Abstract
Several nations have recently begun to relax their public health protocols, particularly regarding the use of face masks when engaging in outdoor activities. This is because there has been a general trend towards fewer cases of coronavirus disease 2019 (COVID‐19). However, new Omicron sub‐variants (designated BA.4 and BA.5) have recently emerged. These two subvariants are thought to be the cause of an increase in COVID‐19 cases in South Africa, the United States, and Europe. They have also begun to spread throughout Asia. They evolved from the Omicron lineage with characteristics that make them even more contagious and which allow them to circumvent immunity from a previous infection or vaccination. This article reviews a number of scientific considerations about these new variants, including their apparently reduced clinical severity.
Keywords: BA.4 and BA.5, COVID‐19, evolution, omicron sub‐variants, SARS‐CoV‐2
Abbreviations
- rACE2
angiotensin‐converting enzyme 2
- COVID‐19
Coronavirus Disease 2019
- ECDC
European Centre for Disease Prevention and Control
- FCS
furin cleavage site
- GISAID
Global Initiative on Sharing All Influenza Data
- HCoV
human coronavirus
- PCR
polymerase chain reaction
- RBD
receptor‐binding domain
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TCID50
median tissue culture infectious dose
- WGS
whole genome sequencing
- WHO
World Health Organisation
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) pandemic is still ongoing. However, there are signs that it is making the transition to an endemic status. The causative agent of this disease, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has claimed many victims. The World Health OrganizationOrganisation reports that as of 18 July 2022, there have been 559,469,605 confirmed cases of COVID‐19, with 6,361,157 deaths. However, vaccination programs continue, with a total of 12,130,881,147 vaccine doses administered as of 11 July 2022 (https://covid19.who.int; accessed on 18 July 2022).
The significant number of mutations in the newly emerged, highly transmissible Omicron variants of SARS‐CoV‐2 has aroused concern. The exceptional capacity of the more than 50 mutations overall to avoid antibodies may considerably compromise the existing immunisation effort. 1 , 2 About 30 of these are mutations in the spike protein. 3 The Omicron variant has been shown to improve its transmissibility and ability to evade the immune system, 4 but there are also reports that it reduces hospitalisation rates and causes milder symptoms, such as in vaccinated individuals. 5 , 6 This high transmission ability is partly due to the variant's very high affinity for angiotensin‐converting enzyme 2 (ACE2), compared to the wild‐type (Wuhan‐Hu‐1) strain. 7 Moreover, one group comprised of three mutations adjacent to the S1/S2 furin cleavage site was detected in Omicron. This mutation is believed to increase spike protein cleavage and also fusion with host cells, which is thought to contribute to enhanced transmissibility. 8
Molecular dynamics simulations have revealed that the receptor‐binding domain (RBD) of the Omicron variant's spike shows a more diffuse network of interactions caused by the mutations, leading to an increased number of hydrophobic and hydrogen bonding interactions with ACE2. 9 Mathematical modelling supports the use of in silico data to determine the affinity of mutations in the spike protein for ACE2 for predicting the infectivity of SARS‐CoV‐2, where the evolutionary distance of the S gene correlates with the infectivity of this virus. 10
Considering that SARS‐CoV‐2 continues to change, resulting in diverse variants, endemic projections are predicated. Although the global incidence of COVID‐19 continues to decline, the world must remain vigilant for the emergence of new variants. Newly emerged sub‐variants of Omicron have recently been detected and they are on the rise. 11 They are Pango lineages BA.4 (22A) and BA.5 (22B), respectively, which are distinct from Pango lineages BA.2.12.1 (22C) (Figure 1). One study found that the Omicron variant was absent from an intermediate evolutionary branch, which leads one to believe that it could have originated in a species other than humans. Their findings point to the possibility that Omicron evolved in a mouse host because the virus contains five mutation sites that are adapted to mice. 12 Evidence for the evolution of Omicron is critical for future research since it pertains to the behaviour of this variant and its sub‐variants. Therefore, this article gives an update on the occurrence of Omicron BA.4 and BA.5, which appear to predominate in most current COVID‐19 infections around the world.
FIGURE 1.

Three new clades of SARS‐CoV‐2 Omicron evolution. 22A corresponds to Pango lineage BA.4, 22B corresponds to Pango lineage BA.5, and 22C corresponds to Pango lineage BA.2.12.1. (Adapted from Nextstrain; https://nextstrain.org/ncov/gisaid/global/6m)
2. DIFFERENCES OF SUB‐VARIANTS BA.4 AND BA.5 FROM OTHER OMICRON SUBVARIANTS
The international virus classification group advocates the separation of BA.4 and BA.5 from other Omicron sub‐variants. According to the sequencing results for BA.4 and BA.5, the spike (S) protein region contains six mutations that distinguish them from BA.2. BA.4 and BA.5 share the same amino acid mutation, F486V, which is intriguing. The polymerase chain reaction (PCR) test, which is currently the gold standard for detecting the presence of the virus in a person, may fail due to the high number of mutations in the spike of the Omicron variant. As a result, this must be taken into account so that a definitive diagnosis of COVID‐19 can be made. 13
Important antibodies generated in response to the COVID‐19 vaccine and previous SARS‐CoV‐2 infections neutralise the virus by binding to spike proteins. Furthermore, other areas of the variants' genomes contain a variety of mutations. In comparison to BA.2, the spike proteins of BA.4 and BA.5 have a number of distinguishing mutations, including Δ69‐70, L452R, F486V, and R493Q (a reverse mutation, R493Q, is found in the SARS‐CoV‐2 wild‐type strain). 11 Most likely, the presence of a mutation in the S protein is associated with antibody evasion, as well as enhanced viral fitness and ACE2 receptor binding. 7 , 14 , 15 For this reason, vulnerable individuals require a booster to keep their antibody levels high. 16 Figure 2 presents a phylogeny that illustrates the evolutionary relationships among SARS‐CoV‐2 variants that emerged during the pandemic. The branch length of the Omicron group is extremely long. 12 The trends indicate that the Omicron subvariant will continue to predominate in the future. In addition, some findings indicate that Omicron's ability to evade antibody neutralisation is the result of several mutations in the spike protein. Therefore, it is believed that Omicron infections will recur as a result of the clade's ongoing development. 11 , 17
FIGURE 2.
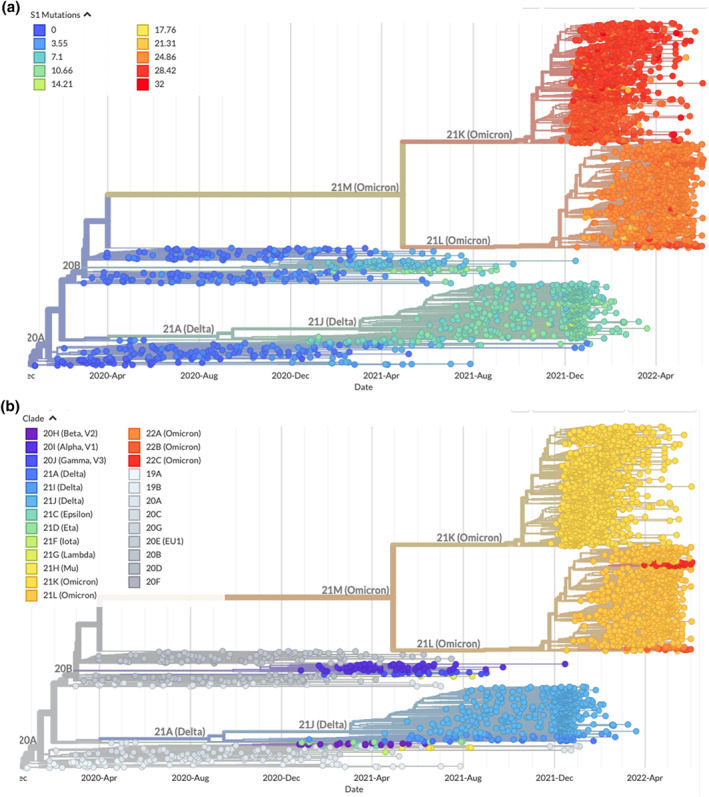
Phylogenies based on mutations at spike protein (a) (available at https://nextstrain.org/ncov/gisaid/global/6m?c=S1_mutations) and clade (b) (available at https://nextstrain.org/ncov/gisaid/global/6m) show evolutionary relationships of SARS‐CoV‐2 from the ongoing COVID‐19 pandemic. The phylogenies are relatively rooted in early Wuhan samples. Temporal resolution assumes that there is a nucleotide substitution rate of 8 × 10−4 substitutions per site per year. The results of Obermeyer 18 are used as the basis for the calculation of mutational fitness
These mutations could provide SARS‐CoV‐2 with a genetic advantage. 19 , 20 Mutations in the Delta variant cause more severe illness than the wild‐type strain, 21 , 22 whereas the omicron variant is more contagious. 23 Chances are that BA.5 will overtake BA.4 because it has increased by around 85% in the US, while in South Africa it has seen an increase of 74% of all COVID‐19 cases. When compared to BA.2, the resistance of BA.2.12.1 to sera from individuals who have been vaccinated and boosted is only slightly increased (1.8‐fold). On the other hand, BA.4/5 is significantly (4.2‐fold) more resistant, and as a result, it has a greater potential to cause vaccine breakthrough infections (an infection of a fully vaccinated person). 24
3. THE EMERGENCE OF THE OMICRON SUB‐VARIANTS BA.4 AND BA.5
According to reports, BA.4 first appeared around the middle of December 2021, whereas BA.5 first appeared around the beginning of 20 January22. 25 In addition, it was claimed that these two sub‐variants spread slightly more rapidly than the preceding Omicron sub‐variants. 26 Therefore, on 22 May 2022, the ECDC (European Centre for Disease Prevention and Control) reclassified each from ‘variant of interest’ to ‘variant of concern.’ Furthermore, BA.4 can only be distinguished from BA.5 by assays targeted at discordant sites that are outside of the spike gene or by whole genome sequencing. An additional finding from a phylogenetic investigation was that the Omicron variants form a monophyletic group, with the Gamma variant serving as its sister lineage. In addition to this, it was hypothesised that the Omicron and Gamma lineages would probably split sometime during the first half of 2020. 12
However, there are currently no indications that these two variants are more clinically severe than previous Omicron lineages. Preliminary in vitro data revealed that both viruses are antigenically distinct from their ancestor. 11 , 27 In addition, when compared to BA.1 and BA.2, they are neutralised by the sera of individuals who have received three doses of the COVID‐19 vaccine with a lower level of effectiveness (Vaxzevria or Comirnaty), or with sera derived from the BA.1 vaccine. 11 , 28 Epidemiologically, it is necessary to re‐evaluate the risk posed by sub‐variants BA.4 and BA.5. The question has been raised as to why there is immune escape by these two sub‐variants in individuals who have previously contracted SARS‐CoV‐2 with other variants, including the previous Omicron sub‐variants, as well as in vaccinated individuals.
4. THE COVID‐19 WAVE
It has been reported that SARS‐CoV‐2 mutations are becoming increasingly rare, thus indicating that there has been inhibition of viral mutations in various ways. The mutation is present in less than 50 of the nearly 10 million SARS‐CoV‐2 sequence samples on Global Initiative on Sharing Avian Influenza Data. 29 This demonstrates that vaccination will continue to be an effective means of prevention, even though there are several types or subtypes of escape antibodies that neutralise them. Hospitalizations are thought to be significantly reduced by complete mass vaccination with booster shots, especially in vulnerable and immunocompromised populations. These actions will undoubtedly significantly lessen the harm that the Omicron variant and its subvariants cause. 30 However, the presence of these variants could still lead to a significant increase in the number of COVID‐19 cases in certain regions, particularly in Europe and the United States, in the coming weeks and months, despite the fact that the overall proportion of BA.4 and BA.5 is still relatively low at this time. It is anticipated that the number of hospital patients will rise if there is a widespread outbreak of these variants. It is believed, however, that it will not be as high as during the Omicron BA.1 and BA.2 waves. Epidemiologists are still investigating whether these two Omicron sub‐variants will be able to displace the dominant Omicron sub‐variants BA.2.12.1 and BA.2, which accounted for 62% and 25% of infections, respectively, in the previous week. With the appearance of this new wave peak of the Omicron sub‐variant, it appears that the BA.4 and BA.5 sub‐variants are not the end of new Omicron sub‐variants.
In approximately 6 months, it is anticipated that additional sub‐variants will appear. However, the peak of the waves will eventually decrease as the amplitude and frequency of the waves decrease over time, although as with the seasonal flu, they will not disappear completely. There appears to be a new wave every 5–7 months (Figure 3), and it is possible that there will only be one wave per year with a lower peak in the future. For instance, the human coronavirus OC43 (HCoV‐OC43) once caused a pandemic about 120 years ago but has since become endemic, and now a new wave of infection occurs only once every year. The clinical manifestations of HCoV‐OC43 and SARS‐COV‐2 share a significant degree of similarity with one another. 31 In 1967, the HCoV‐OC43 strain was discovered in the nasopharynx of a patient who was experiencing cold‐like symptoms. 32
FIGURE 3.

New waves of COVID‐19 appear periodically and are triggered by the emergence of new variants (adapted from World Health Organization (WHO) COVID‐19 Dashboard; https://covid19.who.int)
The question remains as to why an individual who has been exposed to COVID‐19 can contract it again, causing a new wave of infections with each new variant. This may be analogous to seasonal flu, in which vaccinated individuals can contract the virus multiple times. A person who was infected with the virus that causes COVID‐19, made a full recovery, and then later became infected again is said to have been reinfected with the virus. Although reinfected individuals do not show symptoms of severe disease, there is an extraordinary proportion of those naturally infected or who have been vaccinated who are subsequently reinfected with newly emerged variants. 33 In the end, COVID‐19 will develop into this state, which indicates that it has already become endemic. The emergence of new infectious diseases that can be transmitted from animals to humans is the current threat that the world needs to be aware of and anticipate.
Reinfection occurred after at least 6 months (twice with HCoV‐229E and once with HCoV‐OC43) and 9 months (once with HCoV‐NL63), in some cases. Reinfections, on the other hand, were common at 12 months. This is due to the fact that seasonal coronavirus protective immunity is transient. 34
It appears that BA.4 and BA.5 are more infectious than BA.2, which was infectious to a greater extent than BA.1, the original Omicron strain. The inability of some spike gene targeting PCR assays to detect Omicron variants is due to a small deletion in this gene. 25 This small deletion was the signature of the original Omicron strain, but not of BA.2 or BA.3. BA.4 and BA.5 are closely related and should exhibit comparable behaviours. However, care should be taken to avoid the possibility that a particular mutation in the RBD will result in antibody neutralisation escape. This is due to the fact that previous infections with Omicron BA.1 have been proved insufficient to prevent a second infection with BA.4 or BA.5. 28 Although sufficient immunity exists to protect against severe disease, it is insufficient to protect against symptomatic infection.
5. THE CRUCIAL ROLE THAT HEALTH PROTOCOLS PLAY IN PUTTING AN END TO THE COVID‐19 PANDEMIC
When health protocols were implemented, very few people contracted SARS‐CoV‐2, resulting in a significant decline in COVID‐19 cases. Logic dictates that the virus is extremely unlikely to breach masks. Even though it is capable of penetrating, the amount of virus is then so low that it cannot cause illness, as the ability of the virus to cause illness is proportional to the amount of virus that enters the body. The majority of respiratory viruses appear to be as infectious in humans as they are in tissue culture. It was reported that doses of less than one TCID50 of influenza virus, rhinovirus, and adenovirus were sufficient to infect 50% of the tested population. 35
Various factors can affect the level of infectivity of a virus, including age, sex, genetic makeup, social lifestyle factors, population density, number of elderly care facilities, 36 individual immune regulation, population age, 37 air pollution, temperature, and other environmental factors. 38 One example is rhinovirus, the primary cause of the common cold. Although the underlying mechanism is unknown, most strains of this virus replicate more effectively at the lower temperatures found in the nasal cavity than they do at temperatures found in the lungs. 39 Due to the highly infectious nature of SARS‐CoV‐2, 36 masks and social distancing are effective in preventing airborne transmission. 40
The human immune system provides two fundamental lines of defence: innate and adaptive immunities. 41 With each wave that passes, our resistance increases, allowing us to be better prepared for the waves that follow. Young children's immune systems are still maturing, 42 , 43 so it is not surprizing that they fall ill with infection more frequently than do older children and adults. 42
In the end, the magnitude and frequency of COVID‐19 waves will decrease after the human immune layer is formed by several waves. Layered immunity will emerge as a result of reinfection. 44 At this time, it is most likely that COVID‐19 has started to become an endemic disease like the seasonal flu. In the end, it is predicted that the Omicron waves will not disappear, but will become smaller and less frequent over time.
6. CONCLUSIONS
As SARS‐CoV‐2 continues to evolve, it is likely that this virus will not completely vanish. However, as the Omicron sub‐variants continue to predominate, it is predicted that COVID‐19 will eventually become comparable to seasonal influenza, where the disease persists with milder symptoms. Additionally, with layered immune protection acquired by the majority of the population, herd immunity is likely to be achieved. As a result, COVID‐19 will have to be integrated into everyday life. Expectations that the COVID‐19 pandemic will become endemic have been bolstered by the appearance of the Omicron sub‐variants, which cause mild symptoms despite being significantly more contagious than the other variants. The community should be equipped with the knowledge necessary to coexist with COVID‐19 so that it does not interfere with daily life. Data suggests that vaccination can lower the risk of long‐term COVID‐19 infection. Therefore, there is still a need to encourage vaccination advocacy in order to protect individuals who are particularly susceptible to the disease.
AUTHOR CONTRIBUTIONS
Trina Ekawati Tallei: Conceptualisation, Investigation, Formal analysis, Writing ‐ original draft, Writing ‐ review and editing. Saad Alhumaid: Investigation, Formal analysis, Writing ‐ review and editing. Zainab AlMusa: Investigation, Formal analysis. Fatimawali: Investigation, Validation, Writing ‐ review and editing. Diah Kusumawaty: Investigation, Validation, Writing ‐ original draft. Ahlam Alynbiawi: Validation, Writing ‐ original draft. Abeer N. Alshukairi: Conceptualisation, Validation, Writing ‐ original draft. Ali A. Rabaan: Supervision, Project administration, Writing ‐ review and editing.
CONFLICT OF INTEREST
The author declares that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
STATEMENTS AND DECLARATIONS
The authors received no funding for the work they submitted. Aside from that, there are no known financial or personal relationships between the authors that might have influenced the findings reported in this article.
Tallei TE, Alhumaid S, AlMusa Z, et al. Update on the omicron sub‐variants BA.4 and BA.5. Rev Med Virol. 2022;e2391. 10.1002/rmv.2391
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are included within the article.
References
- 1. Celik I, Abdellattif MH, Tallei TE. An Insight based on computational analysis of the interaction between the receptor‐binding domain of the Omicron variants and human angiotensin‐converting enzyme 2. Biology. 2022;11(5):797. 10.3390/biology11050797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khandia R, Singhal S, Alqahtani T, et al. Emergence of SARS‐CoV‐2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID‐19 pandemic. Environ Res. 2022;209:112816. 10.1016/j.envres.2022.112816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin DP, Lytras S, Lucaci AG, et al. Selection analysis identifies unusual clustered mutational changes in Omicron lineage BA.1 that likely impact Spike function. bioRxiv Prepr Serv Biol. Published online January. 2022. 10.1101/2022.01.14.476382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. SARS‐CoV‐2 Omicron variant: recent progress and future perspectives. Signal Transduct Targeted Ther. 2022;7(1):141. 10.1038/s41392-022-00997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abas AH, Marfuah S, Idroes R, et al. Can the SARS‐CoV‐2 Omicron variant confer natural immunity against COVID‐19? Molecules. 2022;27(7):2221. 10.3390/molecules27072221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christie B. Covid‐19: early studies give hope omicron is milder than other variants. BMJ. 2021;375:n3144. 10.1136/bmj.n3144 [DOI] [PubMed] [Google Scholar]
- 7. Mannar D, Saville JW, Zhu X, et al. SARS‐CoV‐2 Omicron variant: antibody evasion and cryo‐EM structure of spike protein\&\#x2013;ACE2 complex. Science. 2022;375(6582):760‐764. 10.1126/science.abn7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS‐CoV‐2 Omicron variant in southern Africa. Nature. 2022;603(7902):679‐686. 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shishir TA, Jannat T, Naser IB. An in‐silico study of the mutation‐associated effects on the spike protein of SARS‐CoV‐2, Omicron variant. PLoS One. 2022;17(4):e0266844. 10.1371/journal.pone.0266844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takaoka Y, Sugano A, Morinaga Y, et al. Prediction of infectivity of SARS‐CoV2: mathematical model with analysis of docking simulation for spike proteins and angiotensin‐converting enzyme 2. Microb Risk Anal. Published online. 2022:100227. 10.1016/j.mran.2022.100227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tuekprakhon A, Huo J, Nutalai R, et al. Further antibody escape by Omicron BA.4 and BA.5 from vaccine and BA.1 serum. bioRxiv; 2022. Published online. 10.1101/2022.05.21.492554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun Y, Lin W, Dong W, Xu J. Origin and evolutionary analysis of the SARS‐CoV‐2 Omicron variant. J Biosaf Biosecurity. 2022;4(1):33‐37. 10.1016/j.jobb.2021.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Islam F, Dhawan M, Nafady MH, et al. Understanding the omicron variant (B.1.1.529) of SARS‐CoV‐2: mutational impacts, concerns, and the possible solutions. Ann Med Surg. 2022;78:103737. 10.1016/j.amsu.2022.103737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saville JW, Mannar D, Zhu X, et al. Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS‐CoV‐2 variants. Nat Commun. 2022;13(1):742. 10.1038/s41467-022-28324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mannar D, Saville JW, Zhu X, et al. Structural analysis of receptor binding domain mutations in SARS‐CoV‐2 variants of concern that modulate ACE2 and antibody binding. Cell Rep 2021;37(12):110156. 10.1016/j.celrep.2021.110156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Juno JA, Wheatley AK. Boosting immunity to COVID‐19 vaccines. Nat Med. 2021;27(11):1874‐1875. 10.1038/s41591-021-01560-x [DOI] [PubMed] [Google Scholar]
- 17. Hachmann NP, Miller J, Collier AY, et al. Neutralization escape by the SARS‐CoV‐2 Omicron variants BA.2.12.1 and BA.4/BA.5. medRxiv. Published online 2022. 10.1101/2022.05.16.22275151 [DOI] [Google Scholar]
- 18. Obermeyer F, Schaffner SF, Jankowiak M, et al. Analysis of 2.1 million SARS‐CoV‐2 genomes identifies mutations associated with transmissibility. medRxiv; 2021. Published online. 10.1101/2021.09.07.21263228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dearlove B, Lewitus E, Bai H, et al. A SARS‐CoV‐2 vaccine candidate would likely match all currently circulating variants. Proc Natl Acad Sci. 2020;117(38):23652‐23662. 10.1073/pnas.2008281117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao S, Lou J, Cao L, et al. Quantifying the transmission advantage associated with N501Y substitution of SARS‐CoV‐2 in the UK: an early data‐driven analysis. J Trav Med. 2021;28(2). 10.1093/jtm/taab011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Z, Huang X, Zhang J, Fu S, Ding D, Tao Z. Differences in clinical characteristics between delta variant and wild‐type SARS‐CoV‐2 infected patients. Front Med. 2022;8. 10.3389/fmed.2021.792135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Islam S, Islam T, Islam MR. New coronavirus variants are creating more challenges to global healthcare system: a brief report on the current nnowledge. Clin Pathol. 2022;15:2632010X221075584. 10.1177/2632010X221075584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62(2):412‐422. 10.1021/acs.jcim.1c01451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q, Guo Y, Iketani S, et al. SARS‐CoV‐2 Omicron BA.2.12.1, BA.4, and BA.5 subvariants evolved to extend antibody evasion. bioRxiv. Published online 2022. 10.1101/2022.05.26.493517 [DOI] [Google Scholar]
- 25. Tegally H, Moir M, Everatt J, et al. Continued emergence and evolution of Omicron in South Africa: new BA.4 and BA.5 lineages. medRxiv. Published online 2022. 10.1101/2022.05.01.22274406 [DOI] [Google Scholar]
- 26. Mohapatra RK, Kandi V, Sarangi AK, et al. The recently emerged BA.4 and BA.5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID‐19 pandemic ‐ Correspondence. Int J Surg. 2022;103:106698. 10.1016/j.ijsu.2022.106698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willett BJ, Kurshan A, Thakur N, et al. Distinct antigenic properties of the SARS‐CoV‐2 Omicron lineages BA.4 and BA.5. bioRxiv. Published online 2022. 10.1101/2022.05.25.493397 [DOI] [Google Scholar]
- 28. Khan K, Karim F, Ganga Y, et al. Omicron sub‐lineages BA.4/BA.5 escape BA.1 infection elicited neutralizing immunity. medRxiv; 2022. Published online. 10.1101/2022.04.29.22274477 [DOI] [Google Scholar]
- 29. Maxmen A. Are new Omicron subvariants a threat? Here’s how scientists are keeping watch. Nature. 2022;604(7907):605‐606. 10.1038/d41586-022-01069-4 [DOI] [PubMed] [Google Scholar]
- 30. Dhawan M, Saied AA, Emran TB, Choudhary OP. Emergence of omicron variant’s sublineages BA.4 and BA.5: risks assessment and possible countermeasures. New Microbes New Infect. 2022;48:100997. 10.1016/j.nmni.2022.100997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keshavarz Valian N, Pourakbari B, Asna Ashari K, Hosseinpour Sadeghi R, Mahmoudi S. Evaluation of human coronavirus OC43 and SARS‐COV‐2 in children with respiratory tract infection during the COVID‐19 pandemic. J Med Virol. 2022;94(4):1450‐1456. 10.1002/jmv.27460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24(6):490‐502. 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rahman S, Rahman MM, Miah M, et al. COVID‐19 reinfections among naturally infected and vaccinated individuals. Sci Rep. 2022;12(1):1438. 10.1038/s41598-022-05325-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short‐lasting. Nat Med. 2020;26(11):1691‐1693. 10.1038/s41591-020-1083-1 [DOI] [PubMed] [Google Scholar]
- 35. Yezli S, Otter JA. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ Virol. 2011;3(1):1‐30. 10.1007/s12560-011-9056-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abu‐Hammad O, Alnazzawi A, Borzangy SS, et al. Factors influencing global variations in COVID‐19 cases and fatalities; A review. Healthc. 2020;8(3):216. 10.3390/healthcare8030216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Freitas E Silva R, Pitzurra R. What are the factors influencing the COVID‐19 outbreak in Latin America? Trav Med Infect Dis. 2020;35:101667. 10.1016/j.tmaid.2020.101667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aabed K, Lashin MMA. An analytical study of the factors that influence COVID‐19 spread. Saudi J Biol Sci. 2021;28(2):1177‐1195. 10.1016/j.sjbs.2020.11.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foxman EF, Storer JA, Fitzgerald ME, et al. Temperature‐dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells. Proc Natl Acad Sci. 2015;112(3):827‐832. 10.1073/pnas.1411030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valsamatzi‐Panagiotou A, Penchovsky R. Environmental factors influencing the transmission of the coronavirus 2019: a review. Environ Chem Lett. 2022;20(3):1603‐1610. 10.1007/s10311-022-01418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marshall JS, Warrington R, Watson W, Kim HL. An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol. 2018;14(2):49. 10.1186/s13223-018-0278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proceedings Biol Sci. 2015;282(1821):20143085. 10.1098/rspb.2014.3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kloc M, Ghobrial RM, Kuchar E, Lewicki S, Kubiak JZ. Development of child immunity in the context of COVID‐19 pandemic. Clin Immunol. 2020;217:108510. 10.1016/j.clim.2020.108510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Netea MG, Domínguez‐Andrés J, Barreiro LB, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20(6):375‐388. 10.1038/s41577-020-0285-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


