Abstract
The growth phase-dependent activity profile of the alternate transcription factor ςB and its effects on the expression of sar and agr were examined in three different Staphylococcus aureus strains by Northern blot analyses and by the use of reporter gene fusion experiments. Significant ςB activity was detectable only in the clinical isolates MSSA1112 and Newman, carrying the wild-type rsbU allele, but not in the NCTC8325 derivative BB255, which is defective in rsbU. ςB activity peaked in the late exponential phase and diminished towards the stationary phase when bacteria were grown in Luria-Bertani medium. Transcriptional analysis and a sarP1-sarP2-sarP3 (sarP1-P2-P3)-driven firefly luciferase (luc+) reporter gene fusion demonstrated a strong ςB activity- and growth phase-dependent increase in sar expression that was totally absent in either rsbU or ΔrsbUVWsigB mutants. In contrast, expression of the agr locus, as measured by RNAIII levels and by an hldp::luc+ fusion, was found to be higher in the absence of ςB activity, such as in rsbU or ΔrsbUVWsigB mutants, than in wild-type strains. Overexpression of ςB in BB255 derivatives resulted in a clear increase in sarP1-P2-P3::luc+ expression as well as a strong decrease in hldp::luc+ expression. The data presented here suggest that ςB increases sar expression while simultaneously reducing the RNAIII level in a growth phase-dependent manner.
Staphylococcus aureus is a major human pathogen causing a variety of infections, ranging from minor skin and wound infections to life-threatening diseases (42). Pathogenicity in S. aureus is based on a wide range of cell wall-associated and extracellular proteins that are regulated in a coordinate and growth phase-dependent manner. These virulence determinants are controlled among others by the accessory gene regulator agr and the staphylococcal accessory regulator sar (48). Mutations in either agr or sar result in mutants that are strongly attenuated in virulence compared to their corresponding parental strains (1, 8, 15, 31).
The agr locus regulates the expression of cell wall-associated proteins and secreted exoproteins in response to the density of the bacterial population (37). The proposed function of this regulatory system is to enhance the production of wall-associated adhesins, which interact with the host's matrix proteins, and potential defense factors (protein A) in the early stages of infection. This is followed by the expression of excreted invasion factors, such as hemolysins, proteases, and lipases, that are suggested to be involved in the dissemination of the organism from the primary site of infection once the infection has been established (58). The agr locus comprises two divergent transcriptional units, RNAII and RNAIII, which are transcribed from the agrP2 and agrP3 promoters, respectively (Fig. 1B) (reviewed in reference 46). RNAII encodes a four-gene operon, including a two-component signal transduction system that responds to the concentration of a secreted and processed peptide pheromone, which is encoded within the operon itself. The primary function of the RNAII gene products is to activate the agrP2 and agrP3 promoters, significantly aided by SarA. Transcription from the agrP3 promoter results in a 510-nucleotide RNA molecule (RNAIII), which appears to be the effector molecule of the positive and negative regulation of virulence genes that are controlled by the agr locus (36, 49). RNAIII is thought to regulate most target genes at the level of transcription but has also been shown to influence the translation of some genes (45, 49) and contains a small open reading frame coding for delta hemolysin (hld).
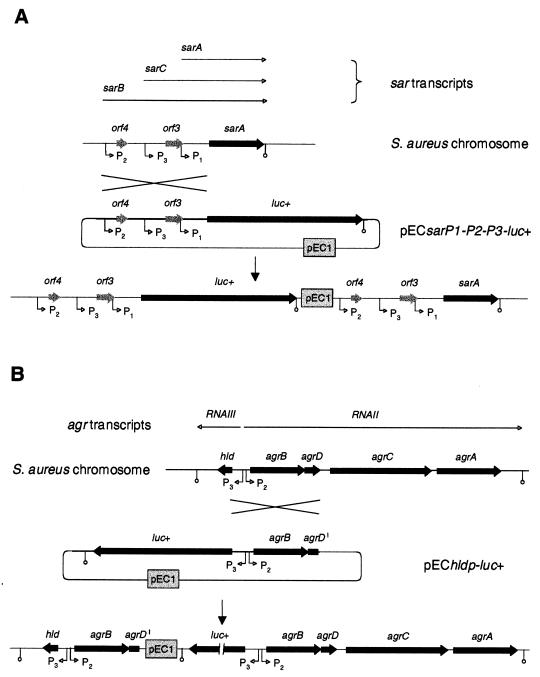
FIG. 1.
Genetic organization of the sar and agr loci of S. aureus. Genetic organization of the sar locus (A) and the agr locus (B) of S. aureus and schematic representation of the integration of sarP1-P2-P3::luc+ or hldp::luc+ fusion constructs into the S. aureus chromosome by single crossover. For a description of construction of the plasmids pECsarP1-P2-P3-luc+ and pEChldp-luc+ and integration of the constructs into the S. aureus chromosome, see Materials and Methods. Open reading frames, promoters, and respective transcripts are indicated.
SarA, the major functional protein encoded by the sar locus, is generally believed to be required for the activation of expression of the agr locus (20, 21, 46) and influences the regulation of several virulence factors independently from agr (7, 11, 14, 19, 41, 60). SarA is essentially involved in the capacity of S. aureus to survive inside of polymorphonuclear neutrophils (33), and the ability of S. aureus to enter mammalian cells and induce apoptosis is supposed to be dependent on factors regulated by sar and agr (58). SarA expression itself is controlled by three different, tandemly arranged promoters (Fig. 1A) in a growth phase-dependent manner (3, 23, 43). Although one of these transcripts, sarC, was shown to be controlled by the alternative transcription factor ςB in vitro (10, 28, 43), the inadvertant use of an rsbU mutant to compare sar expression in a sigB mutant may have wrongly suggested that ςB was involved neither in the transcriptional control of the sar locus nor in agr expression (12, 14, 17). The strains used in those studies were recently shown to possess almost no ςB activity due to a mutation in the rsbU gene, which encodes a positive regulator of ςB (30). The transcription factor ςB itself, organized in the rsbUVWsigB operon, is supposed to be activated by a cascade encompassing RsbU, an RsbV-specific phosphatase, the anti-anti-sigma factor RsbV, and the anti-sigma factor RsbW.
In this study we demonstrate by the use of transcriptional analyses and reporter gene fusion experiments in three different genetic backgrounds that transcription of both the sar and agr loci are clearly influenced by the ςB activity in S. aureus strains harboring a functional sigB operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus was routinely grown in Luria-Bertani (LB) medium at 37°C and at 200 rpm. Antibiotics were used at the following concentrations: for erythromycin and tetracycline, 10 μg ml−1; for ampicillin, 50 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH10B | F− φ80dlacZΔM15 recA1 | Gibco |
| S. aureus | ||
| RN4220 | NCTC8325-4, r− m+ (restriction minus, modification plus), rsbU | 38 |
| BB255 | Essentially the same as NCTC8325, rsbU | 4 |
| Newman | ATCC25904; clinical isolate, high level of clumping factor, rsbU+ | 24 |
| MSSA1112 | Clinical isolate, bla rsbU+ | 25 |
| GP266 | RN4220, rsbU+sigB1(Am) Tcr | 6 |
| GP268 | BB255, rsbU+V+W+sigB+ Tcr | 30 |
| IK181 | BB255, ΔrsbUVWsigB::erm(B) Emr | 40 |
| IK184 | Newman, ΔrsbUVWsigB::erm(B) Emr | 40 |
| MB32 | Newman, asp23+asp23p::pECasp23p-luc+ Emr | 30 |
| MB33 | BB255, rsbU asp23+asp23p::pECasp23p-luc+ Emr | 5 |
| MB39 | MSSA1112, ΔrsbUVWsigB::erm(B) Emr | This study |
| MB49 | GP268, asp23+asp23p::pECasp23p-luc+ Tcr Emr | 30 |
| MB69 | Newman, ΔrsbUVWsigB::erm(B) asp23+asp23p::pECasp23p-luc+ Tcr Emr | 30 |
| MB70 | MSSA1112, ΔrsbUVWsigB::erm(B)asp23+asp23p::pECasp23p-luc+ Tcr Emr | This study |
| MB73 | MSSA1112, asp23+asp23p::pECasp23p-luc+ Emr | This study |
| MB90 | BB255, ΔrsbUVWsigB::erm(B) asp23+asp23p::pECasp23p-luc+ Tcr Emr | 30 |
| MB94 | BB255, rsbU+hld+hldP::luc+ Tcr Emr | This study |
| MB95 | BB255, rsbU hld+hldp::pEChldp-luc+ Emr | This study |
| MB97 | Newman, hld+hldp::pEChldp-luc+ Emr | This study |
| MB98 | BB255, rsbU sar+sar-P1-P2-P3::pECsar-P1-P2-P3-luc+ Emr | This study |
| MB100 | Newman, sar+sar-P1-P2-P3::pECsar-P1-P2-P3-luc+ Emr | This study |
| MB101 | Newman, sigB1(Am) sar+sar-P1-P2-P3::pECsar-P1-P2-P3-luc+ Tcr Emr | This study |
| MB102 | BB255, rsbU+sigB1(Am) sar+sar-P1-P2-P3::pECsar-P1-P2-P3-luc+ Tcr Emr | This study |
| MB103 | Newman, sigB1(Am) hld+hldp::pEChldp-luc+ Tcr Emr | This study |
| MB104 | MSSA1112, hld+hldp::pEChldp-luc+ Emr | This study |
| MB105 | MSSA1112, sar+sar-P1-P2-P3::pECsar-P1-P2-P3-luc+ Emr | This study |
| MB112 | MSSA1112, sigB1(Am) hld+hldp::pEChldp-luc+ Tcr Emr | This study |
| MB113 | MSSA1112, sigB1(Am) sar+sar-P1-P2-P3::pECsar-P1-P2-P3-luc+ Tcr Emr | This study |
| Plasmids | ||
| pSP-luc+ | Apr, firefly luciferase casette vector | Promega |
| pEC1 | Apr Emr, 1.45-kb ClaI erm(B) fragment of Tn551 in pUC18 | 9 |
| pTX15 | TcrPxyl, staphylococcal origin of replication | 50 |
| pIK64 | Tcr, pTX15 derivative, containing sigB under the control of the xylose-inducible promoter Pxyl | 40 |
| pSPsarP1-P2-P3 | Apr, 867-bp PCR fragment of sar promoter from strain RN4220 in pSP-luc+ | This study |
| pSPhldp | Apr, 1-kb PCR fragment of hld promoter from strain RN4220 in pSP-luc+ | This study |
| pECsarP1-P2-P3-luc+ | Emr, 2.5-kb KpnI-EcoRI sarP1-P2-P3-luc+ fragment of pSPsarP1-P2-P3 in pECI, S. aureus integration vector that inserts into the sar promoters (sarP1-P2-P3) | This study |
| pEChldP::luc+ | Emr, 2.6-kb KpnI-EcoRI hldp-luc+ fragment of pSPhldP in pEC1, S. aureus integration vector that inserts into the hld promoter (hldp) | This study |
Abbreviations are as follows: Apr, ampicillin resistant; Emr, erythromycin resistant; Tcr, tetracycline resistant.
General methods.
All DNA manipulations, basic molecular methods, and handling of Escherichia coli were performed in accordance with standard protocols (54). Genetic manipulation of S. aureus was done as described earlier (39). The general transducing phage 80α was used for transductions.
Construction of pECsarP1-P2-P3-luc+ and pEChldp-luc+.
A DNA fragment covering 867 bp of the sar promoter region of S. aureus RN4220 was generated by PCR using an upstream primer (5′-CGGTACCGTTGATTTGGGTAGTATGC-3′) including a KpnI linker (underlined) and a downstream primer (5′-TTGCCATGGTTAAAACCTCCC-3′) including a NcoI site (underlined), with italic nucleotides corresponding to positions 5 to 24 and 852 to 872 of the sequence found under GenBank accession no. U46541, respectively. For hldp::luc+, a DNA fragment covering 1 kb of the agr locus of S. aureus RN4220 was generated by PCR using an upstream primer (5′-GTGCCATGGAAATCACTCCTTCC-3′) including a NcoI site (underlined) and a downstream primer (5′-TGGTACCTCAACTTCATCCATTATG-3′) including a KpnI site (underlined), with italic nucleotides corresponding to positions 397 to 419 and 1348 to 1372 of the sequence found under GenBank accession no. AF230358, respectively. The PCR products obtained were digested with KpnI and NcoI and cloned in frame with the 5′ end of the luciferase gene of plasmid pSP-luc+. Sequence analysis and comparison confirmed the identity of the constructs to the RN6390 sequence or RN4220 sequence, respectively. A 2.5-kb KpnI-EcoRI fragment, including the sar promoter region fused to the luciferase coding region, or a 2.6-kb KpnI-EcoRI fragment, including the hld promoter region fused to the luciferase coding region, was subsequently cloned into the suicide plasmid pEC1 (9) to obtain the plasmids pECsarP1-P2-P3-luc+ (Fig. 1A) and pEChldp-luc+ (Fig. 1B), respectively. The plasmids obtained were transformed by electroporation into RN4220 and transduced into different S. aureus genetic backgrounds.
Northern blot analyses.
Isolation of total RNA was done as described by Cheung et al. (16). Eight micrograms of total RNA of each sample was electrophoresed through a 1.5% agarose–0.66 M formaldehyde gel in morpholinepropanesulfonic acid running buffer (20 mM morpholinepropanesulfonic acid, 10 mM sodium acetate, 2 mM EDTA [pH 7]). Blotting of RNA onto a positively charged nylon membrane (Roche, Basel, Switzerland) was performed with a vacuum blotter (Pharmacia, Uppsala, Sweden). The intensities of the 23S and 16S rRNA bands stained with ethidium bromide were verified to be equivalent in all the samples before transfer. Labeling and hybridization were done by the use of the digoxigenin labeling and detection kits according to the manufacturer's instructions (Roche). The following specific primers were used to generate the digoxigenin-labeled DNA probes by PCR labeling: SasarA+, 5′-AGGGAGGTTTTAAACATGGC-3′; SasarA−, 5′-CTCGACTCAATAATGATTCG-3′ (nucleotides 851 to 870 and 1177 to 1196 of the sequence found under GenBank accession no. U46541); RNAIII+, 5′-GTGATGGAAAATAGTTGATGAG-3′; RNAIII−, 5′-GTGAATTTGTTCACTGTGTCG-3′ (nucleotides 453 to 474 and 333 to 353 of the sequence under GenBank accession no. AF230358).
Luciferase assay.
Bacterial cells from overnight cultures containing the appropriate antibiotic were diluted with fresh LB medium to an optical density at 600 nm (OD600) of 0.01. Freshly diluted cells were incubated without antibiotics at 37°C and at 200 rpm. S. aureus cells, obtained at different growth stages, were harvested by centrifugation at 11,000 × g during 1 min at room temperature, and the cell pellets were resuspended in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.3]) to an OD600 of 10. Luciferase activity was that determined by rapidly mixing PBS-resuspended cells (10 μl) with an equal volume of Luciferase Assay Substrate (Promega, Madison, Wis.). Luminescence was measured on a Turner Designs TD-20/20 Luminometer (Promega) for a time period of 10 s with a delay of 2 s.
Fibronectin binding assay.
Binding of S. aureus to fibronectin was measured quantitatively in microtiter plates, by a slight modification of a previously described method (44). Briefly, 50 μl of human fibronectin (Sigma, Buchs, Switzerland) was serially diluted twofold from a starting concentration of 500 μg/ml in PBS. Bacterial cells grown to exponential or stationary phase were harvested by centrifugation and washed in PBS, and 20 μl of a suspension (5 × 108 CFU) was added to the fibronectin dilutions. The lowest concentration of fibronectin triggering clumping after overnight incubation at 4°C was recorded as the titer.
RESULTS
ςB activity in S. aureus.
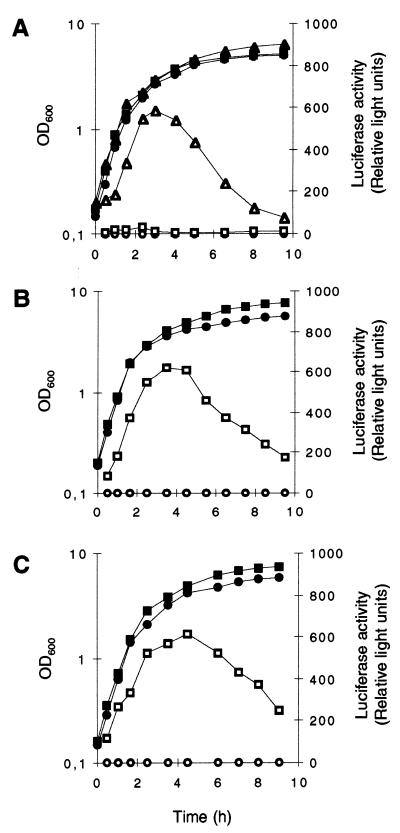
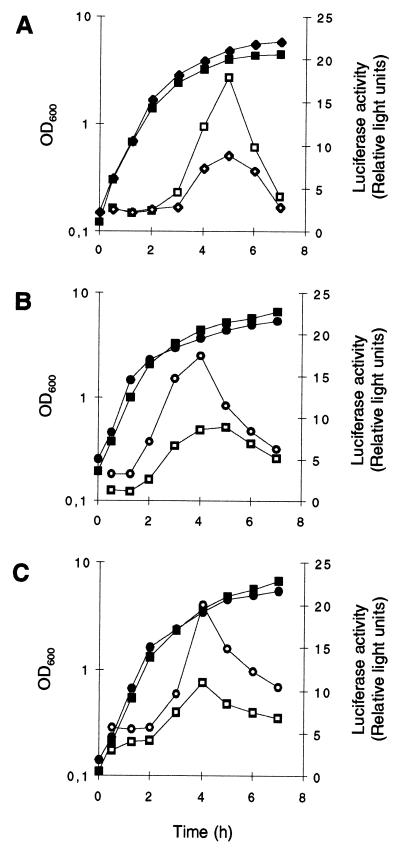
The ςB activities of the two genetically distinct strains Newman and MSSA1112 and their respective ΔrsbUVWsigB mutants, as well as those of the rsbU strain BB255 and its derivative GP268, transformed with the rsbU wild-type allele, were analyzed during growth by the use of the asp23 reporter gene system (30). The two clinical isolates MSSA1112 and Newman possessed quite similar ςB activity profiles in LB medium, with a maximal ςB activity in late exponential growth phase followed by a significant decrease thereafter (Fig. 2B and C). The ςB activity profile obtained for strain GP268 (MB49) was comparable to those found for strains MSSA1112 (MB73) and Newman (MB32), while its parental strain produced almost no ςB activity throughout the whole growth cycle (Fig. 2A). Additionally, all three ΔrsbUVWsigB mutants were unable to produce any ςB-dependent activity at all. The unexpected strong decrease in ςB activity that was observed from the onset of stationary phase was confirmed by monitoring the transcription of the ςB-dependent genes asp23 and sigB in Northern blot analyses. Both transcripts were found to be the most abundant during late exponential growth phase, and their levels were drastically reduced during stationary phase (data not shown).
FIG. 2.
ςB activity during growth of S. aureus. Expression of asp23::luc+ during growth of S. aureus strains BB255 (A), MSSA1112 (B), Newman (C), and their respective sigB mutants. Strains were grown in LB medium at 37°C. Bacterial growth was measured as the OD600 (solid symbols). ςB transcriptional activity was determined by measuring the luciferase activity of Luc+ (open symbols), the product of the luc+ reporter gene fused to the ςB-dependent promoters of asp23 (asp23p). (A) Squares, S. aureus strain MB33 (BB255, asp23p::luc+); triangles, strain MB49 (BB255, rsbU+ asp23p::luc+); circles, strain MB90 (BB255, ΔrsbUVWsigB asp23p::luc+). (B) Squares, S. aureus strain MB73 (MSSA1112, asp23p::luc+); circles, strain MB70 (MSSA1112, ΔrsbUVWsigB asp23p::luc+). (C) Squares, S. aureus strain MB32 (Newman, asp23p::luc+); circles, strain MB69 (Newman, ΔrsbUVWsigB asp23p::luc+).
Influence of ςB on the expression of the sar locus.
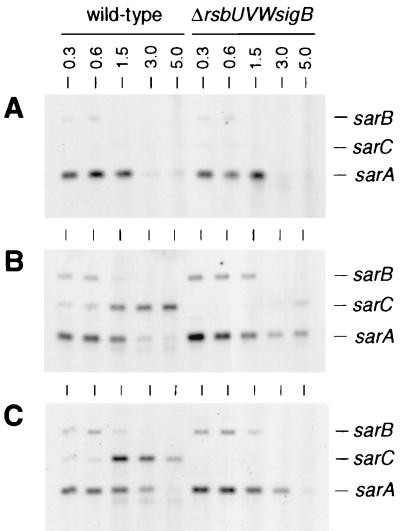
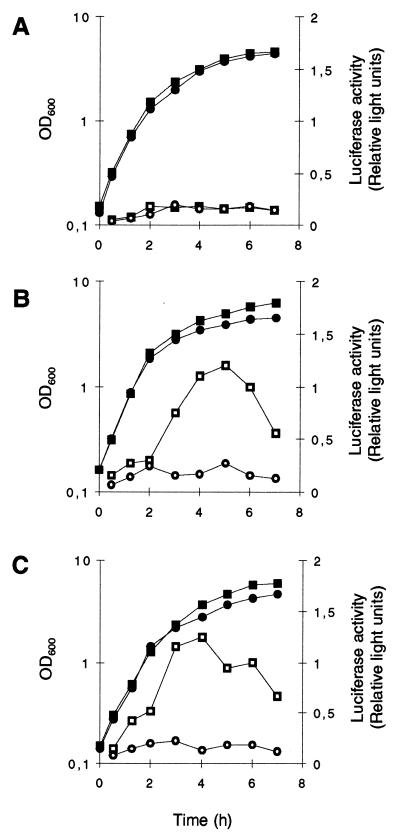
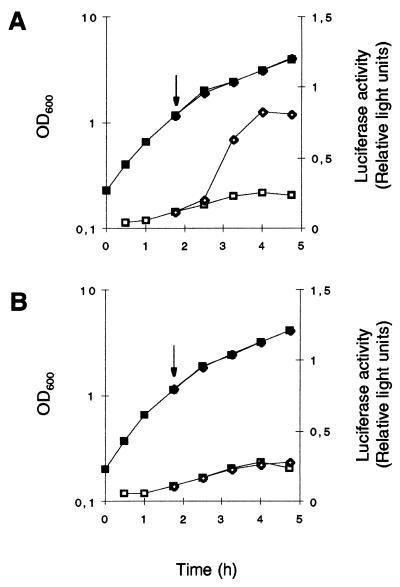
Northern blot analyses with the sarA gene as a probe showed strong sarA transcription, originating from the ςA-dependent sarP1 promoter, during exponential growth (OD600, 0.3 to 1.5) and declining with the onset of stationary phase in all strains analyzed (Fig. 3). A similar time course but much weaker transcription was observed in all strains for sarB, originating from the ςA-dependent sarP2 promoter. In contrast, ςB-dependent transcription of sarC from the sarP3 promoter was detectable only in the wild-type strains MSSA1112 and Newman, not in BB255 or any of the ΔrsbUVWsigB mutants. sarC-specific transcripts were detectable abundantly from late exponential growth phase up to stationary phase (OD600, 1.5 to 5.0). Reporter gene fusion experiments with the luciferase gene luc+ fused to the sarP1, sarP2, and sarP3 (sarP1-P2-P3) promoters suggested an increased SarA production in strains MSSA1112 and Newman with the beginning of late exponential growth phase that paralleled the time course of the overall sar transcripts observed in those strains. No such increase in luciferase activity was detectable with the rsbU mutant BB255 (Fig. 4A) or in any of the ΔrsbUVWsigB mutants. Plasmid pIK64 (Pxyl::sigB)-mediated xylose-induced overexpression of ςB in the BB255 derivative MB98 resulted in a strong increase in sarP1-P2-P3::luc+ expression (Fig. 5A), which was not observed with the control plasmid pTX15 (Fig. 5B), unambiguously proving ςB to influence sar expression directly or indirectly.
FIG. 3.
Northern blot analyses of the sar locus. Total RNAs (8 μg/lane) of S. aureus strains BB255 (A), MSSA1112 (B), Newman (C), and their respective sigB mutants were blotted onto positively charged nylon membranes and subjected to Northern blot analyses. RNAs were obtained from cells grown in LB medium at 37°C and harvested at different growth stages (indicated as OD600 values [numbers above the lanes]). The blotted membranes were hybridized using a digoxigenin-labeled DNA probe specific for sarA (for details on the construction, see Materials and Methods). The RNA molecular weight marker I (Roche) was used as a size marker. Relevant transcript signals are indicated.
FIG. 4.
Role of ςB in the regulation of sarP1-P2-P3::luc+ expression during growth. S. aureus derivatives of strains BB255 (A), MSSA 1112 (B), Newman (C), and their respective sigB mutants were grown in LB medium at 37°C. Bacterial growth was measured by OD600 (closed symbols). sarP1-P2-P3::luc+ expression was determined by measuring the luciferase activity of the reporter gene luc+ (open symbols). (A) Squares, S. aureus strain MB98 (BB255, sarP1-P2-P3::luc+); circles, strain MB102 (BB255, sigB sarP1-P2-P3::luc+). (B) Squares, S. aureus strain MB105 (MSSA1112, sarP1-P2-P3::luc+); circles, strain MB113 (MSSA1112, sigB sarP1-P2-P3::luc+). (C) Squares, S. aureus strain MB100 (Newman, sarP1-P2-P3::luc+); circles, strain MB101 (Newman, sigB sarP1-P2-P3::luc+).
FIG. 5.
Effect of overexpressed ςB on the expression of sarP1-P2-P3::luc+. S. aureus derivatives of strain MB98 (BB255, sarP1-P2-P3::luc+), harboring plasmid pIK64 (Pxyl::sigB) (A) or control plasmid pTX15 (Pxyl) (B) were grown in LB medium at 37°C. Growth was measured as the OD600 (closed symbols). Growing cultures were split into equal parts at the OD600 of 1, and overexpression of ςB was induced in one of the parts by supplementing 0.5% xylose (diamonds), while the control part was left without addition (squares). The arrow denotes the time point of supplementation. sarP1-P2-P3::luc+ expression (open symbols) was determined by measuring the luciferase activity of the reporter gene luc+ as described in Materials and Methods.
Influence of ςB on the expression of the agr locus.
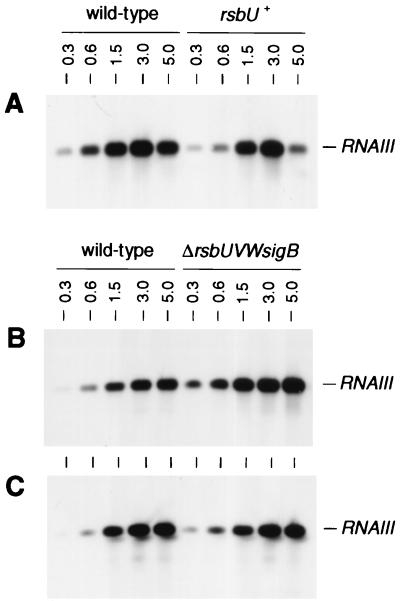
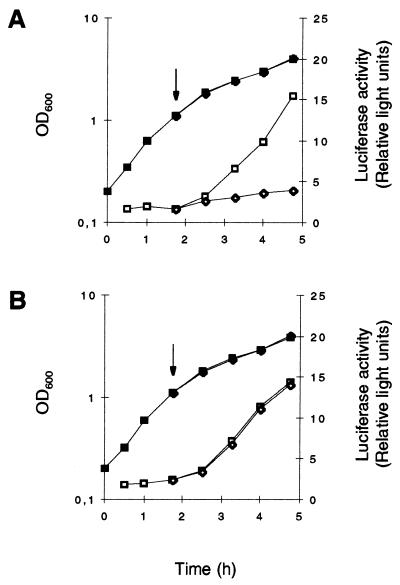
Comparison of the growth phase-dependent transcription of RNAIII in Northern blot analyses revealed low transcription levels and a delayed induction of RNAIII expression in the wild-type strains MSSA1112 and Newman compared to results for their corresponding ΔrsbUVWsigB mutants (Fig. 6B and C). The RNAIII expression profile of strain GP268 (BB255 rsbU+) paralleled the transcription profiles found for strains MSSA1112 and Newman, with expression being significantly delayed and at a lower level than that for BB255 (Fig. 6A). Reporter gene fusion experiments, using the luciferase gene luc+ fused to the hld gene, carried by RNAIII (Fig. 1B), confirmed these data. The RNAIII-representing luciferase activity profiles of strains carrying the wild-type rsbU allele were significantly lower than those for ΔrsbUVWsigB mutants and BB255, respectively (Fig. 7). Overexpression of ςB in the BB255 derivative MB95, harboring plasmid pIK64, resulted in a strong decrease in hldp::luc+ expression from that of the control (Fig. 8), corroborating the negative regulatory effect of ςB on agr expression.
FIG. 6.
Northern blot analyses of RNAIII. Total RNAs (8 μg/lane) of S. aureus strains BB255 and GP268 (BB255, rsbU+) (A), MSSA1112 and MB39 (MSSA1112, ΔrsbUVWsigB) (B), and Newman and IK 184 (Newman, ΔrsbUVWsigB) (C) were blotted onto positively charged nylon membranes and subjected to Northern blot analyses. RNAs were obtained from cells grown in LB medium at 37°C and harvested at different growth stages (indicated as OD600 values above the lanes). The blotted membranes were hybridized using a digoxigenin-labeled DNA probe specific for RNAIII (for details on construction, see Materials and Methods). The RNA molecular weight marker I (Roche) was used as the size marker. Relevant transcript signals are indicated.
FIG. 7.
Role of ςB in the regulation of hldP::luc+ expression during growth. S. aureus derivatives of strains BB255 (A), MSSA1112 (B), and Newman (C) were grown in LB medium at 37°C. Bacterial growth was measured by OD600 (closed symbols). hldp::luc+ expression was determined by measuring the luciferase activity of the reporter gene luc+ (open symbols) as described in Materials and Methods. (A) Squares, S. aureus strain MB95 (BB255, hldp::luc+); diamonds, strain MB94 (BB255, rsbU+ hldp::luc+). (B) Squares, S. aureus strain MB104 (MSSA1112, hldp::luc+); circles, strain MB112 (MSSA1112, sigB hldp::luc+). (C) Squares, S. aureus strain MB97 (Newman, hldp::luc+); circles, strain MB103 (Newman, sigB hldp::luc+).
FIG. 8.
Effect of overexpressed ςB on the expression of hldP::luc+. S. aureus derivatives of strain MB95 (BB255 hldp::luc+), harboring plasmid pIK64 (Pxyl::sigB) (A) or control plasmid pTX15 (Pxyl) (B), were grown in LB medium at 37°C. Growth was measured as the OD600 (closed symbols). Growing cultures were split into equal parts at the OD600 of 1, and overexpression of ςB was induced in one of the parts by supplementing 0.5% xylose (diamonds), while the control part was left without addition (squares). The arrow denotes the time point of supplementation. hldp::luc+ expression (open symbols) was determined by measuring the luciferase activity of the reporter gene luc+ as described in Materials and Methods.
Fibronectin binding activity.
Since sar and agr are known to influence the ability of S. aureus to bind to fibronectin (55, 60), the effects of RsbU on the fibronectin binding capacity were determined with strain BB255 and its derivatives (Table 2). Strain GP268, carrying an intact sigB operon, showed a more than 100-fold-lower fibronectin-clumping titer than its rsbU-defective parent, BB255, irrespective of the growth phase, while no difference was apparent between BB255 and its ΔrsbUVWsigB mutant.
TABLE 2.
Titration of fibronectin clumping of S. aureus
| Strain (genotype) | Fibronectin-clumping titera
|
|
|---|---|---|
| Exponential growth phase | Stationary growth phase | |
| GP268 (rsbU+V+W+sigB+) | 1 | 1 |
| BB255 (rsbU) | 250 | 125 |
| BB255 (ΔrsbUVWsigB) | 125 | 125 |
Clumping titer, lowest fibronectin concentration (in μg/ml) showing clumping. Results are expressed as the median of results from five to nine independent experiments.
DISCUSSION
Most of our knowledge of the regulation of ςB in S. aureus has been adapted from the well-characterized ςB regulon of the closely related soil bacterium Bacillus subtilis (reviewed in reference 34). In this organism, ςB has been shown to function as a stress- and stationary phase-specific transcription factor. In B. subtilis, activation of ςB appears to be basically dependent on the activity of the two RsbV-specific phosphatases, RsbU and RsbP, with the latter being essential for the stationary phase and energy stress-dependent activation of ςB (56). RsbU was found to be of importance only for the environmental stress activation of this sigma factor (57, 59, 61). The data presented here and elsewhere (6, 30) instead suggest ςB of S. aureus to be a transcription factor with the main activity in late exponential growth phase rather than in stationary phase. Additionally, the data clearly demonstrated that the natural rsbU mutant BB255 was almost completely unable to express ςB activity, illustrating the importance of RsbU for the overall activity level of ςB in S. aureus. Accidentally, nearly all studies of the influence of ςB on the expression of sar and agr have been carried out with NCTC8325 isogenic backgrounds, harboring the mutation in rsbU (12, 14, 23, 43). In consequence, the findings presented here call into question the ςB-dependent results obtained from such strains.
Transcriptional data and reporter gene fusions of the sar locus revealed a strong ςB-dependent transcription of sarC in S. aureus strains harboring an intact sigB operon, while no such transcription was detectable in the rsbU-deficient strain BB255 nor in any of the ΔrsbUVWsigB mutants (Fig. 3 and 4). This results are in contrast to the findings of Bayer et al. (3) and Manna et al. (43), who detected significant amounts of sarC transcripts during late exponential phase in the closely related rsbU-deficient strain RN6390. It is noteworthy that the reporter gene fusion experiments of Manna et al. (43) performed with strain RN6390 revealed only little activity for the sarP3 promoter, the level of which was approximately 50-fold lower than that for the sarP1 promoter. Interestingly, the maximum activity obtained in this study for the sarP3 promoter was comparable to that found for the sarP2 promoter. A comparison of the reporter gene data obtained from Manna et al. (43) with the intensities of the different sar transcripts presented here in Fig. 3 led us to the conclusion that the RN6390 strain used by Manna et al. also possessed almost no ςB activity. This conclusion is further strengthened by the findings of Cheung et al. (17), who reported neither in vitro nor in vivo activity of the sarP3 promoter in strain RN6390, in accordance with our finding that the luciferase activity profile of BB255 was indistinguishable from that of its ΔrsbUVWsigB mutant. Both findings fit well with our deduction that normal levels of ςB-dependent transcription of sarC occur only in the presence of a functional RsbU phosphatase. Our data demonstrate that expression of the sar locus is significantly upregulated by ςB in a growth phase-dependent manner in S. aureus strains harboring an intact sigB operon. Thus, we postulate that ςB positively contributes to the overall level of SarA in S. aureus. This hypothesis is strengthened by recent findings of Gertz et al. (29), who reported significantly lower SarA levels in a ΔrsbUVWsigB mutant of the rsbU+ strain COL.
The influence of SarA on the expression of the agr locus has been the topic of several studies (13, 20, 21, 35, 46, 53). Even though a factor(s) other than SarA (e.g., ORF3, RAP, and RIP) is suggested to participate in controlling agr-related transcription (2, 13, 20, 21), SarA is currently believed to stimulate the expression from both the agrP2 and agrP3 promoters, leading ultimately to the upregulation of RNAIII (21; for a review, see reference 48). The ςB-dependent upregulation of sar expression observed in rsbU+ strains would be expected therefore to result in an increase in agr expression. However, the data presented here revealed a weaker RNAIII transcription for the rsbU+ strains MSSA1112 and Newman than for their ΔrsbUVWsigB mutants, suggesting that agr expression is negatively influenced by ςB activity, irrespective of the positive effect of ςB on sar expression. This finding is supported by a recent study of Chakrabarti and Misra (10), demonstrating an inhibitory influence of SarA on transcription from both the agrP2 and agrP3 promoters in vitro. The authors suggest that either SarA, together with an as-yet-uncharacterized cellular factor(s), activates transcription of the agr operon, or SarA regulates expression of one or more factors which then activate agr expression. In line with this hypothesis, this as-yet-uncharacterized cellular factor(s) involved in the activation of agr expression may be positively regulated by SarA but dominated negatively by ςB activity.
Many potential virulence factors have been shown to be regulated by SarA and RNAIII in a cooperative way (11, 13, 18, 27, 52), but the expression of several virulence factors was found to be upregulated by one regulator but repressed by the other (55, 60). Additionally, some virulence factors are influenced by one of the two loci but unaffected by the other one (7, 26, 32). Thus, the ςB-mediated increase in SarA, accompanied by the decrease in RNAIII, is very likely to enhance these phenomena, resulting in severe growth phase-dependent differences in the expression profiles of some virulence factors. One possible role of ςB in this scenario is to prolong the production of cell surface proteins, such as fibronectin binding proteins, that are positively influenced by SarA and negatively influenced by agr (55, 60). Simultaneously, ςB may down-regulate RNAIII-specific activities, i.e., the repression of protein A and upregulation of exoproteins (36, 49) or the production of capsular polysaccharides (22, 51). Consistent with this hypothesis is our finding that the rsbU+ derivative GP268 possessed a significantly lower fibronectin-clumping titer than its rsbU-defective parent, BB255, signalling the presence of larger quantities of fibronectin binding proteins in GP268 than in BB255.
The impact of either SarA and/or RNAIII on the expression of virulence factors in S. aureus is well documented. On account of the studies performed with NCTC8325 derivatives, it is unquestionable that sigB mutants are still able to produce sufficient amounts of those two global regulators to be virulent, which led to the conclusion that ςB has no essential function in the virulence and pathogenicity of S. aureus (12). This conclusion is further strengthened by the findings of Nicholas et al. (47), who observed no differences between the clinical isolate WCUH29 and its isogenic ΔsigB mutant in their ability to cause infections in three distinct animal infection models. However, we agree with Gertz et al. (29), who question whether the infection models analyzed so far really reflect the natural situation in the host. The findings that both agr and sar expression are significantly influenced by ςB in an rsbU+ genetic background should be reason to reevaluate if and how ςB is involved in the virulence and pathogenicity of S. aureus.
ACKNOWLEDGMENTS
We are grateful to B. Berger-Bächi for critically reading and commenting on the manuscript. Preliminary sequence data was obtained from The Institute of Genomic Research (TIGR) through the website at http://www.tigr.org. Sequencing of Staphylococcus aureus COL was accomplished with support from National Institute of Allergy and Infectious Diseases (NIAID) and the Merck Genome Research Institute (MGRI).
This work was supported by the Swiss National Science Foundation grant NF 31-46762.96 to F. H. Kayser.
REFERENCES
- 1.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban N, Goldkorn T, Nhan R T, Dang L B, Scott S, Ridgley R M, Rasooly A, Wright S C, Larrick J W, Rasooly R, Carlson J R. Autoinducer of virulence as a target for vaccine and therapy against Staphylococcus aureus. Science. 1998;280:438–440. doi: 10.1126/science.280.5362.438. [DOI] [PubMed] [Google Scholar]
- 3.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger-Bächi B. Increase in transduction efficiency of Tn551 mediated by the methicillin resistance marker. J Bacteriol. 1983;154:533–535. doi: 10.1128/jb.154.1.533-535.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff M, Roos M, Putnik J, Wada A, Glanzmann P, Giachino P, Vaudaux P, Berger-Bächi B. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol Lett. 2001;194:77–82. doi: 10.1111/j.1574-6968.2001.tb09449.x. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff M, Berger-Bächi B. Teicoplanin stress-selected mutations increasing ςB activity in Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1714–1720. doi: 10.1128/AAC.45.6.1714-1720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins J S, Gillaspy A F, Rechtin T M, Hurlburt B K, Smeltzer M S. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol Microbiol. 1999;33:317–326. doi: 10.1046/j.1365-2958.1999.01475.x. [DOI] [PubMed] [Google Scholar]
- 8.Booth M C, Cheung A L, Hatter K L, Jett B D, Callegan M C, Gilmore M S. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65:1550–1556. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brücker R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti S K, Misra T K. SarA represses agr operon expression in a purified in vitro Staphylococcus aureus transcription system. J Bacteriol. 2000;182:5893–5897. doi: 10.1128/jb.182.20.5893-5897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan P F, Foster S J, Ingham E, Clements M O. The Staphylococcus aureus alternative sigma factor ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung A L, Bayer M G, Heinrichs J H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung A L, Chien Y T, Bayer A S. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect Immun. 1999;67:1331–1337. doi: 10.1128/iai.67.3.1331-1337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar-agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 17.Cheung A L, Nast C C, Bayer A S. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect Immun. 1998;66:5988–5993. doi: 10.1128/iai.66.12.5988-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung A L, Ying P. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien Y, Manna A C, Projan S J, Cheung A L. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 20.Chien Y T, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;273:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 21.Chien Y T, Manna A C, Cheung A L. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol Microbiol. 1998;30:991–1001. doi: 10.1046/j.1365-2958.1998.01126.x. [DOI] [PubMed] [Google Scholar]
- 22.Dassy B, Hogan T, Foster T J, Fournier J M. Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide. J Gen Microbiol. 1993;139:1301–1306. doi: 10.1099/00221287-139-6-1301. [DOI] [PubMed] [Google Scholar]
- 23.Deora R, Tseng T, Misra T K. Alternative transcription factor ςSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J Bacteriol. 1997;179:6355–6359. doi: 10.1128/jb.179.20.6355-6359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duthie E S, Lorenz L L. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 25.Entenza J M, Vouillamoz J, Glauser M P, Moreillon P. Levofloxacin versus ciprofloxacin, flucloxacillin, or vancomycin for treatment of experimental endocarditis due to methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1662–1667. doi: 10.1128/aac.41.8.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto D F, Bayles K W. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol. 1998;180:3724–3726. doi: 10.1128/jb.180.14.3724-3726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto D F, Brunskill E W, Bayles K W. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J Bacteriol. 2000;182:4822–4828. doi: 10.1128/jb.182.17.4822-4828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gertz S, Engelmann S, Schmid R, Ohlsen K, Hacker J, Hecker M. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol Gen Genet. 2000;261:558–566. doi: 10.1007/s004380051001. [DOI] [PubMed] [Google Scholar]
- 29.Gertz S, Engelmann S, Schmid R, Ziebandt A-K, Tischer K, Scharf C, Hacker J, Hecker M. Characterization of the ςB regulon in Staphylococcus aureus. J Bacteriol. 2000;182:6983–6991. doi: 10.1128/jb.182.24.6983-6991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giachino P, Engelmann S, Bischoff M. ςB activity depends on RsbU in Staphylococcus aureus. J Bacteriol. 2001;183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillaspy A F, Hickmon S G, Skinner R A, Thomas J R, Nelson C L, Smeltzer M S. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillaspy A F, Patti J M, Pratt F L J, Iandolo J J, Smeltzer M S. The Staphylococcus aureus collagen adhesin-encoding gene (cna) is within a discrete genetic element. Gene. 1997;96:239–248. doi: 10.1016/s0378-1119(97)00256-4. [DOI] [PubMed] [Google Scholar]
- 33.Gresham H D, Lowrance J H, Caver T E, Wilson B S, Cheung A L, Lindberg F P. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol. 2000;164:3713–3722. doi: 10.4049/jimmunol.164.7.3713. [DOI] [PubMed] [Google Scholar]
- 34.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 35.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreiswirth B N, Löfdahl S, Bentley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectable transmitted by a prophage. Nature. 1983;305:680–685. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 39.Kullik I, Giachino P. The alternative sigma factor sigB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 40.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindsay J A, Foster S J. Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol Gen Genet. 1999;262:323–331. doi: 10.1007/s004380051090. [DOI] [PubMed] [Google Scholar]
- 42.Lowy F. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 43.Manna A C, Bayer M G, Cheung A L. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol. 1998;180:3828–3836. doi: 10.1128/jb.180.15.3828-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDevitt D, Vaudaux P, Foster T J. Genetic evidence that bound coagulase of Staphylococcus aureus is not clumping factor. Infect Immun. 1992;60:1514–1523. doi: 10.1128/iai.60.4.1514-1523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;15:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 47.Nicholas R O, Li T, McDevitt D, Marra A, Sucoloski S, Demarsh P L, Gentry D R. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect Immun. 1999;67:3667–3669. doi: 10.1128/iai.67.7.3667-3669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novick R P. Pathogenicity factors and their regulation. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 392–407. [Google Scholar]
- 49.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peschel A, Ottenwälder B, Götz F. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiQ using an improved staphylococcal expression system. FEMS Microbiol Lett. 1996;137:279–284. doi: 10.1111/j.1574-6968.1996.tb08119.x. [DOI] [PubMed] [Google Scholar]
- 51.Pöhlmann-Dietze P, Ulrich M, Kiser K B, Döring G, Lee J C, Fournier J M, Botzenhardt K, Wolz C. Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect Immun. 2000;68:4865–4871. doi: 10.1128/iai.68.9.4865-4871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramadurai L, Lockwood K J, Nadakavukaren M J, Jayaswal R K. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology. 1999;145:801–808. doi: 10.1099/13500872-145-4-801. [DOI] [PubMed] [Google Scholar]
- 53.Rechtin T M, Gillaspy A F, Schumacher M A, Brennan R G, Smeltzer M S, Hurlburt B K. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol Microbiol. 1999;33:307–316. doi: 10.1046/j.1365-2958.1999.01474.x. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Laboratory Press; 1989. [Google Scholar]
- 55.Saravia-Otten P, Muller H P, Arvidson S. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J Bacteriol. 1997;179:5259–5263. doi: 10.1128/jb.179.17.5259-5263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 57.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wesson C A, Liou L E, Todd K M, Bohach G A, Trumble W R, Bayles K W. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infect Immun. 1998;66:5238–5243. doi: 10.1128/iai.66.11.5238-5243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolz C, Pöhlmann-Dietze P, Steinhuber A, Chien Y T, Manna A, van Wamel W, Cheung A L. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol Microbiol. 2000;36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]
- 61.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine proteine kinase and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]