Abstract
The diversity of prokaryotic symbionts in Ciliophora and other protists is fascinatingly rich; they may even include some potentially pathogenic bacteria. In this review, we summarize currently available data on biodiversity and some morphological and biological peculiarities of prokaryotic symbionts mainly within the genera Paramecium and Euplotes. Another direction of ciliate symbiology, neglected for a long time and now re‐discovered, is the study of epibionts of ciliates. This promises a variety of interesting outcomes. Last, but not least, we stress the new technologies, such as next generation sequencing and the use of genomics data, which all can clarify many new aspects of relevance. For this reason, a brief overview of achievements in genomic studies on ciliate's symbionts is provided. Summing up the results of numerous scientific contributions, we systematically update current knowledge and outline the prospects as to how symbiology of Ciliophora may develop in the near future.
Keywords: Ciliophora, endosymbiosis, episymbiosis, Euplotes, Holospora, Holospora‐like bacteria, morphology, Paramecium, phylogeny, symbionts
ESTIMATES of the number of species of ciliates vary from 3000 to 30,000 (Foissner, 2008; Fokin, 2012). Numerous descriptions of new ciliate taxa from exotic habitats and scantily investigated regions and sometimes places not yet investigated by professional taxonomists (Foissner, 2016; Foissner & Berger, 2021; Krenek et al., 2015; Rossi et al., 2016) underpin the argument that a total of 30,000 species for all Ciliophora is not unrealistic. Ciliates occupy highly diverse ecological niches. Most are free‐living, some are epi and endocommensals, parasites of other unicellular organisms and metazoans, and serve as hosts for bacterial epi and endosymbionts, fungi, algae, and other protists (Dziallas et al., 2012; Fokin et al., 2014; Görtz, 2006). Ecological and trophic preferences (e.g. being bacterivores) of ciliates seem to favor endosymbioses with bacteria and a diversity of symbiotic relationships. The biodiversity of prokaryotic symbionts in Ciliophora is fascinatingly rich; and it is not by chance they might harbor potentially pathogenic bacteria.
Most of the studies published over the past decade in the field of symbiosis in ciliated protists address newly discovered or re‐investigated systems of ciliate bacteria or have summarized such results (Bright et al., 2014; Fokin, 2012; Görtz, 2014; Potekhin et al., 2021; Schrallhammer & Potekhin, 2020; Serra et al., 2016; Szokoli et al., 2016; Vannini et al., 2014), and some of the investigations were devoted to the molecular aspects of such systems (Boscaro et al., 2017; Dohra et al., 2014; Fujishima, 2009; Garushyants et al., 2018).
The ability to gain and retain symbionts varies among the different ciliate groups, as it is in the case of other protistan taxa. Most of the detected prokaryotic symbionts belong to Alphaproteobacteria, but step‐by‐step symbiotic abilities have been demonstrated in other bacterial groups (Boscaro et al., 2012; Bright et al., 2014; Fokin, 2004, 2012; Rosati et al., 1998; Schrallhammer & Potekhin, 2020).
Holospora and Holospora‐like bacteria (HLB) are non‐motile Alphaproteobacteria, showing a characteristic life cycle with infectious and reproductive forms, found mainly in the nuclear apparatus of representatives of Oligohymenophorea, Heterotrichea, Armophorea, Phyllopharyngea, and Prostomatea (Fokin, 2012; Fokin & Görtz, 2009). The majority of such cases have not been investigated in depth, and representatives of the Holospora genus were confirmed only in Paramecium. Bacteria associated with bacteriophages were found in species of Heterotrichea and Oligohymenophorea. A rare type of endosymbiotic motile bacteria was found in ciliates of the two different classes: in the cytoplasm of heterotrichs or in the macronucleus and its perinuclear space of Oligohymenophora (Fokin, 2012; Vannini et al., 2014). Some other non‐motile endosymbionts were discovered in the perinuclear space of both nuclei of ciliates (Fokin & Karpov, 1995). Episymbionts are more common in Heterotrichea, Armophorea, and Plagiopylea, but recently such bacteria were recorded for Oligohymenophorea (Bright et al., 2014; Castelli et al., 2019).
Apparently, Oligohymenophorea and Spirotrichea are the most promising groups of Ciliophora for the investigation of prokaryotic symbioses. A remarkable wealth of symbioses has been observed so far in Paramecium, a member of the first group. They include an infectious type of bacterial symbiosis (parasitism). A similar kind was detected in the related genus Frontonia (Fokin et al., 2019; Schrallhammer & Potekhin, 2020). In this review, particular attention will be given to the HLB group—“Candidatus Gortzia” (from here on, we will use symbiotic bacterial taxa names with abbreviation of the prefix “Candidatus”—a term that indicates that the taxon name does not yet meet all requirements for nomenclature of prokaryotes), “Ca. Hafkinia”, and Preeria as well as to endosymbionts of Euplotes species.
Among spirotrichs, Euplotes is certainly one of the most studied genera for its proneness to establish symbiotic relations with bacterial organisms of different natures. It often harbors, in its cytoplasm, true microbial consortia (i.e. more than one bacterial species stably present inside the host cell). The best known Euplotes symbiont is Polynucleobacter necessarius, a betaproteobacterium which establishes an obligate mutualistic relationship with the host (Heckmann & Schmidt, 1987; Vannini et al., 2005, 2012). Many other species of bacteria have been found in different species of Euplotes, ranging from Alphaproteobacteria to Gammaproteobacteria and even to Verrucomicrobia (Boscaro et al., 2019; Schrallhammer et al., 2013; Serra et al., 2020). It is still unclear which is the role played by the symbiont in these cases.
Another direction of ciliate's symbiology, neglected for long time and now re‐discovered, is the study of epibionts of ciliates. The topic promises diverse and interesting insights, as those achieved with epixenosomes in Euplotidium spp. (Rosati et al., 1997, 1998, 1999), and more recently with “Ca. Deianiraea vastatrix” in Paramecium (Castelli et al., 2019), and the episymbionts of Parablepharisma spp. (Campello‐Nunes et al., 2020) and Zoothamnium niveum (Bright et al., 2019; Rinke et al., 2006).
Endosymbionts can occupy almost all cellular compartments of ciliated protists, but not much is known about the way in which colonization takes place and how the symbiont communicates with host ciliate cell (Boscaro et al., 2017; Fujishima, 2009; Garushyants et al., 2018; Sabaneyeva et al., 2010; Schrallhammer & Potekhin, 2020). Many groups of ciliates other than Oligohymenophorea and Spirotrichea have been shown to contain symbionts, but they have generally been little investigated. Many of those endosymbionts were found and described in the “pre‐molecular era”. It is valuable to find them again, analyze them using molecular approaches, and investigate how they interact with the host cell. In this perspective, new technologies, such as next‐generation sequencing and the use of genomics data, can provide essential information to clarify many aspects of the subject. For this reason, also a brief overview of achievements in genomics studies on ciliate's symbionts is provided.
HOLOSPORACEAE SYMBIONTS IN OLIGOHYMENOPHOREAN CILIATES
The genus Paramecium (Ciliophora, Oligohymenophorea), which comprises around 20 morphospecies (Krenek et al., 2015; Przyboś & Tarcz, 2018), is one of the most studied ciliate genera. Therefore, it is not surprising that the largest number of bacterial symbionts has been reported for these ciliates, some of them having been precisely described from the middle of the 20th century (Fokin & Görtz, 2009; Potekhin et al., 2021).
Most endosymbionts of Paramecium belong to two orders within Alphaproteobacteria, namely, Rickettsiales and Holosporales (Fokin et al., 2019; Schrallhammer & Potekhin, 2020; Serra et al., 2016; Szokoli et al., 2016). We focus on representatives of the Holosporaceae family as they are the most studied in ciliate symbiosis.
Bacteria from the family Holosporaceae are known as obligate endosymbionts of eukaryotes, mostly ciliates. In the last decade, new members of the family were found in various hosts from different environments (Boscaro, Fokin, et al., 2013; Fokin, 2012; Fokin et al., 2019; Lanzoni et al., 2015; Serra et al., 2016).
The biology of most Holospora and HLB, as well as their morphological peculiarities, have been described in detail in several reviews (Fokin & Görtz, 2009; Fokin & Sera, 2014; Görtz, 2006, 2014; Schrallhammer & Potekhin, 2020). They have been characterized as immobile, generally host‐ and compartment‐specific infectious bacteria, with reproductive and infectious forms in their life cycles. Members of Holospora have been traditionally defined as bacteria able to invade nuclei of certain Paramecium species only.
Among Paramecia, two species, P. bursaria and P. chlorelligerum, contain autotrophic eukaryotic symbionts, that is, Chlorella or Chlorella‐like green algae. We described such a symbiotic complex in P. chlorelligerum, in which the unicellular algae belong to Meyerella genus and inhabit its cytoplasm, while a new endosymbiont, “Ca. Holospora parva” colonizes the host macronucleus (Lanzoni et al., 2015). “Ca. Holospora parva” is a basally branching member of the Holospora but shares almost all characteristics of the genus, except connecting piece formation during the nuclear division. The connecting piece is a median body of the infected macronucleus or of the micronucleus formed during the host cell division process, in which most of the infective forms concentrate (Figure 1a): this peculiar formation is thought to improve the release of the infectious symbionts to the environment (Fokin & Sabaneyeva, 1997). This phenomenon could be considered an adaptation of bacteria that exploit the host cell division machinery to accomplish complex life cycles (Fokin, 2015; Fokin & Sabaneyeva, 1997). This feature has been found only in some HLB species retrieved in the macronuclei of representatives of Paramecium (Figure 1b) and Frontonia species (Beliavskaia et al., 2020; Boscaro, Fokin, et al., 2013; Fokin et al., 2019; Potekhin et al., 2018; Serra et al., 2016), more in detail, in Holospora species infecting P. caudatum and P. bursaria, “H. bacillata” from P. nephridiautum or P. calkinsi, and “H. curvata” from P. calkinsi (Fokin, 1989a, 2015; Fokin & Sabaneyeva, 1993, 1997). In other words, it was shown that the connecting piece is characteristic for all Holospora species, except “Ca. Holospora parva”. Therefore, the formation of a distinctive connecting piece during host cell division was proposed as a key feature to discriminate between classical Holospora and other HLB (Fokin et al., 1996).
FIGURE 1.
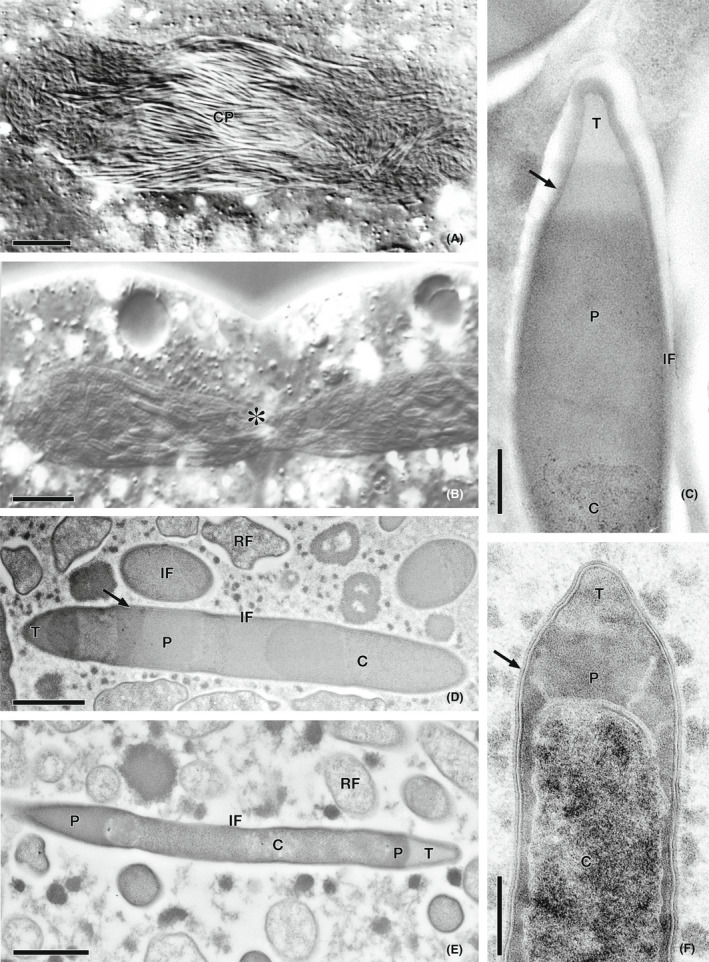
Morphology of Holospora and Holospora‐like bacteria. (A) Equatorial connecting piece (CP) of a dividing Paramecium caudatum macronucleus infected with H. obtusa. (B) Macronucleus of P. putrinum infected with “Ca. Gortzia yakutica” undergoing division shows the absence of the CP (asterisk). (C) Ultrastructure of the infectious form (IF) of “Ca. Holospora parva”; recognition tip (T); the arrowhead indicates the subdivision of periplasmic space (P); bacterial cytoplasm (C). (D) “Ca. Gortzia infectiva” reproductive form (RF), other abbreviations are the same. (E) IF and RF of Preeria caryophila. IF with two recognition tips, located at the opposite ends of the bacterial cell. (F) IF of “Ca. Gortzia shahrazadis”. All abbreviations are the same as in (C–E). (A, B) differential interference contrast microscopy; (C–F) transmission electron microscopy. Scale bars represent 20 μm (A), 10 μm (B), 0.3 μm (C), 1.0 μm (D), 0.6 μm (E), 0.5 μm (F)
Classical characteristics of Holospora infectious forms are the straight rod shape with differentiated cytoplasmic and periplasmic parts and a recognition tip‐like structure. In case of “Ca. Holospora parva” the periplasmic region of the cell always has two different regions, a larger and denser one and another more electronically transparent (Figure 1c). The latter characteristic was never recorded in other Holosporas, but is common for HLB (Figure 1d–f).
Considering both morphological and molecular data obtained for “Ca. H. parva”, we do not claim the presence of the connecting piece as a purely apomorphic feature for all Holospora species. However, apparently, this adaptation originated during co‐evolution between Holosporas and Paramecia at a rather early step, being present in five Holospora species and absent only from one, “Ca. H. parva” (Figure 2).
FIGURE 2.
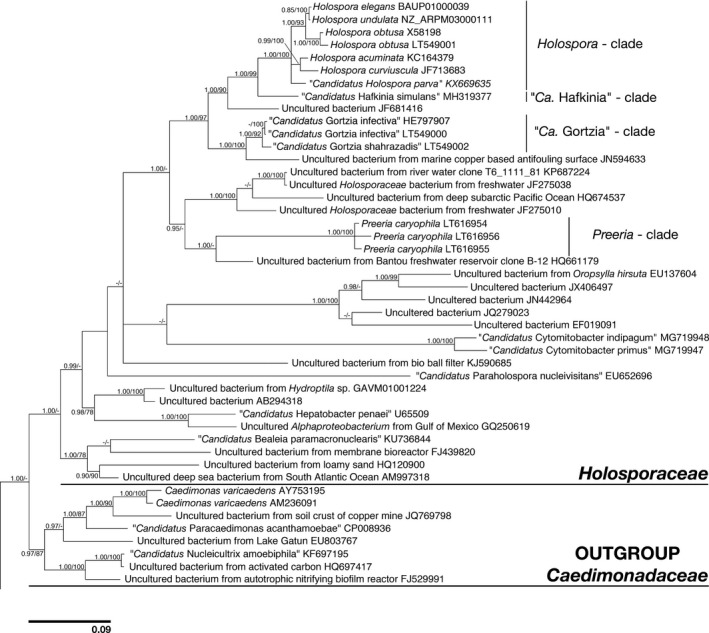
Bayesian inference tree of the family Holosporaceae based on 16S rRNA gene sequences (1288 character matrix). Numbers on nodes represent posterior probabilities and maximum likelihood bootstrap values, respectively (only values above 0.85–75 are shown). Bar stands for an estimated sequence divergence of 9%
Recently the former Holospora member Holospora caryophila (Preer & Preer, 1982) was reinvestigated (Potekhin et al., 2018). The authors wrote “surprisingly, they are only distantly related to other Holospora species, suggesting that they belong to a new genus within the family Holosporaceae, here described as Preeria caryophila comb. nov.” (Potekhin et al., 2018). Indeed, according to the 16S rRNA gene sequence comparison made, this HLB species is located in a different branch separated from all classical Holosporas and even other investigated HLB (Fokin et al., 2019; Potekhin et al., 2018). Two and a half decades ago, this conclusion had been reached on the basis of the absence of the connecting piece in the life cycle of the endosymbiont (Fokin et al., 1996).
Another feature that could generally separate Holospora from HLB is the relatively low host specificity of the latter. In the case of Preeria caryophila, the endosymbiont could not only infect but also survive in three ciliates of P. aurelia complex: P. biaurelia, P. octaurelia, and P. novaurelia as well in P. caudatum and P. polycaryum (Fokin, 2004; Fokin & Görtz, 2009; Lebedeva, Potekhin and Fokin, unpubl. data; Potekhin et al., 2018). “Ca. Gortzia” has the same tendency: “Ca. G. infectiva” (Boscaro, Fokin, et al., 2013) could infect the macronucleus of P. jenningsi (main host) and P. quadecaurelia (another native host); “Ca. G. yakutica” (Beliavskaia et al., 2020) infected the macronucleus in P. putrinum and P. nephridiatum (Fokin, Lebedeva, Serra, unpubl. data); “Holospora bacillata” (Fokin, 1989a) infected the macronucleus in P. nephridiatum and P. calkinsi. “Ca. Hafkinia simulans” from Frontonia salmastra and “Ca. G. shahrazadis” from P. multimicronucleatum have only been observed once (Fokin et al., 2019; Serra et al., 2016).
Regarding symbionts other than in the genus Holospora, the first complete description of “Ca. Gortzia” (Boscaro, Fokin, et al., 2013) was from P. jenningsi (main host) and from P. quadecaurelia from Thailand (Figure 1d). It is a member of the Holospora—“Hafkinia”—“Gortzia” cluster (Figure 2). This bacterium has low host specificity, and it has been identified again in India from P. jenningsi (Serra et al., 2016).
Holospora phylogeny suggests that this genus has co‐evolved with Paramecium, occurring in P. caudatum, P. chlorelligerum, and P. bursaria (Fokin et al., 2019; Potekhin et al., 2021). Three other genera of HLB closely related to Holospora include infectious bacteria manifesting similar life cycles and inhabiting the macronuclei of peniculine ciliates. They are: “Ca. Gortzia” (Boscaro, Fokin, et al., 2013) and Preeria (Potekhin et al., 2018), which colonize different Paramecium species, and “Ca. Hafkinia” which is so far known only from Frontonia salmastra. Preeria seems to be more divergent from Holospora in molecular analyses (Figure 2).
The number of HLBs might be much higher than the 12 or so members referred to in the literature (see: Fokin et al., 2019). Several species, described in pre‐molecular era, such as “H. bacillata”, “H. curvata”, and “Holospora sp.” from the macronucleus of P. putrinum (Fokin, 1989a; Fokin et al., 1999; Fokin & Görtz, 2009; Fokin & Sabaneyeva, 1993), probably belong to the “Ca. Gortzia” genus, considering the type of host and morphological traits (Figure 1b).
The infectious forms of “Ca. Hafkinia simulans”—HLB of the F. salmastra macronucleus have a distinctive appearance. They are present in the nucleus as large (up to 30 × 3.5 µm) spindle‐shaped or even skittle‐like infectious forms, quite similar in shape with the diatom Phaeodactylum tricornutum—a common prey of the ciliate. Some forms (probably the developing infectious forms) have many periplasmic zones and lack the recognition tip—an otherwise distinctive trait of the infectious form. Conversely, some infectious forms had two recognition tips located at opposite ends of the bacterial cell (Fokin et al., 2019). The last feature possibly improves the probability of infection. Double tips were reported in Preeria caryophila as well (Figure 1e).
In phylogenetic trees, “Ca. Hafkinia simulans” fell inside the Holosporaceae as a sister taxon to the Holospora clade, with strong statistical support (Figure 2). Thus, the biology and morphology of “Ca. Hafkinia simulans” differs more that of from Holospora, than other HLB, but phylogenetically it is more similar to classical holosporas than the “Ca. Gortzia” genus representatives (Fokin et al., 2019). The opposite argument could be made for Preeria caryophila—it is morphologically most similar (among HLB) to classical holosporas, but on phylogenetic reconstruction trees, it is far from Holospora and other HLB (Figure 2; Fokin et al., 2019; Potekhin et al., 2018). Thus, morphological and molecular results in these cases are considerably different.
One representative of the “Ca. Gortzia” genus, “Ca. G. shahrazadis”, detected in P. multimicronucleatum in India (Serra et al., 2016), has an unusual and surprising ability to reproduce in the host cytoplasm as well as in the macronucleus. This pattern has never been reported for any species of Holospora nor for members of “Ca. Gortzia”, Preeria, or “Ca. Hafkinia” (Fokin et al., 2019; Potekhin et al., 2018; Serra et al., 2016). Double infection of the cytoplasm and the macronucleus occurs in “Ca. Paraholospora nuclevisitans” (Eschbach et al., 2009), which is phylogenetically distant from the “Ca. Gortzia” clade (Figure 2; Fokin et al., 2019). As in the case of “Ca. Gortzia shahrazadis”, the endosymbiont is mainly located in the cytoplasm and morphologically does not resemble HLB; we conclude that this life cycle arose independently in these lineages of Holosporaceae.
Currently, the closest relative of “Ca. P. nucleovisitans” is “Ca. Mystax nordicus”, a cytoplasmic symbiont of P. nephridiatum (Korotaev et al., 2020). This endosymbiont has a remarkable appearance, suggesting it is the same as described in the 1980s (Fokin, 1989b). The endosymbiont was never observed inside the macronucleus; instead, it aggregated in clusters close to mitochondria.
New findings of bacteria belonging to major HLB groups and other representatives of Holosporaceae will definitely continue to occur. Some of endosymbionts described in the pre‐molecular period have been recollected from nature, and the bacteria were identified according to the current rules of bacterial nomenclature. This applies to “Ca. M. nordicus” (Korotaev et al., 2020) and probably to “Ca. G. yakutica”, even though the re‐description of the endosymbiont was made without transmission electron microscope investigation (Beliavskaia et al., 2020). The finding of the endosymbiont in the macronucleus of F. leucas that resemble Preeria (Fokin, unpubl. data) also suggests that more HLB biodiversity will be reported.
As last remarks: first, we recommend that we avoid the practice of naming symbiotic bacteria by the geographic place of their occurrence (Beliavskaia et al., 2020; Korotaev et al., 2020) because in our opinion, the same endosymbiont may be found in other host populations far from the place of the first description. As an example, “Ca. Gortzia yakutica” (from Yakutia, Sakha Republic, Russia) was first found in Germany (Fokin et al., 1999) and more recently in another host species, P. nephridiatum, on the Baltic Sea coast (Fokin, Lebedeva, Serra, unpubl. data).
The second remark concerns the proposal of H. undulata as the type species of the genus Holospora some time ago (Gromov & Ossipov, 1981). Indeed, it is now clear that this species has considerable morphological plasticity, and its 16S rRNA gene sequence is almost identical to the one of H. elegans and H. recta (Potekhin et al., 2021; Serra et al., 2016; Wackerow‐Kouzova & Myagkov, 2021). A recent attempt to solve this issue was made by Wackerow‐Kouzova and Myagkov (2021), who proposed to introduce a subspecies level for those three holosporas, namely, Holospora undulata subsp. elegans, H. undulata subsp. recta, and Holospora undulata subsp. undulata. This proposal has, unfortunately, been made based on few molecular data, not sufficient in our opinion to correctly address the problem, and moreover, we would discourage the use of subspecies level for bacterial species.
ENDOSYMBIONTS IN EUPLOTES
Euplotes is one of the most studied genera of Spirotrichea (Ciliophora). An increasing number of species is being described (Serra et al., 2020; Syberg‐Olsen et al., 2016), with currently more than 40 free‐living species. Members of the genus typically feed on bacteria, microalgae, and smaller protists. They are worldwide in distribution and widely present in marine, brackish, freshwater, and terrestrial habitats.
Generally, symbiotic events in Euplotes appear to be common and widespread, especially involving bacteria (Boscaro et al., 2019; Görtz, 2006; Heckmann & Schmidt, 1987; Serra et al., 2020; Vannini et al., 2005). Most studies have focused on freshwater or brackish water Euplotes, and little is known about endosymbionts in marine species (Görtz, 2006; Vannini et al., 2004).
An important event in the evolutionary history of the genus was the origin of the obligate symbiosis among some phylogenetically close species of Euplotes and Polynucleobacter necessarius or Polynucleobacter spp. (Betaproteobacteria, Burkholderiales), previously known as omicron or omicron‐like particles (Heckmann & Schmidt, 1987). This kind of obligate endosymbionts occurs in all Euplotes species from brackish or freshwater habitats, belonging to a monophyletic group known as “clade B” (Syberg‐Olsen et al., 2016). The obligate relationship with Polynucleobacter, essential for Euplotes, has been deeply explored, although not all mechanisms or benefits are understood. Thanks to genomic studies, it is clear that Euplotes acquired Polynucleobacter several times, with a process of free‐living bacterial strains taking over existing symbiotic systems and replacing the former symbiotic bacteria (Boscaro et al., 2017).
This kind of plasticity in symbiont replacement was demonstrated also by laboratory experiments, in which endosymbiotic bacteria have been transferred between different Euplotes species by microinjection (Fujishima & Heckmann, 1984; Vannini et al., 2017).
Interestingly, if P. necessarius is not present in the cytoplasm of the host, another less common Burkholderiales bacterium, namely, “Ca. Protistobacter hekmanni” can take its place (Boscaro, Petroni, et al., 2013; Vannini et al., 2007, 2012) or, in the case E. platysoma/harpa, distantly related endosymbionts, such as “Ca. Devosia symbiotica” (Alphaproteobacteria, Hyphomicrobiales; Boscaro et al., 2019). The presence of this bacterium in a “clade B” Euplotes has led to alternative scenarios that explain the initial symbiotic event between this clade of Euplotes and their modern symbionts. “Ca. Devosia spp.” (Alphaproteobacteria, Hyphomicrobiales) have been detected in two “clade A”—Euplotes (E. magnicirratus, E. enigma; Boscaro et al., 2019), suggesting that the first symbiosis event involved a common ancestor of clades A and B and an alphaproteobacterium, and the symbiosis with a betaproteobacterium would have arisen later. Further studies and Euplotes screening for symbionts are required to solve the issue.
Beside Betaproteobacteria endosymbionts, Euplotes (from “clades A”, “C”, and “E”) can host a wide range of phylogenetic far‐related bacteria, spanning from Alphaproteobacteria to Gammaproteobacteria and Verrucomicrobia (Boscaro et al., 2019; Schrallhammer et al., 2013; Serra et al., 2020).
Alphaproteobacteria symbionts are among the most frequently found endosymbionts in the cytoplasm of Euplotes spp. Most belong to the Rickettsiales (Boscaro, Petroni, et al., 2013; Senra et al., 2016; Vannini et al., 2010) or Holosporales (Boscaro et al., 2019) or Hyphomicrobiales (Boscaro et al., 2019; Vannini et al., 2004).
Representatives of Gammaproteobacteria are found in few species of Euplotes, e.g. “Ca. Nebulobacter yamunensis” (Boscaro et al., 2012), Francisella endociliophora (Schrallhammer et al., 2011), F. adeliensis (Vallesi et al., 2019), and “Ca. Endonucleariobacter sp.” (Boscaro et al., 2019).
The first verrucomicrobium found as an endosymbiont of a ciliate is represented by “Ca. Pinguicoccus supinus” (Verrucomicrobia, Opitutae), endosymbiont of E. vanleeuwenhoeki, a “clade A” Euplotes (Serra et al., 2020). This roundish prokaryote is about 1 µm in size and is a very peculiar endosymbiont from morphological and molecular points of view. It has distinctive invaginations of the inner membrane and an ultra‐reduced genome, one of the smallest found in symbionts of ciliates, being only 163,218 bp, encoding 205 genes (Serra et al., 2020). The most striking and fascinating feature of this bacterium is that it does not possess any gene for the catalytic subunit of the DNA polymerase. Unfortunately, the role of this symbiont in its host is still unclear.
Another verrucomicrobial endosymbiont was recently found in association with one Euplotes sp. from the Organic lake in Antarctica: “Ca. Organicella extenuata”, phylogenetically close to “Ca. Pinguicoccus supinus” (Williams et al., 2021). The genome of this endosymbiont is even smaller: 158,228 bp (encoding 194 genes). It lacks any capacity for the biosynthesis of amino acids or vitamins. It has a certain capacity for replication, transcription, translation, and protein‐folding, but no dedicated DNA polymerase for DNA replication was detected, as in “Ca. Pinguicoccus supinus”. Unfortunately, for “Ca. Organicella extenuata”, no ultrastructural analyses of the endosymbiont nor fluorescence in situ hybridization (FISH) experiments for localization inside the host have been reported.
Generally speaking, Euplotes species have a tendency to harbor bacteria and even true microbial consortia (i.e. two or more species of different prokaryotic species stably associated to the host.). Similar bacterial associations were detected and characterized from different Euplotes species. For example, one E. octocarinatus strain from Italy has been found infected by six different endosymbionts (including P. necessarius and four Alphaproteobacteria; Boscaro et al., 2019); one freshwater E. aediculatus strain from India harbored P. necessarius together with two Alpha‐ and one Gammaproteobacteria species in a stable association (Boscaro, Petroni, et al., 2013; Boscaro et al., 2012; Vannini et al., 2012, 2014). One brackish E. woodruffi strain from Brazil was reported to host the Alphaproteobacteria “Ca. Bandiella woodruffii” and P. necessarius (Senra et al., 2016). In all these cases, all the endosymbionts occupied the cytoplasm of the host, and P. necessarius was present. As they were found in different Euplotes species, from different habitats and very distant locations, it suggests that these kinds of associations could be more common than one might expect. The dynamics among these prokaryotic endosymbionts is still far from being understood, although it was hypothesized that P. necessarius might play a crucial role in facilitating the establishment of other symbionts in the same ciliate host (Senra et al., 2016).
As general considerations, all the endosymbionts of Euplotes were found in the cytoplasm (with or without a symbiosome; i.e. a specialized subcellular compartment of the host that houses an endosymbiont in a symbiotic relationship) and not in organelles, such as the macronucleus, but the reason for this is still unclear (Boscaro et al., 2019). Possibly, it is connected with the particular way that the macronucleus reorganizes before cell division of the ciliate. Moreover, none seems to be provided of any motile structures (Boscaro et al., 2019). Despite a considerable amount of knowledge on Euplotes’ symbionts having been recently acquired, a lot of work has still to be done to understand how and why these kinds of symbioses take place in Euplotes (so often!) and which cellular metabolic pathways are involved.
EPISYMBIONTS IN CILIATES
The episymbiosis phenomenon in ciliates is a tremendously interesting and diverse topic. Episymbioses have been observed in many ciliate species belonging to different classes of the phylum, involving many kinds of bacteria from Verrucomicrobia to Rickettsiales to Gammaproteobacteria and many others. The nature of the relationship can differ greatly from case to case. The prokaryotic symbiont may be parasitic and highly damaging for the host, as is the case of the episymbiont “Ca. Deianiraea vastatrix” (Alphaproteobacteria, Rickettsiales) in P. primaurelia as it causes loss of cilia and the death of the host (Castelli et al., 2019).
Relationships may be mutualistic, as in the case of the mouthless karyorelictean Kentrophoros and its microbial “kitchen garden” (Fenchel & Finlay, 1989), from which, from time to time the ciliate gains energy as food, phagocytizing a number of bacteria. The symbionts of Kentrophoros are phagocytosed by the ciliates along the whole cell body. The bacteria are sulfur oxidizers (thiotrophs) and assigned to the so called “Ca. Kentron” clade within the Gammaproteobacteria (Seah et al., 2017).
One of the most interesting and peculiar cases of mutualistic symbiosis is the relationship among Euplotidium species with the so‐called epixenosomes. These are Verrucomicrobia bacteria distributed along the dorsal surface of the ciliate. They protect the ciliate from predation with an extrusive apparatus that is triggered by external signals which are mediated by membrane receptors (Rosati et al., 1997, 1998, 1999).
Episymbionts can influence the lifestyle of their ciliate hosts, and even their morphology, as demonstrated for Zoothamnium niveum and its thiotrophic ectosymbiont “Ca. Thiobios zoothamnicoli” (Bright et al., 2014; Rinke et al., 2006), another thiotrophic bacterium belonging to the Gammaproteobacteria. Bright et al. (2019) demonstrated that Z. niveum is capable of a striking polymorphism connected to the presence of these ectosymbionts. Symbiotic and aposymbiotic organisms differed significantly in colony growth, form, and fitness (Bright et al., 2019).
Episymbionts of ciliates can also be employed as diagnostic features, such as in case of members of Parablepharisma (Campello‐Nunes et al., 2020): P. bacteriophora and P. brasiliensis possess rod‐shaped ectosymbiotic bacteria transversally attached to the cortex, while P. granulata shows episymbionts longitudinally attached to the cortex. No molecular studies have been carried out yet on these organisms, and we still do not know if there is a species‐specificity among Parablepharisma species and their ectosymbionts.
A fascinating aspect is of episymbionts of ciliates living in anoxic or semi‐anoxic conditions, such as members of Plagiopylea and Armophorea, for example, Plagiopyla ramani, P. nasuta (Nitla et al., 2019), Metopus contortus, and Caenomorpha levanderi (Fenchel & Ramsing, 1992). Molecular data are not available for these cases, but preliminary FISH experiments suggest the presence of sulfate‐reducing microorganisms (Fenchel & Ramsing, 1992).
The examples above reflect a small part of the literature dedicated to episymbionts of ciliates. Despite the remarkable observations, the field is still poorly investigated. Episymbioses is a topic that deserves extensive efforts from the scientific community as they may provide the keys to understanding: (1) how eukaryote cells are colonized by prokaryotes which could later become endosymbionts in a second evolutionary step after the prolonged cell to cell contact of an episymbiotic relationship and/or (2) how prokaryotes evolved a separate adaption path to another different kind of niche (extra vs. intracellular).
GENOMES OF CILIATE SYMBIONTS
Second‐ and third‐generation sequencing techniques allow us to obtain genomes for uncultivable organisms, such as the symbionts in ciliates. To date, around 20 complete genomes of such symbionts have been sequenced and annotated (Table 1). Most belong to the Proteobacteria, in particular the Alphaproteobacteria and Gammaproteobacteria (Table 1), but recently, the first genomes of two methanogenic archaeal endosymbionts have been provided (Lind et al., 2018).
TABLE 1.
Available genomes of symbionts of ciliates
| Host | Symbiont | Symbiont classification | Localization | GC content (%) | Genome size (Mb) | References |
|---|---|---|---|---|---|---|
| Paramecium caudatum | Holospora obtusa | Bacteria; Alphaproteobacteria; Holosporales; Holosporaceae | Macronucleus | 35.2 | 1.33 | Dohra et al. (2014) |
| Paramecium caudatum | Holospora elegans | Bacteria; Alphaproteobacteria; Holosporales; Holosporaceae | Macronucleus | 36 | 1.27 | Dohra et al. (2014) |
| Paramecium caudatum | Holospora undulata | Bacteria; Alphaproteobacteria; Holosporales; Holosporaceae | Macronucleus | 36.1 | 1.40 | Dohra et al. (2013, 2014) |
| Paramecium bursaria | Holospora curviuscula | Bacteria; Alphaproteobacteria; Holosporales; Holosporaceae | Macronucleus | 37.6 | 1.70 | Garushyants et al. (2018) |
| Paramecium biaurelia | "Caedimonas varicaedens" | Bacteria; Alphaproteobacteria; Holosporales; Caedimonadaceae | Cytoplasm or macronucleus | 42.1 | 1.68 | Suzuki et al. (2015) |
| Paramecium sp. | "Candidatus Fokinia solitaria" | Bacteria; Alphaproteobacteria; Rickettsiales; Midichloriaceae | Cytoplasm | 35.8 | 0.83 | Floriano et al. (2018) |
| Paramecium primaurelia | "Candidatus Deianiraea vastatrix" | Bacteria; Alphaproteobacteria; Rickettsiales; Deianiraeaceae | External cell surface | 32.9 | 1.2 | Castelli et al. (2019) |
| Paramecium tredecaurelia | "Candidatus Sarmatiella mevalonica" | Bacteria; Alphaproteobacteria; Rickettsiales; Rickettsiaceae | Cytoplasm | 38 | 1.27 | Castelli et al. (2021b) |
| Paramecium polycaryum | "Candidatus Gromoviella agglomerans" | Bacteria; Alphaproteobacteria; Holosporales; Holosporaceae | Cytoplasm | 32.2 | 0.589 | Castelli et al. (2021a) |
| Euplotes petzi | Francisella adeliensis | Bacteria; Gammaproteobacteria; Thiotrichales; Francisellaceae | Cytoplasm | 32.6 | 2.05 | Vallesi et al. (2019) |
| Undescribed Plagiopylea | "Candidatus Azoamicus ciliaticola" | Bacteria; Gammaproteobacteria | Cytoplasm | 24.4 | 0.29 | Graf et al. (2021) |
| Kentrophoros sp. | "Candidatus Kentron" clade | Bacteria; Gammaproteobacteria | External cell surface | Approximately 50 | 3.3–5.0 | Seah et al. (2019) |
| Paramecium tetraurelia | Caedibacter taeniospiralis | Bacteria; Gammaproteobacteria; Thiotrichales; Fastidiosibacteraceae | Cytoplasm | 41.3 | 1.32 | Pirritano et al. (2020) |
| Pseudoblepharisma tenue | "Candidatus Thiodictyon intracellulare" | Bacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae | Cytoplasm | 64.2 | 2.9 | Muñoz‐Gómez et al. (2021) |
| Trimyema compressum | Unnamed, strain TC1 | Bacteria; Firmicutes | Cytoplasm | 32.8 | 1.59 | Shinzato et al. (2016) |
| All Euplotes spp. belonging to clade B | Polynucleobacter spp., including Polynucleobacter necessarius | Bacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae | Cytoplasm | 45.6 | 1.5–1.9 | Boscaro, Felletti, et al. (2013) |
| Euplotes sp. (AntOrgLke) | "Candidatus Organicella extenuata" | Bacteria; Verrucomicrobia | Unknown | 32 | 0.158 | Williams et al. (2021) |
| Euplotes vanleeuwenhoeki (clade A) | "Candidatus Pinguicoccus supinus" | Bacteria; Verrucomicrobia | Cytoplasm | 25.1 | 0.163 | Serra et al. (2020) |
| Metopus contortus | Methanocorpusculum sp. MCE | Archaea; Methanomicrobia; Methanomicrobiales | Cytoplasm | 50.1 | 1.69 | Lind et al. (2018) |
| Nyctotherus ovalis | Methanobrevibacter sp. NOE | Archaea; Methanobacteria; Methanobacteriales | Cytoplasm | 25.4 | 1.92 | Lind et al. (2018) |
In terms of genome size, symbionts of ciliates range from 158 and 163 kbp of “Ca. Organicella extenuata” and of the E. vanleeuwenhoeki endosymbiont, “Ca. Pinguicoccus supinus” (Serra et al., 2020), to the genomes of 3.31 and 5.02 Mbp long from the symbionts "Ca. Kentron" clade from Kentrophoros (Seah et al., 2019). The latter are unique among thiotrophic symbionts because they do not encode canonical pathways for autotrophic carbon fixation but have a variety of heterotrophic features. They have the potential to oxidize sulfur to provide energy for assimilating organic carbon as the main carbon source for growth (Seah et al., 2019).
Complete genome assembly of “Ca. Gromoviella agglomerans” is the smallest reported genome among the order Holosporales (589.967 bp) and presents a severely reduced metabolism, both for what concerns biosynthetic pathways and for energy production and conversion (Castelli et al., 2021a).
Some obligate intracellular symbionts are subject to genomic reduction that may be accompanied by extensive gene loss, pseudogenization, a high rate of mutations, and low GC content (Floriano et al., 2018; Garushyants et al., 2018; Lind et al., 2018). The highly reduced genome of “Ca. Azoamicus ciliaticola”, from an undescribed plagiopylid, has presumably preserved traits that are beneficial to the host and that provide energy from anerobic respiration. Indeed, this symbiont is an obligate endosymbiont that has retained cellular functions that are markedly similar to those of mitochondria, although it did not originate from the mitochondrial line of descent (Graf et al., 2021).
In other cases, genome reduction is much less extreme in P. necessarius, the betaproteobacterial endosymbiont of the ciliate Euplotes has a genome (1.56 Mbp long) that is approximately as large as its free‐living counterparts (Boscaro et al., 2017). Genomic analyses show that intracellular bacteria use strategies to interact with, invade, and exploit their host cell, including secretion systems and effector, such as Type IV and Type VI secretion systems (Castelli et al., 2019), proteins with repeat motifs such as ankyrin repeat motifs (Floriano et al., 2018) and ADP/ATP translocase that directly import ATP from the host (Castelli et al., 2021b; Dohra et al., 2014; Garushyants et al., 2018; Vallesi et al., 2019). This potential genomic adaptation for an intracellular lifestyle is not always so marked. This is shown by the genomes of the two methanogenic endosymbionts of the anerobic ciliates, Metopus contortus and Nyctotherus ovalis. Their genomes are in an early stage of adaptation toward endosymbiosis, as evidenced by the large number of genes undergoing pseudogenization (Lind et al., 2018).
PERSPECTIVES
Genomic studies of ciliate symbionts give excellent opportunities to deepen our knowledge on the processes of symbioses. Currently, this line of research is poorly explored because of the absence of extensive molecular data. Hence, a fundamental challenge will be to increase the number of complete genomes which, in parallel with comparative genomics analyses, will shed light on specific adaptations to symbiosis. A better understanding of host–symbiont interactions will depend on more targeted genomic studies.
ACKNOWLEDGMENTS
Michele Giovannini is acknowledged for his support in “Genomes of ciliates’ symbionts” part preparation. Alessandro Allievi is acknowledged for his support in English revision. Simone Gabrielli is acknowledged for his assistance in tree and photo editing. This study was supported by the European Community’s H2020 Programme H2020‐MSCA‐RISE 2019 (grant agreement no. 872767), and by the Fondazione Cassa di Risparmio di Pistoia e Pescia ‐ giovani@ricercascientifica2019 fellowship (no. 2019.0380). Open Access Funding provided by Universita degli Studi di Pisa within the CRUI‐CARE Agreement.
Fokin, S.I. & Serra, V. (2022) Bacterial symbiosis in ciliates (Alveolata, Ciliophora): Roads traveled and those still to be taken. Journal of Eukaryotic Microbiology, 69, e12886. 10.1111/jeu.12886
[Correction added on 12 May 2022, after first online publication: CRUI funding statement has been added.]
REFERENCES
- Beliavskaia, A.Y. , Predeus, F.V. , Garushyants, S.K. , Logacheva, M.D. , Gong, J. , Zou, S. et al. (2020) New intranuclear symbiotic bacteria from macronucleus of Paramecium putrinum—Candidatus Gortzia yakutica. Diversity, 12, 198. 10.3390/d12050198 [DOI] [Google Scholar]
- Boscaro, V. , Felletti, M. , Vannini, C. , Ackerman, M.S. , Chain, P.S. , Malfatti, S. et al. (2013) Polynucleobacter necessarius, a model for genome reduction in both free‐living and symbiotic bacteria. Proceedings of the National Academy of Sciences of the United States of America, 110, 18590–18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscaro, V. , Fokin, S.I. , Schrallhammer, M. , Schweikert, M. & Petroni, G. (2013) Revised systematic of Holospora‐like bacteria and characterization of “Candidatus Gortzia infectiva”, a novel macronuclear symbiont of Paramecium jenningsi . Microbial Ecology, 65, 255–267. [DOI] [PubMed] [Google Scholar]
- Boscaro, V. , Husnik, F. , Vannini, C. & Keeling, P.J. (2019) Symbionts of the ciliate Euplotes: diversity, patterns and potential as models for bacteria–eukaryote endosymbioses. Proceedings of the Royal Society B: Biological Sciences, 286, 20190693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscaro, V. , Kolisko, M. , Felletti, M. , Vannini, C. , Lynn, D.H. & Keeling, P.J. (2017) Parallel genome reduction in symbionts descended from closely related free‐living bacteria. Nature Ecology & Evolution, 1, 1160–1167. [DOI] [PubMed] [Google Scholar]
- Boscaro, V. , Petroni, G. , Ristori, A. , Verni, F. & Vannini, C. (2013) “Candidatus Defluviella procrastinata” and “Candidatus Cyrtobacter zanobii”, two novel ciliate endosymbionts belonging to the “Midichloria clade. Microbial Ecology, 65, 302–310. [DOI] [PubMed] [Google Scholar]
- Boscaro, V. , Vannini, C. , Fokin, S.I. , Verni, F. & Petroni, G. (2012) Characterization of “Candidatus Nebulobacter yamunensis” from the cytoplasm of Euplotes aediculatus (Ciliophora, Spirotrichea) and emended description of the family Francisellaceae . Systematic and Applied Microbiology, 35, 432–440. [DOI] [PubMed] [Google Scholar]
- Bright, M. , Espada‐Hinojosa, S. , Lagkouvardos, I. & Volland, J.‐M. (2014) The giant ciliate Zoothamnium niveum and its thiotrophic epibiont Candidatus Thiobios zoothamnicoli: a model system to study interspecies cooperation. Frontiers in Microbiology, 5, 145. 10.3389/fmicb.2014.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright, M. , Espada‐Hinojosa, S. , Volland, J.‐M. , Drexel, J. , Kesting, J. , Kolar, I. et al. (2019) Thiotrophic bacterial symbiont induces polyphenism in giant ciliate host Zoothamnium niveum . Scientific Reports, 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello‐Nunes, P.H. , Fernandes, N.M. , Szokoli, F. , Fokin, S.I. , Serra, V. , Modeo, L. et al. (2020) Parablepharisma (Ciliophora) is not a heterotrich: a phylogenetic and morphological study with the proposal of new taxa. Protist, 17, 125716. [DOI] [PubMed] [Google Scholar]
- Castelli, M. , Lanzoni, O. , Giovannini, M. , Lebedeva, N. , Gammuto, L. , Sassera, D. et al. (2021a) ‘Candidatus Gromoviella agglomerans’, a novel intracellular Holosporaceae parasite of the ciliate Paramecium showing marked genome reduction. Environmental Microbiology Reports, 1–16. 10.1111/1758-2229.13021 [DOI] [PubMed] [Google Scholar]
- Castelli, M. , Lanzoni, O. , Nardi, T. , Lometto, S. , Modeo, L. , Potekhin, A. et al. (2021b) ‘Candidatus Sarmatiella mevalonica’ endosymbiont of the ciliate Paramecium provides insights on evolutionary plasticity among Rickettsiales . Environmental Microbiology, 23, 1684–1701. 10.1111/1462-2920.15396 [DOI] [PubMed] [Google Scholar]
- Castelli, M. , Sabaneyeva, E. , Lanzoni, O. , Lebedeva, N. , Floriano, A.M. , Gaiarsa, S. et al. (2019) Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales . ISME Journal, 13, 2280–2294. 10.1038/s41396-019-0433-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohra, H. , Tanaka, K. , Suzuki, T. , Fujishima, M. & Suzuki, H. (2014) Draft genome sequences of three Holospora species (Holospora obtusa, Holospora undulata, and Holospora elegans), endonuclear symbiotic bacteria of the ciliate Paramecium caudatum . FEMS Microbiology Letters, 359, 16–18. 10.1111/1574-6968.12577 [DOI] [PubMed] [Google Scholar]
- Dziallas, C. , Allgaier, M. , Monaghan, M. T. & Grossart, H. P. (2012) Act together—implications of symbioses in aquatic ciliates. Frontiers in microbiology, 3, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach, E. , Pfannkuchen, M. , Schweikert, M. , Drutschmann, D. , Brümmer, F. , Fokin, S. et al. (2009) Candidatus Paraholospora nucleivisitans”, an intracellular bacterium in Paramecium sexaurelia shuttles between the cytoplasm and the nucleus of its host. Systematic and applied microbiology, 32(7), 490–500. [DOI] [PubMed] [Google Scholar]
- Fenchel, T. & Finlay, B.J. (1989) Kentrophoros: a mouthless ciliate with a symbiotic kitchen garden. Ophelia, 30, 75–93. [Google Scholar]
- Fenchel, T. & Ramsing, N.B. (1992) Identification of sulphate‐reducing ectosymbiotic bacteria from anaerobic ciliates using 16S rRNA binding oligonucleotide probes. Archives of Microbiology, 158, 394–397. [DOI] [PubMed] [Google Scholar]
- Floriano, A.M. , Castelli, M. , Krenek, S. , Berendonk, T.U. , Bazzocchi, C. , Petroni, G. et al. (2018) The genome sequence of “Candidatus Fokinia solitaria”: insights on reductive evolution in Rickettsiales . Genome Biology and Evolution, 10, 1120–1126. 10.1093/gbe/evy072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner, W. (2008) Protist diversity and distribution: some basic considerations. Biodiversity and Conservation, 17, 235–242. [Google Scholar]
- Foissner, W. (2016) Terrestrial and semiterrestrial ciliate (Protozoa, Ciliophora) from Venezuela and Galapogos. Denisia, 35, 1–912. [Google Scholar]
- Foissner, W. & Berger, H. (2021) Terrestrial ciliates (Protista, Ciliophora) from Australia and some other parts of the world. Ser. Monogrphiae Ciliophorae, 5, 1–380. [Google Scholar]
- Fokin, S.I. (1989a) Bacterial endobionts of the ciliate Paramecium woodruffi. I. Endobionts of the macronucleus. Tsitologiya, 31, 839–844. [Google Scholar]
- Fokin, S.I. (1989b) Bacterial endobionts of the ciliate Paramecium woodruffi. III. Endobionts of the cytoplasm. Tsitologia, 31, 964–969. [Google Scholar]
- Fokin, S.I. (2004) Bacterial endocytobionts of Ciliophora and their interactions with the host cell. International Review of Cytology, 236, 181–249. [DOI] [PubMed] [Google Scholar]
- Fokin, S.I. (2012) Frequency and biodiversity of symbionts in representatives of the main classes of Ciliophora. European Journal of Protistology, 48, 138–148. [DOI] [PubMed] [Google Scholar]
- Fokin, S.I. (2015) Release of Holospora‐like bacteria in different ciliate species. Mat. VII European Congress of Protistology. 5–10 September, Seville. p. 279.
- Fokin, S.I. , Brigge, T. , Brenner, J. & Gȍrtz, H.‐D. (1996) Holospora species infecting the nuclei of Paramecium appear to belong to two groups of bacteria. European Journal of Protistology, 32(Suppl. 1), 19–24. [Google Scholar]
- Fokin, S. , Brigge, T. & Gȍrtz, H.‐D. (1999) An infectious bacterium inhabiting the macronucleus of Paramecium putrinum . Journal of Eukaryotic Microbiology, 46, 11. [Google Scholar]
- Fokin, S.I. & Görtz, H.‐D. (2009) Diversity of Holospora bacteria in Paramecium and their characterization. In: Fujishima, M. (Ed.) Microbiology monographs 12. Berlin, Heidelberg: Springer‐Verlag, pp. 161–199. [Google Scholar]
- Fokin, S.I. & Karpov, S.A. (1995) Bacterial endocytobionts inhabiting the perinuclear space of Protista. Endocytosis Cell Research, 11, 81–94. [Google Scholar]
- Fokin, S.I. & Sabaneyeva, E.V. (1993) Bacterial endocytobionts of the ciliate Paramecium calkinsi . European Journal of Protistology, 29, 390–395. [DOI] [PubMed] [Google Scholar]
- Fokin, S. & Sabaneyeva, E. (1997) Release of endonucleobiotic bacteria Holospora bacillata and Holospora curvata from the macronucleus of their host cells Paramecium woodruffi and Paramecium calkinsi . Endocytobiosis and Cell Research, 12, 49–56. [Google Scholar]
- Fokin, S.I. , Schrallhammer, M. , Chiellini, C. , Verni, F. & Petroni, G. (2014) Free‐living ciliates as potential reservoirs for eukaryotic parasites: occurrence of a trypanosomatid in the macronucleus of Euplotes encysticus . Parasites Vectors, 7, 203. 10.1186/1756-3305-7-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin, S.I. & Sera, V. (2014) The hidden biodiversity of ciliate‐endosymbionts systems. JSM Microbiology, 2, 1015–1018. [Google Scholar]
- Fokin, S.I. , Serra, V. , Ferrantini, F. , Modeo, L. & Petroni, G. (2019) “Candidatus Hafkinia simulans” gen. nov., sp. nov., a novel Holospora‐like bacterium from the macronucleus of the rare brackish water ciliate Frontonia salmastra (Oligohymenophorea, Ciliophora): multidisciplinary characterization of the new endosymbiont and its host. Microbial Ecology, 77, 1092–1106. 10.1007/s00248-018-1311-0 [DOI] [PubMed] [Google Scholar]
- Fujishima, M. (2009) Infection and maintenance of Holospora species in Paramecium caudatum . In: Fujishima, M. (Ed.) Microbiology monographs 12. Berlin, Heidelberg: Springer‐Verlag, pp. 201–225. [Google Scholar]
- Fujishima, M. & Heckmann, K. (1984) Intra‐and interspecies transfer of endosymbionts in Euplotes . Journal of Experimental Zoology, 230, 339–345. [Google Scholar]
- Garushyants, S. , Beliavskaia, A.Y. , Malko, D.B. , Logacheva, M.D. , Rautian, M.S. & Gelfand, M.S. (2018) Comparative genomic analysis of Holospora spp., intranuclear symbiont of paramecia. Frontiers in Microbiology, 9(738), 1–11. 10.3389/fmicb.2018.00738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görtz, H.‐D. (2006) Symbiotic associations between ciliates and prokaryotes. In: Dworkin, M. , Falkow, S. , Rosenberg, E. , Schleifer, K.H. & Stackebrandt, E. (Eds.) The prokaryotes. New York, NY: Springer, pp. 364–402. [Google Scholar]
- Görtz, H.‐D. (2014) Prokaryotic endosymbionts in ciliates. In: Hausmann, K. & Radek, R. (Eds.) Cilia and flagella ciliates and flagellates. Ultrastructure and cell biology, function and systematic, symbiosis and biodiversity. Tübingen: Schweizerbart Science Publishers, pp. 229–238. [Google Scholar]
- Graf, J.S. , Schorn, S. , Kitzinger, K. , Ahmerkamp, S. , Woehle, C. , Huettel, B. et al. (2021) Anaerobic endosymbiont generates energy for ciliate host by denitrification. Nature, 591, 445–450. 10.1038/s41586-021-03297-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromov, B.V. & Ossipov, D.V. (1981) Holospora (ex Hafkine 1890) nom.rev., a genus of bacteria inhabiting the nuclei of paramecia. International Journal of Systematic Bacteriology, 31, 348–352. [Google Scholar]
- Heckmann, K. & Schmidt, H.J. (1987) Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus . International Journal of Systematic and Evolutionary Microbiology, 37, 456–457. [Google Scholar]
- Korotaev, A. , Benken, K. & Sabaneyeva, E. (2020) “Candidatus Mystax nordicus” aggregates with mitochondria of its host, the ciliate Paramecium nephridiatum . Diversity, 12, 251. 10.3390/d12060251 [DOI] [Google Scholar]
- Krenek, S. , Berendonk, T.U. & Fokin, S.I. (2015) New Paramecium (Ciliophora, Oligohymenophorea) congeners shape our view on its biodiversity. Organisms, Diversity, and Evolution, 15, 215–233. 10.1007/s13127-015-0207-9 [DOI] [Google Scholar]
- Lanzoni, O. , Fokin, S.I. , Lebedeva, N. , Migunova, A. , Petroni, G. & Potekhin, A. (2015) Rare freshwater ciliate Paramecium chlorelligerum Kahl, 1935 and its macronuclear symbiotic bacterium “Candidatus Holospora parva”. PLoS One, 11, e0167928. 10.1371/journal.pone.0167928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind, A.E. , Lewis, W.H. , Spang, A. , Guy, L. , Embley, T.M. & Ettema, T.J.G. (2018) Genomes of two archaeal endosymbionts show convergent adaptations to an intracellular lifestyle. ISME Journal, 12, 2655–2667. 10.1038/s41396-018-0207-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz‐Gómez, S. A. , Kreutz, M. & Hess, S. (2021) A microbial eukaryote with a unique combination of purple bacteria and green algae as endosymbionts. Science Advances, 7(24), eabg4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitla, V. , Serra, V. , Fokin, S.I. , Modeo, L. , Verni, F. , Sandeep, B.V. et al. (2019) Critical revision of the family Plagiopylidae (Ciliophora: Plagiopylea), including the description of two novel species, Plagiopyla ramani and Plagiopyla narasimhamurtii, and redescription of Plagiopyla nasuta Stein, 1860 from India. Zoological Journal of the Linnean Society London, 186, 1–45. [Google Scholar]
- Pirritano, M. , Zaburannyi, N. , Grosser, K. , Gasparoni, G. , Müller, R. , Simon, M. & Schrallhammer, M. (2020) Dual‐Seq reveals genome and transcriptome of Caedibacter taeniospiralis, obligate endosymbiont of Paramecium . Scientific reports, 10(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potekhin, A. , Nekrasova, I. & Flemming, F.E. (2021) In shadow of Holospora—the continuous quest for new Holosporaceae members. Protistology, 15, 127–141. [Google Scholar]
- Potekhin, A. , Schweikert, M. , Nekrasova, I. , Vitali, V. , Schwarzer, S. , Anikina, A. et al. (2018) Complex life cycle, broad host range and adaptation strategy of the intranuclear Paramecium symbiont Preeria caryophila comb. nov. FEMS . Microbial Ecology, 94, 2018, fiy076. [DOI] [PubMed] [Google Scholar]
- Preer, J.R. & Preer, L.B. (1982) Revival of names of protozoan endosymbionts and proposal of Holospora caryophila nom. nov. International Journal of Systematic Bacteriology, 32, 140–141. [Google Scholar]
- Przyboś, E. & Tarcz, S. (2018) Paramecium (Protista, Ciliophora, Oligohymenophorea) as a model organism in biological studies, especially concerning speciation process. Krakow: ISEA PAS, pp. 1–68. [Google Scholar]
- Rinke, C. , Schmitz‐Esser, S. , Stoecker, K. , Nussbaumer, A.D. , Molnár, D.A. , Vanura, K. et al. (2006) “Candidatus Thiobios zoothamnicoli”, an ectosymbiotic bacterium covering the giant marine ciliate Zoothamnium niveum . Applied and Environment Microbiology, 72, 2014–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati, G. , Giambelluca, M.A. , Grossi, M. & Morelli, A. (1997) Epixenosomes, peculiar epibionts of the ciliate Euplotidium itoi: involvement of membrane receptors and the adenylate cyclase‐cyclic AMP system in the ejecting process. Protoplasma, 197, 57–63. [Google Scholar]
- Rosati, G. , Petroni, G. , Quochi, S. , Modeo, L. & Verni, F. (1999) Epixenosomes: peculiar epibionts of the hypotrich ciliate Euplotidium itoi defend their host against predators. Journal of Eukaryotic Microbiology, 46, 278–282. [Google Scholar]
- Rosati, G. , Verni, F. , Lenzi, P. , Giambelluca, M.A. , Sironi, M. & Bandi, C. (1998) Epixenosomes, peculiar epibionts of the ciliated protozoon Euplotidium itoi: what kind of organisms are they? Protoplasma, 201, 38–44. [Google Scholar]
- Rossi, A. , Boscaro, V. , Carducci, D. , Serra, V. , Modeo, L. , Verni, F. et al. (2016) Ciliate communities and hidden biodiversity in freshwater biotopes of the Pistoia province (Tuscany, Italy). European Journal of Protistology, 53, 11–19. [DOI] [PubMed] [Google Scholar]
- Sabaneyeva, E.V. , Derkacheva, M.E. , Benken, K.A. , Fokin, S.I. , Vainio, S. & Skovorodkin, I.N. (2010) Actin‐based mechanism of Holospora obtusa trafficking in Paramecium caudatum . Protist, 160, 205–219. [DOI] [PubMed] [Google Scholar]
- Schrallhammer, M. , Ferrantini, F. , Vannini, C. , Galati, S. , Schweikert, M. , Görtz, H.D. et al. (2013) ‘Candidatus Megaira polyxenophila’ gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent Rickettsiae . PLoS One, 8, e72581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrallhammer, M. & Potekhin, A. (2020) Epidemiology of nucleus‐dwelling Holospora: infection, transmission, adaptation, and interaction with Paramecium . In: Kloc, M. (Ed.) Symbiosis: cellular, molecular, medical and evolutionary aspects. Results and problems in cell differentiation, vol. 69, Cham: Springer, 4, pp. 106–135. 10.1007/978-3-030-51849-3_4 [DOI] [PubMed] [Google Scholar]
- Schrallhammer, M. , Schweikert, M. , Vallesi, A. , Verni, F. & Petroni, G. (2011) Detection of a novel subspecies of Francisella noatunensis as endosymbiont of the ciliate Euplotes raikovi . Microbial Ecology, 61, 455–464. [DOI] [PubMed] [Google Scholar]
- Seah, B.K.B. , Antony, C.P. , Huettel, B. , Zarzycki, J. , Schada von Borzyskowski, L. , Erb, T.J. et al. (2019) Sulfur‐oxidizing symbionts without canonical genes for autotrophic CO2 fixation. MBio, 10(3), 1–18. 10.1128/mBio.01112-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah, B.K. , Schwaha, T. , Volland, J.M. , Huettel, B. , Dubilier, N. & Gruber‐Vodicka, H.R. (2017) Specificity in diversity: single origin of a widespread ciliate‐bacteria symbiosis. Proceedings of the Royal Society London B: Biological Sciences, 284, 20170764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senra, M.V. , Dias, R.J. , Castelli, M. , Silva‐Neto, I.D. , Verni, F. , Soares, C.A. et al. (2016) A house for two—double bacterial infection in Euplotes woodruffi Sq1 (Ciliophora, Euplotia) sampled in Southeastern Brazil. Microbial Ecology, 71, 505–517. [DOI] [PubMed] [Google Scholar]
- Serra, V. , Fokin, S.I. , Castelli, M. , Basuri, C.K. , Nitla, V. , Verni, F. et al. (2016) “Candidatus Gortzia shahrazadis”, a novel endosymbiont of Paramecium multimicronucleatum and a revision of the biogeographical distribution of Holospora‐like bacteria. Frontiers in Microbiology, 7, 1704. 10.3389/fmicb.2016.01704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra, V. , Gammuto, L. , Nitla, V. , Castelli, M. , Lanzoni, O. , Sassera, D. et al. (2020) Morphology, ultrastructure, genomics, and phylogeny of Euplotes vanleeuwenhoeki sp. nov. and its ultra‐reduced endosymbiont “Candidatus Pinguicoccus supinus” sp. nov. Scientific Reports, 10, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato, N. , Aoyama, H. , Saitoh, S. , Nikoh, N. , Nakano, K. & Shimoji, M. (2016) Complete genome sequence of the intracellular bacterial symbiont TC1 in the anaerobic ciliate Trimyema compressum. Genome announcements, 4(5), e01032‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H. , Dapper, A. L. , Jackson, C. E. , Lee, H. , Pejaver, V. & Doak, T. G. et al. (2015) Draft genome sequence of Caedibacter varicaedens, a kappa killer endosymbiont bacterium of the ciliate Paramecium biaurelia . Genome announcements, 3(6), e01310‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syberg‐Olsen, M.J. , Irwin, N.A. , Vannini, C. , Erra, F. , Di Giuseppe, G. , Boscaro, V. et al. (2016) Biogeography and character evolution of the ciliate genus Euplotes (Spirotrichea, Euplotia), with description of Euplotes curdsi sp. nov. PLoS One, 11, e0165442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szokoli, F. , Castelli, M. , Sabaneyeva, E. , Schrallhammer, M. , Krenek, S. , Doak, T.G. et al. (2016) Disentangling the taxonomy of Rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) sharing the cytoplasm of the ciliate protist Paramecium biaurelia . Applied and Environment Microbiology, 82, 7236–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallesi, A. , Sjödin, A. , Petrelli, D. , Luporini, P. , Taddei, A.R. , Thelaus, J. et al. (2019) A new species of the γ‐Proteobacterium Francisella, F. adeliensis sp. nov., endocytobiont in an Antarctic marine ciliate and potential evolutionary forerunner of pathogenic species. Microbial Ecology, 77, 587–596. [DOI] [PubMed] [Google Scholar]
- Vannini, C. , Boscaro, V. , Ferrantini, F. , Benken, K. , Mironov, T.I. , Schweikert, M. et al. (2014) Flagellar movement in two bacteria of the family Rickettsiaceae: a re‐evaluation of motility in an evolutionary perspective. PLoS One, 9, e87718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini, C. , Ferrantini, F. , Schleifer, K.H. , Ludwig, W. , Verni, F. & Petroni, G. (2010) “Candidatus Anadelfobacter veles” and “Candidatus Cyrtobacter comes”, two new Rickettsiales species hosted by the protist ciliate Euplotes harpa (Ciliophora, Spirotrichea). Applied and Environment Microbiology, 76, 4047–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini, C. , Ferrantini, F. , Ristori, A. , Verni, F. & Petroni, G. (2012) Betaproteobacterial symbionts of the ciliate Euplotes: origin and tangled evolutionary path of an obligate microbial association. Environmental Microbiology, 14, 2553–2563. [DOI] [PubMed] [Google Scholar]
- Vannini, C. , Petroni, G. , Verni, F. & Rosati, G. (2005) Polynucleobacter bacteria in the brackish‐water species Euplotes harpa (Ciliata Hypotrichia). Journal of Eukaryotic Microbiology, 52, 116–122. [DOI] [PubMed] [Google Scholar]
- Vannini, C. , Pöckl, M. , Petroni, G. , Wu, Q.L. , Lang, E. , Stackebrandt, E. et al. (2007) Endosymbiosis in statu nascendi: close phylogenetic relationship between obligately endosymbiotic and obligately free living Polynucleobacter strains (Betaproteobacteria). Environmental Microbiology, 9, 347–359. [DOI] [PubMed] [Google Scholar]
- Vannini, C. , Rosati, G. , Verni, F. & Petroni, G. (2004) Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of ‘ Candidatus Devosia euplotis'. International Journal of Systematic and Evolutionary Microbiology, 54, 1151–1156. [DOI] [PubMed] [Google Scholar]
- Vannini, C. , Sigona, C. , Hahn, M. , Petroni, G. & Fujishima, M. (2017) High degree of specificity in the association between symbiotic betaproteobacteria and the host Euplotes (Ciliophora, Euplotia). European Journal of Protistology, 59, 124–132. 10.1016/j.ejop.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Wackerow‐Kouzova, N. D. & Myagkov, D. V. (2021) Clarification of the taxonomic position of Paramecium caudatum micronucleus symbionts. Current microbiology, 78(12), 4098–4102. [DOI] [PubMed] [Google Scholar]
- Williams, T.J. , Allen, M.A. , Ivanova, N. , Huntemann, M. , Haque, S. , Hancock, A.M. et al. (2021) Genome analysis of a verrucomicrobial endosymbiont with a tiny genome discovered in an Antarctic Lake. Frontiers in Microbiology, 12, 674758. 10.3389/fmicb.2021.674758 [DOI] [PMC free article] [PubMed] [Google Scholar]


