Abstract
The small heat shock protein (sHSP) from the hyperthermophile Pyrococcus furiosus was specifically induced at the level of transcription by heat shock at 105°C. The gene encoding this protein was cloned and overexpressed in Escherichia coli. The recombinant sHSP prevented the majority of E. coli proteins from aggregating in vitro for up to 40 min at 105°C. The sHSP also prevented bovine glutamate dehydrogenase from aggregating at 56°C. Survivability of E. coli overexpressing the sHSP was enhanced approximately sixfold during exposure to 50°C for 2 h compared with the control culture, which did not express the sHSP. Apparently, the sHSP confers a survival advantage on mesophilic bacteria by preventing protein aggregation at supraoptimal temperatures.
Hyperthermophilic microorganisms that can grow at or above 100°C are now fairly well known. However, little is known about their adaptive responses to extremely high temperatures. Ubiquitous heat shock responses are observed when organisms are confronted by near-lethal temperatures. Sets of proteins with diverse functions known as heat shock proteins (HSPs) are induced in response to abrupt temperature changes (11). The synthesis of these proteins can lead to acquired thermotolerance, allowing the organisms to survive at even higher temperatures (9, 21, 33, 36). HSPs have been divided into five classes based on their molecular weights: HSP100, HSP90, HSP70, HSP60, and small HSPs (sHSPs) (35). Most HSPs function as molecular chaperones, catalyzing refolding of denatured proteins, assisting maturation of newly synthesized proteins, or suppressing protein aggregation (7, 8, 10, 22).
sHSPs are a common class of HSPs; their molecular masses range from 15 to 42 kDa, and they normally form multisubunit complexes of 200 to 800 kDa (13). Some sHSPs are abundant in nonstressed cells (18), but others are stress induced (20, 29). Most sHSPs share a conserved motif with α-crystallin proteins, which are ubiquitous proteins found in vertebrate eye lenses and which are known to function as molecular chaperones (10, 15, 34). The in vivo functions of sHSPs remain unclear. It has been proposed that they confer thermotolerance or tolerance to chemical challenges such as superoxide (17). Survival of an HSP30-deficient mutant of Neurospora crassa under high-temperature, carbohydrate-limited conditions was shown to be reduced dramatically compared to survival of the wild type (26). However, mechanisms of action have not been defined.
In particular, the functions and regulation of archaeal sHSPs remain unclear, although the sHSP from the hyperthermophilic methanogenic archaeon Methanococcus jannaschii has been cloned and expressed and its crystal structure has been reported (14). Based on its crystal structure, the M. jannaschii sHSP forms oligomeric structures having 24 subunits of the 16.5-kDa monomer. We have studied the sHSP from Pyrococcus furiosus (Pfu-sHSP), a hyperthermophilic archaeon that grows optimally at about 100°C (4). We report the cloning and regulation of Pfu-sHSP, the effects of its overexpression in Escherichia coli, and the mechanism of its action in stabilizing a mesophilic protein at elevated temperatures.
Sequence comparison and phylogenetic analysis.
The Pfu-sHSP gene was identified from the genome sequence (27) based on its similarity to other α-crystallin-like sHSP genes. The protein is composed of 167 amino acids encoded by an open reading frame of 504 nucleotides (GenBank accession no. AF256212) and has a predicted pI of 5.25. The low-molecular-weight sHSPs have significantly higher diversities in size and amino acid sequence than do the higher-molecular-weight HSPs (24). The amino acid sequence of Pfu-sHSP showed low similarity to the sHSP of Clostridium acetobutylicum and to other bacterial and eukaryotic sHSPs (31). Typically, a nonconserved region occurs at the amino terminus (14).
Comparison of the amino acid sequences of Pfu-sHSP with other sHSPs by BestFit (Wisconsin Package, version 10.0; Genetics Computer Group, Madison, Wis.) reveals, not surprisingly, that the sHSP of the marine hyperthermophilic archaeon Pyrococcus horikoshii (5) has the highest percent identity and similarity to Pfu-sHSP. Interestingly, the sHSP from the bacterium Aquifex aeolicus is more similar to that of P. furiosus than are sHSPs from archaea with lower optimal growth temperatures. The sHSP from the mesophilic bacterium C. acetobutylicum shows the lowest percent identity to Pfu-sHSP (Table 1).
TABLE 1.
Amino acid similarity of Pfu-sHSP to corresponding proteins of other organisms
| Organism | Comparison to Pfu-sHSPa
|
Reference | |
|---|---|---|---|
| % Identity | % Similarity | ||
| Pyrococcus horikoshii | 87.3 | 91.6 | 12 |
| Aquifex aeolicus | 43.0 | 59.8 | 2 |
| Archaeoglobus fulgidus | 39.2 | 51.5 | 19 |
| Methanobacterium thermoautotrophicum | 37.3 | 52.7 | 33 |
| Oryza sativa | 34.0 | 47.9 | 38 |
| Methanococcus jannaschii | 33.8 | 40.1 | 14 |
| Clostridium acetobutylicum | 31.0 | 45.0 | 31 |
Percent identity and percent similarity were calculated from an alignment of the Pfu-sHSP amino acid sequence with other sHSPs by BestFit (Genetics Computer Group, Madison, Wis.).
Native Pfu-sHSP and its mRNA are induced by heat shock.
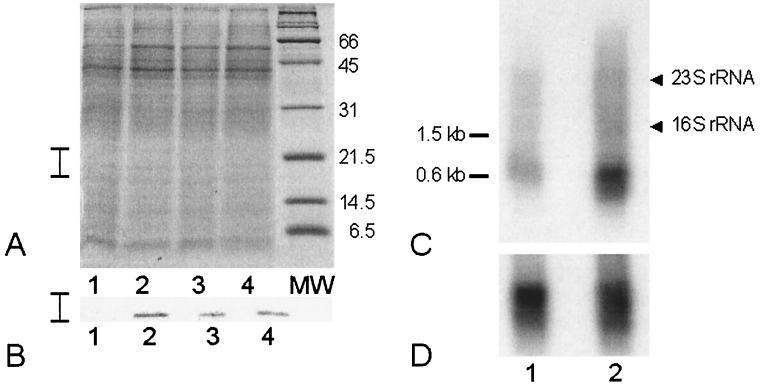
In order to determine whether Pfu-sHSP is regulated in response to heat shock, we obtained polyclonal antiserum against Pfu-sHSP by immunization of a rabbit with purified, recombinant Pfu-sHSP (BioWorld, Dublin, Ohio). Western blot analysis using this antiserum demonstrated that Pfu-sHSP is indeed heat shock inducible. P. furiosus was cultured in a modified 20-liter fermentor (New Brunswick) under standard conditions with 5 g of S0/liter (1). The cultures were incubated at 95°C for 4 h and then shifted to 105°C for 0, 30, 60, or 120 min before being chilled on ice and harvested by centrifugation at 7,500 × g for 15 min. Heat shock was carried out at 105°C because the maximum temperature for growth of P. furiosus is 103°C (4). The total protein levels of the cell extracts were measured, and equal amounts of protein were loaded onto each lane of a sodium dodecyl sulfate (SDS)-polyacrylamide gel (Fig. 1A). Western blot analysis was done as described elsewhere (30). A strong signal was observed at 20 kDa (Fig. 1B, lanes 2 to 4), corresponding to 30, 60, and 120 min after the onset of heat shock, whereas no signal was observed for the non-heat-shocked control culture (Fig. 1B, lane 1). This indicates that native Pfu-sHSP is strictly heat inducible and that Pfu-sHSP is apparently not required for rapid growth at the optimal growth temperature.
FIG. 1.
Expression of native Pfu-sHSP. Protein expression is shown by SDS–15% PAGE and Western blot analysis in panels A and B, respectively. (A and B) Lanes 1, culture growing at 95°C; lanes 2, 3, and 4, P. furiosus cultures after heat shock treatment at 105°C for 30, 60, and 120 min, respectively. The vertical bars on the left show the section of the gel represented in the Western blot analysis. Northern blot analyses of Pfu-sHSP gene mRNA expression and P. furiosus GDH gene mRNA (control) expression are shown in panels C and D, respectively. (C and D) Lanes 1, autoradiogram of a blot of mRNA from non-heat-shocked cells; lanes 2, corresponding mRNA signals from heat-shocked cells.
We measured the expression of mRNA from the gene encoding Pfu-sHSP under heat shock conditions by Northern blot analysis (30). Total RNA was isolated from P. furiosus after exposure to 105°C for 120 min and compared to a non-heat-shocked control culture. Total RNA (4 μg) was electrophoresed on a 1.5% agarose gel for Northern blot analysis with a radiolabeled PCR probe from the Pfu-sHSP gene generated by PCR amplification using [32P]dCTP. Hybridization with this probe revealed a transcript of 600 nucleotides, corresponding to the size of the putative Pfu-sHSP gene (Fig. 1C). A radiolabeled probe from the gene encoding P. furiosus glutamate dehydrogenase (GDH), which is expressed constitutively (3, 32), was used as a control (Fig. 1D).
Cloning, overexpression, and purification of Pfu-sHSP in E. coli.
The region encoding the 504-nucleotide Pfu-sHSP gene was amplified from P. furiosus genomic DNA by PCR using primers Pfu-shspN, containing an NcoI site (C′CATG_G [underlined in the full primer sequence]) (5′GCCATGGTGAGGAGAATAAGAAGATGG), and Pfu-shspC, containing an XhoI site (C′TCGA_G [underlined in the full primer sequence]) (5′ACTCGAGCTATTCAACTTTAACTTCGAATCCTTC). The gene fragment was cloned into the pCR Zero Blunt vector (Invitrogen, Carlsbad, Calif.). The insert was digested by NcoI and XhoI and then subcloned into the IPTG (isopropyl-1-thio-β-d-galactopyranoside)-inducible pET19b expression vector (Novagen, Madison, Wis.). The construct was designated pPfu-shsp. E. coli BL21(DE3) (Novagen), carrying pSJS1240, which encodes the rare E. coli Arg-tRNAAGA and Ile-tRNAATA (16), was used as an expression host. E. coli cultures were grown in Luria-Bertani broth in the presence of 50 μg each of ampicillin and spectinomycin per ml to an A595 of 0.6. Pfu-sHSP expression was induced by the addition of 1 mM IPTG for 3 h. The same strain carrying pET19b and pSJS1240 was the negative control. SDS-polyacrylamide gel electrophoresis (PAGE) of E. coli overexpressing Pfu-sHSP crude extract revealed an additional protein of 20 kDa, which corresponds to the protein molecular weight deduced from the predicted amino acid sequence. After induction, E. coli cells overexpressing Pfu-sHSP were harvested and resuspended in 25 mM potassium phosphate buffer (pH 7.0)–2 mM dithiothreitol–1 mM EDTA (buffer A). The cells were disrupted using a French press (SLM Instruments, Urbana, Ill.) at 16,000 lb/in2, and the extract was centrifuged at 5,000 × g for 15 min. Pfu-sHSP appeared in the particulate fraction as indicated by SDS-PAGE. The pellet was washed and dissolved in buffer A by heating at 85°C for 20 min. The dissolved pellet was then filtered and loaded onto an anion-exchange column (MonoQ; Pharmacia Biotech, Uppsala, Sweden) previously equilibrated with buffer A. Pfu-sHSP was eluted as a single peak at 0.35 M NaCl by using a linear gradient from 0 to 1 M NaCl. Fractions were subsequently pooled and concentrated.
Protection of E. coli cell extracts by Pfu-sHSP at 105°C.
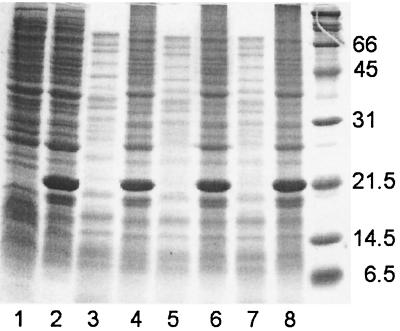
Several sHSPs are known to prevent aggregation of proteins during heating in vitro (15, 23). We examined the effect of Pfu-sHSP expressed in E. coli. The cells were harvested and prepared as described above. The total protein concentration was determined using the Bradford protein assay kit (Bio-Rad, Hercules, Calif.). The cell extracts were diluted in buffer A to a uniform protein concentration of 4 mg/ml. Diluted cell extracts were covered with mineral oil and heated at 105°C for 0, 20, 30, and 40 min in 1.5-ml microcentrifuge tubes. After being cooled to room temperature, the samples were centrifuged at 10,000 × g for 5 min at 25°C and the supernatants were collected. The residual proteins were visualized by SDS-PAGE and subjected to the Bradford protein concentration assay. At least 90% of the proteins in the E. coli cell extracts containing overexpressed Pfu-sHSP remained soluble after heat treatment for 40 min and appeared in the supernatant fractions (Fig. 2). Approximately 30% of the soluble proteins remained in the control supernatants after heat treatment. SDS-PAGE showed that high-molecular-weight proteins of E. coli were protected by Pfu-sHSP, whereas those in the control were rapidly aggregated at 105°C (Fig. 2). This finding indicates that sHSP prevents aggregation of many different proteins at or above their denaturing temperatures.
FIG. 2.
SDS–15% PAGE analysis of thermal protection of E. coli crude extracts by Pfu-sHSP at 105°C. Lanes 1 (E. coli crude extracts without Pfu-sHSP) and 2 (E. coli with overexpressed Pfu-sHSP) represent control experiments without heat treatment. Lanes 3, 5, and 7 show E. coli crude extracts without overexpressed Pfu-sHSP heated to 105°C for 20, 30, and 40 min, respectively; lanes 4, 6, and 8 show E. coli crude extracts with overexpressed Pfu-sHSP heated to 105°C for 20, 30, and 40 min, respectively.
Pfu-sHSP prevents aggregation of mesophilic GDH as a result of heat inactivation.
Since Pfu-sHSP can prevent aggregation of other proteins in response to heat stress, we addressed the question of whether or not it could extend the half-life (t1/2) of a purified enzyme in vitro. Bovine glutamate dehydrogenase (boGDH) (Sigma, Milwaukee, Wis.) was used as a model. boGDH is a mesophilic enzyme with an optimal assay temperature of 25°C that is inactivated rapidly at 56°C. Purified Pfu-sHSP was added to solutions of boGDH during heat treatment at 56°C in EPPS (N-2-hydroxyethylpiperazine-N′-3-propanesulfonic acid) buffer, pH 8.0, to a boGDH concentration of 0.9 mg/ml, with 2.25 mg of purified Pfu-sHSP/ml. Samples were removed and assayed at 0, 2, 4, and 8 min and centrifuged at 10,000 × g for 2 min. The residual activities of boGDH in the supernatant samples were assayed as described previously (28) using a Beckman DU640 spectrophotometer with the temperature controlled at the optimum, 25°C. The assay mixture contained 100 mM EPPS (pH 8.0), 65 mM glutamic acid, and 16.25 mM NADP. There was no detectable GDH activity in the purified Pfu-sHSP (data not shown).
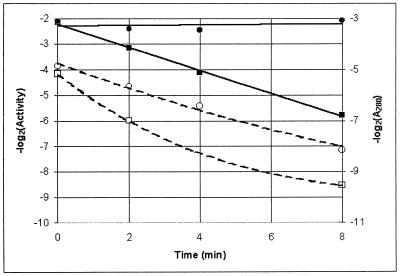
The t1/2 for boGDH precipitation was measured by centrifugation and recording of the A280 of the supernatant. In the experiment in which boGDH was incubated alone, the apparent t1/2 was approximately 2 min, whereas the boGDH to which sHSP was added did not precipitate at all during the course of the experiment. The activity of boGDH in the supernatants, on the other hand, declined in both cases (Fig. 3). Thus, much of the boGDH that remained in solution was soluble but inactive. In this case, the enzyme was maintained in solution but not preserved from denaturation. This is an important result, indicating that the probable mode of action of Pfu-sHSP is toward aggregation of nonnative proteins, thus allowing them to be recruited to either refolding or protein turnover pathways.
FIG. 3.
Effects of Pfu-sHSP on boGDH incubated at 56°C for up to 8 min. Solubility is shown as log2 A280 of boGDH in the presence (●) and absence (▪) of Pfu-sHSP (solid lines). Activity of boGDH is shown in the presence (○) and absence (□) of Pfu-sHSP (dashed lines).
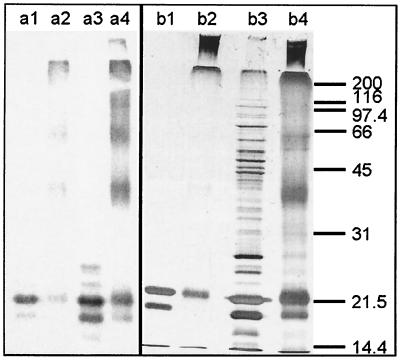
We addressed the question of the subunit structure of Pfu-sHSP by incubating purified Pfu-sHSP and E. coli extracts containing expressed Pfu-sHSP in 0.01% (wt/vol) glutaraldehyde to cross-link the proteins. The control and cross-linked preparations were subjected to SDS-PAGE and visualized by silver staining and Western blotting (Fig. 4). In contrast to M. jannaschii sHSP, which is reported to occur mostly as 24-mers (14), Pfu-sHSP appears to be polydisperse. This characteristic has been reported for several other α-crystallin homologs (6). The polydisperse patterns of the Pfu-sHSP polymer from purified Pfu-sHSP and from E. coli crude extracts are indistinguishable.
FIG. 4.
Cross-linking of purified Pfu-sHSP and Pfu-sHSP in unpurified E. coli extracts as visualized by Western blot analysis (a) and silver-stained SDS–12% PAGE (b). Lanes 1, un-cross-linked purified Pfu-sHSP; lanes 2, cross-linked purified Pfu-sHSP; lanes 3, un-cross-linked E. coli extracts; lanes 4, cross-linked E. coli extracts.
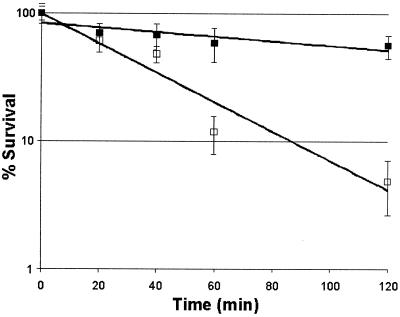
Survivability at 50°C of E. coli overexpressing Pfu-sHSP.
sHSPs from the chestnut (Castanea sativa) and from murine and human αβ-crystallin have been shown to confer slight thermotolerance at 50°C on E. coli (23, 25, 34). We show here that an archaeal sHSP can also confer very significant thermotolerance on a typical mesophilic bacterium. E. coli cultures containing pPfu-shsp/pSJS1240 were induced as described above, and the cells were diluted in Luria-Bertani broth containing 50 μg each of ampicillin and spectinomycin per ml to an A595 of 0.6. The cultures were rapidly shifted to 50°C in a water bath shaker. Samples were removed at 0, 20, 40, 60, and 120 min, diluted appropriately, and plated on Luria-Bertani agar containing 50 μg each of ampicillin and spectinomycin per ml, and the plates were incubated at 37°C overnight. Cell viability was determined by counting of CFU after overnight incubation. The first-order rate of decline of viability of E. coli overexpressing Pfu-sHSP was significantly higher, approximately six- to sevenfold, than that of the culture transformed with pET19b and pSJS1240 (Fig. 5). As a result, the difference in viability between protected and unprotected cells after a 120-min exposure at 50°C was approximately 50-fold.
FIG. 5.
Effect of recombinant Pfu-sHSP expression on E. coli viability at 50°C. The reduction of viability as an exponential function of time is shown. □, pET19b/pSJS1240; ▪, pPfu-shsp/pSJS1240.
We conclude that Pfu-sHSP increases the thermotolerance of E. coli by preventing aggregation of heat-compromised proteins. Because many proteins from hyperthermophiles are nonfunctional at the optimal growth temperatures of mesophiles, it is intriguing that a component of the adaptive response of an archaeon growing at 100°C can enhance the heat resistance in E. coli cells growing at “mesophilic” temperatures. However, Trent et al. found that the chaperonin TF55 from Sulfolobus shibatae can bind a denatured protein at 25°C as efficiently as at 70°C (37). These findings will enable us to assess the biological activity of sHSP mutants by using E. coli as a surrogate genetic system for P. furiosus, which lacks systems for mutation selection and recombinant gene expression.
Acknowledgments
We thank Juan M. Gonzalez for providing P. furiosus genomic DNA, Jocelyne DiRuggiero and Robert Belas for critical discussions, and Anchalee Jiemjit for technical assistance and assistance with the manuscript.
This work was supported by grants from the National Science Foundation (grant BES-9632554 to F.T.R.) and the Knut and Alice Wallenberg Foundation.
Footnotes
Contribution 524 from the Center of Marine Biotechnology.
REFERENCES
- 1.Adams M W W. Large-scale growth of hyperthermophiles. In: Robb F T, Place A R, editors. Archaea: a laboratory manual. 3. Thermophiles. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 47–49. [Google Scholar]
- 2.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 3.DiRuggiero J, Robb F T. Enzymes of central nitrogen metabolism from hyperthermophiles: characterization, thermostability, and genetics. Adv Protein Chem. 1996;48:311–339. doi: 10.1016/s0065-3233(08)60365-4. [DOI] [PubMed] [Google Scholar]
- 4.Fiala G, Stetter K O. P. furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 5.Gonzalez J M, Masuchi Y, Robb F T, Ammerman J W, Maeder D L, Yanagibayashi M, Tamaoka J, Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- 6.Haley D A, Bova M P, Huang Q-L, Mchaourab H S, Stewart P L. Small heat-shock protein structures reveal a continuum from symmetric to variable assemblies. J Mol Biol. 2000;298:261–272. doi: 10.1006/jmbi.2000.3657. [DOI] [PubMed] [Google Scholar]
- 7.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 8.Hayes S A, Dice J F. Roles of molecular chaperones in protein degradation. J Cell Biol. 1996;132:255–258. doi: 10.1083/jcb.132.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hightower L E. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 10.Horwitz J. α-Crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kagawa H K, Osipiuk J, Maltsev N, Overbeek R, Quaite-Randall E, Joachimiak A, Trent J D. The 60 kDa heat shock proteins in the hyperthermophilic archaeon Sulfolobus shibatae. J Mol Biol. 1995;253:712–725. doi: 10.1006/jmbi.1995.0585. [DOI] [PubMed] [Google Scholar]
- 12.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Yoshizawa T, Nakamura Y, Robb F T, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 13.Kim K K, Yakota H, Santoso S, Lerner D, Kim R, Kim S-H. Purification, crystallization and preliminary X-ray crystallographic data analysis of a small heat shock protein homolog from Methanococcus jannaschii, a hyperthermophile. J Struct Biol. 1998;121:76–80. doi: 10.1006/jsbi.1998.3969. [DOI] [PubMed] [Google Scholar]
- 14.Kim K K, Kim R, Kim S-H. Crystal structure of a small heat shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 15.Kim R, Kim K K, Yokota H, Kim S-H. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc Natl Acad Sci USA. 1998;95:9129–9133. doi: 10.1073/pnas.95.16.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim R, Sandler S J, Goldman S, Yokota H, Clark A J, Kim S-H. Overexpression of archaeal proteins in Escherichia coli. Biotechnol Lett. 1998;20:207–210. [Google Scholar]
- 17.Kitagawa M, Matsumura Y, Tsuchido T. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stress in Escherichia coli. FEMS Microbiol Lett. 2000;184:165–171. doi: 10.1111/j.1574-6968.2000.tb09009.x. [DOI] [PubMed] [Google Scholar]
- 18.Klemenz R, Andres A C, Frohli E, Schafer R, Aoyama A. Expression of the murine small heat shock proteins hsp25 and αB-crystallin in the absence of stress. J Cell Biol. 1993;120:639–645. doi: 10.1083/jcb.120.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D'Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 20.Leroux M R, Melki R, Gordon B, Batelier D, Candido E P M. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J Biol Chem. 1997;272:24646–24656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- 21.Lindquist S. Heat shock proteins and stress tolerance in microorganisms. Curr Opin Genet. 1992;2:748–755. doi: 10.1016/s0959-437x(05)80135-2. [DOI] [PubMed] [Google Scholar]
- 22.Martin J, Hartl F U. Chaperone-assisted protein folding. Curr Opin Struct Biol. 1997;7:41–52. doi: 10.1016/s0959-440x(97)80006-1. [DOI] [PubMed] [Google Scholar]
- 23.Muchowski P, Clark J I. ATP-enhanced molecular chaperone functions of the small heat shock protein human αB-crystallin. Proc Natl Acad Sci USA. 1998;95:1004–1009. doi: 10.1073/pnas.95.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagao R T, Czarnecka E, Gurley W B, Schoffl F, Key J L. Genes for low-molecular-weight heat shock proteins of soybeans: sequence analysis of a multigene family. Mol Cell Biol. 1985;5:3417–3428. doi: 10.1128/mcb.5.12.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plater M L, Goode D, Crabbe M J C. Effects of site-directed mutations on the chaperone-like activity of αB-crystallin. J Biol Chem. 1996;271:28558–28566. doi: 10.1074/jbc.271.45.28558. [DOI] [PubMed] [Google Scholar]
- 26.Plesofsky-Vig N, Brambl R. Disruption of the gene for hsp30, an α-crystallin-related heat shock protein of Neurospora crassa, causes defects in thermotolerance. Proc Natl Acad Sci USA. 1995;92:5032–5036. doi: 10.1073/pnas.92.11.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robb F T, Maeder D L, Brown J R, DiRuggiero J, Stump M D, Yeh R K, Weiss R B, Dunn D M. Genomic sequence of hyperthermophile Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–157. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- 28.Robb F T, Park J B, Adams M W W. Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Biochim Biophys Acta. 1992;1120:267–272. doi: 10.1016/0167-4838(92)90247-b. [DOI] [PubMed] [Google Scholar]
- 29.Roy S K, Hiyama T, Nakamoto H. Purification and characterization of the 16-KDa heat-shock-responsive protein from the thermophilic cyanobacterium Synechococcus vulcanus, which is an α-crystallin-related, small heat shock protein. Eur J Biochem. 1999;262:406–416. doi: 10.1046/j.1432-1327.1999.00380.x. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sauer U, Durre P. Sequence and molecular characterization of a DNA region encoding a small heat shock protein of Clostridium acetobutylicum. J Bacteriol. 1993;175:3394–3400. doi: 10.1128/jb.175.11.3394-3400.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreier H J, Robinson-Bidle K A, Romashko A M, Patel G V. Heterologous expression in the archaea: transcription from Pyrococcus furiosus gdh and mlrA promoters in Haloferax volcanii. Extremophiles. 1999;3:11–19. doi: 10.1007/s007920050094. [DOI] [PubMed] [Google Scholar]
- 33.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soto A, Allona I, Collada C, Guevara M-A, Casado R, Rodriguez-Cerezo E, Aragoncillo C, Gomez L. Heterologous expression of a plant small heat-shock protein enhances Eschericia coli viability under heat and cold stress. Plant Physiol. 1999;120:521–528. doi: 10.1104/pp.120.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trent J D. A review of acquired thermotolerance, heat-shock proteins and molecular chaperones in archaea. FEMS Microbiol Rev. 1996;18:249–258. [Google Scholar]
- 36.Trent J D, Gabrielsen M, Jensen B, Neuhard J, Olsen J. Acquired thermotolerance and heat shock proteins in thermophiles from the three phylogenetic domains. J Bacteriol. 1994;176:6148–6152. doi: 10.1128/jb.176.19.6148-6152.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trent J D, Nimmesgern E, Wall J S, Hartl F U, Horwich A L. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature. 1991;354:490–493. doi: 10.1038/354490a0. [DOI] [PubMed] [Google Scholar]
- 38.Tzeng S-S, Yeh K W, Chen Y M, Lin C-Y. Two Oryza sativa genomic DNA clone 16.9-kilodalton heat shock proteins. Plant Physiol. 1992;99:1723–1725. doi: 10.1104/pp.99.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]