Abstract
It is thought that in the gram-negative soil bacterium Sinorhizobium meliloti the protein ExoP is involved in biosynthesis of the acidic exopolysaccharide succinoglycan (EPS I). The amounts and compositions of EPS I produced by mutants expressing ExoP proteins characterized by specific amino acid substitutions in the C-terminal cytoplasmic domain were analyzed. The cytoplasmic domain of the ExoP protein was shown to have ATPase activity. Mutations in the highly conserved Walker A ATP-binding motif prevented ATPase activity of the ExoP protein. Phenotypically, these mutations resulted in much lower levels of succinoglycan which consisted only of monomers of the octasaccharide repeating unit. The ExoP protein has similarities to proteins with autophosphorylating protein tyrosine kinase activity. We found that ExoP was phosphorylated on tyrosine and that site-directed mutagenesis of specific tyrosine residues in the cytoplasmic domain of ExoP resulted in an altered ratio of low-molecular-weight succinoglycan to high-molecular-weight succinoglycan.
The soil bacterium Sinorhizobium meliloti (Rhizobium meliloti) has the ability to produce the acidic exopolysaccharide (EPS) succinoglycan (EPS I), which is required for invasion of Medicago sativa root nodules by S. meliloti (2, 19, 30, 31, 32, 37, 54). Succinoglycan is composed of octasaccharide subunits, which consist of one galactose and seven glucose residues, joined by β-glycosidic linkages (1). It can be modified by acetyl, succinyl, and pyruvyl groups (41). S. meliloti produces a high-molecular-weight (HMW) form and a low-molecular-weight (LMW) form of succinoglycan (30). LMW succinoglycan comprises monomers, dimers, and trimers of the octasaccharide subunit, and it has been shown that the trimer is the symbiotically active species (2, 20, 53). The production of succinoglycan is influenced by the osmolarity of the growth medium. An increase in osmotic pressure results in enhanced production of HMW succinoglycan at the expense of LMW succinoglycan (10).
The biosynthesis of succinoglycan is directed by 21 exo and exs genes, located in a 30-kb gene cluster on megaplasmid 2 (3–6, 8, 11, 17, 18, 36, 42). The octasaccharide repeating unit is synthesized on an undecaprenyl lipid carrier located in the cytoplasmic membrane (48). In a study of the roles of the various exo gene products in succinoglycan biosynthesis, membrane-associated proteins ExoP, ExoQ, and ExoT were determined to be involved in polymerization and secretion of succinoglycan (20). ExoQ was found to be required for production of HMW succinoglycan, and it was suggested that ExoT is involved in synthesis or secretion of the LMW succinoglycan dimers and trimers but not the monomers. A mutation in exoP blocked polymerization of succinoglycan octasaccharide subunits (20), indicating that ExoP plays an important role in the polymerization of succinoglycan.
Analysis of the membrane topology of the ExoP protein showed that this protein can be divided into an N-terminal domain located mainly in the periplasmic space and a C-terminal domain located in the cytoplasm (8). S. meliloti strains carrying a mutated exoP∗ gene, expressing only the N-terminal domain, produced a reduced amount of succinoglycan with an increased ratio of the LMW form to the HMW form. These exoP mutants were still able to invade root nodules (8). Specific amino acid substitutions in the proline-rich motif, which is located near the second transmembrane region in the N-terminal domain, also affected the ratio of HMW succinoglycan to LMW succinoglycan to the benefit of LMW succinoglycan (7). This led to the conclusion that the cytoplasmic C-terminal domain is not essential for production and export of succinoglycan but may have a regulatory function.
The ExoP protein has similarities to proteins involved in polysaccharide chain length determination (8). Related proteins involved in the biosynthesis of lipopolysaccharides (LPS), O-antigen polysaccharides, capsular polysaccharides (CPS), and EPS can be distinguished on the basis of their coiled-coil prediction profiles and other characteristics used for classification, such as size, type of polysaccharide synthesized, sequence similarity, location of transmembrane regions, and the presence of ATP-binding domains (35, 38). ExoP, with a periplasmic domain flanked by two transmembrane regions and an additional cytoplasmic domain, was placed in the PCP2a (polysaccharide copolymerase) family. Like the Ptk protein of Acinetobacter johnsonii or the Wzc protein of Escherichia coli K-12, the C-terminal domain of ExoP contains Walker A and B ATP-binding motifs (15, 24, 52). In several gram-positive bacteria (e.g., Staphylococcus aureus and Streptococcus pneumoniae) the cytoplasmic domain is encoded by a separate gene in CPS biosynthesis operons (34).
Several members of the PCP2a family have been shown to be autophosphorylating protein tyrosine kinases. In the case of Ptk of A. johnsonii, the ATP-binding motif is required for this activity (15). Morona et al. (34) obtained evidence that protein tyrosine phosphorylation negatively regulates CPS production in S. pneumoniae. A similar finding for E. coli was recently described by Vincent et al. (51), although these authors hypothesized that the processes of protein autophosphorylation are different in gram-positive and gram-negative bacteria. The role of autophosphorylation in EPS biosynthesis has not been determined yet. In this context we focused on the biochemical activity of ExoP, particularly with regard to the C-terminal cytoplasmic domain, and demonstrated that ATPase activity and tyrosine phosphorylation occur. The possible role of the ATP-binding motif and individual tyrosine residues in biosynthesis of succinoglycan was investigated.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference and/ or source |
|---|---|---|
| S. meliloti strains | ||
| Rm2011 | Wild type, Nod+ Fix+ Inf+ Cfw+ Nxr Smr | 13 |
| RmΔexoP | Rm2011 carrying a Sper cassette replacing the 40-bp exoN 3′ portion, the intergenic region, the complete exoP region, and the 262 bp downstream of exoP | 7 |
| RmΔPII15 | RmΔexoP, expA1#1015-lacZ-aaC1 | 7 |
| E. coli strains | ||
| XL1-Blue | recA 1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]thi | 12 |
| S17-1 | E. coli 294, thi RP4-2-Tc::Mu-Km::Tn7 integrated into the chromosome | 45 |
| BL21 | F′ ompT rB− mB− | 47 |
| XLmutS | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 relA1 lac mutS::Tn10 (Tetr) [F′ proAB lacIqZΔM15 Tn5 (Kmr)]thi | Stratagene |
| Plasmids | ||
| pK18 | lacZα Kmr | 40 |
| pUK21 | lacZα Kmr | 50 |
| pUC21 | lacZα Apr | 50 |
| pHIP1 | lacZα Gmr Kms, pK18 derivative | This study |
| pHIP1-EB | pHIP derivative carrying the 1.77-kb EcoRI-BamHI fragment of the exoP-thi coding region | This study |
| pExoP | pK19mdGE derivative carrying the fragment deleted from the exoP-exoN region of RmΔexoP | 7 |
| pExoP.ΔESt.Gm | pExoP derivative carrying a Gmr cassette instead of the 665-bp EcoRI-StuI fragment of exoP | This study |
| pExoP.A583D | pExoP with a nucleotide substitution resulting in the amino acid change A583D | This study |
| pExoP.A583P | pExoP with a nucleotide substitution resulting in the amino acid change A583P | This study |
| pExoP.G588E | pExoP with a nucleotide substitution resulting in the amino acid change G588E | This study |
| pExoP.G588V | pExoP with a nucleotide substitution resulting in the amino acid change G588V | This study |
| pExoP.K5891 | pExoP with a nucleotide substitution resulting in the amino acid change K5891 | This study |
| pExoP.Y477G | pExoP with a nucleotide substitution resulting in the amino acid change Y477G | This study |
| pExoP.Y505S | pExoP with a nucleotide substitution resulting in the amino acid change Y505S | This study |
| pExoP.Y758S | pExoP with a nucleotide substitution resulting in the amino acid change Y758S | This study |
| pExoP.Y775S | pExoP with a nucleotide substitution resulting in the amino acid change Y775S | This study |
| pExoP.Y505S/Y775S | pExoP with nucleotide substitutions resulting in the amino acid changes Y505S and Y775S | This study |
| pGEX-5x-1 | GST expression vector | 46; Pharmacia Biotech |
| pGEX-exoPc | pGEX-5x-1 carrying the 925-bp exoP 3′ portion and 70 bp of the intergeneic region | This study |
Culture media and growth conditions.
E. coli strains were grown in Pennassay broth or in Luria-Bertani broth (43) at 37°C. For overexpression the strains were grown in Superbroth (43) at 37 or 30°C. S. meliloti strains were grown in TY (9) or in Luria-Bertani medium. For succinoglycan production S. meliloti strains were grown at 30°C in glutamate–d-mannitol–salts (GMS) medium (pH 7.0) supplemented with 0.24 M sodium chloride, biotin, thiamine, and trace elements (55).
Antibiotics were added as required at the following concentrations: for E. coli, 100 μg of ampicillin per ml, 50 μg of kanamycin per ml, 50 μg of gentamicin per ml, and 10 μg of tetracycline per ml; and for S. meliloti, 200 μg of spectinomycin per ml, 600 μg of streptomycin per ml, 8 μg of nalidixic acid per ml, 8 μg of tetracycline per ml, 40 μg of gentamicin per ml, and 120 μg of neomycin per ml.
DNA and protein biochemistry.
Preparation of plasmid DNA, DNA restriction, agarose gel electrophoresis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), cloning procedures, and transformations of E. coli cells were carried out by using previously described protocols (28, 43). Southern hybridizations were performed as described by Kessler (25). Total DNA was isolated from rhizobia as described by Meade et al. (33).
DNA sequencing.
DNA sequencing to verify new plasmid constructs or mutations was carried out by the Institut für Innovationstransfer an der Universität Bielefeld (IIT Biotech) with an ABI PRISM 377 DNA sequencer (Perkin-Elmer). Sequence data were obtained and processed by using the ABI software according to the manufacturer's instructions.
Construction of the exoP expression plasmid.
Plasmid pExoP (6, 7) (exoP sequence EMBL/GenBank/DDBJ accession number Z22636) served as the template for PCR amplification of the exoPC gene flanked by a BamHI restriction site and a SmaI restriction site. The sequences of the two primers were 5′-GCC CGG ATC CTT GCC TTC CTC GAA TTC CGC G-3′ at the N terminus and 5′-GCA ACC CGG GTC GAT CGC CGC AAG GCT TGA C-3′ at the C terminus. The BamHI-SmaI fragment comprising 925 bp of the 3′ portion of exoP and 70 bp downstream of the exoP coding region was ligated into vector pGEX-5x-1 (Pharmacia Biotech), resulting in plasmid pGEX-exoPC. The sequence of the PCR fragment containing exoPC was verified by DNA sequencing.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by using a Chameleon double-stranded site-directed mutagenesis kit from Stratagene according to the manufacturer's protocol. The target mutagenic primers and the selective primer which were used to generate site-specific mutations are shown in Table 2. The mutations were introduced into plasmid pHIP1-EB, which resulted from insertion of the 1.77-kb EcoRI-BamHI fragment of the exoP-thi gene into the vector pHIP1. All mutations were verified by sequencing.
TABLE 2.
Selective and mutagenic primers
| Primer | Sequencea |
|---|---|
| Selective primer | CAC TTT GAC ATC GAC CCA AGT ACC |
| Mutagenic primers | |
| A583D | ATT GCA TCG GAC CTT CCG GAC |
| A583P | ATT GCA TCG CCC CTT CCG GAC |
| G588E | CCG GAC GAG GAA AAA TCG ATC |
| G588V | CCG GAC GAG GTA AAA TCG ATC |
| K589I | GAC GAG GGA ATA TCG ATC ATT |
| Y477G | GGC GGC GCT GGC GCG GCC TTC |
| Y505S | TCG CTC GGC TCC GTT CCG CTG |
| Y758S | CTC GGC AAA TCC AGC GAC TT |
| Y755S | GGC AAA TAT TCC GTC GAG AAT |
Substitutions in the selective and mutagenic primers are underlined.
Purification of the ExoPC protein.
E. coli BL21 cells were transformed with plasmid pGEX-exoPC. Cells from an overnight culture of this strain were used to inoculate 1 liter of Superbroth supplemented with ampicillin. Cultures were incubated at 37°C with shaking until the A600 was 0.6. Then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM. Incubation was continued at 30°C for 2.5 h with shaking. Cells were harvested by centrifugation at 4,200 × g for 10 min at 4°C and suspended in 40 ml of buffer 1 (pH 7.4) (10 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA, 10% glycerol) containing 20 μg of RNase A per ml and 10 μg of DNase 1 per ml. The cells were disrupted with a French pressure cell at 20,000 lb/in2 two or three times. The state of the cells was checked by light microscopy. Each cell suspension was supplemented with Triton X-100 at a final concentration of 1% and centrifuged at 12,500 × g for 10 min at 4°C.
A column was packed with glutathione-Sepharose 4B matrix as recommended by the manufacturer (bulk glutathione-S-transferase [GST] purification module; Pharmacia Biotech) and equilibrated with buffer 1. Before loading, each GST-ExoPC fusion protein solution was centrifuged at 150,000 × g for 50 min at 4°C. The fusion protein-resin complex in the column was washed three times with 10 ml of buffer 1 containing 1% Triton X-100. Protein was eluted with buffer 2 (pH 8.0) (50 mM Tris-HCl, 5 mM MgCl, 10% glycerol) containing 0.1% Triton X-100 and 10 mM glutathione. After 0.5 ml of elution buffer was loaded, the column was incubated for 0.5 h at 4°C. This process was repeated up to five times. The eluted fractions were collected separately, analyzed by SDS-PAGE, and stored at −20°C.
Immunoblot analysis.
Samples were subjected to SDS-PAGE and transferred to a nitrocellulose membrane by using a semidry electrophoretic transfer cell (Trans-Blot SD-Dry Transfer Cell; Bio-Rad) and the procedure of Towbin et al. (49).
Polyclonal anti-GST antibody (goat; Pharmacia Biotech) was diluted in phosphate-buffered saline supplemented with 0.3% Tween 20 and 10% (wt/vol) nonfat dry milk. Binding of the secondary anti goat immunoglobulin G (IgG)–alkaline phosphatase conjugate (Sigma) was detected with 4 nitroblue tetrazolium chloride (Sigma) and BCIP (5-bromo-4-chloro-3-indolylphosphate) (Sigma).
Monoclonal anti-phosphotyrosine antibody PT-66 (mouse; Sigma) was diluted in Tris-buffered saline supplemented with 0.1% Tween 20 and 0.3% (wt/vol) nonfat dry milk. Binding of the biotinylated secondary anti mouse IgG (Amersham) was detected with streptavidin-biotinylated horseradish peroxidase complex (Amersham) and diaminobenzidine (DAB)-H2O2 (Sigma).
ExoP-peptide antibody (rabbit; Eurogentec) raised with peptide EWGRTPSRLVR was diluted in Tris-buffered saline supplemented with 0.1% Tween 20 and 0.3% (wt/vol) nonfat dry milk. Binding of the secondary biotinylated anti-rabbit IgG (Amersham) was detected with streptavidin-biotinylated horseradish peroxidase complex (Amersham) and DAB-H2O2 (Sigma).
ATPase activity.
For in situ demonstration of ATPase activity by detection of released inorganic phosphate (Pi), the affinity-purified fusion proteins were separated on nondenaturing acrylamide gel as described by Koronakis et al. (26, 27). The gels were either stained with Coomassie brilliant blue or incubated with ATP buffer (40 mM Tris-HCl, 4 mM ATP, 4 mM MgCl2, 5% glycerol) for 20 min at 37°C. After incubation the reaction was stopped with color reagent (0.034% malachite green, 0.1% Triton X-100, and 10.5 g of ammonium molybdate per liter in 1 M HCl) and 34% citric acid (29).
Production of succinoglycan.
S. meliloti strains were grown at 30°C for 10 days in GMS medium as described by Zevenhuizen and van Neerven (55). Cells were removed by centrifugation (11,200 × g, 1 h, 10°C), and the clear culture supernatants, containing the secreted EPS, were lyophilized. After suspension in water (20% of the primary volume), carbohydrates were precipitated with 10 volumes of ethanol and pelleted by centrifugation. The carbohydrates were resuspended and desalted by dialysis (Spectra/Por membrane; molecular weight cutoff, 1,000; Roth) against water for 4 days, and this was followed by concentration of EPS by lyophilization.
Analysis of extracellular carbohydrates by high-performance liquid chromatography (HPLC)–gel permeation chromatography.
Succinoglycan fractions were separated by gel permeation chromatography on Nucleogel columns (2× GFC 4000-8, 1× GFC 300-8; 300 by 7.7 mm; Macherey-Nagel, Düren, Germany) by using a flow rate of 0.8 ml min−1 as described by Becker and Pühler (7); the eluent was 200 mM sodium chloride–200 mM sodium phosphate buffer (pH 7.0).
Analysis of extracellular carbohydrates by gel filtration chromatography and HPAEC-PAD.
Cyclic glucans and HMW and LMW succinoglycan fractions were separated by gel filtration chromatography (Bio-Gel P6, fine mesh; Bio-Rad) by using the procedure of Wang et al. (53); the size of the column (Merck) was 1.6 by 120 cm, 1 ml was loaded, the flow rate was 0.2 ml min−1 and the buffer was pyridine–0.1 M acetic acid (pH 5.0). Ninety 1.5-ml fractions were collected and analyzed for total carbohydrates by the HCl–l-cysteine method (14).
LMW succinoglycan fractions were lyophilized, resolved in water, and analyzed by HPLC–anion-exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) (Dionex Corp.) by using a CarboPac PA-100 column (4 by 250 mm; Dionex Corp.) with a gradient of sodium nitrate in sodium hydroxide buffer (eluent A was 500 mM NaOH, and eluent B was 500 mM NaOH–100 mM NaNO3) and a flow rate of 1 ml min−1. The pulsed amperometric detector (Dionex Corp.) was operated at a sensitivity of 0.1 μC by using the following wave forms (potentials and durations): E1, 0.05 V and 240 ms; E2, 0.75 V and 180 ms; and E3, −0.6 V and 360 ms. The resulting chromatographic data were integrated by using a Merck/Hitachi Chromato-Integrator D 2000.
RESULTS
The C-terminal domain of ExoP fused to GST is phosphorylated on tyrosine.
To investigate the function of the C-terminal domain of ExoP independent from the N-terminal domain, an exoP gene lacking the coding region for the N-terminal domain was synthesized by PCR and inserted into vector pGEX-5x-1. The resulting plasmid, termed pGEX-exoPC, expressed a 57-kDa fusion protein consisting of the C-terminal domain of ExoP with the GST at its N terminus.
The presence of autophosphorylating protein tyrosine kinases in prokaryotic organisms was recently reported by Grangeasse et al. (21), Vincent et al. (51, 52), and Morona et al. (34). The proteins from organisms like A. johnsonii, E. coli, and S. pneumoniae have structural similarities to ExoP. Thus, we examined the overexpressed GST-ExoPC fusion protein for the presence of tyrosine phosphorylation by Western immunoblotting, using a mouse anti-phosphotyrosine monoclonal antibody. Phosphorylation of tyrosine was detected in the purified fusion protein and in the fusion protein in the E. coli cell extract. This result was indicated by inhibition by phosphotyrosine. The possibility that phosphorylation of other amino acids occurred was eliminated by the results of immunoblot analysis with specific antibodies against phosphoserine and phosphothreonine (data not shown).
GST-ExoPC fusion protein possesses ATPase activity.
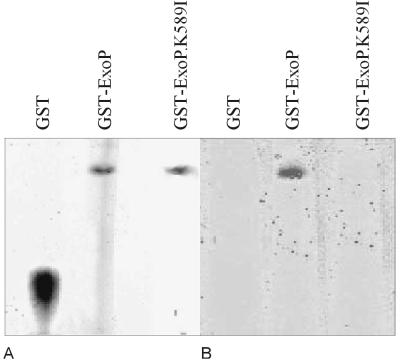
The ExoP protein contains two conserved sequences in the cytoplasmic domain, S582ALPDEGKS590 and V691VVD694, which are similar to the Walker A motif ([AG]X4GK[ST]) and the Walker B motif ([hhhD]), respectively (X indicates any amino acid; h indicates a hydrophobic amino acid; and alternative residues are enclosed in brackets). These motifs were also identified in ExoP homologues like Ptk from A. johnsonii and CpsD from S. pneumoniae (15, 34). Doublet et al. showed that these conserved features are involved in binding of ATP by the Ptk protein kinase of A. johnsonii. Hence, the affinity-purified GST-ExoPC protein was assayed for ATPase activity in nondenaturing polyacrylamide gels. When ATP was provided, the GST-ExoPC protein produced free inorganic phosphate, while GST alone did not (Fig. 1). If ATP was not added to the incubation buffer, free inorganic phosphate was not observed. This result confirmed that the cytoplasmic domain of ExoP is able to hydrolyze ATP.
FIG. 1.
In situ demonstration of GST-ExoP ATPase activity. Affinity-purified GST-ExoP, GST-ExoP. K589I, and control GST protein were separated on a nondenaturing acrylamide gel and either stained with Coomassie brilliant blue (A) or incubated with ATP. A dark green precipitate was formed in the gel upon reaction of released Pi with malachite green (B).
Substitution of amino acids in the Walker A ATP-binding motif blocks ATPase activity and phosphorylation of tyrosine residues.
To assess the relevance of the ATP-binding motif in ExoP in general and with respect to protein tyrosine kinase activity, GST-ExoPC fusion proteins characterized by specific amino acid substitutions in the conserved segment were constructed by site-directed mutagenesis of the exoPC gene.
Constructs for overexpression of the GST-ExoPC mutant proteins were obtained by replacing the EcoRI-SstI wild-type fragments with the corresponding fragments of mutagenized plasmid pHIP1-EB (Table 1) carrying base pair substitutions. After affinity chromatography and electrophoresis of the mutant proteins in nondenaturing polyacrylamide gels, released inorganic phosphate was not detected in any of the mutant protein lanes. An examination of the intensities of the fusion protein bands in the Coomassie blue-stained gel indicated that comparable amounts of these proteins were present. Figure 1 shows the results obtained for mutant GST-ExoPC.K589I, a representative of the mutants with mutations in the ATP-binding motif. It has been found that in all of the eukaryotic ATP-binding proteins examined except protein kinases, the conserved lysine residue is essential for nucleotide binding (44). In this study every single-amino-acid substitution in the ATP-binding motif of the cytoplasmic ExoP domain resulted in a loss of ATPase activity (Table 3).
TABLE 3.
ATPase activities of affinity-purified GST-ExoP wild-type and mutant proteins
| Protein | Mutation | ATPase activity |
|---|---|---|
| GST-ExoP | None (wild type) | + |
| GST-ExoP.A583D | exoP, A583D | − |
| GST-ExoP.A583P | exoP, A583P | − |
| GST-ExoP.G588E | exoP, G588E | − |
| GST-ExoP.G588V | exoP, G588V | − |
| GST-ExoP.K589I | exoP, K589I | − |
| GST-ExoP.Y505S | exoP, Y505S | + |
| GST-ExoP.Y758S | exoP, Y758S | + |
| GST-ExoP.Y775S | exoP, Y775S | + |
| GST-ExoP.Y505S/Y775S | exoP, Y505S/Y775S | + |
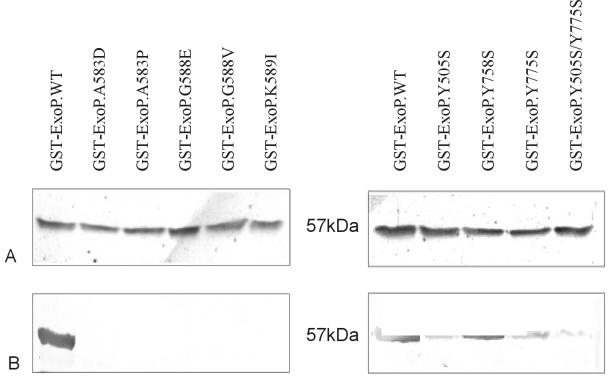
To compare the mutant proteins with the wild-type protein with respect to tyrosine phosphorylation, immunoblot analysis with the monoclonal anti-phosphotyrosine antibody was carried out. Compared to the wild-type GST-ExoPC protein, the mutant proteins did not display phosphorylation signals (Fig. 2). Thus, the mutations in the ATP-binding motif blocked phosphorylation of tyrosine in the cytoplasmic ExoP domain.
FIG. 2.
Presence of the 57-kDa GST-ExoP wild-type and mutant proteins, overexpressed in E. coli BL21. Western immunoblots were probed with the ExoP peptide antibody (A) and with anti-phosphotyrosine antibody PT-66 (B).
Replacement of specific tyrosine residues results in a reduced phosphorylation state in the ExoP protein.
To investigate the role of tyrosine phosphorylation in ExoP, single tyrosine residues were replaced in the ExoPC protein. Site-directed mutagenesis was performed as described above. Grangeasse et al. (21) suggested that there are three putative autophosphorylation tyrosine sites in ExoP. The sequence flanking these tyrosine residues often includes arginine or lysine residues, like the consensus autophosphorylation motifs present in various eukaryotic kinases (39). On the basis of this information, we replaced residue Y477, which is situated in the transmembrane region and is followed by an arginine residue at position +7 (R484). The second tyrosine residue that was replaced was Y505, which is located between the second transmembrane region and the amphiphilic helix at the C terminus of the cytoplasmic domain. This tyrosine was also followed by an arginine residue at position +7 (R512).
The other substitutions were made in the tyrosine-rich region at the C terminus of the protein. We replaced the highly conserved residue Y758, which is flanked by lysine residues at positions −1 (K757) and +8 (K767). The last tyrosine replaced was Y775. As Grangeasse et al. (21) observed that the Ptk protein of A. johnsonii was phosphorylated by preference at multiple tyrosine residues, we also combined the tyrosine substitutions at positions 505 and 775 in one exoPC gene.
The purified mutant proteins were subjected to immunoblot analysis with the monoclonal anti-phosphotyrosine antibody. Protein bands at the expected molecular size positions but with variable intensities were produced (Fig. 2). Except for mutant protein ExoPC.Y758S, which exhibited slightly decreased signal intensity compared to the wild type, replacement of one tyrosine residue resulted in a significant decrease in protein band intensity in the mutants. The protein band of the double mutant was even less intense. Every tyrosine substitution resulted in a modified immunoblot pattern compared to the wild-type ExoPC fusion protein pattern.
The assay for ATPase activity of purified mutant proteins after electrophoretic separation in nondenaturing acrylamide gels showed that none of the mutations affected the ability of the mutants to hydrolyze ATP.
Mutations in the ATP-binding motif result in much lower levels of succinoglycan which consists only of monomers of the repeating unit.
To investigate the phenotypic effect of mutations on the biosynthesis of succinoglycan, S. meliloti Rm2011 mutant strains expressing the exoP mutant genes were constructed. The 665-bp EcoRI-StuI fragment of pHIP1-EB carrying the mutations was inserted into plasmid pExoP, replacing the wild-type fragment. The exoP mutant genes were expressed in S. meliloti exoP deletion mutant RmΔPII15 in order to eliminate interference from ExoP proteins encoded by the endogenous gene and ExoP proteins encoded by the exoP mutant genes, as described by Becker and Pühler (7). Integration of wild-type and mutant pExoP plasmids into the genome of RmΔPII15 by homologous recombination occurred upstream of the deletion site, thereby restoring the native genomic structure, which was verified by Southern hybridization.
When cultured in GMS medium, S. meliloti Rm2011 produced both HMW succinoglycan and LMW succinoglycan, which has been reported to consist of monomers, dimers, and trimers of the octasaccharide subunit (53). Since the mutants with mutations in the ATP-binding motif produced very small amounts of succinoglycan, we increased the osmolarity of the medium to 0.24 M NaCl in order to obtain larger amounts of HMW succinoglycan (10). Fractionation of the supernatants of S. meliloti wild-type and mutant strains on a gel filtration column resulted in three major carbohydrate peaks. The first peak, which eluted in the void volume of the column, was the HMW succinoglycan fraction, the second peak contained the cyclic glucan fraction, and the third peak represented the LMW succinoglycan.
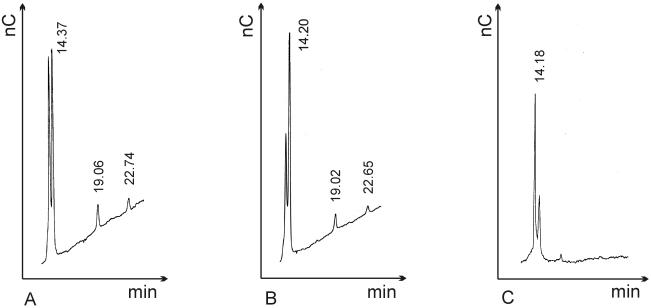
Compared to the wild-type strain, the tyrosine mutant strains showed exactly the same fractionation of succinoglycan. Analysis of the composition of the LMW succinoglycan by HPAEC-PAD resulted in similar chromatograms for the wild-type and tyrosine mutant strains (Fig. 3). The LMW succinoglycan fractions separated into a large monomer peak and two smaller di- and trimer peaks.
FIG. 3.
Chromatographic separation of LMW succinoglycan. (A) Wild type; (B) tyrosine mutant RmΔPII15.pExoP-Y505S; (C) ATP-binding motif mutant RmΔPII15.pExoP-K589I. The monomer fraction eluted after 14 min, the dimer fraction eluted after 19 min, and the trimer fraction eluted after 22 min.
The results obtained for the ATP-binding motif mutants were different from the results for the wild-type strain. In addition to a reduced total succinoglycan yield (Table 4), di- and trimers were not detected in the LMW succinoglycan fractions (Fig. 3). Only the octasaccharide subunit of succinoglycan was found in the supernatants of the ATP-binding motif mutants, which indicates that the mutants were able to synthesize the monomeric unit but were not able to completely polymerize it. Hence, these mutants had the same phenotypic characteristics as exoP deletion mutants (20).
TABLE 4.
Production of HMW and LMW succinoglycans by S. meliloti Rm201I mutants characterized by alterations in the C-terminal domain of the ExoP proteina
| Strain | Mutation | EPS production (mg of Glc equivalents liter−1) | % HMW succinoglycan | % LMW succinoglycan |
|---|---|---|---|---|
| Rm2011 | None (wild type) | 810 | 70 | 30 |
| RmΔPII15/pExoP | None (wild type) | 790 | 71 | 29 |
| RmΔPII15/pExoP.A583D | exoP, A583D | 60 | NDb | 100 |
| RmΔPII15/pExoP.A583P | exoP, A583P | 65 | ND | 100 |
| RmΔPII15/pExoP.G588E | exoP, G588E | 62 | ND | 100 |
| RmΔPII15/pExoP.G588V | exoP, G588V | 59 | ND | 100 |
| RmΔPII15/pExoP.K589I | exoP, K589I | 60 | ND | 100 |
| RmΔPII15/pExoP.Y505S | exoP, Y505S | 360 | 10 | 90 |
| RmΔPII15/pExoP.Y758S | exoP, Y758S | 685 | 30 | 70 |
| RmΔPII15/pExoP.Y775S | exoP, Y775S | 783 | 74 | 26 |
| RmΔPII15/pExoP.Y505S/Y775S | exoP, Y505S/Y775S | 425 | 24 | 76 |
The standard deviations were equal to or less than 10% and equal to or less than 5% for determinations of the total amount of EPS as expressed in glucose equivalents and the ratio of HMW succinoglycan to LMW succinoglycan, respectively.
ND, not detectable.
Tyrosine mutants produce modified ratios of LMW and HMW succinoglycans.
Gel filtration chromatography indicated that some of the tyrosine mutants differed from the wild type in terms of the ratio of HMW succinoglycan to LMW succinoglycan. This observation was verified by gel permeation HPLC on Nucleogel columns. Mutant RmΔPII15.pExoP-Y477G produced the same peak areas for LMW and HMW succinoglycans as the wild type. This was not surprising because the mutation at position 477 was located in the putative transmembrane region and therefore was probably not phosphorylated. Mutant RmΔPII15.pExoP-Y505S produced a completely different result (Table 4). The mutation of this mutant resulted in drastically enhanced production of LMW succinoglycan at the expense of HMW succinoglycan. A ratio of HMW succinoglycan to LMW succinoglycan of 10:90 was obtained. The total amount of EPS was less than 50% of the amount of wild-type EPS produced. Mutant RmΔPII15.pExoP-Y775S acted like the wild type. Strain RmΔPII15.pExoP-Y505S/Y775S carrying the double mutation also produced more LMW succinoglycan than the wild type, but the double mutation did not result in as drastic an alteration as that observed with mutant RmΔPII15.pExoP-Y505S. The ratio of HMW succinoglycan to LMW succinoglycan determined for the double mutant was 24:76. Finally, the mutation at position 758 (RmΔPII15.pExoP-Y758S) resulted in a ratio of HMW succinoglycan to LMW succinoglycan of 30:70 and slightly decreased production of total EPS (Table 4).
DISCUSSION
In this study the C-terminal cytoplasmic domain of ExoP from S. meliloti was shown to influence the polymerization and export of succinoglycan. The function of ExoP is affected by its ATPase activity and the phosphorylation state of its tyrosine residues in the cytoplasmic domain.
Doublet et al. (15) showed that the ATP-binding motif in the C-terminal domain of A. johnsonii Ptk was required for phosphorylation activity. The ATP molecule which serves as the phosphoryl donor binds to highly conserved Walker A and B motif protein sites. The presence of Walker motifs A and B has been described for a wide variety of prokaryotic and eukaryotic ATP-or GTP-binding proteins (22), and these motifs were also found in the protein sequence of the cytoplasmic ExoP domain. Previously (8), we reported a potential ATP- and GTP-binding motif in ExoP and local homology to prokaryotic ATPases. In this study we verified the ATPase activity of ExoP. On the basis of strong structural similarities between Ptk of A. johnsonii (21) and ExoP, it seems very likely that tyrosine phosphorylation in ExoP also occurs due to an autophosphorylating activity, such as that described for Ptk.
In accordance with this assumption, the ExoPC proteins of the ATP-binding motif mutants were not phosphorylated on tyrosine. This finding might be an indication that ATP is the phosphoryl donor, as in Ptk. It also supports the observation of Doublet et al. (15) that binding and hydrolysis of ATP occur at a site different from the phosphorylation site. We found that binding and/or hydrolysis of ATP is essential for the function of ExoP, as the phenotypes of the ATP-binding mutants did not differ from those of exoP deletion mutants with regard to succinoglycan biosynthesis.
Phosphorylation of the CpsD protein in the gram-positive bacterium S. pneumoniae required the presence of another protein, CpsC, which enabled ATP to bind to CpsD (34). However, in contrast to CpsD, no additional protein was necessary to catalyze phosphorylation of Wzc, a protein tyrosine kinase in E. coli comprising a C-terminal cytoplasmic domain and an N-terminal periplasmic domain (51). CpsD corresponds to the C-terminal domain and CpsC is similar to the N-terminal domain of proteins belonging to the PCP2a family, like Wzc and ExoP. However, Vincent et al. (51) reported that even the C-terminal domain of Wzc alone could be phosphorylated. These results supported our hypothesis that the tyrosine kinase activity and a regulatory function in biosynthesis of succinoglycan should be assigned to the C-terminal domain of the ExoP protein of S. meliloti.
In contrast to tyrosine phosphorylation in eukaryotic cells, in which a high number of protein tyrosine kinases and the essential roles of these molecules in the control of various cellular functions, including signal transduction, growth control, and metabolism, are well known (16, 23), tyrosine phosphorylation in prokaryotic organisms is still poorly understood. IIan et al. (24) observed that protein tyrosine kinases in bacterial pathogens are commonly associated with production of EPS and virulence. A direct influence of tyrosine phosphorylation on a phenotype was demonstrated only for the CpsD protein. Morona et al. (34) reported that tyrosine phosphorylation negatively regulated CPS biosynthesis. Recently, Vincent et al. (51) demonstrated that synthesis of the CPS colanic acid in E. coli is modulated by reversible tyrosine phosphorylation of protein Wzc, which has to be dephosphorylated to be active in colanic acid synthesis.
The presence of proteins which catalyze dephosphorylation of proteins at tyrosine residues has also been demonstrated for A. johnsonii and E. coli (51, 52). In these organisms phosphotyrosine protein phosphatases have been identified which are encoded by genes genetically linked to protein tyrosine kinase-encoding genes ptk of A. johnsonii and wzc and etk of E. coli. Interestingly, a phosphatase-encoding gene was not found to be linked to exoP in S. meliloti. The gene cluster involved in biosynthesis of EPS does not encode a gene product that is similar to phosphotyrosine protein phosphatases. A Blast search of the genome of S. meliloti (http://sequence.toulouse.inra.fr/rhime /complete/doc/complete.html) revealed only two potential phosphatases. One of these phosphatases, encoded on pSymA, exhibited 30% identity and 40% similarity to the phosphatase encoded by wzb in E. coli. The other phosphatase, identified an arsenate reductase, is encoded on the chromosome and exhibited 30% identity and 43% similarity to the wzb-encoded enzyme.
Our results also imply that the composition of succinoglycan in S. meliloti is influenced by the phosphorylation state of tyrosine residues in ExoPC. The levels of succinoglycan produced by the tyrosine substitution mutants differed significantly, although the mutant proteins still displayed ATPase activity, indicating that the mutation did not eliminate the function of the proteins. In particular, the tyrosine substitution at amino acid position 505 resulted in a significant decrease in the amount of HMW succinoglycan. When the phosphotyrosine immunoblot analysis procedure was used, the signal intensity did not necessarily indicate that the succinoglycan composition was modified. The signal of the double mutant indicated that we did not replace every tyrosine residue which could be phosphorylated in the cytoplasmic domain. Our results led to the inference that phosphorylation of one tyrosine residue might be influenced by the phosphorylation state of other tyrosine residues. It may be that one phosphorylated tyrosine residue promotes phosphorylation of other residues, independent of its direct contribution to the function of the ExoP protein. This would explain the presence of the intense, almost wild-type-like protein band of mutant RmΔPII15.pExoP-Y758S on the phosphotyrosine immunoblot and the strongly modified phenotype of this mutant.
Mutant RmΔPII15.pExoP-Y505S exhibited phenotypic similarities to mutant RmP∗Δ1, which expressed a truncated ExoP protein lacking the whole C-terminal domain (8). The LMW succinoglycan fraction of RmΔPII15.pExoP-Y505S contained normal amounts of mono-, di-, and trimers, which was not surprising because like all tyrosine mutants, RmΔPII15.pExoP-Y505S was still able to produce at least small amounts of HMW succinoglycan. It is thought that ExoP is required for production of dimers of succinoglycan (20). Since mutant RmΔPII15.pExoP-Y505S produced dimers of the succinoglycan repeating unit, the substitution at position 505 did not destroy this possible function of ExoP. However, the data indicated that an ExoP protein not phosphorylated at position 505 influences the biosynthetic pathway. As the ExoQ protein of S. meliloti is thought to be necessary for production of HMW succinoglycan (20), the mutated ExoP protein might have negative regulatory effects on ExoQ. Otherwise, it is assumed that the ExoT protein is involved in the synthesis of LMW oligosaccharides of succinoglycan (20). An ExoP protein not phosphorylated at position 505 might also be a positive regulator of ExoT activity. Both possibilities are consistent with the hypothetical model which states that ExoP regulates the degree of succinoglycan polymerization by controlling polymerization activities of other proteins, most likely ExoQ and ExoT (20). Whether these proteins also contribute to the phosphorylation process and how the mechanism of tyrosine phosphorylation itself is involved remain to be investigated.
ACKNOWLEDGMENTS
This work was supported by grants Be2121/1-2 and Be2121/1-3 from Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Aman P, McNeil M, Franzen L, Darvill A G, Albersheim P. Structural elucidation using HPLC-MS of the acidic polysaccharide secreted by Rhizobium melilioti strain Rm1021. Carbohydr Res. 1981;95:263–282. [Google Scholar]
- 2.Battisti L, Lara L, Leigh J A. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci USA. 1992;89:5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker A, Kleickmann A, Küster H, Keller M, Arnold W, Pühler A. Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltranferases. Mol Plant-Microbe Interact. 1993;6:735–744. doi: 10.1094/mpmi-6-735. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, Kleickmann A, Arnold W, Pühler A. Analysis of the Rhizobium meliloti exoH/exoK/exoL fragment: ExoK shows homology to excreted endo-β-1,3-1,4-glucanases and ExoH resembles membrane proteins. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00279541. [DOI] [PubMed] [Google Scholar]
- 5.Becker A, Küster H, Niehaus K, Pühler A. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol Gen Genet. 1995;249:487–497. doi: 10.1007/BF00290574. [DOI] [PubMed] [Google Scholar]
- 6.Becker A, Kleickmann A, Keller M, Arnold W, Pühler A. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 7.Becker A, Pühler A. Specific amino acid substitutions in the proline-rich motif of the Sinorhizobium meliloti ExoP protein result in enhanced production of low-molecular-weight succinoglycan at the expense of high-molecular-weight succinoglycan. J Bacteriol. 1998;180:395–399. doi: 10.1128/jb.180.2.395-399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker A, Niehaus K, Pühler A. Low molecular weight succinoglycan is predominantly produced by Rhizobium meliloti strains carrying a mutated ExoP protein characterized by a periplasmic N-terminal and a missing C-terminal domain. Mol Microbiol. 1995;16:191–203. doi: 10.1111/j.1365-2958.1995.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 9.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 10.Breedveld M W, Zevenhuisen L P T M, Zehnder A J B. Osmotically induced oligo- and polysaccharide synthesis by Rhizobium meliloti SU47. J Gen Microbiol. 1990;136:2511–2519. [Google Scholar]
- 11.Buendia A M, Enenkel B, Köplin R, Niehaus K, Arnold W, Pühler A. The Rhizobium meliloti exoZ/exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose-4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol Microbiol. 1991;5:1519–1530. doi: 10.1111/j.1365-2958.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 12.Bullock W C, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactoside selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 13.Casse F, Boucher C, Hulliot J S, Michel M, Denarie J. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J Gen Microbiol. 1979;113:229–242. [Google Scholar]
- 14.Chaplin M F. Reducing sugar-neocuprine assay. In: Chaplin M F, Kennedy J F, editors. Carbohydrate analysis—a practical approach. Oxford, United Kingdom: IRL; 1986. p. 3. [Google Scholar]
- 15.Doublet P, Vincent C, Grangeasse A J, Cozzone B, Duclos B. On the binding of ATP to the autophosphorylating protein, Ptk, of the baterium Acinetobacter johnsonii. FEBS Lett. 1999;445:137–143. doi: 10.1016/s0014-5793(99)00111-8. [DOI] [PubMed] [Google Scholar]
- 16.Fantl W J, Johnson D E, Williams L T. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 17.Glucksmann M A, Reuber T L, Walker G C. Family of glycosyltransferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glucksmann M A, Reuber T L, Walker G C. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González J E, Reuhs B L, Walker G C. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González J E, Semino C E, Wang L, Castellano-Torres L E, Walker G C. Biosynthetic control of molecular weight in the polymerization of the octasaccharide subunits of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Proc Natl Acad Sci USA. 1998;95:13477–13482. doi: 10.1073/pnas.95.23.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grangeasse C, Doublet P, Vaganay E, Vincent C, Deleage G, Duclos B, Cozzone A J. Characterization of a bacterial gene encoding an autophosphorylating protein tyrosine kinase. Gene. 1997;204:259–265. doi: 10.1016/s0378-1119(97)00554-4. [DOI] [PubMed] [Google Scholar]
- 22.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 23.Hunter T. Protein kinases and phosphatases: the Yin and Yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 24.Ilan O, Bloch Y, Frankel G, Ullrich H, Geider K, Rosenshine I. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 1999;18:3241–3248. doi: 10.1093/emboj/18.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler C. Nonradioactive labeling and detection of biomolecules. Berlin, Germany: Springer; 1992. [Google Scholar]
- 26.Koronakis E, Hughes C, Milisav I, Koronakis V. Protein exporter function and in vitro ATPase activity are correlated in ABC-domain mutants of HlyB. Mol Microbiol. 1995;16:87–96. doi: 10.1111/j.1365-2958.1995.tb02394.x. [DOI] [PubMed] [Google Scholar]
- 27.Koronakis V, Hughes C, Koronakis E. ATPase activity and ATP/ADP-induced conformational change in the soluble domain of the bacterial protein translocator HlyB. Mol Microbiol. 1993;8:1163–1175. doi: 10.1111/j.1365-2958.1993.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lanzetta P A, Alvarez L, Reinach P, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 30.Leigh J A, Lee C C. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J Bacteriol. 1988;170:3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 32.Leigh J A, Signer E R, Walker G C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morona J K, Paton J C, Miller D C, Morona R. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol Microbiol. 2000;35:1431–1442. doi: 10.1046/j.1365-2958.2000.01808.x. [DOI] [PubMed] [Google Scholar]
- 35.Morona R, Van Den Bosch L, Daniels C. Evaluation of Wzz/MPA I/MPA 2 proteins based on the presence of coiled-coil regions: Microbiology. 2000;146:1–3. doi: 10.1099/00221287-146-1-1. [DOI] [PubMed] [Google Scholar]
- 36.Müller P, Keller M, Weng W M, Quandt J, Arnold W, Pühler A. Genetic analysis of Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol Plant-Microbe Interact. 1993;6:55–65. doi: 10.1094/mpmi-6-055. [DOI] [PubMed] [Google Scholar]
- 37.Niehaus K, Becker A. The role of microbial surface polysaccharides in the Rhizobium-legume interaction. In: Biswas B B, Das H K, editors. Subcellular biochemistry. Plant-microbe interactions. New York, N.Y: Plenum Press; 1998. pp. 73–116. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen I T, Beness A M, Saier M H. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 39.Pearson R B, Kemp B E. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 40.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 41.Reinhold B B, Chan S Y, Reuber T L, Marra A, Walker G C, Reinhold V N. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J Bacteriol. 1994;176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reuber T L, Walker G C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/technology. 1983;1:784–791. [Google Scholar]
- 46.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-tranferase. Gene. 1988;67:31–41. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 47.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 48.Tolmasky M E, Staneloni R J, Ugalde R A, Leloir L F. Lipid-bound sugars in Rhizobium meliloti. Arch Biochem Biophys. 1980;203:358–364. doi: 10.1016/0003-9861(80)90187-3. [DOI] [PubMed] [Google Scholar]
- 49.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 51.Vincent C, Duclos B, Grangeasse C, Vaganay E, Riberty M, Cozzone A J, Doublet P. Relationship between exopolysaccharide production and protein-tyrosine phopsphorylation in Gram-negative bacteria. J Mol Biol. 2000;304:311–321. doi: 10.1006/jmbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 52.Vincent C, Doublet P, Grangeasse C, Vaganay E, Cozzone A J, Duclos B. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol. 1999;181:3472–3477. doi: 10.1128/jb.181.11.3472-3477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Wang Y, Pellock B, Walker G C. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J Bacteriol. 1999;181:6788–6796. doi: 10.1128/jb.181.21.6788-6796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.York G M, González J E, Walker G C. Exopolysaccharides and their role in nodule invasion. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul. Minn: International Society for Molecular Plant-Microbe Interactions; 1996. pp. 325–330. [Google Scholar]
- 55.Zevenhuisen L P T M, van Neerven A R W. (1,2)-β-d-Glucan and acidic oligosaccharides produced by Rhizobium meliloti. Carbohydr Res. 1983;118:127–134. [Google Scholar]