Abstract
Little is known about the immune environment of ovarian clear cell carcinoma (OCCC) and its impact on various ethnic backgrounds. The aim of this OCCC immune‐related gene expression signatures (irGES) study was to address the interaction between tumour and immune environment of ethnically‐diverse Asian and Caucasian populations and to identify relevant molecular subsets of biological and clinical importance. Our study included 264 women from three different countries (Singapore, Japan, and the UK) and identified four novel immune subtypes (PD1‐high, CTLA4‐high, antigen‐presentation, and pro‐angiogenic subtype) with differentially expressed pathways, and gene ontologies using the NanoString nCounter PanCancer Immune Profiling Panel. The PD1‐high and CTLA4‐high subtypes demonstrated significantly higher PD1, PDL1, and CTLA4 expression, and were associated with poorer clinical outcomes. Mismatch repair (MMR) protein expression, assessed by immunohistochemistry, revealed that about 5% of OCCCs had deficient MMR expression. The prevalence was similar across the three countries and appeared to cluster in the CTLA4‐high subtype. Our results suggest that OCCC from women of Asian and Caucasian descent shares significant clinical and molecular similarities. To our knowledge, our study is the first study to include both Asian and Caucasian women with OCCC and helps to shine light on the impact of ethnic differences on the immune microenvironment of OCCC. © 2021 The Authors. The Journal of Pathology published by John Wiley & Sons, Ltd. on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: ovarian cancer, clear cell cancer, gene expression signatures, immune microenvironment, microsatellite instability, mismatch repair protein, immune subtypes, RNA expression and ethnicity
Introduction
The prevalence of ovarian clear cell carcinoma (OCCC) varies across ethnicity. In Europe and North America, OCCC accounts for 3–12% of epithelial ovarian cancer (EOC) cases [1, 2, 3], while in East Asia, the rates are significantly higher, approaching 25% of EOC cases [4, 5]. However, OCCC is still considered ‘rare’ by ESGO‐GCIG, defined as an annual incidence of <6 per 100 000. This subset of EOC typically has a poorer prognosis and much lower rates of response to platinum‐based chemotherapy compared with patients with high‐grade serous ovarian carcinoma (HGSOC) [6].
In recurrent OCCC, responses to chemotherapy are uniformly low. A study in the Japanese population observed response rates to second‐line chemotherapy of 8% in ‘platinum‐sensitive’ patients and only 6% in the ‘platinum‐resistant’ cohort [7]. In another retrospective study of 39 OCCC patients from Australia, patients who had a partial response or no response to initial chemotherapy failed to respond to second‐line chemotherapy [8]. A retrospective study of response rates in recurrent OCCC patients from Canada and the UK revealed an overall response rate of 9%, confirming the limited activity of chemotherapy in recurrent disease [9]. Typically, response rates of recurrent OCCC are less than 10% [7], with a median progression‐free survival (PFS) of around 11 weeks and a median overall survival (OS) of around 40 weeks [9].
As a consequence of the rare nature of this disease, large‐scale international trials are required in order to change the standard of care. To date, these have only rarely been completed. One of the first large trials conducted specifically for OCCC was JGOG3017 [10], a randomized phase III trial that compared the efficacy and safety of paclitaxel plus carboplatin (TC) versus irinotecan plus cisplatin (CPT‐P) in patients with OCCC. This JGOG/GCIG trial was an international collaboration that accrued 667 patients, mainly from Japan and the remainder from Korea, France, and the UK. The main objective was to evaluate if the CPT‐P regimen was superior to TC. The trial observed no differences in PFS or OS between the two study regimens and suggested that TC therapy remain the standard chemotherapy for patients with OCCC. There is a clear need for new therapeutic strategies for OCCC patients. A recent phase II trial of pembrolizumab for recurrent ovarian cancer (KEYNOTE‐100) showed a low overall response (~8%), although the response rate in patients with clear cell histology was 16% [11], suggesting that certain OCCCs may elicit anti‐tumour immune responses following PD‐1 checkpoint inhibition.
Several reports have demonstrated subsets of OCCC which have immune‐active phenotypes. Using exome‐sequencing and microarray data, Matsushita et al demonstrated that OCCC with high immunoediting, based on the number of neoantigens per somatic mutation, was associated with a T‐cell‐inflamed phenotype, and a more favourable prognosis [12]. Willis et al reported that PD‐L1 expression was found in 43% of OCCCs, particularly in 67% of OCCCs with mismatch repair (MMR) defects [13]. Howitt et al showed that OCCC with MSI exhibited a high number of CD8+ tumour‐infiltrating lymphocytes (TILs) and a higher number of PD‐1‐expressing TILs compared with microsatellite‐stable (MSS) OCCC. PD‐L1 expression in tumour cells or immune cells was also noted in all cases of OCCCs with MSI [14]. In addition, Tan et al [15] demonstrated a subset of OCCCs enriched for genes associated with extracellular matrix organization, adhesion, and collagen binding similar to renal cell cancers that revealed preferential response to anti‐angiogenic agents, emphasizing the complex pathology of this rare subset of EOC and the importance of understanding the underlying biology of this disease.
Cytotoxic T‐lymphocyte‐associated antigen 4 (CTLA‐4) and programmed cell death protein 1 (PD‐1) are both immune inhibitory checkpoints commonly seen on activated T‐cells that make up the tumour‐immune environment and regulate immune responses. In our study, we analysed the tumour‐immune environment of OCCC from women of different ethnic backgrounds who were treated in Singapore, Japan, or the UK, based on a panel of immune‐related genes using NanoString nCounter® technology, and correlated their histopathological characteristics with clinical outcomes.
Materials and methods
Patients and collection of specimens
A cohort of 264 women with ovarian clear cell carcinoma of the ovary, primary peritoneal, or Fallopian tube, diagnosed between 1984 and 2016 (Singapore: between 2000 and 2015; Japan: between 2010 and 2016; UK: between 1984 and 2016), was identified through the National University Cancer Institute (NCIS) Singapore (n = 96); Saitama Medical University International Medical Center, Hidaka, Saitama, Japan (n = 84); and Edinburgh Cancer Research UK Centre, UK (n = 84). A summary of the clinical details is provided in Table 1. All procedures were conducted in accordance with the approved protocols. Eligible patients were at least 18 years of age (21 years of age for patients from Singapore), newly diagnosed with invasive epithelial ovarian, peritoneal, or Fallopian tube cancer, and had a histological diagnosis of clear cell carcinoma verified by two independent pathologists (Singapore: DL, JW; Japan: MY, M Yano; UK: CSH, DL) based on established diagnostic criteria [16, 17]. This study was approved by the Institutional Domain Specific Review Board from all participating sites.
Table 1.
Demographics of patients and their clinicopathological characteristics from the three ovarian clear cell carcinoma cohorts.
| Parameter | Combined | Singapore | Japan | UK |
|---|---|---|---|---|
| n | 255 (100%) | 96 (100%) | 76 (100%) | 83 (100%) |
| Age (median, years) | 56 | 52 | 57 | 61 |
| Follow‐up (median, months) | 41 | 33.5 | 49 | 41.6 |
| Stage | ||||
| I | 139 (54.6%) | 46 (48%) | 50 (65.8%) | 43 (51.8%) |
| IA/IB | 40 (15.7%) | 16 (16.7%) | 9 (11.8%) | 15 (18.1%) |
| IC | 99 (38.9%) | 30 (31.3%) | 41 (54%) | 28 (33.7%) |
| IC‐1 | 27 (10.6%) | NA | 20 (26.3%) | 7 (8.4%) |
| IC‐2 | 8 (3.1%) | NA | 5 (6.6%) | 3 (3.6%) |
| IC‐3 | 34 (13.3%) | NA | 16 (21.1%) | 18 (21.7%) |
| II, III, IV | 113 (44.3%) | 47 (49%) | 26 (34.2%) | 40 (48.2%) |
| NA | 3 (1.2%) | 3 (3.1%) | 0 | 0 |
| Clinical outcomes | ||||
| Overall median survival (months) | 48.4 | 60.5 | 49 | 41.6 |
| Disease‐free median survival (months) | 36.6 | 35 | 29.2 | 31.7 |
| Debulking | ||||
| No residual disease | 186 (72.9%) | 60 (62.5%) | 66 (86.8%) | 60 (72.3%) |
| Optimal (< 1 cm) | 23 (9.2%) | 7 (7.3%) | 2 (2.6%) | 14 (16.9 |
| Suboptimal (≥ 1 cm) | 22 (8.6%) | 5 (5.2%) | 8 (10.5%) | 9 (10.8%) |
| NA | 24 (9.4%) | 24 (25%) | 0 | 0 |
| Adjuvant therapy | ||||
| Yes | 197 (77.3%) | 66 (68.8%) | 64 (84.2%) | 67 (80.7%) |
| No | 38 (14.9%) | 10 (10.4%) | 12 (15.8%) | 16 (19.3%) |
| NA | 20 (7.8%) | 20 (20.8%) | 0 | 0 |
| Therapy | ||||
| Platinum‐containing | 184 (72.2%) | 66 (68.8%) | 62 (81.6%) | 61 (73.5%) |
| Taxol‐containing | 137 (53.7%) | 59 (61.5%) | 45 (59.2%) | 33 (39.8%) |
| Radiotherapy | 6 (2.4%) | 0 | 0 | 6 (7.2%) |
NA, not available.
Clinical and pathological data
Patients' medical records were comprehensively reviewed. Median follow‐up for the combined group analysis was 41 months (Table 1). Outcome analyses were restricted to binary comparisons for two reasons: (1) OCCC is considered a rare cancer as defined by an annual incidence of <6 per 100 000 based on classification from ESGO‐GCIG and RARECAREnet [18]; and (2) OCCC tends to present as early‐stage disease resulting in low event rates, which precluded the ability to observe significant differences between the subtypes analysed. Disease‐free survival (DFS) was defined as the interval between histological diagnosis and first progression, death as a result of disease, or last follow‐up. Death as a result of non‐disease‐related causes was not considered in the calculation of DFS. Overall survival (OS) was defined as the interval between histological diagnosis and the date of death as a result of disease, or last follow‐up. Histopathology data were assessed by expert pathologists based on classical morphology and IHC analysis for positive HNF1β and napsin A, as well as negative WT1.
Sample processing and gene expression profiling
RNA was extracted from 10‐μm formalin‐fixed, paraffin‐embedded (FFPE) sections using QIAGEN RNeasy FFPE Kits (Qiagen N.V., Hilden, Germany) following the manufacturer's protocol (Cat No/ID: 73504). An Agilent Bioanalyzer 2100 (Agilent Technologies, Inc, Santa Clara, CA, USA) was used to check the quality and quantity of RNA extracted from Singapore samples. Only RNA with length greater than 300 nt was considered for quantity calculation for downstream application. Unamplified total FFPE RNA (100 ng) was hybridized to NanoString® Pan‐Cancer Immune Panel Reporter CodeSets and Capture ProbeSets at 65 °C for at least 16 h but not more than 48 h in a thermal cycler. The hybridized RNA samples were then loaded onto a NanoString nCounter system (NanoString Technologies Inc, Seattle, WA, USA) for gene expression analysis.
Gene expression data processing
Gene expression profiling of the OCCC samples on the Pan‐Cancer Immune Panel was retrieved. The NanoString Pan‐Cancer Immune Panel analysed the expression levels of 730 genes related to immune cell types, CT antigens, responses, and functions. Gene expression normalization of NanoString data was performed using nSolver analysis software version 3.0 (NanoString Technologies Inc). Only samples that passed all quality metrics were retained for analysis (supplementary material, Table S1). The raw NanoString counts were subjected to background subtraction, positive control normalization, and 40‐reference gene normalization. The normalized counts were then log2‐transformed prior to downstream analysis.
Immune molecular subtype identification
ConsensusClusterPlus v1.44.0 (with default parameter settings except Euclidean distance, max K = 20, and 1000 permutations) in R v3.5.1 Bioconductor v3.8 was employed to identify subtypes in the Singapore cohort using all 730 immune‐related genes [19]. Silhouette analysis was used to select core samples (silhouette width greater than 0.065, first quantile of silhouette width). A total of 519 differentially expressed immune‐related genes were identified from the 72 core samples from the Singapore cohort (ANOVA p < 0.05 and expressed in more than 10% of the sample) and were used as the gene expression signature (supplementary material, Table s2 ).
Consensus clustering was subsequently applied to the Japan and UK cohorts individually using the differentially expressed immune‐related genes identified. Distinct clusters identified in the Japan and UK cohorts were assigned to the immune subtype that shared the highest similarity to that of the Singapore cohort.
Immunohistochemistry (IHC)
FFPE sections (4 μm) from all three cohorts were immunostained for mismatch repair (MMR) proteins using pre‐diluted antibodies from Ventana (Roche Tissue Diagnostics, Oro Valley, AZ, USA): MLH1 (mouse clone M1); MSH2 (mouse clone G219‐1129); PMS2 (mouse clone A16‐4), and MSH6 (rabbit clone SP93). Deparaffinization was performed using EZ Prep and antigen retrieval was performed using CC1 retrieval solution at 100 °C for 40–92 min. Endogenous peroxidase was blocked using 3% hydrogen peroxide. All steps were performed using a Roche Ventana Ultra Automated IHC machine (Roche Tissue Diagnostics). Tissue sections were counterstained with haematoxylin, dehydrated through a graded ethanol series, and coverslipped.
Statistical analyses
Statistical analyses were conducted using Matlab® R2012a 7.14.0.739 (MathWorks, Natick, MA, USA). Statistical significance of differential expression was evaluated using unpaired t‐tests. Associations were evaluated using Fisher's exact test. A Spearman correlation coefficient test was applied to assess significance of correlation. Kaplan–Meier analyses were conducted using GraphPad Prism® 5.04 (GraphPad Software, San Diego, CA, USA). Statistical significance of Kaplan–Meier analyses was calculated using a log‐rank test. Pathway enrichment scoring was based on Kolmogorov–Smirnov testing as described previously [20].
Results
Patient demographics
A total of 264 women with clear cell carcinoma of the ovary, primary peritoneum, or Fallopian tube from three international sites (Singapore, Japan, and the UK) were included in this study. Nine samples did not pass the quality check of NanoString gene expression profiling and were removed, resulting in a combined dataset of 255 (Figure 1). All samples from the UK, Singapore, and Japan were reviewed by two independent expert gynaecology pathologists and deemed to be histologically consistent with a diagnosis of OCCC based on classical morphology and/or IHC analysis of HNF1β, napsin A, and WT1 [16, 17] (Table 1). Subsequently, eligible histologically confirmed OCCC archival samples from all three countries underwent immune‐related gene expression profiling using the NanoString platform and their associated clinicopathological outcomes were correlated accordingly.
Figure 1.
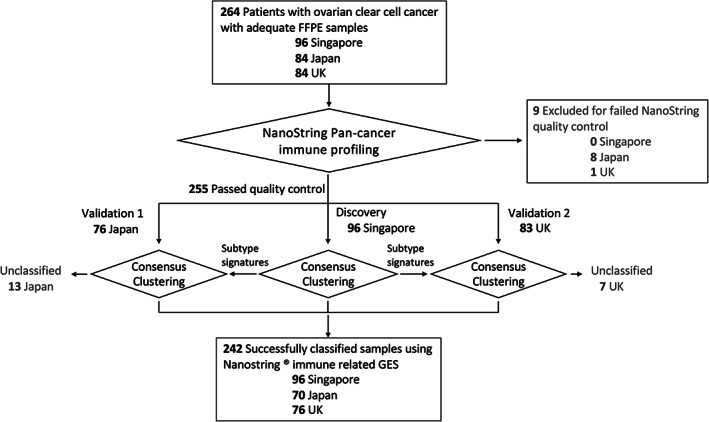
Workflow of the sample processing pipeline.
Median age at diagnosis across the three centres was 56 years of age (Singapore: 52 years; Japan: 57 years; and the UK: 61 years) (Table 1). As expected, early‐stage disease predominated in each cohort, with the proportions relatively similar across the three countries. Altogether, 139 (54.6%) women had stage I disease, while 113 (44.3%) had stage II–IV disease. Japan, with 65.8% (n = 50), had the highest proportion of patients with stage I disease, followed by the UK with 51.8% (n = 43) and Singapore with 48% (n = 46). Information about the initial disease stage was unavailable for three patients (Table 1). Amongst the 139 stage I patients, 28.7% (40/139) had stage IA/IB disease and 71.2% (99/139) had stage IC disease. Within the patients with stage IC, 20 patients from the Japanese cohort and seven patients from the UK cohort were stage IC due to iatrogenic rupture (stage IC‐1), while 21 patients from both the Japan and the UK cohort had surface involvement or positive cytology (stage IC‐2, IC‐3; Table 1). Further information on stage IC disease was not available for the Singapore cohort.
The proportions of women diagnosed with late‐stage disease were similar in the UK and Singapore cohorts, at 48.2% and 49%, respectively, and lower in the Japanese cohort (34.2%). Optimal debulking surgery, defined as no or less than 1 cm residual disease after cytoreductive surgery [21], was achieved in more than 80% of patients from all three cohorts (72.9% with no residual disease; 9.2% with optimal debulking). A total of 24 samples (25%) from Singapore did not have accessible surgical data at the time of analysis (Table 1). Treatment practices appeared to be similar across the three countries. In total, 197 (77.3%) patients received chemotherapy in the adjuvant setting (Table 1), mostly with a platinum‐based regimen (189 of 197 patients; 95.9%), while 137 of 197 (69.5%) received a combination of a platinum‐agent and taxane chemotherapy (Table 1). A total of 38 (14.9%) women did not receive any chemotherapy. Patterns of first‐line chemotherapy administration were similar across the three sites. Six patients from the UK cohort received radiotherapy as part of the adjuvant treatment (7.2%) (Table 1). The median OS for the Singapore, Japan, and UK cohorts were 60.5, 49, and 41.6 months, respectively (Table 1).
Immune‐related gene expression signatures (irGES) identify four novel molecular subtypes of ovarian clear cell carcinoma
Whole tumour immune‐related gene expression profiling was conducted on archival FFPE samples using the NanoString platform. Nine samples did not pass quality control and were excluded from the analysis (Figure 1 and supplementary material, Table S1). To exclude batch artefacts from the three different cohorts due to FFPE storage, specimen age, and processing, the Singapore cohort was used as the discovery set. Unsupervised clustering of the Singapore cohort using gene expression data was performed using the consensus clustering, which revealed four distinct clusters (Figure 2 and supplementary material, Figure S1). The four clusters were named Pro‐angiogenic (ProA), Antigen‐presentation (AP), CTLA4‐high, and PD1‐high relative to the dominant genes or pathways; we termed these as the irGES subtypes. In the ProA subtype, we observed increased overexpression of genes associated with stromal cell types relative to the other subtypes. The stromal markers in ProA include those of activated myofibroblasts (MFGE8, MCAM), vascular endothelial cells (VEGFC), and pericytes (PDGFRB), as well as enrichment of pathways and gene ontology groups defining extracellular matrix production/remodelling (FN1, COL3A1), and immunosuppression (TGFB2) (supplementary material, Figure S1). The AP subtype was characterized by overexpression of cell–cell adhesion markers (CD58, CD164) that enhance the binding of antigen‐presenting cells to T cells, thereby regulating T‐cell activation. Other genes highly expressed in this subtype included CD46, which encodes a protein that regulates the complement pathway, as well as NF‐κB1 and ATF1. Evidence of adaptive immune responses was observed among the genes significantly overexpressed within the CTLA4‐high subtype, including overexpression of HLA class II markers (HLA‐DPA, HLA‐DPB1) and T cells and granulocyte trafficking (CXCL12, CX3CL1, CCL11, CCL13). Overexpressed genes in the PD1‐high subtype not only indicated upregulation of PD‐1 (PDCD1) but also identified overexpressed NK cell markers that orchestrate cell migration and homing in the form of chemokines (CR2, CXCR1, XCL2, CCR4) (supplementary material, Figure S1).
Figure 2.

Clustering of immune‐related gene expression signatures (irGES) from Singapore, Japan, and the UK into four immune subtypes. Heatmap shows the clustering expression data derived from 96 Singapore samples and validated in the Japan and UK cohort. OCCCs were robustly clustered or classified into four k‐means groups: PD1‐high, CTLA4‐high, Antigen‐presentation (AP), and Pro‐angiogenic (ProA). Average linkage hierarchical clustering using a Pearson correlation metric was used to cluster genes based on relative expression.
To ascertain the reproducibility of the identified subtypes, we validated the subtypes in two independent cohorts – Japan (n = 76) and the UK (n = 83) (Figure 2). Core samples from the Singapore cohort (72 with the least ambiguous subtype classification) were used to generate the immune‐related signatures (Figures 1 and 2) to classify the OCCC samples from the two validation cohorts. In the validation cohorts, 91.8% of the OCCC samples could be assigned to one of the four subtypes, with 13 samples (Japan: 6; UK: 7) remaining unclassified (Figure 1). In total, 242 samples from Singapore (n = 96), Japan (n = 70), and the UK (n = 76) were successfully classified (Figures 1 and 2). Among them, 42.5% of women were classified as AP (n = 103), 28.9% as CTLA4‐high (n = 70), 17.7% as ProA (n = 43), and 10.7% as PD1‐high (n = 26) (supplementary material, Table S3). Unclassified samples (Figure 2, grey samples) were subsequently removed from the downstream analysis, yielding a final analysis of 242 irGES profiles with their related clinical and prognostic outcomes.
irGES profiling correlates with distinct clinical features
Correlation of irGES profiles with patient demographics
Information on patient characteristics was subsequently correlated with irGES profiles and we observed no discernible differences in age of disease onset and stage of disease based on irGES profiles between the three cohorts (supplementary material, Figure S2).
Clinical outcomes based on irGES profiles
Kaplan–Meier analyses of overall survival (OS) and disease‐free survival (DFS) were generated using the corresponding survival information available. The PD1‐high and CTLA4‐high subtypes had poorer outcomes compared with the other subtypes (Figure 3). The 5‐year DFS for the PD1‐high patients was almost half that of the other subtypes at 28% and 50.5%, respectively (hazard ratio, HR = 1.93; p = 0.06) (Figure 3). In the CTLA4‐high subtype, 5‐year DFS was 40.8% compared with 51.6% in the other subtypes (HR = 1.49; p = 0.063). Conversely, women classified to the AP subtype had significantly better outcomes compared with the other subtypes, with a median DFS that was more than twice as long as that for the other subtypes (76.7 versus 34.2 months, p = 0.019, supplementary material, Figure S3). There was no observable difference in the OS and DFS outcomes of women classified to the ProA subtype compared with the other subtypes (Figure 3 and supplementary material, Figure S3).
Figure 3.

Five‐year clinical outcomes of women with OCCC in each irGES subset compared with the rest of the cohort. Top row: 5‐year overall survival and lower row: 5‐year disease‐free survival of PD‐1 high (blue) compared with the non‐PD‐1 high subset (black), CTLA4‐high (green) compared with the non‐CTLA4‐high (black), Antigen‐presentation subset (red) compared with the non‐Antigen‐presentation (black), and Pro‐angiogenic subset (brown) compared with the non‐Pro‐angiogenic (black).
In a univariate analysis (supplementary material, Table S4), stage I tumours were associated with better survival (p < 0.001), consistent with previous reports [3]. In multivariate analyses, the PD1‐high subtype was an independent predictor of poorer OS, even after adjusting for age and stage, with HR = 2.1 (95% CI 1.14–3.88; p < 0.018) (supplementary material, Table S5). Five‐year OS and DFS rates in women of the PD1‐high subtype were around two times poorer than those of the other subtypes (5‐year OS: HR = 2.1, p = 0.07 and 5‐year DFS: HR = 1.9; p = 0.06, respectively; Figure 3). Both AP and ProA subtypes showed a non‐significant trend towards improved OS after accounting for age and stage, with an HR of 0.71 (p = 0.10) and an HR of 0.74 (p = 0.27), respectively (supplementary material, Table S5).
Of the 33 women with stage IA/IB disease, 21 (63.6%) women received adjuvant chemotherapy. Of these, ten women (30.3%) were classified to the AP, four (12.1%) to the CTLA4‐high, seven (21.2%) to the ProA, and none to the PD1‐high subtypes (Figure 4 and supplementary material, Table S6).
Figure 4.

Clinical outcomes of stage 1 OCCC women based on their irGES profiles and receipt of adjuvant treatment. Kaplan–Meier plots showing overall survival (upper row) and disease‐free survival (lower row) outcomes of women with stage IA/IB OCCC and stage IC–IV based on their irGES and whether or not they received adjuvant chemotherapy.
The rates of first‐line chemotherapy were higher in patients with stage IC–IV disease, with 164 (88.2%) receiving adjuvant chemotherapy while only 22 (11.8%) patients did not. 72/186 (38.7%) women were in AP, 51/186 (27.4%) women in CTLA4‐high, 15/186 (8.1%) women in PD1‐high, and 26/186 (14.0%) women in ProA subtype groups (supplementary material, Table S6). For the 186 stage IC–IV OCCCs, both the poor prognosis PD1‐high and CTLA‐4 high subtypes appeared to derive significant OS benefit from adjuvant chemotherapy compared with the other subtypes (HR = 0.09, p = 0.027; and HR = 0.08, p = 0.0005 respectively; Figure 4). DFS showed a similar benefit following adjuvant therapy for PD1‐high (HR = 0.13, p = 0.048) and CTLA4‐high (HR = 0.21, p = 0.01; Figure 4).
MMR status based on irGES profiles
It is recommended that all women with OCCC, endometrioid or mucinous ovarian cancer should be offered somatic tumour testing for mismatch repair deficiency (dMMR) [22]. IHC for four mismatch repair (MMR) proteins (MSH2, MSH6, MLH1, PMS2) was performed successfully on 240 out of 242 (99.2%) OCCC samples (Figure 5A). When compared within each cohort, the proportions of MMR‐deficient patients were similar across the three cohorts, with 6% (n = 6) from Singapore, 6% (n = 4) from Japan, and 5% (n = 4) from the UK (Figure 5A,B). The patterns of MMR protein loss were variable in our study. Two patients had loss of both MLH1 and PMS2, whereas one had loss of only PMS2. Loss of both MSH2 and MSH6 occurred in five patients, while the remaining five patients had loss of MSH6 protein only (supplementary material, Table S7). A total of 14 women (5.8%) from Singapore, Japan, and the UK were observed to have loss of MMR proteins with all subtypes included except for ProA, which showed 100% retained MMR expression across the three cohorts (Figure 5C). Strikingly, our data showed a significant enrichment of MMR‐deficient tumours in the CTLA4‐high subtype (p = 0.016) compared with the other subtypes (Figure 5D). Eight (57.1%) MMR‐deficient patients were categorized as CTLA‐4 high, three (21.4%) as AP, and three (21.4%) as PD1‐high (supplementary material, Table S7).
Figure 5.
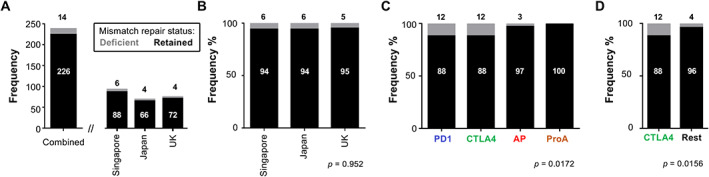
Distribution of retained and deficient MMR proteins in OCCC. (A) Bar chart showing frequency (y‐axis) of combined, and of Singapore, Japan, and UK ovarian clear cell carcinoma cohorts (x‐axis). (B) Bar chart showing frequency percentage (y‐axis) of retained and deficient mismatch repair (MMR) proteins' status assessed using IHC in individual cohorts (x‐axis). (C) Bar chart depicting the combined analysis of the MMR protein status based on their irGES profiles. (D) Comparison of the MMR protein status specifically in the CTLA4‐high subtype relative to the rest of the other subtypes in the combined cohort analysis. Numbers above or within bars indicate the number of samples or the percentage.
Discussion
Of our 255 women with available clinical data, more than half the patients (54.6%) had stage I disease (n = 139), consistent with prior studies [23, 24]. Despite the small numbers, our study demonstrated overall good survival outcomes in stage I OCCC of 83% 3‐year OS (supplementary material, Figure S4), similar to studies published previously [5, 24].
Several studies have reported significant variation in the prognosis of stage I disease [25, 26]. Our study identified that the PD1‐high subtype conferred a less favourable prognosis, with 3‐year DFS of 64.2% and 3‐year OS of 75% despite presenting with stage I disease (supplementary material, Figure S4). The outcomes of stage I PD1‐high women appeared to be comparable to those of stage IC patients with surface involvement, ascites or positive cytology, which was shown to confer a 3‐year DFS and OS rate of 56.4% and 71.9%, respectively, in one study [25]. Historically, due to concerns relating to prognosis and limited data on outcomes with surgery alone, patients with low‐stage OCCC beyond FIGO stage IA/C1 would be recommended post‐operative chemotherapy. Our data demonstrate that women with stage IC–IV OCCC who fall into either the PD1‐high or the CTLA‐4‐high subtype would derive significant survival benefit with the addition of adjuvant chemotherapy (Figure 4). However, we do not know whether women with stage IC disease with surface involvement or positive cytology classified to the other irGES subtypes would derive benefit from adjuvant chemotherapy.
Few reports have investigated associations between expression of immune‐related molecules and clinical features, particularly for OCCC. Mariya et al reported HLA class I expression and its association with T‐cell infiltration and prognosis of EOC. They concluded that high expression of HLA class I was a good prognostic marker of EOC. However, they also reported that OCCC had lower CD3 and CD8 T cells, which did not impact on patients' survival outcome [27]. In addition, Tan et al categorized OCCC into two gene expression subtypes in relation to the individual epithelial–mesenchymal transition states, namely epithelial and mesenchymal OCCC. The latter had a poorer prognosis and was enriched in immune‐related genes such as genes associated with antigen processing and presentation [15]. Our study is the first to include both Asian and Caucasian women with OCCC, which could shine a light on the impact of racial differences in the immune microenvironment of OCCC. In our data, the PD1‐high and CTLA4‐high subtypes were associated with poorer 5‐year DFS and OS outcomes (Figure 4).
In a case report by Bellone et al, an exceptional complete response to pembrolizumab was described in a woman with OCCC following disease refractory to several prior lines of treatment. This patient had increased aberrant expression of PD‐L1 following a gain‐of‐function structural variant disrupting the 3′‐region of the PD‐L1 (CD274) gene [28]. This was consistent with a separate study which found marked elevation of aberrant PD‐L1 transcripts secondary to truncation of their 3′‐UTR [29]. A different study found that OCCC patients with a better prognosis had higher expression of HLA class I genes, and those with poorer outcomes had higher ratios of PD‐1/CD8 or CTLA‐4/CD8 [14]. These data support our results suggesting that expression of antigen presentation genes may confer a better prognosis and that overexpression of immune checkpoint molecules, particularly PD‐1 and CTLA‐4, may confer a poorer outcome in women with OCCC.
Our study found that dMMR was infrequent, with 14 out of 240 (5.8%) OCCCs demonstrating dMMR, similar to other reports [30]. Leskela et al demonstrated that the loss of both MLH1 and PMS2 proteins was the most frequently observed pattern of dMMR across all histological types of EOC [30]. However, this pattern was not observed in our OCCC cohorts, where patterns of MMR protein loss were spread out with no particular pattern standing out. We also observed that the CTLA4‐high subtype had the most women with deficient MMR status (Figure 5 and supplementary material, Table S7). OCCCs with microsatellite instability molecular profiles have been shown to be associated with a high number of CD8+ TILs with higher PD‐1 expression compared with microsatellite‐stable (MSS) tumours [14], suggesting that perhaps patients of the CTLA4‐high subtype may derive additional benefit from immune checkpoint inhibition.
Finally, our collaborative analyses from different ethnic backgrounds revealed similarities between OCCC from the Asian and Caucasian populations. Considering transcriptomes, there were great similarities, with the majority of tumours clustering within the four irGES subtypes (Figure 1). Clinically, the demographics of the women who presented with OCCC were fairly similar, with patients in the UK observed to be slightly older, with a median age at diagnosis of 61 years compared with the Asian population (52 in Singapore; 57 in Japan; Table 1). The stage of disease at diagnosis was also similar between the three cohorts, although the Japanese population had a slightly higher proportion of patients with early‐stage disease (Table 1). These data are reassuring as we aim to understand the drivers and susceptibilities of this rare disease; we are only able to do so through extensive collaborative efforts from around the globe. Despite their similarities, the irGES subtypes across the three cohorts were not fully concordant; in particular, we observed a higher incidence of the PD1‐high subtype in our Singapore cohort compared with the rest. While this difference could be attributed to population differences, it could also arise from the difference in patient demographics or sample handling and processing. Nevertheless, the addition of our data to the taxonomy of OCCC warrants further exploration as it allows classification of women based on irGES into distinct prognostic groups and has the potential to predict treatment responses especially with immunotherapeutic strategies. However, the utility of irGES clustering in OCCC will require further prospective validation.
In conclusion, our data affirm that OCCCs are molecularly and clinically similar in the Asian and Caucasian populations. We also confirm that early‐stage OCCC has a favourable prognosis, but women who fall into the PD1‐high subtype have prognostic outcomes similar to those of women with stage IC‐2/3 disease. Future OCCC‐specific studies are crucial and will require multi‐group collaboration to translate our growing knowledge of OCCC molecular features into intelligently designed clinical trials.
Conflict of interest statement
VH received research funding from AstraZeneca and honoraria from AstraZeneca, Novartis, Eli Lily, and Pfizer. NYLN received honoraria from AstraZeneca and Janssen. DSPT received research funding from AstraZeneca, Bayer, and Karyopharm, and honoraria from AstraZeneca, MSD, Tessa Therapeutics, Novartis, Bayer, and Genmab. CG received research funding from AstraZeneca, Novartis, Aprea, Nucana, and Tesaro, and honoraria/personal interest from Roche, AstraZeneca, Tesaro, Nucana, Clovis, Foundation One, Sierra Oncology, Cor2Ed, and Takeda. RYJH is currently supported by the Yushan Scholar Program by the Ministry of Education, Taiwan (NTU‐110V0402) and NTU Core Consortiums (NTUCC‐110L891101). CSH is the Editor‐in‐Chief of The Journal of Pathology and had no involvement with review or decision‐making for this manuscript. No other potential conflicts of interest were declared.
Author contributions statement
RYH, KH, CG, DSPT and AO supervised the study. RYH, TZT, DSPT, KH, CG and VH designed and conceptualized the study. JY performed sample collection and experiments. NN, MM and YI participated in data collection and methodology of the study. DL, CSH, M Yasuda, M Yano and JW processed and reviewed samples and FFPE. TZT performed the bioinformatics analysis. RYH, TZT, VH, DSPT, KH, CG, CSH, JW and MM analysed the data, interpreted the results and participated in writing, reviewing, and editing of the manuscript. All the authors approved the final version of the manuscript.
Supporting information
Figure S1. Consensus clustering of ovarian clear cell carcinoma in the Singapore cohort
Figure S2. Association of clinical pathological parameters with the immune subtypes by cohort
Figure S3. Binary survival analyses of immune subtypes
Figure S4. Stage‐stratified analyses of overall and disease‐free survival
Table S1. Quality check metrics of NanoString gene expression profiling
Table S2. Gene expression signature for the immune subtype of ovarian clear cell carcinoma
Table S3. Immune subtype distribution
Table S4. Univariate Cox regression analysis of overall survival in the combined data of ovarian clear cell carcinoma patients
Table S5. Multivariate Cox regression analysis of overall survival in the combined data of ovarian clear cell carcinoma patients
Table S6. Subtype distribution in different stages and with/without adjuvant therapy
Table S7. Immune‐related gene expression signature (irGES) subtype, stage, and patterns of MMR protein expression on IHC in patients with deficient MMR protein
Acknowledgements
This study was supported by the National Medical Research Council (NMRC) Singapore, National Research Foundation Singapore, and the Singapore Ministry of Education under its Research Centres of Excellence initiative to RYJH and DSPT; and the National University Cancer Centre of Singapore (NCIS) Research Grant, Theme: EMT in Cancer to RYJH. VH was supported by the Yong Loo Lin fellowship grant and the NHG‐LKC Medicine Clinician Scientist Career Scheme grant (CSCS/20001). CG is supported by the Nicola Murray Foundation. DSPT is supported by the Singapore Ministry of Health's National Medical Research Council Transition Award (NMRC/TA/0019/2013) and Clinician Scientist Award (NMR/CSA‐INV/0016/2017). RYJH is currently supported by the Yushan Scholar Program by the Ministry of Education, Taiwan (NTU‐109V0402).
Contributor Information
David SP Tan, Email: david_sp_tan@nuhs.edu.sg.
Ruby YJ Huang, Email: rubyhuang@ntu.edu.tw.
Data availability statement
The NanoString Pan Cancer Immune Panel profiling of the ovarian clear cell carcinomas has been deposited in the Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE128990.
References
- 1. Mackay HJ, Brady MF, Oza AM, et al. Prognostic relevance of uncommon ovarian histology in women with stage III/IV epithelial ovarian cancer. Int J Gynecol Cancer 2010; 20: 945–952. [DOI] [PubMed] [Google Scholar]
- 2. Köbel M, Kalloger SE, Huntsman DG, et al. Differences in tumor type in low‐stage versus high‐stage ovarian carcinomas. Int J Gynecol Pathol 2010; 29: 203–211. [DOI] [PubMed] [Google Scholar]
- 3. Anglesio MS, Carey MS, Köbel M, et al. Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol Oncol 2011; 121: 407–415. [DOI] [PubMed] [Google Scholar]
- 4. Yamagami W, Aoki D. Annual report of the Committee on Gynecologic Oncology, the Japan Society of Obstetrics and Gynecology. J Obstet Gynaecol Res 2015; 41: 167–177. [DOI] [PubMed] [Google Scholar]
- 5. Okamoto A, Glasspool RM, Mabuchi S, et al. Gynecologic Cancer InterGroup (GCIG) consensus review for clear cell carcinoma of the ovary. Int J Gynecol Cancer 2014; 24(9 Suppl 3): S20–S25. [DOI] [PubMed] [Google Scholar]
- 6. Chan JK, Teoh D, Hu JM, et al. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecol Oncol 2008; 109: 370–376. [DOI] [PubMed] [Google Scholar]
- 7. Takano M, Sugiyama T, Yaegashi N, et al. Low response rate of second‐line chemotherapy for recurrent or refractory clear cell carcinoma of the ovary: a retrospective Japan Clear Cell Carcinoma Study. Int J Gynecol Cancer 2008; 18: 937–942. [DOI] [PubMed] [Google Scholar]
- 8. Pather S, Quinn MA. Clear‐cell cancer of the ovary – is it chemosensitive? Int J Gynecol Cancer 2005; 15: 432–437. [DOI] [PubMed] [Google Scholar]
- 9. Tan DSP, Rye T, Barrie C, et al. Analysis of outcomes in patients (pts) with recurrent ovarian clear cell carcinoma (ROCCC): time to rethink our approach to treatment. J Clin Oncol 2014; 32(15_suppl): 5548. [Google Scholar]
- 10. Sugiyama T, Okamoto A, Enomoto T, et al. Randomized phase III trial of irinotecan plus cisplatin compared with paclitaxel plus carboplatin as first‐line chemotherapy for ovarian clear cell carcinoma: JGOG3017/GCIG Trial. J Clin Oncol 2016; 34: 2881–2887. [DOI] [PubMed] [Google Scholar]
- 11. Matulonis UA, Shapira‐Frommer R, Santin AD, et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE‐100 study. Ann Oncol 2019; 30: 1080–1087. [DOI] [PubMed] [Google Scholar]
- 12. Matsushita H, Hasegawa K, Oda K, et al. The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma. Oncoimmunology 2017; 6: e1338996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Willis BC, Sloan EA, Atkins KA, et al. Mismatch repair status and PD‐L1 expression in clear cell carcinomas of the ovary and endometrium. Mod Pathol 2017; 30: 1622–1632. [DOI] [PubMed] [Google Scholar]
- 14. Howitt BE, Strickland KC, Sholl LM, et al. Clear cell ovarian cancers with microsatellite instability: a unique subset of ovarian cancers with increased tumor‐infiltrating lymphocytes and PD‐1/PD‐L1 expression. Oncoimmunology 2017; 6: e1277308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan TZ, Ye J, Yee CV, et al. Analysis of gene expression signatures identifies prognostic and functionally distinct ovarian clear cell carcinoma subtypes. EBioMedicine 2019; 50: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peres LC, Cushing‐Haugen KL, Anglesio M, et al. Histotype classification of ovarian carcinoma: a comparison of approaches. Gynecol Oncol 2018; 151: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iida Y, Okamoto A, Hollis RL, et al. Clear cell carcinoma of the ovary: a clinical and molecular perspective. Int J Gynecol Cancer 2021; 31: 605–616. [DOI] [PubMed] [Google Scholar]
- 18. Ray‐Coquard I, Trama A, Seckl MJ, et al. Rare ovarian tumours: epidemiology, treatment challenges in and outside a network setting. Eur J Surg Oncol 2019; 45: 67–74. [DOI] [PubMed] [Google Scholar]
- 19. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010; 26: 1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan TZ, Miow QH, Miki Y, et al. Epithelial–mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 2014; 6: 1279–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salani R, Axtell A, Gerardi M, et al. Limited utility of conventional criteria for predicting unresectable disease in patients with advanced stage epithelial ovarian cancer. Gynecol Oncol 2008; 108: 271–275. [DOI] [PubMed] [Google Scholar]
- 22. Konstantinopoulos PA, Norquist B, Lacchetti C, et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol 2020; 38: 1222–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skírnisdóttir I, Seidal T, Karlsson MG, et al. Clinical and biological characteristics of clear cell carcinomas of the ovary in FIGO stages I–II. Int J Oncol 2005; 26: 177–183. [PubMed] [Google Scholar]
- 24. Leskela S, Romero I, Cristobal E, et al. The frequency and prognostic significance of the histologic type in early‐stage ovarian carcinoma: a reclassification study by the Spanish Group for Ovarian Cancer Research (GEICO). Am J Surg Pathol 2020; 44: 149–161. [DOI] [PubMed] [Google Scholar]
- 25. Shu CA, Zhou Q, Jotwani AR, et al. Ovarian clear cell carcinoma, outcomes by stage: the MSK experience. Gynecol Oncol 2015; 139: 236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mizuno M, Kikkawa F, Shibata K, et al. Long‐term prognosis of stage I ovarian carcinoma. Prognostic importance of intraoperative rupture. Oncology 2003; 65: 29–36. [DOI] [PubMed] [Google Scholar]
- 27. Mariya T, Hirohashi Y, Torigoe T, et al. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol Res 2014; 2: 1220–1229. [DOI] [PubMed] [Google Scholar]
- 28. Bellone S, Buza N, Choi J, et al. Exceptional response to pembrolizumab in a metastatic, chemotherapy/radiation‐resistant ovarian cancer patient harboring a PD‐L1‐genetic rearrangement. Clin Cancer Res 2018; 24: 3282–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD‐L1 expression through 3'‐UTR disruption in multiple cancers. Nature 2016; 534: 402–406. [DOI] [PubMed] [Google Scholar]
- 30. Leskela S, Romero I, Cristobal E, et al. Mismatch repair deficiency in ovarian carcinoma: frequency, causes, and consequences. Am J Surg Pathol 2020; 44: 649–656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Consensus clustering of ovarian clear cell carcinoma in the Singapore cohort
Figure S2. Association of clinical pathological parameters with the immune subtypes by cohort
Figure S3. Binary survival analyses of immune subtypes
Figure S4. Stage‐stratified analyses of overall and disease‐free survival
Table S1. Quality check metrics of NanoString gene expression profiling
Table S2. Gene expression signature for the immune subtype of ovarian clear cell carcinoma
Table S3. Immune subtype distribution
Table S4. Univariate Cox regression analysis of overall survival in the combined data of ovarian clear cell carcinoma patients
Table S5. Multivariate Cox regression analysis of overall survival in the combined data of ovarian clear cell carcinoma patients
Table S6. Subtype distribution in different stages and with/without adjuvant therapy
Table S7. Immune‐related gene expression signature (irGES) subtype, stage, and patterns of MMR protein expression on IHC in patients with deficient MMR protein
Data Availability Statement
The NanoString Pan Cancer Immune Panel profiling of the ovarian clear cell carcinomas has been deposited in the Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE128990.


