxA inactivation alone.
Different protocols were used by Kwon et al. and ourselves to assess MTZ susceptibility or resistance. Their experiments relied on en masse growth responses after spotting aliquots of dense cultures (21), with resistance defined operationally as growth on the selective MTZ-containing medium (a traditional method for determining the minimal inhibitory concentrations [MIC] of antibacterial agents). Because up to about 0.01% of cells in fresh culture may form Mtzr colonies on MTZ-containing medium (due to the mutagenic effects of MTZ, followed by selection for resistance [16, 29, and results of this work]), we considered a strain to be resistant only if the efficiencies of colony formation were unaffected by inclusion of MTZ in the medium. That is, cultures of interest were diluted, aliquots of each dilution were spotted, and the numbers of colonies formed at appropriate dilutions were scored on MTZ-containing and on MTZ-free media (12, 13).
The experiments presented here tested several possible explanations for the apparently different results and interpretations of Kwon et al. and ourselves, including (i) regulation of FrxA synthesis or activity by a quorum-sensing (cell density-dependent) mechanism, (ii) use of fundamentally different types of strains by our two groups, and (iii) use of dense rather than dilute cultures to score MTZ resistance, where interpretation of results with dense cultures might have been complicated by the mutagenicity of products of MTZ activation (21). Northern blot hybridization analyses were also carried out and showed that frxA mRNA and perhaps also rdxA mRNA were more abundant in type II than in type I strains.
MATERIALS AND METHODS
Bacterial strains and general methods.
The Mtzs H. pylori strains used here and their origins are listed in Table 1. H. pylori strains were grown on brain heart infusion agar (Difco) supplemented with 7% horse blood, 0.4% Isovitalex, and the antibiotics amphotericin B (8 μg/ml), trimethoprim (5 μg/ml), and vancomycin (6 μg/ml) (referred to here as BHI agar), essentially as described previously (16, 17). MTZ was added when needed at concentrations appropriate to each experiment, as detailed below. Rifampin-resistant mutants were selected on medium with 5 μg of rifampin per ml. H. pylori cultures were incubated at 37°C under microaerobic conditions (5% O2, 10% CO2, 85% N2).
TABLE 1.
H. pylori strains
| Straina | Origin | Source and/or reference |
|---|---|---|
| Type I strains | ||
| CAN9A | Peru | R. H. Gilman, 18 |
| HK192 | Hong Kong | B. Wong, 18 |
| HUP57 | Spain | M. Lopez Brea, 18 |
| 26695 | United Kingdom | K. Eaton, 3, 31 |
| Type II strains | ||
| R10 | South Africa | I. Segal, 18 |
| SS1 | Australia | A. Lee, 17, 22 |
| X47b | United States | H. Kleanthous, 19 |
| 2600 | Texas | D. H. Kwon, 21 |
| 2667 | Texas | D. H. Kwon, 21 |
| 2714 | Texas | D. H. Kwon, 21 |
| A219 | Alaska (Cordova) | A. J. Parkinson |
| A381 | Alaska (Nome) | A. J. Parkinson |
| A1099 | Alaska (Kipnuk) | A. J. Parkinson |
| A10103 | Alaska (Kotlik) | A. J. Parkinson |
Derivatives of these Mtzs strains with null mutant alleles of the rdxA and frxA nitroreductase genes were constructed for the present experiments by electroporation, using standard rdxA::cat and frxA::aph or frxA::cat insertion mutant DNAs, with selection on BHI agar medium containing 15 μg of chloramphenicol per ml or 20 μg of kanamycin per ml, as described previously (16, 17). The genetic structures of transformants were checked by PCR to verify that they had resulted from allelic replacement using primers rdxA-F and rdxA-R or frxA-F1 and frxA-R1, as appropriate (see Table 2 for primer sequences). Mtzr and Rifr mutant frequencies were measured in at least four separate cultures, each derived from a different single transformant colony and each growing exponentially. Numbers or frequencies of resistant colonies reported are averages of four or more separate determinations. H. pylori genomic DNA isolation, PCR, and automated dye terminator cycle sequencing with rdxA-specific primers were carried out as described previously (16–18).
TABLE 2.
Primers used
| Primer | Sequence | Product size (bp)a |
|---|---|---|
| rdxA-F | 5′-GCAGGAGCATCAGATAGTTCT | 886 |
| rdxA-R | 5′-GGGATTTTATTGTATGCTACAA | |
| rdxA-F11 | 5′-CGCCATTCTTGCAAGATGTTTGA | 542 |
| rdxA-R11 | 5′-CTCGCTTCTGCCACCCTCTT | |
| rdxRT-F | 5′-GCATGCTGTGGTTGAATCTCAC | 347 |
| rdxRT-R | 5′-CGAGCGCCATTCTTGCAAGATGT | |
| rdxRT-F | 5′-GCATGCTGTGGTTGAATCTCAC | 238 |
| rdxRT-R2 | 5′-CCATGGCATTTTGTGATGGTTACT | |
| frxA-F | 5′-GGATATGGCAGCCGTTTATCATT | 780 |
| frxA-R | 5′-GAATAGGCATCATTTAAGAGATTA | |
| frxA-F11 | 5′-GCTTTACAGCACCAACGATTTG | 617 |
| frxA-R16 | 5′-TAAATAACTTCTGTCTTCCAGCGG | |
| frxRT-F | 5′-GGACAGAGAACAAGTGGTTGCTT | 341 |
| frxRT-R | 5′-GCGAACCTAGAATTAGTGTCAT | |
| frxRT-F2 | 5′-CTTCAATCGGGCTTGAACCATGGA | 334 |
| frxRT-R2 | 5′-GCTGCCATCATCATGTTCGCCAT |
Size of product from wild-type gene.
Determination of MTZ susceptibility and resistance.
Frozen bacterial cultures were streaked on MTZ-free BHI agar and incubated for 3 days. Cells from one or a few colonies from these initial plates were then restreaked on fresh MTZ-free BHI agar and incubated for one more day. The resulting exponentially growing cells were suspended in phosphate-buffed saline (PBS) buffer; a series of 10-fold dilutions of these cell suspensions was prepared, and 10 μl of each dilution was spotted on freshly prepared BHI agar containing various concentrations of MTZ (0, 0.2, 0.5, 1.5, 3, 5, 8, 16, 32, or 64 μg/ml). A strain was considered to be susceptible to concentrations of MTZ that caused at least a 10-fold decrease in the efficiency of colony formation by individual cells (efficiency of plating, or EOP). In our hands this use of aliquots from dilute bacterial cultures was more sensitive and reliable for scoring MTZ susceptibility and resistance of H. pylori than the traditional agar dilution method (16), which relies on growth versus nongrowth after spotting suspensions of at least >105 cells on MTZ-containing medium. The results of a large clinical trial have led others to also conclude that neither agar dilution nor the popular Etest method is reliable for determining MTZ susceptibility or resistance in H. pylori (despite prior approval of agar dilution by the National Committee for Clinical Laboratory Standards) (27).
Much of the failure of these traditional methods can be attributed to the mutagenicity of products of MTZ activation: growth of H. pylori at concentrations that are close to the MIC for H. pylori can cause a more than 100-fold increase in new mutants among survivors (29 and Table 3). In practice, the spotting of 105 Mtzs cells on such MTZ-containing media often results in growth of new MTZ-induced Mtzr mutants that could be misread as indicating resistance of all bacteria in the spot (16). Based on this understanding, we also did not attempt to assay MTZ susceptibility in liquid culture; any inference as to whether observed growth in liquid is due to all cells in the population or to just a subset of them is made tenuous by the combination of slow H. pylori growth and mutagenicity of products of MTZ activation.
TABLE 3.
Frequency of rifampin-resistant mutants in H. pylori culturesa
| Strain | Pregrowth with MTZ (μg/ml)b | Survivalc
|
No. of Rifr colonies/108 cells
|
||
|---|---|---|---|---|---|
| Wild type | frxA null mutant | Wild type | frxA null mutant | ||
| 26695 | 0 | 1 | 1 | 10 ± 1 | 7 ± 2 |
| 1 | 1 | 1 | 27 ± 10 | 6 ± 2 | |
| 2 | 10−2 | 10−2 | 93 ± 9 | 42 ± 10 | |
| 3 | 10−4 | 10−4 | 233 ± 37 | 61 ± 9 | |
| HUP57 | 0 | 1 | 1 | 6 ± 2 | 5 ± 1 |
| 1 | 1 | 1 | 27 ± 6 | 5 ± 2 | |
| 2 | 10−1 | 10−1 | 64 ± 7 | 29 ± 6 | |
| 3 | 10−3 | 10−3 | 114 ± 43 | 38 ± 5 | |
| CAN9A | 0 | 1 | 1 | 2 ± 1 | 2 ± 1 |
| 1 | 1 | 1 | 7 ± 2 | 5 ± 2 | |
| 2 | 10−3 | 10−3 | 40 ± 19 | 25 ± 6 | |
| 3 | 10−4 | 10−4 | 49 ± 20 | 27 ± 6 | |
| HK192 | 0 | 1 | 1 | 4 ± 3 | 3 ± 1 |
| 1 | 1 | 1 | 4 ± 2 | 3 ± 1 | |
| 2 | 10−2 | 10−2 | 19 ± 5 | 24 ± 3 | |
| 3 | 10−4 | 10−4 | 55 ± 9 | 28 ± 3 | |
| R10 | 0 | 1 | 1 | 2 ± 1 | 5 ± 2 |
| 0.5 | 1 | 1 | 8 ± 3 | 16 ± 3 | |
| 1 | 10−2 | 10−2 | 106 ± 9 | 50 ± 9 | |
| 2 | 10−6 | 10−4 | 197 ± 32 | 58 ± 14 | |
Data are the mean values and standard deviations of four independent cultures.
Pregrowth on medium with MTZ for 48 h before spreading on medium with rifampin.
Survival is defined as efficiency of colony formation on BHI agar with the indicated Mtz concentration.
When MTZ-resistant mutants were rare (<10−6) and accurate estimates of frequencies of resistant mutants were needed, culture aliquots were spread directly on the surface of an entire plate of MTZ-containing BHI agar rather than spotting smaller culture aliquots in a small area.
Throughout this report, the phenotype of a given strain or its mutant derivative is generally referred to with an R and S designation (e.g., 1.5R 3S) to refer to the maximum tolerated concentration of MTZ (in micrograms per milliliter) (R value) and the minimum MTZ concentration that will kill at least 99% of the culture (S value). The S value would generally correspond to MIC or minimal bactericidal concentration values that would be reported in traditional susceptibility testing of agents that are not mutagenic.
Viability and emergence of Mtzr mutants in high-density cultures.
Exponentially growing wild-type and frxA mutant H. pylori cells (1 × 109 to 2 × 109 cells) were suspended in PBS buffer and spread in 4- to 5-cm-diameter circular areas on MTZ-containing (8 μg/ml) BHI medium. Each day cells were scraped from an untouched area within (not at the edges of) these large patches of bacterial growth and were suspended in PBS buffer at an optical density at 600 nm (OD600) of 1.0. Ten-microliter volumes of suspension were then spotted directly or after serial dilution on MTZ-free or MTZ-containing BHI plates (0 and 8 μg of MTZ/ml) to measure overall bacterial titer (survival) and titer of resistant mutants. Mtzr bacteria used for rdxA sequencing were obtained as pools or after purification of single colonies, as appropriate, on BHI agar with 8 μg of MTZ per ml.
mRNA analyses.
To prepare RNA for reverse transcriptase (RT)-PCR (Fig. 1) or Northern blots (Fig. 2 and 3), cells growing exponentially on BHI agar were subcultured and grown for another 16 h, collected, and washed three times with 10% glycerol in distilled water at 0°C. Total RNA was isolated and prepared for RT-PCR and Northern hybridization essentially as described previously (2, 17) using a Qiagen RNeasy kit and treatment of RNA preparations with RNase-free DNase I (RT-PCR) or by phenol-chloroform and isoamyl extraction and ethanol precipitation (Northern hybridization). To check RNA quality, a few micrograms from each sample was electrophoresed in agarose, and the ethidium bromide-stained gel was inspected to make sure that the ratio of intensities of 23S and 16S rRNAs was about 2:1 and that there was little, if any, accumulation of the small RNA fragments (probably degradation products) that run just ahead of the 0.2-kb RNA size marker (Perfect RNA Markers, 0.2 to 10 kb; Novagen, Madison, Wis.).
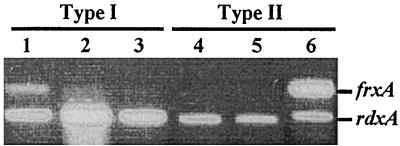
FIG. 1.
RT-PCR amplification of rdxA and frxA gene segments from total RNA of type I and type II strains. The strains tested are as follows: lane 1, HK192; lane 2, 26695; lane 3, HUP57; lane 4, R10; lane 5, X47; lane 6, SS1. The primers frxRT-F2 and frxART-R2 were used to amplify frxA gene segments (334 bp). The primers rdxRT-F and rdxRT-R2 were used to amplify rdxA gene segments (238 bp).
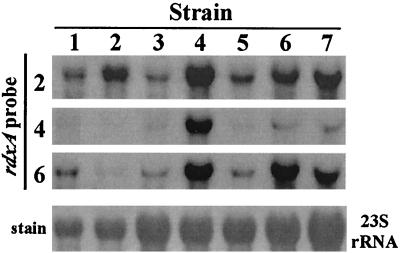
FIG. 2.
DNA sequence of probe affects efficiency of detection of transcripts. RNAs from seven strains were electrophoresed and blotted for Northern analysis in triplicate and probed with rdxA probe DNAs prepared from each of three different strains. The strains used for RNA (and in three cases, DNA probe) preparation were as follows: lane 1, 26695; lane 2, HK192; lane 3, HUP57; lane 4, SS1; lane 5, R10; lane 6, X47; lane 7, 2667 (strains in lanes 1 to 3 are type I and strains in lanes 4 to 7 are type II). 23S and 16S rRNA were visualized after being stained with methylene blue (23S rRNA shown in bottom panel) to ensure that the RNA was of good quality and that equal amounts were loaded in each lane.
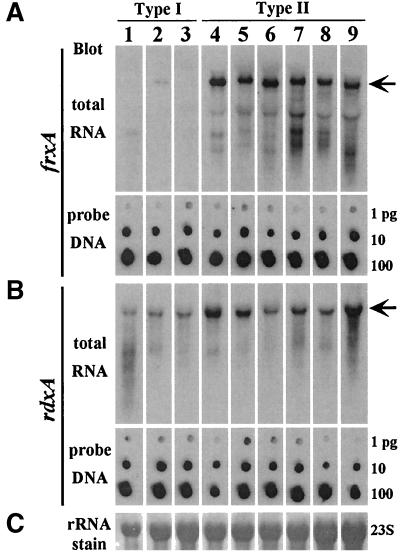
FIG. 3.
frxA and rdxA mRNA transcript levels in type I and type II strains detected with strain-specific probes. Each of the 18 RNA transcripts (rdxA and frxA in each of nine strains) was detected with a probe generated by PCR from genomic DNA from the corresponding strain and DIG labeling. Small differences in amounts or activity of labeled probe were compensated for by using the probe DNA itself as an internal standard and varying exposure for times, ranging from 1 to 5 s, in order to allow direct comparison of amounts of frxA and of rdxA mRNA among the strains tested. The three type I strains tested in this way were as follows: lane 1, 26695; lane 2, HK192; lane 3, HUP57. The six type II stains tested were as follows: lane 4, SS1; lane 5, R10; lane 6, X47; lane 7, 2600; lane 8, 2667; lane 9, 2714.
For RT-PCR experiments, an absence of genomic DNA contamination was verified by PCR using Taq DNA polymerase without reverse transcriptase. Then RT-PCR was carried out with 10 ng of RNA using the One-Step RT-PCR kit (Gibco BRL) with primers frxRT-F and frxRT-R (for frxA mRNA) and rdxRT-F and rdxRT-R (for rdxA mRNA).
For Northern blot analyses 10 μg of RNA was separated by electrophoresis on a 1.5% agarose–6% formaldehyde morpholinepropanesulfonic (MOPS) gel, along with RNA size markers, as above. RNA in the gel was blotted to a Hybond N+ nylon membrane (Amersham) by capillary transfer. DNA fragments used both as probes and as hybridization standards (described below) were generated by PCR from genomic DNA using primer pairs frxA-F11 and frxA-R16 for frxA probes (617 bp) and rdxA-F11 and rdxA-R11 for rdxA probes (541 bp) (Table 1). With each probe, 1 μg of PCR product was labeled with digoxigenin–11 dUTP (DIG) using the DIG High Prime kit (Roche Diagnostics Corporation, Indianapolis, Ind.), and the concentration of labeled product was estimated as recommended by the manufacturer. To avoid effects of base sequence divergence on efficiency of RNA-DNA hybridization (see Fig. 2 and Results), each probe used in the experiment summarized in Fig. 3 was constructed to be strain specific. That is, the probe was generated by PCR from genomic DNA of the strain whose RNA transcripts were to be studied. Eighteen separate probes were prepared in this way, one for rdxA and the other for frxA from each of nine strains.
To prepare hybridization standards, aliquots of rdxA and frxA PCR products from each strain were adjusted to 1 ng/μl, and then aliquots containing 1, 10, and 100 pg of PCR product were spotted as hybridization standards on nylon membranes. Each series of standards was then included in the hybridization sack with its cognate Northern RNA blot and DIG-labeled hybridization probe (concentration of 100 to 200 ng of probe per μl). Hybridization was carried out at 50°C using DIG Easy Hyb buffer (Roche Diagnostics), the filters were washed twice for 15 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 53°C, and hybridization was detected using a DIG Luminescent Detection kit with CDP-star chemiluminescent substrate (Roche Diagnostics). The hybridized Northern Blot and DNA standards were arrayed and exposed for times ranging from 1 to 5 s. Figure 3 was prepared as a composite from various hybridization exposure times, chosen based on giving approximately equal intensities in hybridization standards, thereby allowing direct comparison of frxA and rdxA mRNA levels among the strains tested.
RESULTS
We tested several possible explanations for the apparently different results and proposed mechanisms of MTZ resistance: frxA inactivation is a major cause of MTZ resistance (21) versus frxA inactivation by itself rarely, if ever, confers resistance (16).
Effect of frxA on mutation frequency.
One possible explanation of previous results (20, 21) holds that frxA inactivation rendered H. pylori highly mutable when grown at high cell density, conceivably by perturbing metabolite balances that, in turn, decreased the fidelity of DNA replication or potency of DNA repair (see, e.g., reference 11). This was tested by measuring frequencies of rifampin-resistant mutants (which generally result from point mutations in an RNA polymerase gene [13]) in cultures of five wild-type MTZ-susceptible strains and in their isogenic frxA knockout mutant derivatives. Table 3 shows that frxA inactivation had no significant effect on the frequency of spontaneous mutants, and furthermore that it decreased the yield of MTZ-induced mutants several-fold. Thus, the postulated development of a Mtzr phenotype by frxA inactivation (21), if correct, would not be due to H. pylori entry into a hypermutable state.
Kinetics of MTZ killing at high density.
Several MTZ-susceptible wild-type strains and their frxA knockout mutant derivatives were used to test a model in which incubation at high cell density would render H. pylori tolerant or resistant to MTZ phenotypically, without additional mutation. This entailed inoculation of MTZ-susceptible bacteria at high cell density on MTZ-containing medium, scraping them from the agar surface after one or more days of incubation, and determining bacterial titers on media with and without MTZ. The results (Table 4) showed that all cells that would be considered MTZ-susceptible genetically (unable to form isolated colonies on MTZ-containing medium) were also killed eventually during incubation at high density with MTZ. The kinetics of cell killing depended on both background genotype and whether frxA was functional or not. For example, strain 26695 (type I) was killed more rapidly than HUP57 (also type I) and X47 (type II) (compare titers in Table 4 for day 1, 0 μg of MTZ per ml).
TABLE 4.
H. pylori survival and accumulation of Mtzr mutants on MTZ-containing medium
| Mtzs strain | Titers of H. pylori recovered from MTZ-containing medium on the indicated day and concn (in μg/ml) of MTZ (day [concn])a
|
|||||
|---|---|---|---|---|---|---|
| day 1, MTZ (0) | day 1, MTZ (8) | day 2, MTZ (0) | day 2, MTZ (8) | day 3, MTZ (0) | day 3, MTZ (8) | |
| Type I | ||||||
| 26695 WT | 1.5 × 104 | 1.0 × 104 | 3.1 × 104 | 3.2 × 104 | 3.1 × 105 | 2.8 × 105 |
| 26695 frxA | 2.4 × 104 | 3.4 × 104 | 3.6 × 104 | 4.5 × 104 | 5.3 × 105 | 4.2 × 105 |
| HUP57 WT | 5.9 × 105 | 7.0 × 102 | 1.0 × 103 | 5.0 × 102 | 2.3 × 102 | 3.2 × 102 |
| HUP57 frxA | 4.2 × 106 | 2.0 × 103 | 1.2 × 106 | 1.9 × 103 | 2.2 × 104 | 1.6 × 104 |
| Type II | ||||||
| SS1 WT | 1.5 × 103 | None | None | None | None | None |
| SS1 frxA | 5.6 × 106 | 2.0 × 102 | 3.3 × 106 | 3.0 × 102 | 4.0 × 103 | 3.3 × 103 |
| X47 WT | 1.7 × 105 | None | 5.0 × 102 | None | None | None |
| X47 frxA | 4.0 × 105 | 4.8 × 102 | 5.0 × 104 | 1.9 × 102 | 3.9 × 102 | 1.6 × 102 |
Bacterial suspensions were adjusted to an OD600 of 1.0 and the numbers of CFU per 10-μl aliquot were determined. The titers of H. pylori from drug-free plates (controls) were typically 1 × 107 to 2 × 107 cells per 10 μl per OD600. WT, wild type.
The lack of persistent physiologic resistance without further mutation in bacteria incubated at high density was illustrated by finding that the only cells to survive well on MTZ-containing medium also formed individual Mtzr colonies when replated after appropriate dilution (the identification of new rdxA mutations in such derivatives is described below). Such resistant mutants became abundant after only 1 day in the case of strain 26695 and after 2 days in the case of HUP57 (Table 4). Resistant derivatives of strains SS1 and X47 were not found; this probably reflects a need for mutation in two genes if these strains are to become resistant (16, 17, and below). Inactivation of frxA slowed MTZ-induced killing in all cases, thereby weakening the selection for MTZ resistance. Nevertheless, with frxA mutants, 2 (26695 lineage) or 3 days of incubation on MTZ-containing medium was sufficient to kill most or all cells that had not also become MTZ resistant. Thus, these outcomes argue against the idea that resistance is caused by incubation at high cell density.
Mutation in rdxA needed for frxA mutant strains to become Mtzr.
We next tested the suggestion (20) that Mtzr derivatives of frxA knockout mutant strains selected initially at high cell density rarely, if ever, carried new mutations in rdxA. Mtzr mutants were selected by spreading cells from dense cultures of frxA knockout mutant derivatives of four type I lineages and one type II lineage on MTZ-containing agar, as above, and incubating them for three days. Mtzr colonies formed by single cells were obtained later by streaking diluted aliquots of the dense suspensions of Mtzr cells on new MTZ-containing agar plates. rdxA was PCR amplified from cell suspensions incubated on MTZ-containing agar and from single Mtzr colonies, and the products were sequenced. Table 5 shows that each Mtzr single colony isolate contained a sequence change in rdxA, resulting variously in frameshifts, translation stop codons, or amino acid additions or replacements that would also likely inactivate the gene. In contrast, the rdxA sequence profiles obtained after selecting confluent lawns of Mtzr mutant bacteria matched those from the wild-type parent (data not shown). This might be due to the presence of numerous independent Mtzr mutants in each bacterial pool. In this case, the sequence change in any single mutant allele would be obscured by the many other sequences containing unchanged nucleotides at that site (only when cultures contain just one or a few abundant mutants can each allele be detected reliably). Alternatively, it might reflect DNA extraction before new Mtzr mutants had adequately outgrown their Mtzs siblings (e.g., as shown in Table 4). Either interpretation might explain the seeming wild-type rdxA sequence in H. pylori collected from MTZ-containing agar that was reported previously (20).
TABLE 5.
rdxA alleles from frxA mutant H. pylori recovered from MTZ-containing BHI agara
| Strain | Mtzr isolate | Change in:
|
|
|---|---|---|---|
| DNA | Protein | ||
| Type I | |||
| 26695 frxA::cat | 1 | AG insertion | Frameshift from Glu(138) |
| 2 | GAG→TAG | Glu→stop(75) | |
| 3 | CGC→CAC | Arg→His(16) | |
| HUP57 | 1 | C insertion | Frameshift from Cys(159) |
| frxA::aph | 2 | CAG→TAG | Gln→stop(50) |
| 3 | GA deletion | Frameshift from Glu(107) | |
| CAN9A | 1 | CAA→TAA | Gln→stop(102) |
| frxA::aph | 2 | GAAATCGCT insertion | Insertion of 3 amino acids (Glu, Met, and Ala) from 41 |
| 3 | TGG→TGA | Trp→stop(209) | |
| HK192 | 1 | T insertion | Frameshift from Leu(157) |
| frxA::aph | 2 | T insertion | Frameshift from Met(84) |
| 3 | A deletion | Frameshift from Lys(64) | |
| Type II | |||
| R10 frxA::aph | 1 | GAA→TAA | Glu→stop(32) |
| 2 | GAA→TAA | Glu→stop(32) | |
| 3 | GAG→TAG | Glu→stop(75) | |
H. pylori were recovered from BHI agar containing 8 μg of MTZ per ml. The genes aph and cat confer resistance to kanamycin and chloramphenicol, respectively.
Type II Mtzs strains and frxA.
Three of the clinical isolates whose initial studies had led Kwon and colleagues (20) to conclude that frxA inactivation by itself caused MTZ resistance, strains 2600, 2667, and 2714 (kindly provided by D. H. Kwon and D. Y. Graham) were studied and found to each be highly susceptible to MTZ. In particular, as with type II reference strains from elsewhere, Mtzr mutant colonies were found at frequencies of ≤10−8 on medium with 8 μg of Mtz per ml (see Table 6). In contrast, Mtzr mutant colonies were found in cultures of type I strains at frequencies of about 10−4 (16, 17). There was some variability among type II strains in survival on medium with lower levels of MTZ (e.g., with 3 μg of MTZ per ml, ∼10−6 with each of D. H. Kwon's strains and five of the seven other type II strains, but ∼10−8 with the other two; Table 6). This variability is in accord with the considerable genetic diversity among H. pylori strains, as will be detailed below.
TABLE 6.
Survival of Mtzs H. pylori clinical isolates on MTZ-containing agara
| Strain | Survival on agar containing the following concn of MTZ (μg/ml)
|
||||
|---|---|---|---|---|---|
| 1 | 1.5 | 3 | 5 | 8 | |
| Experimental (type II) | |||||
| 2667 | 1 | 10−3 | 10−6 | 10−7 | <10−8 |
| 2714 | 1 | 10−3 | 10−6 | 10−7 | <10−8 |
| 2600 | 1 | 10−4 | 10−6 | 10−7 | <10−8 |
| X47 | 1 | 10−3 | 10−6 | NDb | <10−8 |
| A1099 | 1 | 10−4 | 10−6 | 10−7 | <10−8 |
| A381 | 10−4 | 10−5 | 10−6 | 10−7 | <10−8 |
| A10103 | 10−4 | 10−5 | 10−6 | ND | <10−8 |
| SS1 | 10−4 | 10−6 | <10−8 | <10−8 | <10−8 |
| R10 | 10−4 | 10−6 | 10−6 | ND | <10−8 |
| A219 | 10−5 | 10−6 | <10−8 | ND | <10−8 |
| Control (type I) | |||||
| 26695 | 1 | 1 | 10−4 | 10−4 | 10−4 |
| CAN9A | 1 | 1 | 10−4 | ND | 10−4 |
| HK197 | 1 | 1 | 10−4 | ND | 10−5 |
| HUP57 | 1 | 1 | 10−4 | ND | 10−4 |
EOP, were determined as described in Materials and Methods, at least four times for each strain, each time with a separate culture.
ND, not determined.
To further examine the roles of nitroreductase genes in the three strains from Kwon and colleagues, new derivatives of these strains with rdxA and frxA null mutations were generated and tested for susceptibility or resistance to MTZ (as detailed in Materials and Methods) (Table 7). We found that each strain remained susceptible to 8 μg of MTZ per ml after inactivation of either one of these genes, as did other type II clinical isolates. In contrast, inactivation of both genes conferred high-level resistance (32 μg per ml) in each case. It is also important that inactivation of either nitroreductase gene dramatically increased the EOP on MTZ-containing agar, i.e., the ease of recovery of new Mtzr mutants (from about ≤10−8 to about 10−4) (Table 7). This is in accord with the need for one new gene mutation in derivatives of type II strains that already lack rdxA or frxA function versus the need for two independent gene mutations in their wild-type (type II) parents.
TABLE 7.
Effects of rdxA and frxA inactivation on H. pylori phenotypea
| Strain | R and S values (in μg/ml) for:
|
|||
|---|---|---|---|---|
| Wild type | rdxA::cat | frxA::aph | rdxA::cat frxA::aph | |
| Type II strainb | ||||
| A219 | 0.5R 1S | 1R 1.5S | 0.5R 1S | 32R 64S |
| SS1 | 0.5R 1S | 1R 1.5S | 1R 1.5S | 32R 64S |
| R10 | 0.5R 1S | 1R 1.5S | 1R 1.5S | 32R 64S |
| A10103 | 0.5R 1S | 1.5R 3S | 0.5R 1S | 32R 64S |
| X47 | 1R 1.5S | 1.5R 3S | 1.5R 3S | 32R 64S |
| 2600 (D. K.)c | 1R 1.5S | 1.5R 3S | 1.5R 3S | 32R 64S |
| A381 | 0.5R 1S | 3R 5S | 0.5R 1S | 32R 64S |
| 2667 (D. K.) | 1R 1.5S | 3R 5S | 1.5R 3S | 32R 64S |
| 2714 (D. K.) | 1R 1.5S | 5R 8S | 1R 1.5S | 32R 64S |
| A1099 | 1R 1.5S | 5R 8S | 1R 1.5S | 32R 64S |
| Type I control strain | ||||
| 26695 | 1.5R 3S | 16R 32S | 1.5R 3S | 32R 64S |
R (as in 1R, 1.5R, etc.) refers to maximum concentration of MTZ that the H. pylori strain can tolerate; S (as in 1.5S, 3S, etc.) refers to minimum concentration of MTZ that will kill at least 99% of the culture and would correspond to the MIC value that is reported in traditional susceptibility testing of antimicrobial agents that are not mutagenic.
The type II strains are grouped according to the change in resistance level that resulted from rdxA inactivation. This, we infer, is related inversely to the level of frxA expression.
Strains studied by D. H. Kwon and associates (21) are identified with (D. K.).
We also note that rdxA inactivation caused small increases in the intrinsic low-level resistance to MTZ in 4 of the 10 type II strains studied (resistant to 3 or 5 but not 8 μg of MTZ per ml; see Table 7), whereas frxA inactivation had little, if any, effect on this low-level MTZ resistance. Since inactivation of both rdxA and frxA increased resistance to the same level in each of these strains (to 32 μg of MTZ per ml; see Table 7), we infer that FrxA activity varies among type II strains. For example, it seemed lower in strains such as 2714 and A1099 than in A219 and SS1, since rdxA mutant derivatives of 2714 and A1099 seemed resistant to slightly higher levels of MTZ (5R versus ≤1R in phenotype).
frxA mRNA is more abundant in type II than in type I cells.
Initially we sought to use RT-PCR (as in reference 17) to test the hypothesis that frxA mRNA is more abundant in type II than in type I strains. Although results with one type II and one type I strain (SS1 and 26695) had supported this view (17 and Fig. 1), results with other strains were erratic. In particular, no frxA RT-PCR product was detected from strains R10 and X47, using rdxA mRNA as an internal standard (RT-PCR with frxA and rdxA primers together), despite mutational evidence showing that acquisition of Mtzr in these strains depended on inactivation of frxA as well as rdxA (Table 7). Various ratios of frxA and rdxA RT-PCR products were obtained from several other type II strains, and one type II strain did not seem to yield an rdxA RT-PCR product at all (see Fig. 1). These unexpected results were obtained repeatedly in trials with RNA preparations from different exponentially growing cultures of the same strain. Accordingly, much of the variation among strains in RT-PCR results was ascribed provisionally to effects of sequence (e.g., secondary structure in single-stranded nucleic acids) on efficiency of amplification rather than differences in levels of target mRNAs.
Northern blot hybridization tests were initiated next to avoid complications that might stem from sequence-dependent differences in amplification efficiency. Preliminary experiments indicated, however, that any given probe tended to detect rdxA mRNA from the same strain more efficiently than from other H. pylori strains. For example, the quite abundant rdxA RNA transcript of the strain labeled 2 in Fig. 2 was detected efficiently only with a DNA probe generated from that strain, not with probes generated from the strains labeled 4 or 6. Similarly, rdxA mRNA from strain 6 was detected more efficiently with the probe from strain 6 than that from 2, and the probe from strain 4 was useful only for detecting the rdxA transcript from 4. These outcomes implied that efficiencies of DNA-RNA hybridization were also affected by sequence divergence among H. pylori strains and that interstrain comparisons based on just one gene-specific probe would be unreliable.
To bypass effects of sequence divergence on efficiencies of hybridization, frxA and rdxA probes specific to (generated from the genomic DNA of) each strain of interest were prepared. To allow direct comparisons of hybridization data from each of nine strains tested (18 separate hybridizations, each with a separately prepared probe), fixed amounts of each rdxA or frxA probe DNA were spotted on separate filters and included as internal standards in each hybridization (for details see Materials and Methods and the legend to Fig. 3). We found that frxA-containing transcripts (Fig. 3A, arrow) were abundant in each of the six type II strains studied, including several that had given erratic results in RT-PCR, whereas frxA transcripts were less abundant or were absent from each of the three type I strains tested. rdxA transcripts were found in all strains tested, as expected, although they seemed to be slightly (perhaps twofold) more abundant in type II than in type I strains.
It is also noteworthy that the transcripts were each longer than could be expected of monocistronic transcription units (about 2.2 kb versus 651 bp for the frxA open reading frame and about 1.4 kb versus 630 bp for the rdxA open reading frame), implying that each is polycistronic. Comparison of the two fully sequenced H. pylori genomes revealed a polymorphism for the presence or absence of a ∼1-kb segment just upstream of frxA (present in strain J99; absent from strain 26695 [4, 31]). PCR tests indicated that this 1-kb segment is present in two of the three type I strains and in each type II strain studied here (data not shown). Thus, our results do not suggest that the presence or absence of this upstream region regulates frxA expression and MTZ susceptibility.
DISCUSSION
The present experiments, initiated to test two alternative views of how H. pylori becomes resistant to MTZ, have given new insights into the genetic diversity of this species. Two types of MTZ-susceptible strains were identified by genetic (mutational) and molecular (Northern blot) tests. Those designated type I needed inactivation only of rdxA to become resistant, apparently because only the rdxA nitroreductase gene was well expressed. Type II strains, in contrast, needed inactivation of both rdxA and the related frxA gene to become resistant, apparently because each gene was highly active. These two Mtzs strain types were also distinguished by frequencies of MTZ-resistant mutants: about 10−4 and 10−8 in cultures of type I and type II strains, respectively, reflecting the need to inactivate just one gene (rdxA) versus two genes (rdxA and frxA) to become resistant. Our tests also showed that frxA inactivation slowed the killing by MTZ of type I strains with functional rdxA genes and increased the resistance of type I derivatives that were already mutated in rdxA (although frxA inactivation by itself did not confer MTZ resistance if rdxA remained functional). These subtle effects of frxA inactivation implied that frxA can be expressed in type I strains, although too weakly for detection in our Northern blots. Given the initial aim of our study, it is noteworthy that these results were also obtained with strains used by another group (21) that had interpreted that frxA inactivation by itself could render H. pylori MTZ resistant. The present results suggest that their interpretation is incorrect.
We ascribe the differences in interpretation to technical aspects of how susceptibility versus resistance was scored. We monitored the susceptibility or resistance of individual cells to MTZ by diluting cultures sufficiently to count colonies formed on media with and without MTZ. This quantitative plating method, although common in bacterial genetics, contrasts with traditional practice in clinical microbiology of scoring (i) growth responses of spots of dense cell suspensions on medium containing the antibiotic (as in reference 21) or (ii) zones of growth inhibition near antibiotic-containing disks or Etest strips laid on dense bacterial lawns. The special value of quantitative plating tests when scoring MTZ resistance stems from the mutagenicity of MTZ and consequently the high frequency of MTZ-resistant mutants (10−4) when mutation in only one gene is needed. This high frequency is enough to sometimes give a misleading appearance of resistance in traditional qualitative en masse growth tests (as in reference 21), a problem that may be exacerbated by the delayed killing of strains that are MTZ susceptible but contain frxA null alleles. Such traditional MTZ resistance tests were also deemed unreliable in a recent large clinical trial (27).
A second technical feature of our experiments also merits comment: the need for strain-specific hybridization probes when comparing transcript levels in different strains. Although initial RT-PCR with strains 26695 (type I) and SS1 (type II) had demonstrated frxA mRNA only in SS1 (17) as expected, RT-PCR with other type II strains often revealed little, if any, frxA product. Erratic results were also obtained when Northern hybridization with a single probe was used to monitor levels of rdxA mRNA in several strains (Fig. 2). These problems can be ascribed to sequence diversity among H. pylori strains (typically some 4 to 6% in loci such as rdxA and frxA) and consequent differences in relative efficiencies of amplification and hybridization between imperfectly matched probe and transcript in Northern blots. Accurate Northern blot results were obtained by generating strain-specific gene probes for each strain of interest, labeling each probe individually, and using each in a separate hybridization. Internal DNA calibration standards were included in each hybridization to allow valid quantitative interstrain comparisons.
Much remains to be learned about the metabolic phenomena that affect MTZ susceptibility and resistance, including the mechanisms underlying the diversity among strains in apparent levels of the related nitroreductases studied here and the actual roles of each nitroreductase during human gastric infection. In particular, although strains with highly expressed frxA genes as defined here seem to be uncommon (≤10% of Mtzs isolates) in many populations (16), we have recently found that about half of Mtzs strains from Lithuania are of this type (G. Dailide, D. Kersulyte, D. Dailidiene, J. Miciuleviciene, and D. E. Berg, unpublished data). Neither the factors underlying this geographic difference nor its generality (i.e., in other Baltic or Northern European countries) are known. That the type I versus type II difference might be adaptive and significant physiologically, however, is suggested by a recent finding that frxA mutants of the type II strain SS1 grow less well in mice than does the wild type (J. Y. Jeong, D. Dailidiene, A. K. Mukhopadhyay, and D. E. Berg, unpublished data). We are attracted to the possibility that the observed differences in gene or enzyme activity may often be significant physiologically for H. pylori—perhaps affecting bacterial nutrition and the persistence of infection in various human hosts.
ACKNOWLEDGMENTS
We thank D. H. Kwon, D. Y. Graham, A. J. Parkinson, H. Kleanthous, B. Velapatiño, R. H. Gilman, B. Wong, I. Segal, T. Alarcon, M. Lopez Brea, A. Lee, and K. Eaton for the various Mtzs H. pylori strains studied here. This research was supported by grants from the U. S. Public Health Service (AI38166, AI49161, DK53727, and P30 DK52574) and the Canadian Institutes for Health Research (MT11318 and RP14292).
REFERENCES
- 1.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Akada J K, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol Microbiol. 2000;36:1071–1084. doi: 10.1046/j.1365-2958.2000.01918.x. [DOI] [PubMed] [Google Scholar]
- 3.Akopyants N S, Eaton K A, Berg D E. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect Immun. 1995;63:116–121. doi: 10.1128/iai.63.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 5.Blaser M J, Berg D E. Helicobacter pylori genetic diversity and risk of human disease. J Clin Investig. 2001;107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cover T L, Berg D E, Blaser M J, Mobley H L T. H. pylori pathogenesis. In: Groisman E A, editor. Principles of bacterial pathogensis. New York, N.Y: Academic Press; 2001. pp. 509–558. [Google Scholar]
- 7.Debets-Ossenkopp Y J, Pot R G, van Westerloo D J, Goodwin A, Vandenbroucke-Grauls C M, Berg D E, Hoffman P S, Kusters J G. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob Agents Chemother. 1999;43:2657–2662. doi: 10.1128/aac.43.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glupczynski Y. Antimicrobial resistance in Helicobacter pylori: a global overview. Acta Gastroenterol Belgium. 1998;61:357–366. [PubMed] [Google Scholar]
- 9.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J O, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 10.Graham D Y. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272–1277. doi: 10.1016/s0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 11.Harris R S, Feng G, Ross K J, Sidhu R, Thulin C, Longerich S, Szigety S K, Winkler M E, Rosenberg S M. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastings J W, Greenberg E P. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol. 1999;181:2667–2668. doi: 10.1128/jb.181.9.2667-2668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heep M, Beck D, Bayerdorffer E, Lehn N. Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:1497–1499. doi: 10.1128/aac.43.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenks P J, Ferrero R L, Labigne A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1999a;43:753–758. doi: 10.1093/jac/43.6.753. [DOI] [PubMed] [Google Scholar]
- 15.Jenks P J, Labigne A, Ferrero R L. Exposure to metronidazole in vivo readily induces resistance in Helicobacter pylori and reduces the efficacy of eradication therapy in mice. Antimicrob Agents Chemother. 1999b;43:777–781. doi: 10.1128/aac.43.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong J Y, Mukhopadhyay A K, Dailidiene D, Wang Y, Velapatiño B, Gilman R H, Parkinson A J, Nair G B, Wong B C Y, Lam S K, Mistry R, Segal I, Yuan Y, Gao H, Alarcon T, Brea M L, Ito Y, Kersulyte D, Lee H-K, Gong Y, Goodwin A, Hoffman P S, Berg D E. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes cause moderate and high-level metronidazole resistance in Helicobacter pylori. J Bacteriol. 2000;182:5082–5090. doi: 10.1128/jb.182.18.5082-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong J Y, Berg D E. Mouse-colonizing Helicobacter pylori SS1 is unusually susceptible to metronidazole due to two complementary reductase activities. Antimicrob Agents Chemother. 2000;44:3127–3132. doi: 10.1128/aac.44.11.3127-3132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kersulyte D, Mukhopadhyay A K, Velapatiño B, Su W W, Pan Z J, Garcia C, Hernandez V, Valdez Y, Mistry R S, Gilman R H, Yuan Y, Gao H, Alarcon T, Lopez Brea M, Nair G B, Chowdhury A, Datta S, Shirai M, Nakazawa T, Ally R, Segal I, Wong B C Y, Lam S K, Olfat F, Boren T, Engstrand L, Torres O, Schneider R, Thomas J E, Czinn S, Berg D E. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleanthous H, Myers G A, Georgakopoulos K M, Tibbitts T J, Ingrassia J W, Gray H L, Ding R, Zhang Z Z, Lei W, Nichols R, Lee C K, Ermak T H, Monath T P. Rectal and intranasal immunizations with recombinant urease induce distinct local and serum immune responses in mice and protect against Helicobacter pylori infection. Infect Immun. 1998;66:2879–2886. doi: 10.1128/iai.66.6.2879-2886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon D H, Kato M, El-Zaatari F A, Osato M S, Graham D Y. Frame-shift mutations in NAD(P)H flavin oxidoreductase encoding gene (frxA) from metronidazole resistant Helicobacter pylori ATCC43504 and its involvement in metronidazole resistance. FEMS Microbiol Lett. 2000;188:197–202. doi: 10.1111/j.1574-6968.2000.tb09193.x. [DOI] [PubMed] [Google Scholar]
- 21.Kwon D H, El-Zaatari F A, Kato M, Osato M S, Reddy R, Yamaoka Y, Graham D Y. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133–2142. doi: 10.1128/aac.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee A, O'Rourke J, De Ungria M C, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 23.Megraud F, Lehn N, Lind T, Bayerdorffer E, O'Morain C, Spiller R, Unge P, van Zanten S V, Wrangstadh M, Burman C F. Antimicrobial susceptibility testing of Helicobacter pylori in a large multicenter trial: the MACH 2 study. Antimicrob Agents Chemother. 1999;43:2747–2752. doi: 10.1128/aac.43.11.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Megraud F. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology. 1998;115:1278–1282. doi: 10.1016/s0016-5085(98)70101-5. [DOI] [PubMed] [Google Scholar]
- 25.Miyaji H, Azuma T, Ito S, Suto H, Ito Y, Yamazaki Y, Sato F, Hirai M, Kuriyama M, Kato T, Kohli Y. Susceptibility of Helicobacter pylori isolates to metronidazole, clarithromycin and amoxycillin in vitro and in clinical treatment in Japan. Ailment Pharmacol Ther. 1997;11:1131–1136. doi: 10.1046/j.1365-2036.1997.00258.x. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay A K, Kersulyte D, Jeong J Y, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya S K, Azuma T, Nair G B, Berg D E. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182:3219–3227. doi: 10.1128/jb.182.11.3219-3227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osato M S, Reddy R, Reddy S G, Penland R L, Graham D Y. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int J Antimicrob Agents. 2001;17:39–44. doi: 10.1016/s0924-8579(00)00320-4. [DOI] [PubMed] [Google Scholar]
- 28.Parsonnet J. Helicobacter and gastric adenocarcinoma. In: Parsonnet J, editor. Microbes and malignancy: infection as a cause of human cancers. New York, N.Y: Oxford University Press; 1999. pp. 372–408. [Google Scholar]
- 29.Sisson G, Jeong J Y, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg D E, Hoffman P S. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA (nitroreductase) gene. J Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tankovic J, Lamarque D, Delchier J C, Soussy C J, Labigne A, Jenks P J. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:608–613. doi: 10.1128/aac.44.3.608-613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K, Klenk H, Gill S, Dougherty B, Nelson K, Quackenbush J, Zhou L, Kirkness E, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H, Glodek A, McKenney K, Fitzegerald L, Lee N, Adams M, Hickey E, Berg D, Gocayne J, Utterback T, Peterson J, Kelley J, Cotton M, Weidman J, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W, Borodovsky M, Karp P, Smith H, Fraser C, Venter J. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Westblom, T. U., S. J. Czinn, and J. G. Nedrud. (ed.). 1999. Current topics in microbiology and immunology, vol. 241. Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Springer Press, Berlin, Germany.