Abstract
The adhesion family of G protein–coupled receptors (GPCRs) is defined by an N-terminal large extracellular region that contains various adhesion-related domains and a highly-conserved GPCR-autoproteolysis-inducing (GAIN) domain, the latter of which is located immediately before a canonical seven-transmembrane domain. These receptors are expressed widely and involved in various functions including development, angiogenesis, synapse formation, and tumorigenesis. GPR125 (ADGRA3), an orphan adhesion GPCR, has been shown to modulate planar cell polarity in gastrulating zebrafish, but its biochemical properties and role in mammalian cells have remained largely unknown. Here, we show that human GPR125 likely undergoes cis-autoproteolysis when expressed in canine kidney epithelial MDCK cells and human embryonic kidney HEK293 cells. The cleavage appears to occur at an atypical GPCR proteolysis site within the GAIN domain during an early stage of receptor biosynthesis. The products, i.e., the N-terminal and C-terminal fragments, seem to remain associated after self-proteolysis, as observed in other adhesion GPCRs. Furthermore, in polarized MDCK cells, GPR125 is exclusively recruited to the basolateral domain of the plasma membrane. The recruitment likely requires the C-terminal PDZ-domain–binding motif of GPR125 and its interaction with the cell polarity protein Dlg1. Knockdown of GPR125 as well as that of Dlg1 results in formation of aberrant cysts with multiple lumens in Matrigel 3D culture of MDCK cells. Consistent with the multilumen phenotype, mitotic spindles are incorrectly oriented during cystogenesis in GPR125-KO MDCK cells. Thus, the basolateral protein GPR125, an autocleavable adhesion GPCR, appears to play a crucial role in apicobasal polarization in epithelial cells.
Keywords: ADGRA3, adhesion GPCR, autoproteolysis, cystogenesis, Dlg1, epithelial cell polarity, GAIN domain, GPR125, GPS motif, spindle orientation
Abbreviations: 7TM, seven transmembrane; aGPCR(s), adhesion GPCR(s); BSA, bovine serum albumin; cDNA, complementary DNA; CTF, C-terminal fragment; CTT, C-terminal tail; ECM, extracellular matrix; ECR, extracellular region; FCS, fetal calf serum; GAIN, GPCR-autoproteolysis inducing; GPCR(s), G protein-coupled receptor(s); GPS, GPCR proteolysis site; HBD, hormone-binding domain; IP, immunoprecipitation; LRR, leucine-rich repeat; MDCK, Madin-Darby canine kidney; NTF, N-terminal fragment; PBM, PDZ domain-binding motif; sgRNA, single-guide RNA
G protein–coupled receptors (GPCRs), comprising over 800 members in humans, relay various extracellular signals by triggering intracellular signaling via their seven transmembrane (7TM) segments (1, 2, 3, 4). On the basis of structural comparison of the 7TM domains, human GPCRs are classified into five major families as follows: the rhodopsin-like (class A), secretin receptor-like (class B or B1), adhesion (class B2), metabotropic glutamate receptor-like (class C), and Frizzled-like (class F) families (4, 5, 6). Adhesion GPCRs (aGPCRs) constitute the second largest family with 33 members in the human genome; they are widely expressed in normal and malignant cells from endodermal, ectodermal, and mesodermal origin and play significant roles in development, reproduction, synapse formation, angiogenesis, immunity, and tumorigenesis (6, 7, 8).
Structurally, according to a topology-based tripartite compartmentation, aGPCRs consist of an N-terminal large extracellular region (ECR), a canonical 7TM helix bundle, and a C-terminal tail (CTT) (6, 7, 8) (Fig. 1A). The large ECRs of aGPCRs contain a variety of adhesion-related domains that are often repeated and are linked to the 7TM domain via the highly conserved GPCR autoproteolysis-inducing (GAIN) domain. The GAIN domain constitutively self-cleaves the receptors into two fragments, namely an N-terminal fragment (NTF) and a C-terminal fragment (CTF), contributing to a cleavage-based bipartite structure. The GPCR proteolysis site (GPS) locates near the C terminus of the GAIN domain; as a result, the NTF harbors all external protein domains and the larger part of the autolyzed GAIN domain, whereas the CTF contains a small part of the cleaved GAIN domain, the entire 7TM helix bundle, and the entire CTT. Because of a dense network of hydrogen bonds within the GAIN domain, the NTF and CTF remain normally associated after cis-autoproteolysis.
Figure 1.
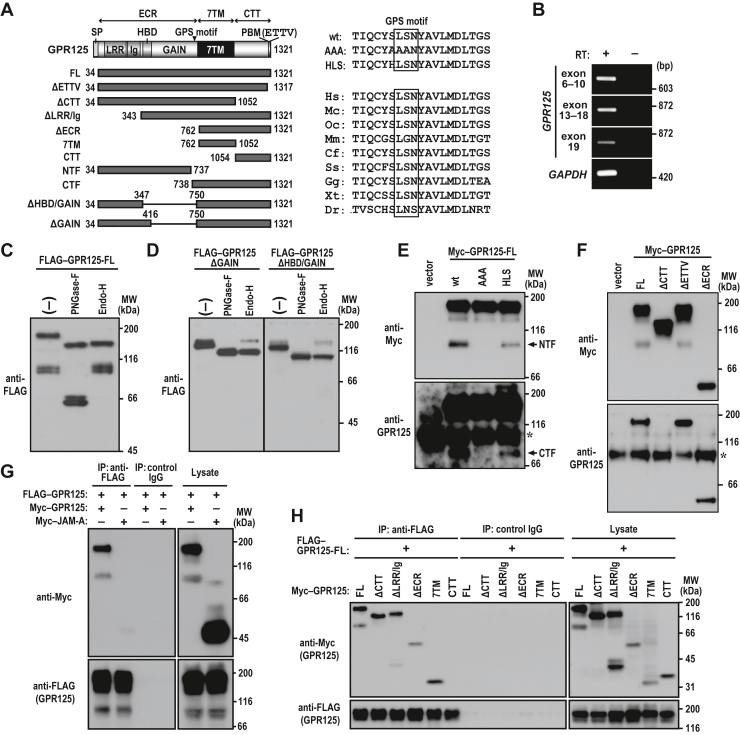
GPR125 is a cleavable aGPCR.A, schematic representation of the domain organization of human GPR125 and its truncated proteins used in the present study (left). ECR, extracellular region; TM, transmembrane; CTT, C-terminal tail; SP, signal peptide; LRR, leucine-rich repeat; Ig, immunoglobulin domain; HBD, hormone-binding domain; GAIN, G protein-coupled receptor (GPCR) autoproteolysis inducing domain; GPS, GPCR proteolysis site; PBM, PDZ domain-binding motif; FL, full-length; NTF, N-terminal fragment; CTF, C-terminal fragment. ETTV indicates the extreme C-terminal tetrapeptide Glu-Thr-Thr-Val, a class I PBM. The amino acid sequence of the flanking regions of a putative cleavage tripeptide in the GPS motif is shown (right). The wild-type (wt) tripeptide Ser-Leu-Ser (SLS) and its mutated tripeptides are boxed; AAA and HLS, the S736A/L737A/S738A and S736H substitutions, respectively. Sequence alignment of regions flanking a putative GPS cleavage site in GPR125 from various species is also shown. Homo sapiens (Hs), Macaca mulatta (Mc), Oryctolagus cuniculus (Oc), Mus musculus (Mm), Canis familiaris (Cf), Sus scrofa (Ss), Gallus gallus (Gg), Xenopus tropicalis (Xt), and Danio rerio (Dr). B, RT-PCR analysis of GPR125 and GAPDH expression in MDCK cells. The amplified region encompassing exons 6 to 10 (exon 6–10), exons 13 to 18 (exon 13–18), or exon 19 of GPR125 gene codes for a part of the ECR, GAIN and 7TM, or CTT of GPR125 protein, respectively. C and D, glycosylation of GPR125. The lysate from MDCK cells expressing FLAG–GPR125 (FL or the indicated truncated protein) was extracted with Triton X-100 and digested with PNGase-F or Endo-H. Proteins were analyzed by immunoblot with the anti-FLAG M2 antibody. E, GPR125 is autoproteolysed at the GPS to produce the NTF and CTF. F, the anti-GPR125 antibody recognizes the CTT of GPR125. An empty vector or the indicated Myc–GPR125 protein was expressed in HEK293 cells, and proteins in the cell lysate were analyzed by immunoblot with the anti-Myc or anti-GPR125 antibody (E and F). The asterisk indicates a nonspecific band present in the cell lysates. G and H, GPR125 forms a homodimer. FLAG–GPR125-FL was coexpressed with the indicated Myc-tagged protein in HEK293 cells, and proteins in the cell lysate were immunoprecipitated (IP) with the anti-FLAG M2 antibody or control IgG, followed by immunoblot analysis with the indicated antibodies. Positions for marker proteins are indicated in kDa. MDCK, Madin-Darby canine kidney.
Although research of aGPCR has been hampered by the orphan status of receptors, recent advancements have revealed a conserved method of aGPCR activation involving a tethered ligand within the GAIN domain (6, 9). NTF–CTF dissociation (for example, mechanically induced dissociation of the fragments) results in the exposed N-terminal peptide of the CTF, i.e., the tethered ligand, which interacts as an agonist intramolecularly with its orthosteric site in the 7TM domain. Thus, cis-autoproteolysis is a prerequisite for tethered peptide agonism for activation of aGPCR, implying that the capability of self-cleavage at least in part renders the receptor out of an orphan state.
GPR125 (a.k.a. ADGRA3), an orphan aGPCR, has a leucine-rich repeat (LRR), an immunoglobulin (Ig) domain, a hormone-binding domain (HBD), and a GAIN domain in the N-terminal ECR (Fig. 1A). In general, the GPS motif in the GAIN domain comprises the tripeptide His-Leu/Ile-Thr/Ser and is cleaved at the peptide bond between Leu/Ile and Thr/Ser. Because the corresponding sequence is Ser-Leu-Ser in GPR125, where the His at position –2 respective to the putative cleavage site is replaced by Ser, this receptor has been predicted noncleavable (6, 7, 8). The possibility, however, has remained to be elucidated. GPR125 was originally identified as a specific marker of adult spermatogonial progenitor cells (10) and has been linked to various tumors (11, 12, 13, 14). Although GPR125 is reported to serve as a modulator of planar cell polarity in gastrulating zebrafish (15), its role in mammalian epithelial cells has remained largely unknown. The previous finding that the CTT of GPR125 is capable of binding to Discs Large 1 (Dlg1), a cell polarity protein (16), may raise its possible role in mammalian epithelial cell polarization via the interaction with Dlg1.
In the present study, we show that human GPR125, expressed in kidney epithelial Madin-Darby canine kidney (MDCK) cells and embryonic kidney HEK293 cells, undergoes cis-autoproteolysis at the atypical GPS during an early stage of receptor biosynthesis. The products NTF and CTF are recruited together to the basolateral domain of the plasma membrane in polarized MDCK cells, which recruitment requires its interaction with Dlg1. GPR125 is indispensable for correct cystogenesis and mitotic spindle orientation in 3D-cultured MDCK cells, indicating its crucial role in epithelial apicobasal polarization.
Results
GPR125 is a cleavable aGPCR
The GPR125 gene is known to be expressed in a variety of tissues and organs including the kidney (16, 17). To know whether the gene is also expressed in canine kidney epithelial MDCK cells, we performed RT-PCR analysis using primer pairs for the regions from exons 6 to 10 (exon 6–10), from exons 13 to 18 (exon 13–18), and within exon 19. As shown in Figure 1B, the three expected PCR products were all obtained, indicating that GPR125 is indeed expressed in MDCK cells.
We next transfected MDCK cells with the complementary DNA (cDNA) for wild-type human GPR125 (FLAG–GPR125-FL), in which the 3 × FLAG-tag was inserted between a signal peptide and the full-length protein (amino acid residues 34–1321) (Fig. 1A). Immunoblot analysis of lysates of the transfected cells with an anti-FLAG antibody revealed that FLAG–GPR125-FL was expressed as two major bands with apparent molecular masses of about 170 and 90 kDa on SDS-PAGE (Fig. 1C). On the extracellular side, GPR125 contains 13 asparagine residues in the putative glycosylation sequence (Asn-Xaa-Ser/Thr; Xaa is any amino acid but proline) (18). Consistent with the identity of GPR125 as a glycoprotein, treatment with peptide:N-glycosidase F (PNGase-F) or endoglycosidase H (Endo-H) each resulted in the appearance of a protein with an about 140 kDa (Fig. 1C), which is similar to the predicted molecular mass of unglycosylated FLAG–GPR125-FL. Thus, the upper band observed in the glycosidase-untreated cell lysate (Fig. 1C) likely represents FLAG–GPR125-FL with high-mannose N-glycans. On the other hand, treatment of the lower band (Fig. 1C) with PNGase-F but not with Endo-H produced a 70 kDa protein, indicating that the lower band is a C-terminally cleaved FLAG–GPR125 with complex-type N-glycans. The cleavage may occur at a position in the GAIN domain in the N-terminal extracellular region, according to a predicted molecular mass of the fragment (about 80 kDa).
GPR125 is autocleaved at the atypical GPS
The above findings raise the possibility that GPR125 is autoproteolysed at the GPS in the GAIN domain, although GPR125 has been predicted to be noncleavable in the GAIN domain because of its atypical GPS motif (Ser736-Leu737-Ser738) but not the canonical one (His-Leu/Ile-Ser/Thr) (6, 7, 8). To test the role of the GAIN domain in GPR125 cleavage, we expressed mutant proteins lacking the GAIN domain alone (amino acid residues 417–749) or both GAIN domain and HBD (amino acid residues 348–749) in MDCK cells. As shown in Figure 1D, the cleavage was not observed in these mutant proteins, suggesting the involvement of the GAIN domain in autoproteolysis of GPR125.
To investigate the significance of the atypical GPS motif in GRP125 cleavage, we expressing an N-terminally Myc-tagged GPR125 protein carrying the S736A/L737A/S738A substitution at the GPS tripeptide (Myc–GPR125-AAA) in HEK293 cells (Fig. 1A). Interestingly, Myc–GPR125-AAA was not cleaved; it failed to produce the NTF of about 90 kDa, which was detected by the anti-Myc antibody (Fig. 1E, upper panel), and the CTF of about 70 kDa (Fig. 1E, lower panel), which was recognized by the antibody against the CTT of GPR125 (Fig. 1F). On the other hand, the efficiency of the cleavage was not enhanced by the substitution of Ser-736 for His (Fig. 1E), a residue conserved in the GPS of most of autocleavable aGPCRs (6, 7, 8). Taken together with the present findings, GPR125 appears to be autocleaved at the atypical GPS motif.
GPR125 is able to self-associate via the 7TM helix bundle
A variety of GPCRs is known to form a homodimer (19, 20, 21, 22). To know whether GPR125 is also able to interact homophilically, we performed an immunoprecipitation (IP) assay using HEK293 cells coexpressing FLAG–GPR125 with Myc-tagged GPR125 or JAM-A, a single-span transmembrane protein with two Ig domains (23). As shown in Fig. 1G, Myc–GPR125 but not Myc–JAM-A was coimmunoprecipitated with FLAG–GPR125, suggestive for self-association of GPR125. When various lengths of GPR125 were coexpressed as Myc-tagged proteins, the 7TM helix bundle alone interacted with FLAG–GPR125 and the interaction was not abrogated by truncation of the ECR or CTT (Fig. 1H). Thus, GPR125 is likely capable of self-associating via the 7TM helix bundle.
GPR125 is recruited to the basolateral membrane
To investigate the role of GPR125 in epithelial cells, we examined its localization in MDCK cells by expressing FLAG–GPR125-FL and subsequent staining with the anti-FLAG antibody. As shown in Figure 2A, FLAG–GPR125 colocalized with β-catenin, a marker protein for the basolateral domain of the plasma membrane, but not with gp135 or ZO-1, which defines the apical membrane or tight junction, respectively. Furthermore, domain-selective cell surface biotinylation analysis revealed that FLAG–GPR125 was detected on the basolateral domain but not the apical surface of the plasma membrane in MDCK cells (Fig. 2B). Thus, GPR125 is recruited to the basolateral membrane in polarized epithelial cells.
Figure 2.
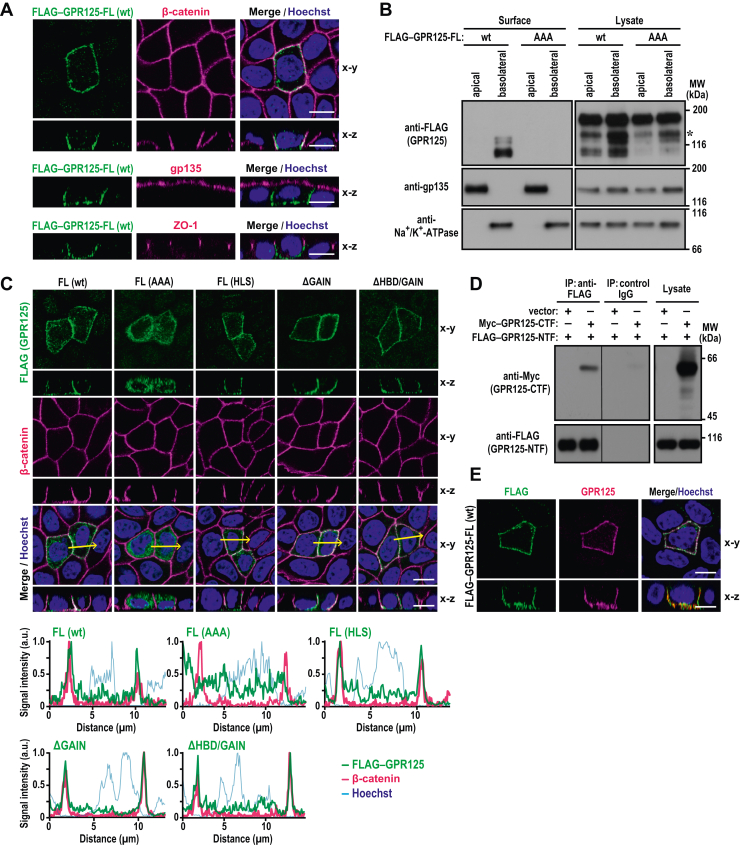
GPR125 localizes to the basolateral membrane.A, representative confocal images of MDCK cells expressing FLAG–GPR125 (wt), visualized with the indicated antibodies and Hoechst. B, domain-selective cell surface biotinylation analysis of GPR125. MDCK cells expressing FLAG–GPR125-wt or FLAG–GPR125-AAA were grown to confluence for 3 days on transwell filters. Proteins at the apical or basolateral surfaces were biotinylated by addition of sulfo-NHS–SS–biotin into the upper or lower reservoir, respectively. The biotinylated proteins were pulled down with streptavidin–Sepharose beads and analyzed by immunoblot with the indicated antibodies. C, representative confocal images of MDCK cells expressing FLAG–GPR125 (wt or the indicated mutant protein), visualized with the indicated antibodies and Hoechst (upper panels). Line plots in the lower panels represent the fluorescent signal intensity along the yellow arrows in the respective upper panels. D, interaction between the NTF and CTF of GPR125. FLAG–GPR125-NTF was coexpressed with or without Myc–GPR125-CTF in HEK293 cells, and proteins in the cell lysate were immunoprecipitated (IP) with the anti-FLAG M2 antibody or control IgG, followed by immunoblot analysis with the indicated antibodies. Positions for marker proteins are indicated in kDa. E, the NTF and CTF of GPR125 remain associated at the basolateral plasma membrane. Representative confocal images of MDCK cells expressing FLAG–GPR125-wt, visualized with the indicated antibodies and Hoechst. The scale bars represents 10 μm. CTF, C-terminal fragment; MDCK, Madin-Darby canine kidney; NTF, N-terminal fragment.
The present finding that the uncleaved GPR125 exclusively contains high-mannose type N-glycans and the NTF of GPR125 solely bears complex type N-glycans (Fig. 1C) suggests that GPR125 is transported to the plasma membrane after cleavage at the GPS. This is because complex type N-glycans are present in mature glycoproteins on the plasma membrane, whereas glycoproteins residing in the endoplasmic reticulum are of the high-mannose type (24). In addition, judging from the molecular mass of about 90 kDa, the NTF is a major form of the basolaterally localized FLAG–GPR125 (Fig. 2B).
To test the possibility that the cleavage of GPR125 is required for its accumulation to the basolateral membrane, we expressed FLAG–GPR125-AAA, a mutant protein with the noncleavable GPS, in MDCK cells and stained with the anti-FLAG antibody. As shown in Figure 2C, this mutant protein was distributed to the cytoplasm but not to the plasma membrane. The failure of the GPR125-AAA to localize to the basolateral membrane was confirmed by domain-selective cell surface biotinylation analysis (Fig. 2B). In contrast, the cleavable mutant protein FLAG–GPR125-HLS as well as the wild-type protein efficiently accumulated at the basolateral membrane of MDCK cells (Fig. 2C). These findings raised a possibility that the uncleaved GAIN domain may have an ability to inhibit basolateral recruitment of GPR125. Indeed, mutant GPR125 proteins lacking the GAIN domain (GPR125-ΔGAIN and GPR125-ΔHBD/GAIN) were both able to localize to the basolateral domain of the plasma membrane (Fig. 2C). Consistent with this, a small but significant amount of these two proteins were resistant to Endo-H, indicative for their localization to the plasma membrane (Fig. 1D). Thus, the uncleaved GAIN domain appears to suppress targeting of GPR125 to the basolateral membrane, which may explain at least partially why the cleavage occurs during an early stage of receptor biosynthesis.
The NTF and CTF of GPR125 remain associated after self-cleavage
GAIN-mediated self-cleavage in aGPCR is generally known to occur constitutively during biosynthesis to produce the two fragments NTF and CTF that remain associated noncovalently (6, 7, 8). To study the association between the NTF and CTF of GPR125, we coexpressed FLAG–GPR125-NTF and Myc–GPR125-CTF in HEK293 cells and performed an IP assay. As shown in Figure 2D, Myc–GPR125-CTF was coprecipitated with FLAG–GPR125-NTF by the anti-FLAG antibody but not by control IgG, suggesting that the separately expressed fragments are able to form a complex. In general, the separately expressed NTF and CTF of aGPCRs do not associate with each other because the N terminus of the isolated CTF, a region that makes direct contacts with the NTF, inserts as a tethered agonist into the 7TM helix bundle and thus fails to associate with the isolated NTF (25, 26, 27, 28, 29). In the present experiment, the insertion into the 7TM helix bundle may be blocked by the 3 × Myc-tag sequence N-terminal to the CTF, which blockade allows the CTF N terminus to bind to FLAG–GPR125-NTF (Fig. 2D). It is also possible that the GPR125 NTF and CTF can associate in a manner distinct from that of other aGPCRs. Furthermore, in MDCK cells transfected with the cDNA for FLAG–GPR125-FL, the basolateral membrane was stained both with the anti-GPR125 antibody, which recognized the CTT, and with the anti-FLAG antibody (Fig. 2E). These observations are consistent with the idea that after the self-cleavage, the NTF and CTF of GPR125 remain associated at the basolateral membrane.
The CTT of GPR125 is required for its basolateral localization
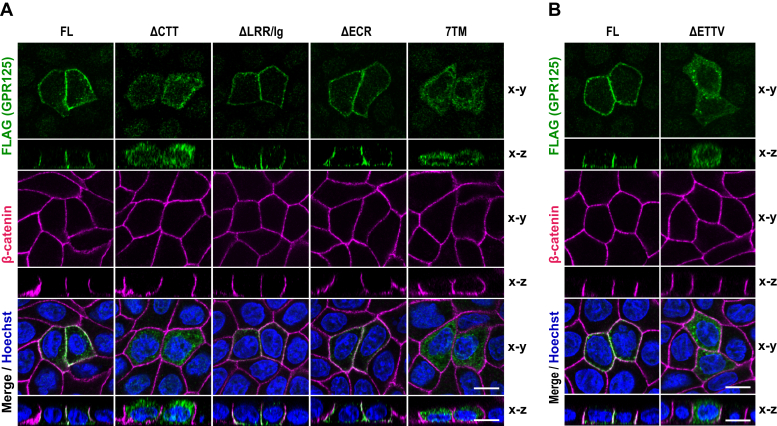
We next clarified the region required for GPR125 localization to the basolateral membrane in polarized epithelial cells. As shown in Figure 3A, not only wild-type GPR125-FL but also mutant proteins without both LRR and Ig-like domain (ΔLRR/Ig) or the entire N-terminal ECR (ΔECR) were capable of localizing to the basolateral membrane in MDCK cells. By contrast, a protein with truncation of the entire CTT (GPR125-ΔCTT) as well as one that contains the 7TM helix bundle but lacks both ECR and CTT (GPR125-7TM) was distributed to the cytoplasm but not to the plasma membrane (Fig. 3A). These findings indicate that the CTT of GPR125 is required for its localization to the basolateral membrane in polarized epithelial cells.
Figure 3.
The C-terminal tail of GPR125 is required for its basolateral localization.A and B, representative confocal images of MDCK cells expressing FLAG–GPR125. The indicated GPR125 proteins (FL or the indicated truncated protein) were expressed by plasmid transfection (A) or by lentivirus infection (B) in MDCK cells, and cells were fixed and stained with the indicated antibodies and Hoechst. The scale bars represents 10 μm. MDCK, Madin-Darby canine kidney.
The CTT of GPR125 contains the extreme C-terminal tetrapeptide Glu-Thr-Thr-Val (amino acid residues 1318–1321), which corresponds to the type I PDZ domain–binding motif (PBM) (30). To investigate the role of the motif in GPR125 localization in polarized epithelial cells, we expressed a mutant protein lacking the last four amino acid residues as a FLAG-tagged protein (FLAG–GPR125-ΔETTV) in MDCK cells. As shown in Figure 3B, the truncation resulted in exclusive localization to the cytoplasm. Thus, the C-terminal PBM is required for recruitment of GPR125 to the basolateral membrane.
GPR125 interacts with Dlg1 via its CTT
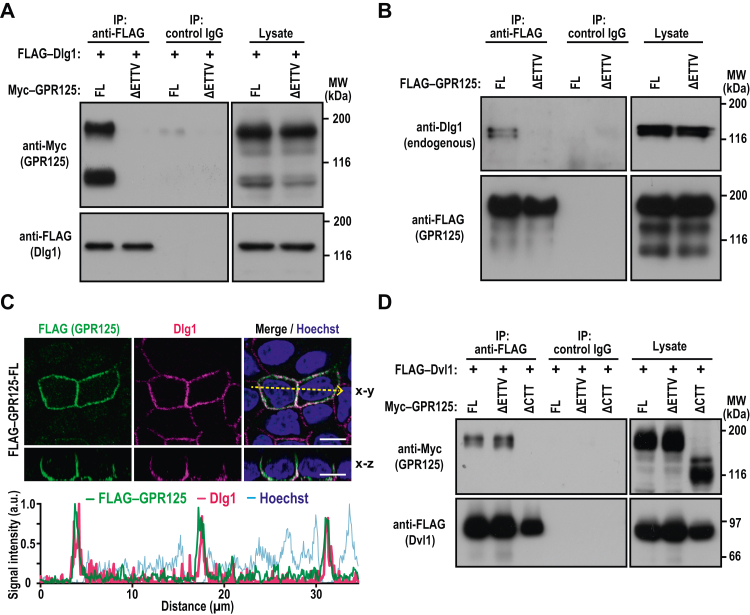
The CTT of bacterially expressed GPR125 has been reported to directly bind to the PDZ domain containing protein Dlg1 (16). To confirm that full-length GPR125 also interacts with Dlg1, we expressed Myc–GPR125-FL and FLAG–Dlg1 in HEK293 cells, followed by a co-IP assay. As shown in Figure 4A, FLAG–Dlg1 interacted with Myc–GPR125-FL but not a mutant protein lacking the C-terminal four amino acid residues of the PBM (Myc–GRP125-ΔETTV). Similarly, endogenous Dlg1 in MDCK cells was coprecipitated with FLAG–GRP125-FL but not with FLAG–GRP125-ΔETTV (Fig. 4B). These findings indicate that GPR125 associates with Dlg1 in a PBM-dependent manner. Consistent with the association, in polarized MDCK cells, FLAG–GPR125 colocalized with endogenous Dlg1 at the basolateral membrane (Fig. 4C).
Figure 4.
GPR125 interacts with Dlg1 via the C-terminal tail.A, FLAG–Dlg1 was coexpressed with Myc–GPR125-FL or Myc–GPR125-ΔETTV in HEK293 cells, and proteins in the cell lysate were immunoprecipitated (IP) with the anti-FLAG M2 antibody or control IgG, followed by immunoblot analysis with the indicated antibodies. B, interaction between endogenous Dlg1 and exogenously expressed FLAG–GPR125. FLAG–GPR125-FL or FLAG–GPR125-ΔETTV was expressed in MDCK cells, and proteins in the cell lysate were immunoprecipitated (IP) as in (A), followed by immunoblot analysis with the indicated antibodies. C, representative confocal images of MDCK cells expressing FLAG–GPR125-FL, visualized with the indicated antibodies and Hoechst (upper panel). Line plots represent the fluorescent signal intensity along the yellow arrow in the upper panel. The scale bars represents 10 μm. D, GPR125 interacts with Dvl1 in a manner independent of its PDZ-binding motif. FLAG–Dvl1 was coexpressed with Myc–GPR125-FL, Myc–GPR125-ΔETTV, or Myc–GPR125-ΔCTT in HEK293 cells, and proteins in the cell lysate were analyzed as in (A). Positions for marker proteins are indicated in kDa. MDCK, Madin-Darby canine kidney.
It has been also reported that the CTT of zebrafish GPR125 interacts with Xenopus Dishevelled (Dvl), another PDZ domain–harboring protein (15). To know whether human Dvl1 also interacts with the PBM of human GPR125, we performed a co-IP assay using HEK293 cells expressing FLAG–Dvl1 and Myc–GPR125-FL, Myc–GPR125-ΔETTV, or Myc–GPR125-ΔCTT. As shown in Figure 4D, FLAG–Dvl1 associated with Myc–GPR125-FL and the association was abolished by truncation of the entire CTT but not by that of the PBM alone. GPR125 appears to also interact with Dvl1 but mainly in a PBM-independent manner, in contrast to the absolute requirement of the PBM for its binding to Dlg1.
Dlg1 is required for basolateral recruitment of GPR125
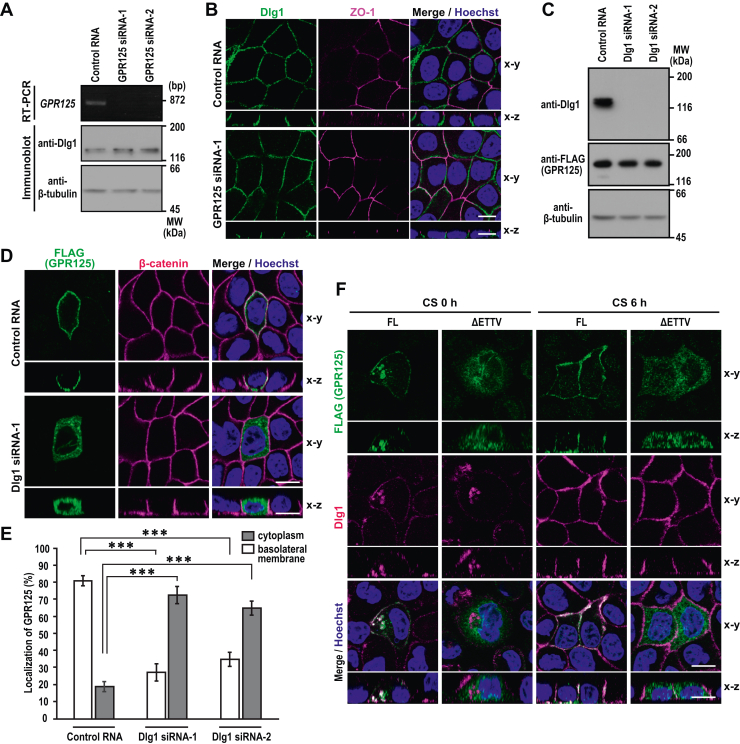
Requirement of the PBM for basolateral recruitment of GPR125 (Fig. 3B) and colocalization of GPR125 with its PBM-binding protein Dlg1 (Fig. 4C) suggest that Dlg1 is required for GPR125 transport to the basolateral domain of the plasma membrane or vice versa. To test the possibility, we first depleted GPR125 in MDCK cells by siRNA-mediated RNAi and performed RT-PCR to confirm specific depletion (Fig. 5A); the anti-GPR125 antibody used in Figure 2 for analysis of the overexpressed protein was not suitable for detection of endogenous GPR125. Depletion of GPR125 did not impair the basolateral localization of Dlg1 (Fig. 5B). We next knocked down Dlg1 (Fig. 5C) and examined its effect on the localization of GPR125 in MDCK cells (Fig. 5D). In Dlg1-depleted cells, GPR125 was distributed in the cytoplasm but not localized to the plasma membrane (Fig. 5, D and E). It is thus likely that Dlg1 is essential for basolateral recruitment of GPR125 in polarized epithelial cells, whereas GPR125 is dispensable for basolateral localization of Dlg1.
Figure 5.
Dlg1 is required for basolateral localization of GPR125.A, RT-PCR analysis of GPR125 and immunoblot analysis of Dlg1 and β-tubulin in MDCK cells transfected with negative control RNA or GPR125 specific siRNA (GPR125 siRNA-1 or siRNA-2). B, representative confocal images of MDCK cells transfected with negative control RNA or GPR125 siRNA-1. Cells were fixed and stained with the indicated antibodies and Hoechst. C, the effect of transfection with Dlg1 siRNA on the protein level of exogenously expressed FLAG–GPR125. MDCK cells were transfected with negative control RNA or Dlg1-specific siRNA (Dlg1 siRNA-1 or siRNA-2) for 24 h, and further transfected with an expression vector for FLAG–GPR125-FL for another 48 h. Proteins in the cell lysates were analyzed by immunoblot with the indicated antibodies. Positions for marker proteins are indicated in kDa. D, representative confocal images of MDCK cells treated as in (C). Cells were fixed and stained with the indicated antibodies and Hoechst. E, quantification of the localization of GPR125. Values are means ± SD from four independent experiments (n ≥ 75 cells/experiment). ∗∗∗p < 0.001 (Dunnett’s test). F, subcellular localization of GPR125 and Dlg1 in depolarized and repolarized MDCK cells. MDCK cells expressing FLAG–GPR125-FL or FLAG–GPR125-ΔETTV were fixed at 0 or 6 h after the calcium switch (CS), followed by staining with the indicated antibodies and Hoechst. The scale bars represent 10 μm. MDCK, Madin-Darby canine kidney.
Dlg1 is implicated in vesicle trafficking, including membrane protein recruitment to the basolateral domain (31). To know a process involved in Dlg1-dependent GPR125 transport, we performed a calcium-switch procedure using MDCK cells; in the procedure, plasma membrane proteins are internalized and thus localized to vesicles in the cytoplasm during culture in low Ca2+ media, and the internalized proteins are trafficked to their appropriate sites by switch to Ca2+-rich media. As shown in Figure 5F, when MDCK cells were cultured in low Ca2+ media, FLAG–GPR125-FL was internalized and accumulated with endogenous Dlg1 at the intracellular compartment. On the other hand, FLAG–GPR125-ΔETTV distributed to the entire cytoplasm without colocalization with Dlg1 (Fig. 5F). After restoration of normal Ca2+ concentration, FLAG–GPR125-FL was transported to the basolateral membrane with Dlg1, whereas FLAG–GPR125-ΔETTV retained in the cytoplasm in spite of basolateral localization of Dlg1 (Fig. 5F). It is thus probable that Dlg1 interacts with GPR125 in a vesicle and delivered the vesicle to the basolateral domain of the plasma membrane.
GPR125 as well as Dlg1 participates in cystogenesis of MDCK cells
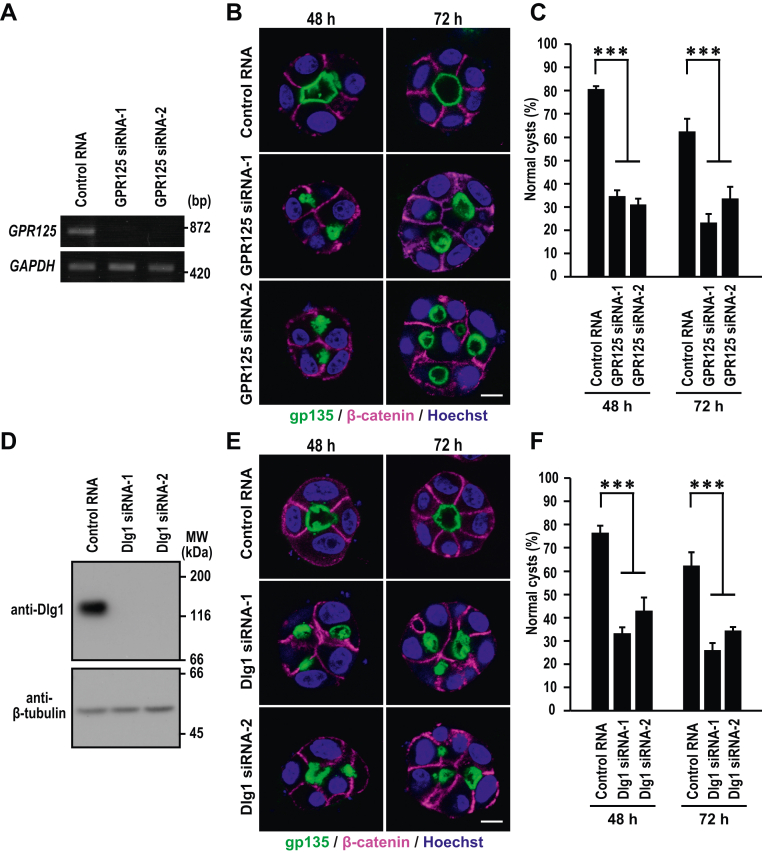
To assess the role for the basolateral membrane–integrated GPR125 in epithelial morphogenesis, we explored cystogenesis in Matrigel 3D culture (32) using MDCK cells depleted of GPR125 by siRNA-mediated RNAi (Fig. 6A). Control RNA-transfected MDCK cells formed normal cysts with a solitary lumen, the surface of which was positive for the apical marker gp135; the basolateral marker β-catenin localized to sites of cell–cell contact and to those facing the extracellular matrix (ECM) (Fig. 6B). By contrast, depletion of GPR125 results in formation of aberrant cysts with multiple lumens (Fig. 6B) and concomitant decrease in normal cystogenesis (Fig. 6C). Thus, GPR125 appears to participate in correct cystogenesis of MDCK cells. In agreement with this conclusion, depletion of Dlg1, a protein required for proper localization of GPR125, also led to abnormal cyst formation (Fig. 6, D–F), as observed in a recent study by Porter et al. (33). It is thus likely that the basolateral recruitment of GPR125 may participate in its regulation of epithelial cystogenesis.
Figure 6.
GPR125 as well as Dlg1 participates in cystogenesis of MDCK cells.A, RT-PCR analysis of GPR125 and GAPDH expression in MDCK cells transfected with negative control RNA or GPR125-specific siRNA (GPR125 siRNA-1 or siRNA-2). B and E, cystogenesis of GPR125-depleted or Dlg1-depleted MDCK cells. Cells were grown for 48 or 72 h in 3D culture and stained with the indicated antibodies and Hoechst. Shown are representative confocal images of cysts of GPR125-depleted (B) or Dlg1-depleted (E) cells. The scale bars represent 10 μm. C and F, quantification of normally oriented cysts with a solitary lumen in 3D culture of MDCK cells transfected with the indicated RNA. Values are means ± SD from four independent experiments (n ≥ 75 cysts/experiment). ∗∗∗p < 0.001 (Dunnett’s test). D, immunoblot analysis of Dlg1 and β-tubulin expression in MDCK cells transfected with negative control RNA or Dlg1 specific siRNA (Dlg1 siRNA-1 or siRNA-2). Positions for marker proteins are indicated in kDa. MDCK, Madin-Darby canine kidney.
GPR125 regulates correct spindle orientation during epithelial cystogenesis
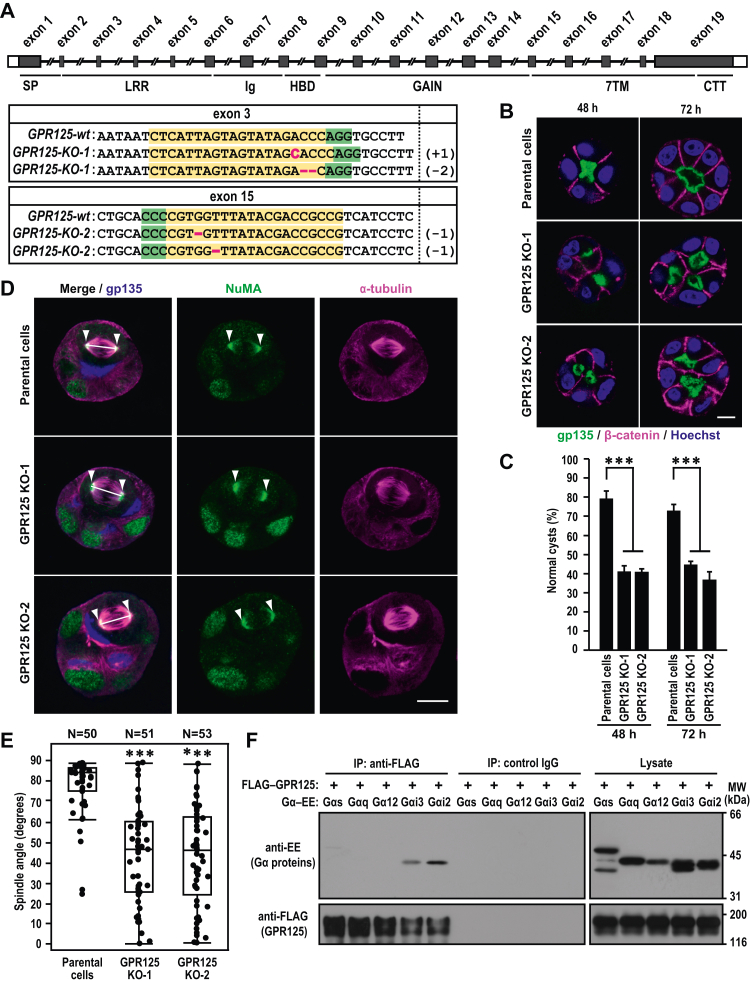
To investigate the mechanism whereby GPR125 regulates epithelial morphogenesis, we used the CRISPR/Cas9 system (34) and generated two independent GPR125-KO MDCK cell lines, namely GPR125 KO-1 and GPR125 KO-2 (Fig. 7A). Successful KO was confirmed by Sanger sequencing: GPR125 KO-1 contains a distinct single frameshift mutation in each allele of the GPR125 gene exon 3, leading to premature termination in the LRR-coding region, and GPR125 KO-2 harbors a distinct one-base deletion in each allele of the GPR125 gene exon 15, leading to premature termination in a region coding the N terminus of the 7TM domain (Figs. 7A and S1). Cystogenesis by these GPR125-deficient cells in Matrigel 3D culture was markedly perturbed with formation of cysts with multiple lumens (Fig. 7, B and C), as observed in GPR125-knockdown cells (Fig. 6). Since apical lumen positioning during epithelial cystogenesis is controlled by the orientation of cell division, misorientation of the mitotic spindle prevents the development of a solitary apical lumen, leading to formation of cysts with multiple lumens in the 3D culture system (35, 36). Hence, we investigated the potential link between GRP125 and mitotic spindle orientation during cystogenesis of MDCK cells. As shown in Figure 7D, the mitotic spindles, defined by immunostaining of spindle microtubules with the anti–α-tubulin antibody and by that of spindle poles with the anti-NuMA antibody, were predominantly perpendicular to the apical–basal axis in control cysts derived from parental MDCK cells. On the other hand, mitotic spindle orientation was almost randomized in GPR125-deficient MDCK cells (Fig. 7, D and E). Thus, GPR125 appears to regulate epithelial cell polarity via correctly orienting mitotic spindles.
Figure 7.
GPR125 regulates mitotic spindle orientation during epithelial cystogenesis.A, structure of human GPR125 gene (upper panel). Exons are denoted by gray boxes (coding region) and open boxes (noncoding region). The lower panel indicates genomic sequence alignment of the wild-type and GPR125-KO alleles. The target and PAM sequences are highlighted in yellow and green, respectively. The inserted or deleted nucleotides in KO cells are indicated in magenta letter or bars, respectively. +1, 1-base insertion; –2, 2-base deletion; –1, 1-base deletion. B, cystogenesis of GPR125-KO MDCK cells. Cells were grown for 48 h or 72 h in 3D culture and stained with the indicated antibodies and Hoechst. Shown are representative confocal images of cysts of GPR125-KO cells. The scale bars represent 10 μm. C, quantification of normally oriented cysts with a solitary lumen in 3D culture of GPR125-KO cells. Values are means ± SD from four independent experiments (n ≥ 75 cysts/experiment). ∗∗∗p < 0.001 (Dunnett’s test). D, representative confocal images of control or GPR125-KO MDCK cells in 3D culture. MDCK cysts grown for 48 h were fixed and stained with the indicated antibodies. Arrowheads and white lines indicate spindle poles and spindle axes, respectively. The scale bars represent 10 μm. E, scatter diagrams and box-and-whisker plots of metaphase spindle angles in parental MDCK cells and GPR125-KO cells. ∗∗∗p < 0.001 (Steel–Dwass test). F, interaction of GPR125 with Gαi proteins. FLAG–GPR125 was coexpressed with the internal EE-tagged Gαs, Gαq, Gα12, Gαi2, or Gαi3 in HEK293 cells. Proteins in the cell lysate were immunoprecipitated (IP) with the anti-FLAG M2 antibody or control IgG, followed by immunoblot analysis with the indicated antibodies. Positions for marker proteins are indicated in kDa. MDCK, Madin-Darby canine kidney.
Correct spindle orientation requires recruitment of the evolutionarily conserved NuMA–LGN protein complex to the lateral cortex, which is mediated via interaction of LGN with GDP-bound Gαi (37, 38, 39), and depletion of Gαi2 or Gαi3 leads to multiple lumen formation in 3D-cultured MDCK cells due to incorrect orientation of mitotic spindles (40, 41). To know whether the lateral membrane protein GPR125 is capable of associating with Gαi, we performed a co-IP assay, which has been utilized for showing the interaction between Gα proteins and GPCRs (42, 43). As shown in Figure 7F, Gαi2 and Gαi3, each with an internal EE-tag, were coprecipitated with FLAG–GPR125 by the anti-FLAG antibody. By contrast, GPR125 did not associate with Gαs, Gαq, or Gα12 (Fig. 7F). These findings suggest that GPR125 interacts with Gαi molecules, although it remains presently unknown whether GPR125 is coupled with Gαi. The interaction with Gαi may be involved in GPR125-mediated control of mitotic spindle orientation.
Discussion
In the present study, we show that the glycoprotein GPR125, a member in the adhesion family of GPCRs, undergoes autoproteolysis at the atypical GPS motif (Fig. 1), and the products NTF and CTF are both recruited to the basolateral domain of the plasma membrane in polarized MDCK cells (Fig. 2). The recruitment requires the C-terminal PBM (Fig. 3), through which GPR125 interacts with the PDZ domain containing protein Dlg1 (Fig. 4). The cell polarity protein Dlg1 is crucial for basolateral localization of GPR125: Dlg1 colocalizes with GPR125 at cytoplasmic vesicles in unpolarized MDCK cells and traffics with GPR125 to the basolateral membrane during cell polarization, and depletion of Dlg1 abrogates plasma membrane localization of GPR125 (Fig. 5). Knockdown of GPR125 as well as that of Dlg1 results in formation of aberrant cysts with multiple lumens in Matrigel 3D culture of MDCK cells (Fig. 6). The multilumen phenotype appears to result from incorrect mitotic spindle orientation during cystogenesis, as observed in GPR125-KO MDCK cells (Fig. 7). Thus, the basolateral membrane–integrated protein GPR125 plays a crucial role in apicobasal polarization in epithelial cells.
It has been previously thought that GPR125 does not undergo autoproteolysis, based on its atypical GPS motif (Ser-Leu-Ser) instead of the canonical tripeptide His-Leu/Ileu-Ser/Thr (6, 7, 8), and thus, the possibility of its autocleavage has not been experimentally tested. Unexpectedly, the present study demonstrates that GPR125 is capable of cleaving an internal peptide bond when expressed in MDCK cells and HEK293 cells. The cleavage likely occurs at the GPS motif because it is abrogated by alanine substitution for the three residues (Fig. 1); the precise site of cleavage, however, needs to be determined in future studies. The uncleaved GPR125 possesses solely high-mannose type N-glycans and all the NTF N-glycans is of the complex type, indicating that the self-cleavage occurs during receptor biosynthesis in an intracellular compartment (Fig. 1). Like most of cleavable aGPCRs, the NTF and CTF remain associated after autoproteolysis (Fig. 2).
The precise mechanism whereby GPR125 self-cleaves at the atypical GPS motif remains presently unknown. Autoproteolysis of aGPCRs requires not only the GPS motif but also the entire GAIN domain, which fine-tunes the chemical environment for peptide bond hydrolysis (44). GAIN-mediated autoproteolysis is thought to proceed via an N–O acyl shift mechanism, in which the hydroxyl group of Thr or Ser at position +1 (respective to the cleavage site) in the GPS motif functions as a nucleophile that attacks into its N-terminal peptide bond (6, 7, 8). His at position –2 likely activates the nucleophile by deprotonating the hydroxyl group as a general base; Leu at position –1 provides a structural anchor to maintain a distorted backbone conformation by fitting its side chain into a hydrophobic pocket (44). As expected from the roles of the three residues in the GPS motif, Thr/Ser at position +1 is absolutely required for GAIN-mediated autoproteolysis; indeed, aGPCRs with replacement of Thr/Ser by Ala or Gly do not undergo autoproteolysis (6, 7, 8). On the other hand, His at position –2 does not seem to be essential as follows. First, the autoproteolytic activity of the GAIN domain in EMR2 (ADGRE2) is severely but not completely suppressed by replacement of the His with various amino acids including Ser (45), although Ala or Gly substitution for the catalytic Thr/Ser results in a complete loss of the activity of various GAIN domain (44, 45, 46). Second, the C-terminal domain of the human nucleoporin Nup98 is a GAIN-related module that has evolved from an ancient autoproteolytic domain with a His-Phe-Ser motif (47), and substitution of Ala or Gln for His-862 in the motif leads to only a slight reduction in autocleavage of Nup98, whereas a mutant protein with Ala substitution for the catalytic Ser-864 does not undergo autoproteolysis (48). Finally, self-cleavage of the FIIND, another GAIN-homologous domain (47), usually occurs at the His-Phe-Ser motif, but the Phe-Ser peptide bond in the atypical sequence Ser-Phe-Ser is cleaved in some proteins such as CARD8 and NLRP1, in which a nearby His is assumed to participate as a general base in electrophile activation instead of the canonical His residue in the tripeptide motif (49, 50). It is thus tempting to postulate that, also in GPR125, a His (or other basic) residue located sterically close to Ser-738 but not at position –2 within the GAIN domain may serve as a general base. This could explain the reason why His substitution for Ser-736 in the GPS motif of GPR125 does not facilitate autoproteolysis any more (Fig. 1).
In addition to a general base, ground state destabilization of the scissile peptide bond via conformational strain is crucial for acceleration of cis-autoproteolysis (51, 52, 53). Self-cleavage of the SEA domain of the transmembrane mucin MUC1 proceeds by applying torsional strain to the scissile bond in the absence of a general base (51). In the GAIN domain, Leu at position –1 in the GPS motif is trapped in a conserved hydrophobic pocket and thus maintains a distorted backbone conformation (44), which contributes to acceleration of GAIN-mediated autoproteolysis. It seems possible that the torsional strain on the Leu-737–Ser-738 peptide bond in GPR125 might be stronger than that in other aGPCRs and thus the His residue as a general base is less required at position –2 in the GPS motif of GPR125. This may also agree with the finding that substitution of His for Ser-736 in the GPS motif does not further enhance autoproteolysis (Fig. 1).
GPR125 is an orphan receptor. The receptor might be constitutively active to some extent; it is known that many GPCRs can signal in the absence of endogenous agonists, a phenomenon known as basal activity, and display different basal signaling activity levels that depend on the individual properties of each receptor (1, 54). Although it remains obscure how most aGPCRs are activated in endogenous tissues, their activation is currently considered to involve the two fundamental modes, i.e., orthosteric agonism and allosteric activation. In the latter models of aGPCR activation, ligand binding to an allosteric site in the NTF (but not to an orthosteric site within the 7TM helix bundle) induces receptor conformational changes, which leads to signal transduction via trimeric G proteins. On the other hand, orthosteric agonism, also referred to NTF shedding, involves NTF–CTF dissociation and subsequent action of the aGPCR tethered peptide, the core of which is the N-terminal heptapeptide of the CTF, sharing the consensus sequence Thr/Ser-Asn/Ser/Ala-Phe-Ala-Val-Leu-Met: the core residues are completely embedded within the GAIN domain in the holoreceptor form comprising both NTF and CTF; after fragment dissociation, which may be induced by a substantial force under appropriate conditions, the tethered peptide binds as an agonist intramolecularly to its orthosteric site in the 7TM domain (6, 9). Thus, autoproteolysis is crucial for tethered peptide agonism but dispensable for allosteric activation of aGPCR. The present finding of self-cleavage of GPR125 at the GPS motif raises a possibility that GPR125 also transduces signals via the mode of tethered peptide agonism. The sequence of the tethered peptide core is well conserved in GPR125: Ser-Asn-Tyr-Ala-Val-Leu-Met (Fig. 1A).
A variety of GPCRs is known to form a homodimer (19, 20), the role of which still remains uncertain especially about those of the rhodopsin-like family (21, 22). The present study shows that GPR125 is able to self-associate via the 7TM helix bundle (Fig. 1H). Although little is known whether self-association of other aGPCRs also occurs, the secretin receptor, belonging to the family (class B1) closest to the aGPCRs (class B2) in the GPCR superfamily (5, 55), forms a homodimer via the fourth transmembrane segment (TM4) (56). Both Gly-243 and Ile-247 in TM4 of the secretin receptor (Phe-Val-Ala-Phe-Gly-Trp-Gly-Ser-Pro-Ala-Ile-Phe-Val-Ala-Leu-Trp-Ala-Ile-Ala) has been shown to play a crucial role in receptor dimerization (57, 58). Intriguingly, the corresponding two residues are conserved in GPR125 TM4: Gly-883 and Ile-887 in Phe-Tyr-Leu-Ile-Gly-Gly-Gly-Ile-Pro-Ile-Ile-Val-Cys-Gly-Ile-Thr-Ala-Ala-Ala. The role for dimerization of GPR125 should be addressed in future studies.
It has been reported that GPS cleavage is necessary for membrane targeting of some aGPCRs (59, 60, 61), whereas others have shown that cleavage-deficient receptors are correctly recruited to the plasma membrane (44, 62, 63). In the case of GPR125, Ala substitution for the three residues in the GPS motif abrogates both autoproteolysis and basolateral localization (Fig. 2), suggesting that the uncleaved GAIN domain inhibits the recruitment of GPR125 to the basolateral domain of the plasma membrane. Consistent with this idea, mutant proteins lacking the GAIN domain are still targeted to the basolateral membrane (Fig. 2). Thus, it seems likely that the GAIN domain has an inhibitory role in basolateral recruitment of GPR125 and the inhibition is released by the cleavage at the GPS. Although it remains possible that Ala substitution in the GPS motif might impair the structural integrity of the GAIN domain for proper membrane trafficking, the GAIN domain per se is dispensable for basolateral recruitment of GPR125 (Fig. 2) as described above.
Basolateral localization of GPR125 requires the C-terminal PBM (Fig. 3) and its binding protein Dlg1 (a.k.a. SAP97) (Figs. 4 and 5). Although GPR124, a closest homolog of GPR125 in the aGPCR family (6, 7, 8), also interacts with Dlg1 via the C-terminal PBM, the interaction is not required for membrane localization of GPR124 (64). On the other hand, Dlg1 is involved in transport of a variety of proteins to the plasma membrane (65, 66, 67). It is also known that Dlg1 is capable of interacting with Sec8, a component of the exocyst (67, 68), which is now widely recognized as a basolateral targeting complex in epithelial cells (69). Dlg1 may recruit GPR125 to the basolateral membrane via the exocyst complex.
In zebrafish, GPR125 regulates distribution of the Dvl1 homolog Dishevelled and planar cell polarity during gastrulation (15); on the other hand, the role of GPR125 in mammalian epithelial cell polarity has not been investigated. As shown in the present study, RNAi-mediated depletion of GPR125 and ablation of the GPR125 gene each lead to formation of cysts with multiple noncentrally located apical lumens in MDCK 3D culture using Matrigel as ECM, indicative of a defect of apicobasal polarity (Figs. 6 and 7). The multilumen phenotype can occur as a result from mitotic spindle misorientation during cystogenesis (70). Simple epithelial cells such as MDCK cells orientate the mitotic spindle and cell division perpendicular to the apicobasal axis toward the central lumen, which secures the expansion of a single apical lumen at the center of the cell mass. Indeed, mitotic spindles are misoriented during cyst formation in GPR125-deficient MDCK cells (Fig. 7).
It is known that the major determinant of mitotic spindle orientation is the evolutionarily conserved NuMA–LGN–Gαi complex, which exerts pulling forces on the spindle poles by anchoring astral microtubules and dynein motors to the cell cortex (37, 38). The coiled-coil protein NuMA directly binds to both microtubules and force-generating dynein motors and is recruited to the cell cortex via its association with the adaptor protein LGN, which specifically interacts with Gαi in the GDP-bound form but not in the GTP-loaded form (37, 38, 39). Indeed, the localization of LGN to the lateral membrane in symmetrically dividing cells depends on its binding to Gαi–GDP (37, 38), and Gαi2 and Gαi3 are both crucial for proper spindle orientation in 3D-cultured MDCK cells, as their depletion results in aberrant lumen formation (40, 41). It has been postulated that specific GPCRs possibly generate a localized pool of Gαi molecules acting as a positional cue that instructs LGN–NuMA recruitment to the lateral cell cortex (38). GPR125 at the lateral membrane may play such a role because it specifically interacts with Gαi2 and Gαi3 but not with Gαs, Gαq, or Gα12 (Fig. 7). Furthermore, since Dlg1 directly binds to LGN (71) and regulates mitotic spindle orientation (33), GPR125 may function via the PBM-dependent interaction with Dlg1 (Fig. 4) for proper anchoring of the LGN–NuMA complex. Given that Dvl1 binds to NuMA in a process of spindle positioning during mitosis (72), the function of GPR125 might be also mediated via Dvl1, which interacts with the extra-PBM region of GPR125 (Fig. 4). Based on these findings, we propose a model, in which GPR125 interacts with Gαi molecules, Dlg1, and Dvl1 and thus serves as a scaffold for accumulation of the NuMA–LGN–Gαi complex at the lateral cortex, contributing to proper orientation of mitotic spindles.
The occurrence of multiple ectopic lumens, which contain surviving cells not in contact with ECM, resembles epithelial tissue phenotypes observed in preinvasive or noninvasive carcinomas (73). The multilumen phenotype induced by a loss of GPR125 in MDCK cells may imply its possible role in regulation of cancer progression. Many aGPCRs including GPR125 are mutated with high frequency in cancer (11). It has been reported that the expression of GPR125 is downregulated in colorectal cancer tissues, and patients with cancer of higher GPR125 expression have a better prognosis, suggesting that GPR125 may function as an antioncogene in colorectal cancer (13); by contrast, GPR125 is regarded as a high-risk gene in thyroid cancer (14). The evaluation of the impact of GPR125 in cancer warrants further investigation.
Experimental procedures
Plasmid construction
The cDNAs encoding human GPR125, JAM-A, Dlg1, and Dvl1 were prepared by PCR using Human Multiple Tissue cDNA panels (BD Biosciences). For expression of the N-terminally tagged transmembrane protein GPR125 or JAM-A in mammalian cells, we utilized pCMV-8 (Sigma–Aldrich) or pLVSIN-CMV-Neo (Takara Bio), in which vectors the preprotrypsin signal peptide sequence precedes the 3 × FLAG-tag or 3 × Myc-tag sequence. The cDNAs for Dlg1 and Dvl1 were ligated to the mammalian expression vectors pcDNA3 (Thermo Fisher Scientific) and pEF-BOS (74), respectively. The cDNAs for human Gαi2, Gαi3, Gαs, Gαq, and Gα12 with an internal Glu-Glu (EE)-tag were prepared and ligated into pcDNA3, as described previously (41, 75). All of the constructs were sequenced for confirmation of their identities.
Antibodies
Anti-FLAG (M2; catalog no. F3165), anti-Myc (9E10; catalog no. 11 667 203 001), and anti-β-tubulin (TUB 2.1; catalog no. T4026) mouse mAbs and anti-FLAG rabbit polyclonal antibody (catalog no. F7425) were purchased from Sigma–Aldrich; anti-GPR125 (catalog no. GTX88735) goat polyclonal antibody from GeneTex; anti-ZO-1 (R40.76; catalog no. sc-33725) rat mAb, anti-β-catenin (H-102; catalog no. sc-7199) rabbit polyclonal antibody, and anti-Dlg1 (2D11; catalog no. sc-9961) mouse mAb from Santa Cruz Biotechnology; anti-NuMA (catalog no. 3888) rabbit polyclonal antibody from Cell Signaling Technology; anti-Na+/K+-ATPase α1 subunit (EP1845Y; catalog no. ab76020) rabbit mAb, and anti-α-tubulin (YOL1/34; catalog no. ab6161) rat mAb from abcam; anti-Glu-Glu (EE) mouse mAb from Covance; and control IgG1 from DakoCytomation (catalog no. X0931). The anti-gp135 (3F2) mouse mAb (76) was generously gifted from G.K. Ojakian (State University of New York, USA).
Cell culture
MDCKII and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS).
RT-PCR
Total RNAs from MDCKII cells were extracted using TRIsure (BIOLINE) according to the instructions of the manufacturer, and 1 μg of the RNA was reverse transcribed by M-MLV Reverse Transcriptase (Thermo Fisher Scientific) using Random Hexamer primer (Sigma–Aldrich). The obtained cDNAs were subjected to PCR analysis using primer pairs for the following three regions of canine GPR125: from exons 6 to 10, 5′-TATCCCAAGTCTCTTCAGG-3′ (sense) and 5′-AGCCAGTAACTGTCGGGCTG-3′ (antisense); from exons 13 to 18, 5′-AGATCACCTGATGACTCT-3′ (sense) and 5′-TGATGCCGCACACTATGAC-3′ (antisense); and in exon 19, 5′-TACTCCATGCAGGTGGACG-3′ (sense) and 5′-CACGGCCGTCTCGTGTTTCC-3′ (antisense). Primer pairs for canine GAPDH were as follows: 5′-AACGGATTTGGCCGTATTGG-3′(sense) and 5′-AGTTGGTGGTGCAGGAGGC-3′ (antisense).
Glycosidase treatment
MDCKII cells expressing FLAG-tagged GPR125 were lysed with a lysis buffer (150 mM NaCl, 5 mM EDTA, 1 mM DTT, 1% Triton X-100, 10% glycerol, and 50 mM Tris–HCl, pH 7.5) containing Protease Inhibitor Cocktail (Sigma–Aldrich), and the lysate was centrifuged for 10 min at 20,000g. The resultant supernatant was subjected to treatment with PNGase-F (New England Biolabs) or Endo-H (New England Biolabs). Proteins were applied to 8% SDS-PAGE, followed by immunoblot analysis with the anti-FLAG (M2) antibody. The blots were developed using ImmunoStar Zeta (FUJIFILM Wako Pure Chemical Corporation) for visualization of the proteins.
Domain-selective cell surface biotinylation assay
The domain-selective cell surface biotinylation was carried out as previously described with minor modifications (77, 78). Briefly, MDCKII cells were transfected with the indicated cDNAs using Lipofectamine 2000 (Thermo Fisher Scientific) and were cultured for 3 days on a 24 mm Transwell filter (0.4 μm pore size; Corning). Cells were then washed with PBS (137 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, and 1.47 mM KH2PO4, pH 7.4) containing 1 mM MgCl2 and 0.1 mM CaCl2. Proteins at the apical or basolateral surfaces were biotinylated by the addition of 0.5 mM EZ-link Sulfo-NHS–SS–biotin (Pierce) into the upper or lower reservoir, respectively. The labeling reaction was quenched with 50 mM glycine in PBS containing 1 mM MgCl2 and 0.1 mM CaCl2, and cells were then scraped with PBS containing 1 mM EDTA and 4 mM EGTA, followed by centrifugation for 3 min at 7300g. The resultant pellets were lysed with a lysis buffer (150 mM NaCl, 5 mM EDTA, 1 mM DTT, 2% Triton X-100, 10% glycerol, and 50 mM Tris–HCl, pH 7.5) containing Protease Inhibitor Cocktail (Sigma–Aldrich), and biotinylated proteins were precipitated with streptavidin–Sepharose (GE Healthcare Biosciences). The precipitants were washed three times with the lysis buffer and analyzed by immunoblot with mAbs against FLAG, gp135, and Na+/K+-ATPase.
IP assay
IP was performed as previously described (79). Briefly, HEK293T cells and MDCKII cells were transfected using X-tremeGENE HP DNA transfection reagent (Sigma–Aldrich) with indicated cDNAs and cultured for 24 or 48 h in DMEM with 10% FCS. Cells were lysed with a lysis buffer (150 mM NaCl, 5 mM EDTA, 1 mM DTT, 1% Triton X-100, 10% glycerol, and 50 mM Tris–HCl, pH 7.5) containing Protease Inhibitor Cocktail (Sigma–Aldrich). The lysates were precipitated with the indicated antibodies in the presence of Protein G-Sepharose (GE Healthcare Biosciences). The precipitants were washed three times with the lysis buffer and analyzed by immunoblot with the indicated antibodies.
Lentiviral transfection
The lentiviral vector pLVSIN-CMV-Neo and Lentiviral High Titer Packaging Mix were obtained from Takara Bio. The lentivirus for expression of FLAG–GPR125 was generated according to the manufacture’s instruction. Briefly, HEK293T cells were cotransfected with pLVSIN-CMV-Neo-FLAG–GPR125 and Lentiviral High Titer Packaging Mix plasmids by using X-tremeGENE HP DNA Transfection Reagent (Sigma–Aldrich). After culture for 48 h, supernatants from transfected HEK293T cells were collected and filtered with a 0.45 μm filter (Millipore). The viral supernatants were added to subconfluent MDCKII cells in the presence of polybrene (2 μg/ml). After 24 h, the virus-containing medium was replaced with fresh DMEM with 10% FCS. The infected cells were cultured for another 24 h and then fixed for immunostaining.
Knockdown with siRNA
As double-strand siRNAs targeting canine GPR125 and Dlg1, the sequences on the sense strand of 25-nucleotide modified synthetic RNAs (Stealth RNAi; Thermo Fisher Scientific) used were as follows: GPR125-siRNA-1, 5′-GGAUGGACGGAUAGUUGAAACUGAU-3′; GPR125-siRNA-2, 5′-GAUUAGCAAAUGGAGCUCAUGUUUA-3′; Dlg1-siRNA-1, 5′-UCAACAGGCUGAGAAGAAGAGUUGG-3′; and Dlg1-siRNA-2, 5′-CCCAGUCAGGGAUUGAACUUCAAAU-3′. Medium GC Duplex of Stealth RNAi Negative Control Duplexes #2 (Thermo Fisher Scientific) was used as a negative control. MDCKII cells plated at 3 × 104/cm2 were transfected with siRNA (7.2 pmol) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) and cultured for 48 to 72 h.
Generation of GPR125-KO MDCK cell lines
Guide RNAs for CRISPR/Cas9-mediated KO of GPR125 were designed using the guide RNA selection tool CRISPOR (34). For expression of the designed single-guide RNA (sgRNA) in MDCKII cells, the following double-stranded synthetic oligomers were ligated to pSpCas9(BB)-2A-Puro (PX459) vector (gift from Feng Zhang, Addgene, catalog no. #48139) (80): GPR125-sgRNA#1; 5′-CACCGCTCATTAGTAGTATAGACCC-3′ (sense) and 5′-AAACGTCATTTTTAGGATTCTGAGC-3′ (antisense); GPR125-sgRNA#2; 5′-CACCGCGGCAGTCGTATAAACCACG-3′ (sense) and 5′-AAACCGTGGTTTATACGACTGCCGC-3′ (antisense). With these plasmids, MDCKII cells were transfected using Amaxa Nucleofector (Lonza) and selected in the presence of puromycin (3 μg/ml; EMD Millipore) for 2 days. Cells were then cloned and cultured for another 2 weeks. Successful gene editing was confirmed by Sanger sequencing of purified PCR products amplifying the sgRNA target sites from edited and wild-type samples (see Fig. S1). The transient expression of GPR125-sgRNA#1 or GPR125-sgRNA#2 established the MDCK clone GPR125-KO-1 or GPR125-KO-2, respectively.
Calcium switch assay
The calcium switch assay was performed as previously described (32). Briefly, MDCKII cells (5.0 × 105) were seeded on a 35 mm glass bottom dish (Matsunami Glass) and grown in DMEM with 10% FCS containing 1.8 mM calcium (the normal calcium medium) for the formation of confluent monolayers. The monolayer cells were cultured for 24 h in the low calcium medium containing 2.1 μM calcium (S-MEM; Thermo Fisher Scientific) supplemented with dialyzed 5% FCS. The calcium switch assay was initiated by incubation of the cells in the normal calcium medium.
Immunofluorescence microscopy
Immunofluorescence microscopy was performed as previously described (32, 41). For staining of β-catenin, ZO-1, gp135, Dlg1, and FLAG-tagged proteins, MDCKII cells were fixed for 10 min in 2% formaldehyde and permeabilized for 30 min with 0.5% Triton X-100 in PBS containing 3% bovine serum albumin (BSA). For 3D culture, MDCKII cells were trypsinized to a single cell suspension of 1.6 × 104 cells/ml in 5% Matrigel, containing laminin, type IV collagen, and entactin (Corning Costar), and 250 μl of the suspension was plated in an 8-well cover glass chamber (Iwaki) precoated with 40 μl of Matrigel. For staining of gp135 and β-catenin, MDCKII cells plated in Matrigel were fixed for 30 min in 4% paraformaldehyde and subsequently permeabilized for 30 min in PBS containing 0.5% Triton X-100 and 3% BSA. For staining of NuMA and α-tubulin, MDCKII cells in Matrigel were fixed for 2 min in 100% methanol at 4 °C and then for 30 min in 4% paraformaldehyde, followed by blocking for 2 h with PBS containing 3% BSA. Indirect immunofluorescence analysis was performed using the following secondary antibodies: Alexa Fluor 488–labeled goat anti-rabbit or anti-mouse IgG antibodies (Thermo Fisher Scientific), Alexa Fluor 594–labeled goat anti-rabbit, anti-mouse, or anti-rat IgG antibodies (Thermo Fisher Scientific), and Alexa Fluor 594–labeled donkey anti-rabbit IgG (Thermo Fisher Scientific). Nuclei were stained with Hoechst 33342 (Thermo Fisher Scientific). Confocal images were captured at room temperature on the confocal microscope LSM700 (Carl Zeiss) and analyzed using ZEN (Carl Zeiss). The microscopes were equipped with a Plan-Apochromat 63 × /1.4 NA oil-immersion objective lens. For the analysis of cyst morphogenesis of MDCKII cells, more than 75 cysts were tested: A normal cyst had a single lumen surrounded by cells that exhibit both intense gp135 staining at the apical surface and β-catenin staining at the surface facing the ECM. To discriminate lumens from cytoplasmic gp135-rich vesicles/vacuoles, we defined a lumen as a gp135-rich structure that makes a contact with β-catenin-containing membranes.
Measurement of mitotic spindle orientation
The measurement of spindle angle was performed as previously described (32, 41). In brief, MDCK cysts grown for 48 h were fixed and stained with anti-NuMA, anti-α-tubulin, and anti-gp135 antibodies. The acute angle between the spindle axis and the line perpendicular to the apical surface was measured by Fiji/ImageJ (version 2.0; NIH).
Statistical analysis
Statistical differences were determined as follows: data for cystogenesis and localization of FLAG–GPR125 in cells transfected with siRNA were analyzed one-way ANOVA with Dunnett’s multiple comparison of mean test, and data for analysis of spindle orientation were analyzed by Steel–Dwass test.
Data availability
All of the data are contained within the article and the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr George K. Ojakian (State University of New York, USA) for the gp135 (3F2) antibody; Namiko Kubo (Kyushu University) for technical assistance; and Hiromi Takeyama (Kyushu University) for secretarial assistance. We also appreciate for technical supports from the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences.
Author contributions
T. S., S. K., and H. S. conceptualization; S. K., J. H., A. K., and H. S. methodology; T. S., S. K., J. H., A. K., and H. S. formal analysis; T. S., S. K., J. H., and A. K. investigation; S. K., J. H., A. K., and H. S. resources; T. S., S. K., J. H., A. K., and H. S. data curation; T. S., S. K., and H. S. writing–original draft; T. S., S. K., J. H., A. K., M. N., and H. S. writing–review and editing; T. S., S. K., J. H. and A. K. visualization; S. K., J. H., A. K., M. N., and H. S. supervision; H. S. project administration; S. K. and H. S. funding acquisition.
Funding and additional information
This work was supported in part by JSPS (Japan Society for the Promotion of Science) KAKENHI Grants JP21H02698 (to H. S.) and JP22K06901 (to S. K.), and by Grant-in-Aid for Transformative Research Areas [A] (21H05267) (to H. S.).
Edited by Henrik Dohlman
Supporting information
References
- 1.Hilger D., Masureel M., Kobilka B.K. Structure and dynamics of GPCR signaling complex. Nat. Struct. Mol. Biol. 2018;25:4–12. doi: 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatakrishnan A.J., Deupi X., Lebon G., Tate C.G., Schertler G.F., Babu M.M. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 3.Sutkeviciute I., Vilardaga J.P. Structural insights into emergent signaling modes of G protein-coupled receptors. J. Biol. Chem. 2020;295:11626–11642. doi: 10.1074/jbc.REV120.009348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wootten D., Christopoulos A., Marti-Solano M., Babu M.M., Sexton P.M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2018;19:638–653. doi: 10.1038/s41580-018-0049-3. [DOI] [PubMed] [Google Scholar]
- 5.Nordström K.J., Sällman Almén M., Edstam M.M., Fredriksson R., Schiöth H.B. Independent HHsearch, Needleman–Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol. Biol. Evol. 2011;28:2471–2480. doi: 10.1093/molbev/msr061. [DOI] [PubMed] [Google Scholar]
- 6.Vizurraga A., Adhikari R., Yeung J., Yu M., Tall G.G. Mechanisms of adhesion G protein-coupled receptor activation. J. Biol. Chem. 2020;295:14065–14083. doi: 10.1074/jbc.REV120.007423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langenhan T., Aust G., Hamann J. Sticky signaling–adhesion class G protein-coupled receptors take the stage. Sci. Signal. 2013;6:re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- 8.Hamann J., Aust G., Araç D., Engel F.B., Formstone C., Fredriksson R., et al. International union of basic and clinical pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosa M., Noel T., Harris M., Ladds G. Emerging roles of adhesion G protein-coupled receptors. Biochem. Soc. Trans. 2021;49:1695–1709. doi: 10.1042/BST20201144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seandel M., James D., Shmelkov S.V., Falciatori I., Kim J., Chavala S., et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu V., Yeerna H., Nohata N., Chiou J., Harismendy O., Raimondi F., et al. Illuminating the Onco-GPCRome: novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019;294:11062–11086. doi: 10.1074/jbc.REV119.005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu J.F., Yen T.H., Chen Y., Huang Y.J., Hsu C.L., Liang D.C., et al. Involvement of Gpr125 in the myeloid sarcoma formation induced by cooperating MLL/AF10(OM-LZ) and oncogenic KRAS in a mouse bone marrow transplantation model. Int. J. Cancer. 2013;133:1792–1802. doi: 10.1002/ijc.28195. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y., Chen W., Gong L., Ke C., Wang H., Cai Y. Elevated G-protein receptor 125 (GPR125) expression predicts good outcomes in colorectal cancer and inhibits Wnt/β-catenin signaling pathway. Med. Sci. Monit. 2018;24:6608–6616. doi: 10.12659/MSM.910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H., Zhang S., Shao S., Fang H. Identification of a prognostic 3-gene risk prediction model for thyroid cancer. Front. Endocrinol. 2020;11:510. doi: 10.3389/fendo.2020.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Roszko I., Sepich D.S., Ni M., Hamm H.E., Marlow F.L., et al. Gpr125 modulates Dishevelled distribution and planar cell polarity signaling. Development. 2013;140:3028–3039. doi: 10.1242/dev.094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto Y., Irie K., Asada M., Mino A., Mandai K., Takai Y. Direct binding of the human homologue of the Drosophila disc large tumor suppressor gene to seven-pass transmembrane proteins, tumor endothelial marker 5 (TEM5), and a novel TEM5-like protein. Oncogene. 2004;23:3889–3897. doi: 10.1038/sj.onc.1207495. [DOI] [PubMed] [Google Scholar]
- 17.Cazorla-Vázquez S., Engel F.B. Adhesion GPCRs in kidney development and disease. Front. Cell Dev. Biol. 2018;6:9. doi: 10.3389/fcell.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breitling J., Aebi M. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013;5:a013359. doi: 10.1101/cshperspect.a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milligan G. The prevalence, maintenance, and relevance of G protein-coupled receptor oligomerization. Mol. Pharmacol. 2013;84:158–169. doi: 10.1124/mol.113.084780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferré S., Casadó V., Devi L.A., Filizola M., Jockers R., Lohse M.J., et al. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurevich V.V., Gurevich E.V. GPCRs and signal transducers: interaction stoichiometry. Trends Pharmacol. Sci. 2018;39:672–684. doi: 10.1016/j.tips.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan G., Ward R.J., Marsango S. GPCR homo-oligomerization. Curr. Opin. Cell Biol. 2019;57:40–47. doi: 10.1016/j.ceb.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuncay H., Ebnet K. Cell adhesion molecule control of planar spindle orientaion. Cell Mol. Life Sci. 2018;73:1195–1207. doi: 10.1007/s00018-015-2116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley P. Golgi glycosylation. Cold Spring Harb. Perspect. Biol. 2011;3:a005199. doi: 10.1101/cshperspect.a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beliu G., Altrichter S., Guixà-González R., Hemberger M., Brauer I., Dahse A.K., et al. Tethered agonist exposure in intact adhesion/class B2 GPCRs through intrinsic structural flexibility of the GAIN domain. Mol. Cell. 2021;81:905–921. doi: 10.1016/j.molcel.2020.12.042. [DOI] [PubMed] [Google Scholar]
- 26.Ping Y.Q., Xiao P., Yang F., Zhao R.J., Guo S.C., Yan X., et al. Structural basis for the tethered peptide activation of adhesion GPCRs. Nature. 2022;604:763–770. doi: 10.1038/s41586-022-04619-y. [DOI] [PubMed] [Google Scholar]
- 27.Xiao P., Guo S., Wen X., He Q.T., Lin H., Huang S.M., et al. Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4. Nature. 2022;604:771–778. doi: 10.1038/s41586-022-04590-8. [DOI] [PubMed] [Google Scholar]
- 28.Barros-Álvarez X., Nwokonko R.M., Vizurraga A., Matzov D., He F., Papasergi-Scott M.M., et al. The tethered peptide activation mechanism of adhesion GPCRs. Nature. 2022;604:757–762. doi: 10.1038/s41586-022-04575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu X., Qiu N., Wang M., Zhang B., Du J., Zhong Z., et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature. 2022;604:779–785. doi: 10.1038/s41586-022-04580-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung A.Y., Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 31.Walch L. Emerging role of the scaffolding protein Dlg1 in vesicle trafficking. Traffic. 2013;14:964–973. doi: 10.1111/tra.12089. [DOI] [PubMed] [Google Scholar]
- 32.Hayase J., Kamakura S., Iwakiri Y., Yamaguchi Y., Izaki T., Ito T., et al. The WD40 protein Morg1 facilitates Par6–aPKC binding to Crb3 for apical identity in epithelial cells. J. Cell Biol. 2013;200:635–650. doi: 10.1083/jcb.201208150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter A.P., White G.R.M., Mack N.A., Malliri A. The interaction between CASK and the tumour suppressor Dlg1 regulates mitotic spindle orientation in mammalian epithelia. J. Cell Sci. 2019;132:jcs230086. doi: 10.1242/jcs.230086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.B., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffe A.B., Kaji N., Durgan J., Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z., Zhu H., Wan Q., Liu J., Xiao Z., Siderovski D.P., et al. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J. Cell Biol. 2010;189:275–288. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.di Pietro F., Echard A., Morin X. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 2016;17:1106–1130. doi: 10.15252/embr.201642292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechler T., Mapelli M. Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat. Rev. Mol. Cell Biol. 2021;22:691–708. doi: 10.1038/s41580-021-00384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takayanagi H., Hayase J., Kamakura S., Miyano K., Chishiki K., Yuzawa S., et al. Intramolecular interaction in LGN, an adaptor protein that regulates mitotic spindle orientation. J. Biol. Chem. 2019;294:19655–19666. doi: 10.1074/jbc.RA119.011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki K., Kakuwa T., Akimoto K., Koga H., Ohno S. Regulation of epithelial cell polarity by PAR-3 depends on Girdin transcription and Girdin–Gαi3 signaling. J. Cell Sci. 2015;128:2244–2258. doi: 10.1242/jcs.160879. [DOI] [PubMed] [Google Scholar]
- 41.Chishiki K., Kamakura S., Hayase J., Sumimoto H. Ric-8A, an activator protein of Gαi, controls mammalian epithelial cell polarity for tight junction assembly and cystogenesis. Genes Cells. 2017;22:293–309. doi: 10.1111/gtc.12477. [DOI] [PubMed] [Google Scholar]
- 42.Antolin-Fontes B., Li K., Ables J.L., Riad M.H., Görlich A., Williams M., et al. The habenular G-protein-coupled receptor 151 regulates synaptic plasticity and nicotine intake. Proc. Natl. Acad. Sci. U. S. A. 2020;117:5502–5509. doi: 10.1073/pnas.1916132117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Gai S., Zhang W., Huang X., Ma S., Huo Y., et al. The GABAB receptor mediates neuroprotection by coupling to G13. Sci. Signal. 2021;14 doi: 10.1126/scisignal.aaz4112. [DOI] [PubMed] [Google Scholar]
- 44.Araç D., Boucard A.A., Bolliger M.F., Nguyen J., Soltis S.M., Südhof T.C., et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H.H., Chang G.W., Davies J.Q., Stacey M., Harris J., Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 46.Wei W., Hackmann K., Xu H., Germino G., Qian F. Characterization of cis-autoproteolysis of polycystin-1, the product of human polycystic kidney disease 1 gene. J. Biol. Chem. 2007;282:21729–21737. doi: 10.1074/jbc.M703218200. [DOI] [PubMed] [Google Scholar]
- 47.Liao Y., Pei J., Cheng H., Grishin N.V. An ancient autoproteolytic domain found in GAIN, ZU5 and Nucleoporin98. J. Mol. Biol. 2014;426:3935–3945. doi: 10.1016/j.jmb.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodel A.E., Hodel M.R., Griffis E.R., Hennig K.A., Ratner G.A., Xu S., et al. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 49.D'Osualdo A., Weichenberger C.X., Wagner R.N., Godzik A., Wooley J., Reed J.C. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finger J.N., Lich J.D., Dare L.C., Cook M.N., Brown K.K., Duraiswami C., et al. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 2012;287:25030–25037. doi: 10.1074/jbc.M112.378323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macao B., Johansson D.G., Hansson G.C., Härd T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 52.Johansson D.G., Wallin G., Sandberg A., Macao B., Aqvist J., Härd T. Protein autoproteolysis: conformational strain linked to the rate of peptide cleavage by the pH dependence of the N → O acyl shift reaction. J. Am. Chem. Soc. 2009;131:9475–9477. doi: 10.1021/ja9010817. [DOI] [PubMed] [Google Scholar]
- 53.Buller A.R., Freeman M.F., Wright N.T., Schildbach J.F., Townsend C.A. Insights into cis-autoproteolysis reveal a reactive state formed through conformational rearrangement. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2308–2313. doi: 10.1073/pnas.1113633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manglik A., Kobilka B. The role of protein dynamics in GPCR function: insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens R.C., Cherezov V., Katritch V., Abagyan R., Kuhn P., Rosen H., et al. The GPCR network: a large-scale collaboration to determine human GPCR structure and function. Nat. Rev. Drug Discov. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harikumar K.G., Happs R.M., Miller L.J. Dimerization in the absence of higher-order oligomerization of the G protein-coupled secretin receptor. Biochim. Biophys. Acta. 2008;1778:2555–2563. doi: 10.1016/j.bbamem.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harikumar K.G., Pinon D.I., Miller L.J. Transmembrane segment IV contributes a functionally important interface for oligomerization of the Class II G protein-coupled secretin receptor. J. Biol. Chem. 2007;282:30363–30372. doi: 10.1074/jbc.M702325200. [DOI] [PubMed] [Google Scholar]
- 58.Ward R.J., Pediani J.D., Harikumar K.G., Miller L.J., Milligan G. Spatial intensity distribution analysis quantifies the extent and regulation of homodimerization of the secretin receptor. Biochem. J. 2017;474:1879–1895. doi: 10.1042/BCJ20170184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krasnoperov V., Lu Y., Buryanovsky L., Neubert T.A., Ichtchenko K., Petrenko A.G. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J. Biol. Chem. 2002;277:46518–46526. doi: 10.1074/jbc.M206415200. [DOI] [PubMed] [Google Scholar]
- 60.Jin Z., Tietjen I., Bu L., Liu-Yesucevitz L., Gaur S.K., Walsh C.A., et al. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum. Mol. Genet. 2007;16:1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- 61.Liebscher I., Schön J., Petersen S.C., Fischer L., Auerbach N., Demberg L.M., et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014;9:2018–2026. doi: 10.1016/j.celrep.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang G.W., Stacey M., Kwakkenbos M.J., Hamann J., Gordon S., Lin H.H. Proteolytic cleavage of the EMR2 receptor requires both the extracellular stalk and the GPS motif. FEBS Lett. 2003;547:145–150. doi: 10.1016/s0014-5793(03)00695-1. [DOI] [PubMed] [Google Scholar]
- 63.Bohnekamp J., Schöneberg T. Cell adhesion receptor GPR133 couples to Gs protein. J. Biol. Chem. 2011;286:41912–41916. doi: 10.1074/jbc.C111.265934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Posokhova E., Shukla A., Seaman S., Volate S., Hilton M.B., Wu B., et al. GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Rep. 2015;10:123–130. doi: 10.1016/j.celrep.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leonoudakis D., Conti L.R., Radeke C.M., McGuire L.M., Vandenberg C.A. A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels. J. Biol. Chem. 2004;279:19051–19063. doi: 10.1074/jbc.M400284200. [DOI] [PubMed] [Google Scholar]
- 66.Underhill S.M., Wheeler D.S., Amara S.G. Differential regulation of two isoforms of the glial glutamate transporter EAAT2 by DLG1 and CaMKII. J. Neurosci. 2015;35:5260–5270. doi: 10.1523/JNEUROSCI.4365-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolis A., Coviello S., Visigalli I., Taveggia C., Bachi A., Chishti A.H., et al. Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J. Neurosci. 2009;29:8858–8870. doi: 10.1523/JNEUROSCI.1423-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inoue M., Chiang S.H., Chang L., Chen X.W., Saltiel A.R. Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol. Biol. Cell. 2006;17:2303–2311. doi: 10.1091/mbc.E06-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Polgar N., Fogelgren B. Regulation of cell polarity by exocyst-mediated trafficking. Cold Spring Harb. Perspect. Biol. 2018;10:a031401. doi: 10.1101/cshperspect.a031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overeem A.W., Bryant D.M., van IJzendoorn S.C. Mechanisms of apical–basal axis orientation and epithelial lumen positioning. Trends Cell Biol. 2015;25:476–485. doi: 10.1016/j.tcb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 71.Saadaoui M., Machicoane M., di Pietro F., Etoc F., Echard A., Morin X. Dlg1 controls planar spindle orientation in the neuroepithelium through direct interaction with LGN. J. Cell Biol. 2014;206:707–717. doi: 10.1083/jcb.201405060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y., Liu M., Li D., Ran J., Gao J., Suo S., et al. CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled-NuMA-dynein/dynactin complex formation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2158–2163. doi: 10.1073/pnas.1319341111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Debnath J., Brugge J.S. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 74.Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucl. Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chishiki K., Kamakura S., Yuzawa S., Hayase J., Sumimoto H. Ubiquitination of the heterotrimeric G protein α subunits Gαi2 and Gαq is prevented by the guanine nucleotide exchange factor Ric-8A. Biochem. Biophys. Res. Commun. 2013;435:414–419. doi: 10.1016/j.bbrc.2013.04.103. [DOI] [PubMed] [Google Scholar]
- 76.Ojakian G.K., Schwimmer R. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 1988;107:2377–2387. doi: 10.1083/jcb.107.6.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lisanti M.P., Le Bivic A., Sargiacomo M., Rodriguez-Boulan E. Steady-state distribution and biogenesis of endogenous Madin-darby canine kidney glycoproteins: evidence for intracellular sorting and polarized cell surface delivery. J. Cell Biol. 1989;109:2117–2127. doi: 10.1083/jcb.109.5.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miyano K., Sumimoto H. N-linked glycosylation of the superoxide-producing NADPH oxidase Nox1. Biochem. Biophys. Res. Commun. 2014;443:1060–1065. doi: 10.1016/j.bbrc.2013.12.086. [DOI] [PubMed] [Google Scholar]
- 79.Kamakura S., Nomura M., Hayase J., Iwakiri Y., Nishikimi A., Takayanagi R., et al. The cell polarity protein mInsc regulates neutrophil chemotaxis via a noncanonical G protein signaling pathway. Dev. Cell. 2013;26:292–302. doi: 10.1016/j.devcel.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 80.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are contained within the article and the supporting information.