Abstract
Cell extracts of Methanococcus jannaschii have been shown to readily convert l-ornithine to l-proline. This cyclization reaction proceeds with the loss of only the C-2 nitrogen, as has been documented for ornithine cyclodeaminase (EC 4.3.1.12). Since no gene homologous to that coding for ornithine cyclodeaminase is present in the genome of M. jannaschii, these results indicate that proline biosynthesis in M. jannaschii is accomplished by a previously unrecognized enzyme.
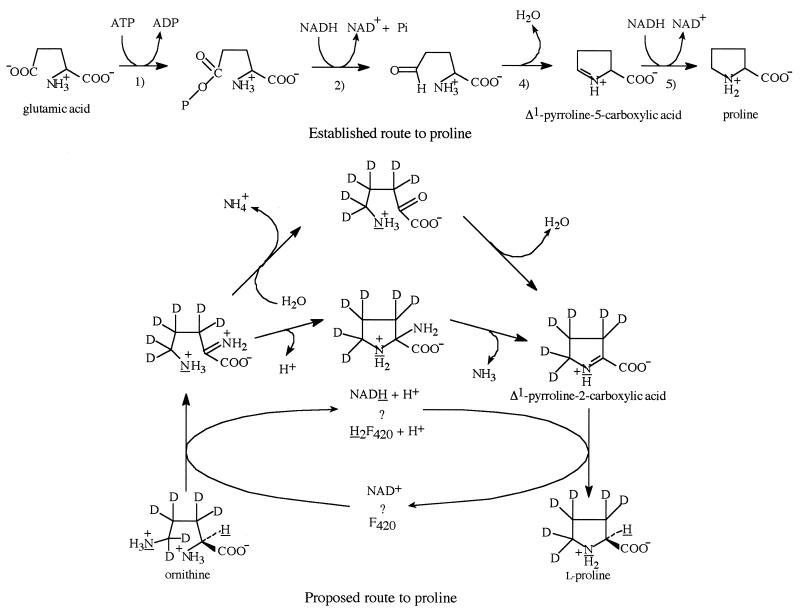
The established pathway for proline biosynthesis in microorganisms is shown in the upper portion of Fig. 1. The reaction sequence involves (i) the phosphorylation of the δ-carboxyl of l-glutamate to form l-glutamyl-5-P, (ii) the NADH-dependent reduction of l-glutamyl-5-P to glutamic acid-γ-semialdehyde, (iii) the cyclization of glutamate-γ-semialdehyde to Δ1-pyrroline-5-carboxylic acid, and (iv) the reduction of Δ1-pyrroline-5-carboxylic acid to l-proline (2, 5, 13). As first pointed out by Selkov et al. for Methanococcus jannaschii (10), and later as a general characteristic of the genomes of most of the Archaea, genes coding for the three enzymes in this pathway are largely absent in the Archaea (4). In contrast, the genes for the biosynthesis of l-ornithine are generally present in all of the archaeal genomes (4). A simple solution to explain the absence of the proline biosynthetic genes in the genomes of some of the Archaea is that proline is derived by the cyclization of l-ornithine. This reaction could be accomplished by ornithine cyclodeaminase (EC 4.3.1.12), an enzyme which is presently considered to have a limited distribution among the bacteria (12). Ornithine cyclodeaminase was first isolated from Clostridium sporogenes, where it functions as the first step in the anaerobic catabolism of l-ornithine via proline to δ-aminovaleric acid (1). The proposed chemical steps for the mechanism of this enzyme involve the oxidative deamination of the α-amino group of ornithine to 2-oxo-5-aminopentanoic acid, which cyclizes to Δ1-pyrroline-2-carboxylic acid, which is subsequently reduced to l-proline. The ornithine cyclodeaminase has been shown to contain 1 mol equivalent of bound NAD+, which is considered to function as a recycling redox carrier in this transformation (7). Mass spectroscopic data showed that [5-15N]ornithine is converted to [15N]proline by ornithine cyclodeaminase, confirming that the initial oxidation of the ornithine is at C-2 (8). Using deuterated ornithine and [15N]ornithine, we have now demonstrated that proline in M. jannaschii is derived from ornithine by a mechanism that is analogous to that demonstrated by the ornithine cyclodeaminase. Since there is no enzyme-encoding gene with a sequence homologous to that coding for the ornithine cyclodeaminase present in the M. jannaschii genome, we propose that a currently unidentified enzyme, functioning with an analogous mechanism, is involved in proline biosynthesis in M. jannaschii.
FIG. 1.
Established and proposed pathways for proline biosynthesis.
Preparation and analysis of cell extracts.
Cell extracts of M. jannaschii, Methanosarcina thermophila strain TM-1, and Methanobacterium thermoautotrophicum strains ΔH and Marburg were prepared as previously described (16). The protein concentrations of the cell extracts used typically ranged from 7 to 26 mg/ml.
Incubation with substrates.
Cell extracts (50 μl) were incubated with millimolar concentrations of the substrates under argon for 2 h at 50°C. l-[2,4,4′-2H3]glutamic acid and l-[3,3′,4,4′,5′-2H6]ornithine were obtained from Cambridge Isotope Laboratories, Inc. [5-15N]ornithine was prepared from potassium [15N]-phthalimide by the following series of reactions. Potassium [15N]-phthalimide (98 atom% 15N) was reacted with dibromopropane in acetone to form 15N-labeled N-(3-bromopropyl)phthalimide (14), which was condensed with ethyl acetamidocyanoacetate in ethanol in the presence of sodium ethoxide. Acid hydrolysis of the condensation product (6 M HCl, 24 h, 110°C) and separation of the resulting products on a Dowex 50-8X (H+) column with an HCl gradient resulted in the isolation of chromatographically pure [5-15N]ornithine. Δ1-Pyrroline-5-carboxylic acid was prepared from the 2,4-dinitrophenylhydrazine derivative as previously described (6).
After incubation, 0.1 M HCl in methanol (200 μl) was added, followed by centrifugation (10 min, 14,000 × g) to remove the precipitated proteins. The resulting clear liquid was evaporated to dryness with a stream of nitrogen gas, and the free amino acids contained within were converted into the methyl ester trifluoroacetyl derivatives and analyzed by gas chromatography-mass spectrometry (GC-MS) as previously described (17). Quantitation of proline and ornithine was determined from the areas of the intensities of their m/z 166 ions, or the m/z 167 or m/z 172 ions for the labeled prolines, using known mixtures of proline and ornithine for calibration. For samples not containing ornithine, the m/z 211 ion from the β-glutamate present in the M. jannaschii cell extracts (9) was used as an internal standard. The establishment of the product of the incubation as l-proline was accomplished by GC-MS of the methyl ester trifluoroacetyl derivative using a type G-TA Chiraldex column as previously described (3).
As can be seen from the data presented in Table 1, incubation of a cell extract of M. jannaschii with [2,4,4′-2H3]glutamic acid, ATP, NADH, and NADPH produced no detectable amount of labeled proline. Likewise, incubation with Δ1-pyrroline-5-carboxylic acid, NADH, and NADPH at the same concentrations failed to produce any detectable amount of proline (data not shown). Incubation of the cell extract with l-ornithine (9.1 mM) resulted in the production of 0.13 μmol of proline, which corresponded to the conversion of 26% of ornithine to proline. To confirm that ornithine was the sole precursor of the proline, we incubated the cell extracts with l-[3,3′,4,4′,5,5′-2H6]ornithine and measured the incorporation of six deuteriums in the generated proline, which indicated that the carbon skeleton of the proline was derived from the ornithine as an intact unit. To establish which of the nitrogens was lost in the cyclization, the experiment was repeated with [5-15N]ornithine, and the recovered proline contained 98% 15N. This result showed that the C-2 nitrogen was the one lost in the cyclization. Since the direct displacement of the C-2 amino group by the C-5 amino group is without enzymatic or chemical precedent, the most likely chemical steps for the reaction would involve the oxidation of the C-2 carbon to an imine, intramolecular cyclic addition of the C-5 nitrogen to the C-2 carbon, loss of ammonia, and reduction of the resulting imine, Δ1-pyrroline-2-carboxylic acid, as shown in lower portion of Fig. 1. Alternately, the imine intermediate generated in the first oxidation could undergo hydrolysis to the keto acid before the cyclization would occur. In this case, elimination of water would produce the Δ1-pyrroline-2-carboxylic acid. Although not directly confirmed by the data, the likely choice for the coenzyme to be involved in this process would be an enzyme-bound NAD+ as occurs in ornithine cyclohydrolase. The possible involvement of coenzyme F420 must also be considered since coenzyme F420 can also effect hydride transfer reactions (15).
TABLE 1.
Proline formation in cell extracts of M. jannaschii
| Expt | Precursor(s) | Proline produced (μmol) | % of substrate converted into proline |
|---|---|---|---|
| 1 | l-[2,4,4′-2H3]glutamic acid (7.7 mM), ATP (7.7 mM), NADH, and NADPH (7.7 mM) | 0.01a | <0.024 |
| 2 | l-Ornithine (9.1 mM) | 0.13 | 26 |
| 3 | l-[3,3′,4,4′,5,5′-2H6]ornithine (9.1 mM) | 0.19b | 38 |
| 4 | [2-15N]ornithine (9.1 mM) | 0.19c | 38 |
| 5 | l-Ornithine (9.1 mM) and 40% 2H2O | 0.13d | 20 |
The proline contained <0.1% of deuterated prolines. The small amount of unlabeled proline observed resulted from the proline which was present in the cell extract.
All proline molecules contained six deuterium atoms.
The recovered proline had the same 15N abundance as the precursor ornithine.
The recovered proline had no detectable deuterium (<1%).
Evidence supporting the involvement of either NAD+ or F420 in the reaction comes from the experiment demonstrating that the hydrogen removed during the C-2 oxidation is reincorporated at C-2 in the proline product during the reduction. Thus, the incubation of a cell extract containing 40% deuterated water with ornithine produced proline with no deuterium (Table 1, data for precursor 5). The conclusion from this experiment is that the hydrogen, which is removed from C-2, is not mixed with the solvent and is the same hydrogen that is incorporated in the reduction. These data are consistent with the idea that the enzyme in M. jannaschii functions with a mechanism analogous to that of ornithine cyclodeaminase from C. sporogenes. Evidence supporting the involvement of NAD+ was the observation that NADH was found to inhibit the conversion presumably by competing with the required NAD+ (data not shown).
Similar data were also obtained using cell extracts of M. thermoautotrophicum strain ΔH and strain Marburg, indicating that these autotrophs generate their proline by the same mechanism as that found in M. jannaschii. Although no gene coding for ornithine cyclodeaminase can be found in the M. jannaschii genome, a gene homologous to one coding for C. sporogenes ornithine cyclodeaminase is present in the M. thermoautotrophicum ΔH (11) and Methanosarcina barkeri (http://www.jgi.doe.gov) genomes. Cell extracts of M. thermophila strain TM-1, on the other hand, were found to produce no proline from ornithine. The M. barkeri genome also has the genes for the biosynthesis of proline from glutamate via Δ1-pyrroline-5-carboxylic acid. These genomic data indicate that several different routes may be operating in the Archaea for the biosynthesis of proline. Although our data cannot rule out the possibility that additional pathways to proline function in vivo, this seems unlikely due to the efficient conversions from ornithine that were observed in vitro.
In total, these observations show that an enzyme with no homology to the known ornithine cyclodeaminases is involved in proline biosynthesis in M. jannaschii. We are presently in the process of isolating the enzyme responsible for carrying out the reaction demonstrated here in order to establish the gene required for its production.
Acknowledgments
We thank Kim Harich for the GC-MS analyses and David Graham for reviewing the manuscript prior to publication. We thank James G. Ferry for the cells of M. thermophila strain TM-1, Ralph S. Wolfe for M. thermoautotrophicum strains ΔH and Marburg, and Biswarup Mukhopadhyay for cells of M. jannaschii.
This work was supported in part by National Science Foundation grant MCB9985712.
REFERENCES
- 1.Costilow R N, Laycock L. Ornithine cyclase (deaminating) J Biol Chem. 1971;246:6655–6660. [PubMed] [Google Scholar]
- 2.Csonka L N, Baich A. Proline biosynthesis. In: Herrmann K M, Somerville R L, editors. Amino acids biosynthesis and genetic regulation. Reading, Mass: Addison-Wesley Publishing Co.; 1983. pp. 35–51. [Google Scholar]
- 3.Graupner M, Xu H, White R H. Identification of an archaeal 2-hydroxy acid dehydrogenase catalyzing reactions involved in coenzyme biosynthesis in methanoarchaea. J Bacteriol. 2000;182:3688–3692. doi: 10.1128/jb.182.13.3688-3692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higuchi S, Kawashima T, Suzuki M. Comparison of pathways for amino acid biosynthesis in archaebacteria using their genomic DNA sequences. Proc Jpn Acad Ser B. 1999;75:241–245. [Google Scholar]
- 5.Meister A. Biochemistry of the amino acids. 2nd ed. II. New York, N.Y: Academic Press, Inc.; 1965. pp. 707–715. [Google Scholar]
- 6.Mezl V A, Knox W E. Properties and analysis of a stable derivative of pyrroline-5-carboxylic acid for use in metabolic studies. Anal Biochem. 1976;74:430–440. doi: 10.1016/0003-2697(76)90223-2. [DOI] [PubMed] [Google Scholar]
- 7.Muth W L, Costilow R N. Ornithine cyclase (deaminating). II. Properties of the homogeneous enzyme. J Biol Chem. 1974;249:7457–7462. [PubMed] [Google Scholar]
- 8.Muth W L, Costilow R N. Ornithine cyclase (deaminating). III. Mechanism of the conversion of ornithine to proline. J Biol Chem. 1974;249:7463–7467. [PubMed] [Google Scholar]
- 9.Robertson D E, Roberts M F, Belay N, Stetter K O, Boone D R. Occurrence of β-glutamate, a novel osmolyte, in marine methanogenic bacteria. Appl Environ Microbiol. 1990;56:1504–1508. doi: 10.1128/aem.56.5.1504-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selkov E, Maltsev N, Olsen G J, Overbeek R, Whitman W B. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–GC26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 11.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soto M, van Dillewijn P, Olivares J, Toro N. Ornithine cyclodeaminase activity in Rhizobium meliloti. FEMS Microbiol Lett. 1994;119:209–214. [Google Scholar]
- 13.Umbarger H E. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:532–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 14.Volford J, Banfi D. Synthesis of 15N-labeled 2-substituted 2-thiasolines and analogous thiazines. J Label Compd. 1975;11:419–426. [Google Scholar]
- 15.Walsh C. Naturally occurring 5-deazaflavin coenzymes: biological redox roles. Acc Chem Res. 1986;19:216–221. [Google Scholar]
- 16.White R H. Methanopterin biosynthesis: methylation of the biosynthetic intermediates. Biochim Biophys Acta. 1998;1380:257–267. doi: 10.1016/s0304-4165(97)00148-7. [DOI] [PubMed] [Google Scholar]
- 17.Zheng L, White R H, Cash V L, Dean D R. Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]