Summary
How mycoheterotrophic plants that obtain carbon and soil nutrients from fungi are integrated in the usually mutualistic arbuscular mycorrhizal networks is unknown. Here, we compare autotrophic and mycoheterotrophic plant associations with arbuscular mycorrhizal fungi and use network analysis to investigate interaction preferences in the tripartite network.
We sequenced root tips from autotrophic and mycoheterotrophic plants to assemble the combined tripartite network between autotrophic plants, mycorrhizal fungi and mycoheterotrophic plants. We compared plant–fungi interactions between mutualistic and antagonist networks, and searched for a diamond‐like module defined by a mycoheterotrophic and an autotrophic plant interacting with the same pair of fungi to investigate whether pairs of fungi simultaneously linked to plant species from each interaction type were overrepresented throughout the network.
Mycoheterotrophic plants as a group interacted with a subset of the fungi detected in autotrophs but are indirectly linked to all autotrophic plants, and fungi with a high overlap in autotrophic partners tended to interact with a similar set of mycoheterotrophs. Moreover, pairs of fungi sharing the same mycoheterotrophic and autotrophic plant species are overrepresented in the network.
We hypothesise that the maintenance of antagonistic interactions is maximised by targeting well linked mutualistic fungi, thereby minimising the risk of carbon supply shortages.
Keywords: antagonism, arbuscular mycorrhizal fungi, mutualism, mycoheterotrophy, mycorrhizal networks
Introduction
Since the early history of life, interspecific mutualisms have been paramount in the functioning of ecosystems (Thompson, 2005; Bascompte & Jordano, 2014). Mutualisms can form complex networks of interdependence between dozens or even hundreds of species. A prime example of this ‘web of life’ is the 450‐million‐yr‐old mutualism between the majority of land plants and arbuscular mycorrhizal (AM) fungi (Strullu‐Derrien et al., 2018). In this interaction, plants supply the AM Glomeromycotina and Mucoromycotina fungi with carbon, essential for fungal survival and growth. In return, the fungi provide their host plants with mineral nutrients and water from the soil (Smith & Read, 2008; Bidartondo et al., 2011). One of the key characteristics of the AM interaction is its low interaction specificity: a mycorrhizal plant typically associates simultaneously with multiple fungi and a mycorrhizal fungus often associates simultaneously with multiple plants (Lee et al., 2013). This creates complex underground networks in which plants of different species are indirectly linked through shared AM fungi (Toju et al., 2015; Chen et al., 2017). Despite this low specificity, there is evidence that networks of plants and AM fungi do not assemble randomly; instead the interactions can be affected by plant functional group (Davison et al., 2011; Sepp et al., 2019), and plant or fungal evolutionary relationships (Montesinos‐Navarro et al., 2015; Chen et al., 2017).
Considerable progress has been made in recent years in dissecting the exchange of resources between plants and AM fungi and its regulation. Experimental work on low‐diversity systems has demonstrated that control is bidirectional, and partners offering the best rate of exchange are rewarded, suggesting that AM networks consist of ‘fair trade’ interactions of carbon‐for‐nutrient exchange (Bever et al., 2009; Kiers et al., 2011). However, the importance of reciprocally regulated resource exchange is questioned, as mycorrhizas also affect plant health, interactions with other soil organisms, host‐defence reactions and suppression of nonmycorrhizal competitor plants (Walder & Van Der Heijden, 2015). Also, strictly reciprocal regulation of carbon‐for‐nutrients exchange does not seem to apply to all AM interactions. For example, some exceptional plants behave as ‘cheaters’ (Selosse & Rousset, 2011; Walder & Van Der Heijden, 2015): mycoheterotrophic plants obtain carbon from root‐associated fungi and some species have replaced photosynthesis with carbon uptake from AM fungi (Leake, 1994; Merckx, 2013). Although the mechanism underpinning carbon transfer from AM fungi to mycoheterotrophic plants remains unclear, mycoheterotrophic plants are often considered cheaters of the mycorrhizal symbiosis because they have evolved from mutualistic ancestors (Merckx et al., 2013) and exploit the AM symbiosis for soil nutrients and carbon without reciprocating, to our current knowledge (Selosse & Rousset, 2011), or without apparently being sanctioned by the fungal partners (Walder & Van Der Heijden, 2015). Moreover, it has been suggested that mycoheterotrophic plants may display a truly biotrophic parasitic mode, digesting the fungus colonising their roots (Imhof et al., 2013). Despite these observations, it is unknown whether mycoheterotrophic plants have a true negative effect on their associated fungi, and we cannot rule out if they provide cryptic benefits to their symbionts.
Within obligate mutualisms, the critical barrier to mutualism breakdown and to the evolutionary stability of the resulting cheater species is thought to be a requirement for three‐species coexistence: a cheater plant relies on a mutualistic partner – a mycorrhizal fungus – which simultaneously interacts with an autotrophic plant (Pellmyr & Leebens‐Mack, 1999). In species‐rich mutualisms, such as the AM symbiosis, for which multispecies coexistence is the rule, a high potential for the occurrence of these tripartite linkages is expected (Merckx & Bidartondo, 2008). Indeed, while evolution of cheating in specialised obligate mutualisms is relatively rare (Sachs & Simms, 2006), cheating in the AM symbiosis has evolved in more than a dozen of plant clades, including over 250 species that together occur in nearly all tropical and subtropical forests (Merckx, 2013; Gomes et al., 2019a).
Previous work has shown that mycoheterotrophic plants target a subset of the mycorrhizal fungi available in the local community (Bidartondo et al., 2002; Gomes et al., 2017a; Sheldrake et al., 2017). Their associated fungal communities can vary in specificity: while many families of mycoheterotrophic plant species associate with fungi that are clustered in the Glomeromycotina phylogeny, large differences in the number of associated fungi and phylogenetic specificity are observed between species of mycoheterotrophic plants (Merckx et al., 2012). This specificity can be shaped by the competitive interactions of the plants. Gomes et al. (2017b) showed that, among mycoheterotrophs, plant species usually associate with more distantly related fungi than expected by chance, and in communities of co‐occurring mycoheterotrophic species, the phylogenetic diversity of the associated fungi increases with the extent of fungal overlap between the mycoheterotrophic species. This pattern may respond to an ecological mechanism driven by maximising co‐occurrence and avoiding competitive exclusion among mycoheterotrophic plants. However, whether partner choice of mycoheterotrophs is affected by the mutualistic interactions of their associated fungi with autotrophic plants is currently unknown.
Here, we hypothesise that mycoheterotrophic plants preferentially associate with ‘keystone’ (Mills & Doak, 1993) fungi that are well connected to many different autotrophic plants simultaneously, as these fungi are potentially more resilient to perturbations (Bascompte & Jordano, 2007) and may be the most reliable source of carbon (Waterman et al., 2013). In addition, as fungal traits play an important role in arbuscular mycorrhizal interactions – phylogenetically related AM fungi (assumed to have similar functional traits), preferentially interact with similar plant species (Chagnon et al., 2015) – we hypothesise that if partner selection in tripartite networks is trait driven we will be able to detect the influence of the phylogenetic relationships of the fungi. We tested these hypotheses on a combined tripartite mycorrhizal network of co‐occurring mycoheterotrophic and surrounding autotrophic plants linked by shared AM fungi compiled by high‐throughput DNA sequencing. To place our results in the context of recent work on comparing different types of ecological interaction networks (Melián et al., 2009; Fontaine et al., 2011; Sauve et al., 2013), we consider autotrophic plants to establish mutualistic interactions with AM fungi and mycoheterotrophic plants to form antagonistic interactions with AM fungi, although it remains unclear whether mycoheterotrophs have a negative impact on their associated fungi.
Materials and Methods
Sampling
As mycoheterotrophic plants are relatively rare and often have patchy distributions (Gomes et al., 2019b), we sampled two 4 × 4 m subplots a few metres apart in a coastal lowland plain rainforest in French Guiana (5°28′25″N, 53°34′51″W) on 28 July 2014, with overlapping mycoheterotrophic species. In both subplots, roots of mycoheterotrophic plants and surrounding autotrophic plants were sampled, cleaned with water, and stored on cetyltrimethylammonium bromide (CTAB) buffer at −20°C until further processing. We found, in total, five mycoheterotrophic plant species: Dictyostega orobanchoides, Gymnosiphon breviflorus (Burmanniaceae), Voyria aphylla, Voyriella parviflora (Gentianaceae) and Soridium spruceanum (Triuridaceae). Complete individuals of mycoheterotrophic plants were dug out and, around each, three root tips of autotrophic plants were collected. In addition, we randomly sampled five autotrophic plant root tips from each quadrant of each plot aiming to better represent the local belowground mutualistic community. In total, we collected 60 root samples of mycoheterotrophic plants and 220 autotrophic plant root samples. For the autotrophic plants, 123 samples could be identified by DNA sequencing (please refer to subsequent paragraphs). Ninety‐nine samples among 28 autotrophic, and 45 samples among the five mycoheterotrophic plant species had Glomeromycotina reads. After removing samples with < 500 Glomeromycotina reads, we retained a total of 77 samples of 21 autotrophic species and 27 samples of the five mycoheterotrophic species. A detailed list of mycoheterotrophic and autotrophic plant species collected in this study can be found in Supporting Information Table S1.
Plant identification and fungal communities sequencing
DNA was extracted from the CTAB‐preserved roots with the NucleoMag96 Plant Kit (Macherey‐Nagel GmbH & Co., Düren, Germany), using the KingFisher Flex Magnetic Particle Processor (Thermo Fisher Scientific, Waltham, MA, USA). Autotrophic plant species were identified by sequencing the markers matK or trnL as described in Gomes et al. (2017a). Plant identification to species, genus, or family level based on Blast against GenBank was reviewed by consulting the checklist of plants in French Guiana (Funk et al., 2007). Fungal communities associated with the roots of mutualistic and antagonistic plants were amplified using the primers fITS7 (Ihrmark et al., 2012) and ITS4 (White et al., 1990) and sequenced using a Personal Genome Machine (Ion Torrent; Life Technologies, Guildford, CT, USA), as described in Gomes et al. (2017b) in two separate runs. Negative controls from the extraction and the PCR reactions were included (and had zero reads). The same bioinformatics methods from Gomes et al. (2017b) were used to process the raw reads from the two runs combined until clustering into 97% operational taxonomic units (OTUs), using Usearch v.7.0 (Edgar, 2010). OTUs that were represented by fewer than six reads in each sample were excluded to avoid spurious OTUs (Lindahl et al., 2013). The taxonomical assignment of each OTU was carried out by querying against the UNITE database (Kõljalg et al., 2013). Because the mycoheterotrophic plants in this study are currently only known to associate with fungi that belong to the subphylum Glomeromycotina (Merckx et al., 2012), we only retained fungal OTUs from this subphylum in the subsequent analysis. In these analyses, we accounted for the phylogenetic relatedness between fungal OTUs, due to the uncertainty of correspondence of individual OTUs with taxon diversity of AM fungi (Flynn et al., 2015), despite the fact that delimitation of OTUs is not likely to interfere with ecological interpretations (Lekberg et al., 2014). We inferred the phylogenetic relationships between the fungal OTUs following the strategy of Gomes et al. (2017b). Briefly, we aligned the OTUs with partial reference sequences and performed a phylogenetic inference in which the relationships between these references were enforced based on Krüger et al. (2012). From this point forwards, we refer to OTUs as ‘fungi’. The fungal communities obtained from the root samples are not necessarily similar representations of the total communities of antagonists and mutualists, because the sampling coverage of their root systems varied: mycoheterotrophic plants are small herbs, and therefore large parts of their root system were collected and extracted, whereas for autotrophic plants, mostly forest trees in our plots (see the Results section), root samples represent only a small fragment of their root system and therefore the detected fungal communities associated with autotrophic plants are likely to be a subset of their total fungal communities. To test for potential bias on the detection of fungal OTUs, due to the differences in root sampling between mycoheterotrophic and autotrophic plants, we generated accumulation curves considering both the number of reads and the individual samples for each plant type (Fig. S1). We observed that, in both cases and for both plant types, the accumulation curves tended to reach an asymptote, and therefore the potential bias of underrepresentation of fungal diversity for both plant types introduced by the uneven sampling is likely to be limited.
Plant–fungi interactions
Fungal communities at the plant individual level were significantly structured by plant species identity with a minor influence of the subplot in which they were collected (Methods S1; Fig. S2). To integrate the interactions of mycoheterotrophic and autotrophic plants, we considered their plant–fungi interactions as a single tripartite network (Fig. 1a) by combining the fungal communities associated with individual plants from the two subplots into overall communities per plant species. By combining the data from the two subplots, we aimed at reconstructing a more robust picture of the interactions in this highly diverse rainforest while compensating for the relatively low sampling intensity (of 200 root tips of autotrophic plants, 123 plant species could be identified). Plant species for which Glomeromycotina fungi were represented by < 500 reads were excluded, resulting in the removal of 11 mutualistic plant species (please refer to details in Table S1). Moreover, rarefying the OTU matrix has been shown to greatly increase the false‐positive rate of OTUs per sample, which can influence the outcome of the analyses (McMurdie & Holmes, 2014). Therefore, we performed the subsequent analyses on 100 matrices rarefied to 844 reads per plant species, based on the lowest number of reads obtained for all plant species in the observed dataset. Results are presented with mean and standard deviation values obtained from running the analyses on the rarefied matrices. To investigate fungal association patterns of mycoheterotrophic and autotrophic plants, we used incidence (binary) data, because our main interest was to determine which interactions could be established and not how abundant they were. Also, due to the difference in root sampling (please refer to preceding paragraphs), read abundance was unlikely to correspond to fungal abundances at the plant species level, in particular between plant types (mycoheterotrophs and autotrophs). We considered the simultaneous presence of a particular fungus in the roots of a mycoheterotrophic and an autotrophic as a potential link between both (Southworth et al., 2005).
Fig. 1.
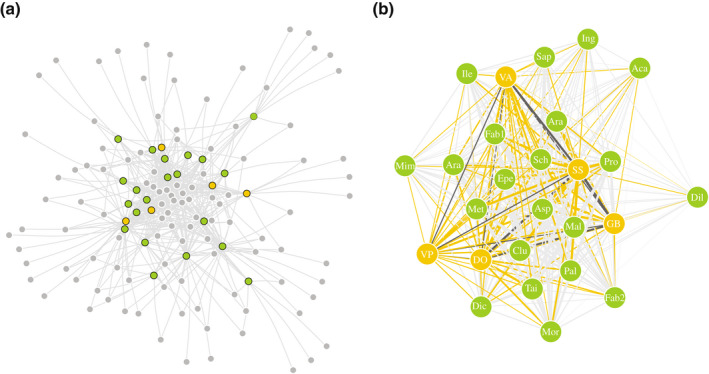
Tripartite arbuscular mycorrhizal interactions. (a) Visualisation of the tripartite network between fungi (grey) and mycoheterotrophic (yellow), and autotrophic (green) plants, in which edges represent a connection between a plant and a fungus. (b) Plant–plant–fungi overlap network, in which edges represent a link between plant species through shared arbuscular mycorrhizal fungus. The thickness of the lines represents the interaction strength between the plants (the thicker the line, the more fungi are shared). Yellow lines link mycoheterotrophic and autotrophic plants; light grey lines link autotrophic to autotrophic plants; and dark grey lines link mycoheterotrophic to mycoheterotrophic plants. Identification of autotrophic plants is indicated by the first three letters of their name (please refer to full names in Fig. 3); mycoheterotrophic plants are Dictyostega orobanchoides (DO), Gymnosiphon breviflorus (GB), Soridium spruceanum (SS), Voyria aphylla (VA), and Voyriella parviflora (VP). In both network representations (a, b), one of the 100 rarefied matrices to a depth of 844 reads was used; and the Fruchterman–Reingold layout was used, in which nodes are evenly distributed through the graph, in which plants that share more connections are closer to each other.
We tested for the effect of plant and fungi phylogenetic relatedness on the observed interactions in both networks. We used the fungal phylogeny described above, and the phylogenetic relationships of the plants as derived from TimeTree (Kumar et al., 2017) (please refer to details in Methods S2). We computed Mantel test correlations between the phylogenetic distance matrix and the community dissimilarity matrix in each instance for autotrophic and mycoheterotrophic species individually using the vegan R package (Oksanen et al., 2015). The phylogenetic distance matrices were extracted from the phylogenetic trees of plants and fungi, and the community dissimilarity matrices were calculated as the Jaccard distance on the binary interaction matrices. Phylogenetic signal was calculated for the 100 rarefied matrices, and consistency of significant results was assessed across multiple rarefaction depths (Fig. S3).
Mutualistic and antagonistic plant–plant interactions
To compare the range of fungal interactions between autotrophic and mycoheterotrophic plants, we calculated their normalised degree and the phylogenetic species variability of their associated fungal communities. The normalised degree of a plant species is the proportion of its associated fungi out of the total possible fungi in the network (Martín González et al., 2010), and was calculated with the ND function of the bipartite R package (Dormann et al., 2018). The phylogenetic species variability (psv) of the fungal community associated with a plant species summarises the level to which the fungi in this community are phylogenetically related (Helmus et al., 2007). When a community consists of unrelated fungi, the index equals 1, indicating maximum phylogenetic variability. As relatedness increases, the index approaches 0, indicating high phylogenetic specificity (Helmus et al., 2007). In addition, we calculated the fungal overlap between each pair of plant species as the number of fungi they share to infer plant–plant interactions (Fig. 1b).
Mutualistic and antagonistic fungal interactions
We compared the ecological similarity of the fungi shared between the two interaction type networks (i.e. mutualistic and antagonistic interactions). The ecological similarity of a pair of fungi represents their similarity in interactions through shared plants. We calculated similarity matrices between the fungi linked to autotrophic and mycoheterotrophic plants separately, using two different measures. The first measure is the Jaccard index, which corresponds to the number of plant species with which both fungi interact divided by the total number of plant species with which they interact, and the second is the overlap measure: C ij /min(d i , d j ), where C ij is the number of shared plants between fungus i and j, and min(d i , d j ) is the smallest number of associated plants between fungus i and j (Saavedra et al., 2013). The overlap measure corresponds to the number of shared plant species relative to the maximum number of plant species that can be shared, while taking into account the possibility of fungi having differential limits in their maximum number of plant partners. We computed the Mantel test correlation between the similarity matrices for the mutualistic and antagonistic interactions using the two different measures. We also computed the partial Mantel test correlation between the similarity matrices calculated using the Jaccard and overlap measures, controlling for the phylogenetic relatedness of the shared fungi.
Network analysis
To investigate how mycoheterotrophic plants are integrated in the mutualistic network of fungi and autotrophic plants, we searched for diamond‐shaped modules in the tripartite network, representing an autotrophic and a mycoheterotrophic plant interacting with the same pair of fungi (Fig. 2a). If the module is overrepresented throughout the entire network, it can be considered a motif (Milo et al., 2002; Bascompte & Melián, 2005; Stouffer et al., 2007). This specific motif does not consider the identity of the partners per se, but searches for this particular topological structure. Because our network consists of a tri‐trophic ‘food chain’ (considering the exchange of carbon), we built on the existing theoretical knowledge of trophic interrelationships and selected the diamond‐shaped module among those that were part of the theoretical research agenda on food webs. This module was described as ‘apparent competition’ and has been found to be overrepresented, and therefore highly relevant, in real food webs (Milo et al., 2002; Bascompte & Melián, 2005). Although the importance of competition in AM interactions is unclear, we expect this module to be effective for testing our main hypothesis: if mycoheterotrophic plant species associate with fungi that are linked to only few autotrophic plant species, the module will be underrepresented (Fig. 2b). By contrast, if mycoheterotrophic plant species associate with fungi that are well connected to autotrophic plants, many of these fungi will be linked to the same autotrophic plant species and therefore the module will be overrepresented throughout the entire network (Fig. 2c) and can be considered a network motif (Milo et al., 2002; Bascompte & Melián, 2005; Stouffer et al., 2007). We did not consider alternative modules, which are potentially also relevant in this context, but this an interesting follow‐up to the present paper.
Fig. 2.
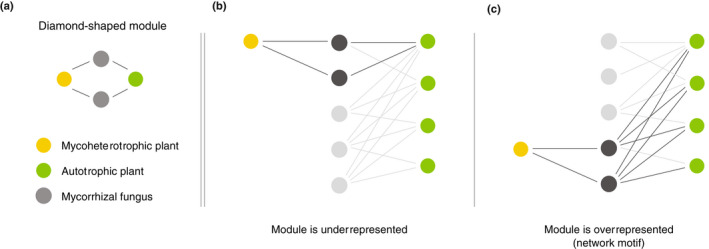
Diamond‐shaped module investigated in this paper. Representation of the module linking pairs of fungi to the same mycoheterotrophic and autotrophic plant species (a). Examples of underrepresentation (b) and overrepresentation (c) of the module in a network. When the module is overrepresented it is considered a network motif.
To assess whether the diamond‐shaped module was overrepresented relative to random expectations, we randomised the original mycoheterotrophic plants – mycorrhizal fungi – autotrophic plants community matrix and calculated the frequency of this diamond‐shaped module in 1000 generated random networks. We used a null model that draws an interaction between a plant and a fungal species that is proportional to the generalisation level of both species. Specifically, this probability is defined as the arithmetic mean of the fraction of interactions of the plant and that of the fungi (null model 2 in Fortuna & Bascompte, 2006; initially proposed in Bascompte et al., 2003). The null models were constructed with two different sets of fungi. First, we generated null models using all fungi in the network. This null model tests whether mycoheterotrophic plants exhibit an overall preference for fungi that are well connected to the autotrophic plants among all fungi present. Next, we generated null models only using fungi that are shared between mycoheterotrophic plants and autotrophic plants in the empirical network. This null model is similar to the previous model, but by restricting the resampling only to overlapping fungi between mycoheterotrophic and autotrophic plants, we assess whether there is a further preference for well connected fungi among those used by the mycoheterotrophs. Because there was an imbalanced number of plant species (five mycoheterotrophic and 21 autotrophic plants), each randomised matrix resulted from randomising the antagonistic and mutualistic interactions separately, and then were combined into a single matrix before the module search. To assess the potential effects of sampling effort both in the number of individuals collected (leading to roughly half of fungal reads belonging to the five mycoheterotrophic species, whereas the other half represent 21 autotrophic species) and in representation of whole (mycoheterotrophic) or partial (autotrophic) root systems, we repeated this procedure for a set of 100 rarefied original matrices. Furthermore, as plant species are represented by an unequal number of samples (Table S1), we also repeated the module search on 1000 matrices created by random resampling three samples per plant species (discarding species for which fewer than three samples were available). When this procedure showed that the number of individuals per species affected the consistency of the results, we alternatively tested the robustness of sampling by repeating the module search at multiple read depths in the rarefaction step to exclude an effect of the chosen sampling depth (please refer to preceding paragraphs). The strategy of resampling only three individuals per species reduced the representation of autotrophs in relation to mycoheterotrophs, from a proportion of 4 : 1 to 2 : 1, drastically deviating from the empirical situation. However, to the best of our knowledge, there was no alternative way to address the potential effect of the sampling bias in our study.
To evaluate whether the diamond‐shaped module was overrepresented throughout the empirical network, we calculated the standard , where obs is the observed network modules and is the mean of the same type of modules across the 1000 random networks using a 95% confidence interval. An empirical result above the null confidence interval indicates that pairs of fungi are shared between particular mycoheterotrophic and autotrophic plants more often than expected by chance, reflecting a preference of mycoheterotrophic plants to link to pairs of fungi that are generalists in their interactions with autotrophic plants. An empirical result below the null confidence interval indicates that mycoheterotrophic plants target pairs of fungi that are specialists in their interactions with autotrophic plants.
Results
Plant–fungi interactions
We successfully identified root tips of 32 autotrophic (123 root tips) and five mycoheterotrophic (60 individuals) plant species (please refer to Table S1 for detailed species list). After retaining samples that contained a minimum of 500 reads identified as Glomeromycotina, we reduced our dataset to 21 autotrophic (77 root tips) and five mycoheterotrophic (27 individuals) plant species, with a total of 365 135 reads. Autotrophic plants belonged to 14 families, and included 16 trees, two shrubs, one herb, one vine and one liana. We obtained 115 AM fungi, identified as Glomeromycotina within three families Gigasporaceae (four fungi), Acaulosporaceae (14) and Glomeraceae (97). Of these, 96 OTUs (49% of total reads) were present in the antagonistic network.
The 100 rarefied binary matrices included 26 plant species (21 autotrophs and five mycoheterotrophs) and 110 ± 1.47 fungi. Of these, mutualistic networks had 100 ± 1.51 fungi (reduced from 115 fungi before rarefaction) and antagonistic networks had 61 ± 3.14 fungi (reduced from 96 fungi before rarefaction). In all the generated matrices, autotrophic and mycoheterotrophic plants shared 51 ± 3.16 fungi, which represents 55% and 92% of total fungi present in autotrophic and mycoheterotrophic, respectively (Fig. 1a).
We measured a relatively low but significant phylogenetic signal of the fungal phylogeny on the antagonistic (r = 0.20 ± 0.06, P = 0.003 ± 0.005) and mutualistic (r = 0.21, P = 0.001) networks (please refer to the effect of rarefaction depth on phylogenetic signal in Fig. S3). We found no significant effects of plant phylogeny in any of the networks.
Mutualistic and antagonistic plant–plant interactions
The number of fungal partners per plant species varied, with Aspidosperma sp. showing the highest normalised degree and Sapindaceae the lowest amongst the autotrophic plants (Fig. 3a). Ranked over all the plants, three mycoheterotrophic species, namely V. aphylla, V. parviflora and D. orobanchoides, presented a normalised degree in the lower half of the spectrum, whereas G. breviflorus and S. spruceanum are amongst the plants with the highest number of associated fungi. We also observed a wide range of phylogenetic species variability (psv) for autotrophic plants, in which Acacia sp. and Aspidosperma sp. had the highest psv and Sapindaceae the lowest, throughout which, the mycoheterotorphic plants are distributed (Fig. 3b).
Fig. 3.
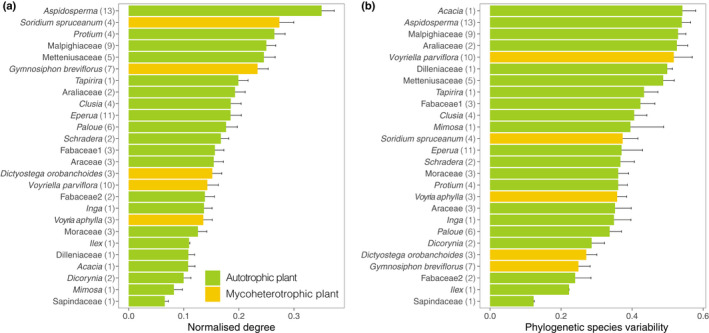
Mean ± SD plant normalised degree (a) and phylogenetic species variability (b) for the 21 autotrophic and five mycoheterotrophic plant species, resulted from the 100 rarefactions to a depth of 844 reads. Values between parentheses represent the number of individual samples per species.
Among the mycoheterotrophic plants, we observed that S. spruceanum shared most fungal interactions with G. breviflorus, followed by S. spruceanum with D. orobanchoides and V. aphylla, and also G. breviflorus with D. orobanchoides. We observed that mycoheterotrophic plants as a group associate with fungi that are simultaneously linked to all autotrophic plants that were retained in our analyses (Fig. 1b). Of all the possible connections between mycoheterotrophic and autotrophic plant species, S. spruceanum had the highest fungal overlap with Clusia sp., and then with Tapirira sp. and Protium sp.; G. breviflorus with Aspidosperma sp. and Protium sp., then with Schadera sp. and Clusia sp.; D. orobanchoides with Tapirira sp. and then with Fabaceae sp. 1; V. parviflora with Aspidosperma sp. and then with Araceae sp., Malpighiaceae sp. and Clusia sp; and V. aphylla with Aspidosperma sp. and then with Paloue sp. These autotrophic plants are also those with highest number of fungal interactions overall (Fig. 3a). Among the autotrophic plants, Aspidosperma sp. had the highest fungal overlap with Protium sp., Malpighiaceae sp. and Metteniusaceae sp., which are also among the highest ranked species in terms of number of associated fungi and phylogenetic diversity (Fig. 3).
Mutualistic and antagonistic plant–fungi interactions
Fungi that present in the mutualistic network but are absent from the antagonistic network were generally characterised by a low normalised degree, except for two Rhizophagus taxa that were present in the majority of autotrophic plants (Fig. 4). Moreover, we found a significant correlation of fungal ecological similarity between the mutualistic and antagonistic interactions (Mantel tests: Jaccard r = 0.20 ± 0.04, P = 0.002 ± 0.002; overlap r = 0.24 ± 0.04, P = 0.001), and also when accounting for the phylogenetic signal of the fungi (partial Mantel tests: Jaccard r = 0.18 ± 0.003, P = 0.018 ± 0.04; overlap r = 0.22 ± 0.04, P = 0.002 ± 0.003). We also observed that the fungi with the highest normalised degrees, both in the mutualistic and antagonistic network, were all members of the Glomeraceae family (Fig. 4).
Fig. 4.
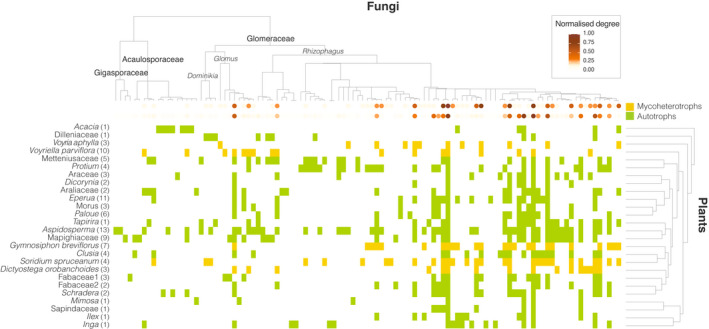
Plant–fungi interactions of autotrophic (green) and mycoheterotrophic (yellow) plants using a matrix rarefied to a depth of 844 reads. Phylogenetic relationships between the fungi are shown at the top. Plant species are listed on the left with the number of individual samples in between parentheses; hierarchy clustered dendrogram based on the Bray–Curtis distance of their fungal communities is shown on the right. The intensity of the orange dots on the tips of the fungal phylogeny depicts the normalised degree (representing their interaction strength) of each fungus in each of the plant–fungi networks (i.e. in the mycoheterotrophic plants–fungi and autotrophic plants–fungi networks). Values between parentheses represent the number of individual samples per species.
Network analysis
The network analysis indicated the presence of 2620.53 ± 271.23 instances of the diamond‐shaped module in the empirical networks across the rarefied matrices. The module was overrepresented in relation to random expectations for all the 100 rarefied matrices, both when including all the fungi in the dataset (z‐score = 7.88 ± 1.06, P = 0), and also when pruning the dataset to only include the overlapping fungi (z‐score = 3.43 ± 0.45, P = 0.003 ± 0.006). Repeating the analysis on resampled matrices that drew three random individuals per species resulted in an overrepresentation of the module in 99% of the cases when including all the fungi in the dataset (modules: 1129.17 ± 280.98, z‐score = 3.00 ± 0.49, P = 0.007 ± 0.010), and in an overrepresentation of the module in 24.4% of the cases when including only the overlapping fungi in the dataset (modules: 1164.29 ± 305.39, z‐score = 1.49 ± 0.42, P = 0.089 ± 0.060). Using the overlapping fungi dataset, the repeated analysis for multiple rarefaction depths indicates that, despite the number of diamond‐shaped modules increases with increasing rarefaction depth, it does not affect the result that pairs of fungi share a mycoheterotrophic and an autotrophic plant more often than expected by chance (Fig. S4).
Discussion
We found that mycoheterotrophic plants as a group target with a subset of the fungi that are potentially available, however this subset of fungi is associated with all autotrophic plants detected in this study. The results of the network analysis indicate that this pattern is produced by a preference of the mycoheterotrophs for well connected fungi. Therefore, despite associating only with a subset of the local pool of fungi, the mycoheterotrophs indirectly reach a wide range of autotrophic plants through their shared fungi, potentially obtaining carbon from any of the autotrophic plants at the study site. Fungi not detected in the roots of mycoheterotrophs were generally connected to few autotrophic plants. Within the fungi that are shared between the mutualistic and antagonistic networks, we detected a significant ecological symmetry between the mutualistic and antagonistic interactions of the fungi: pairs of fungi that interact with overlapping sets of autotrophic plants also interact with overlapping mycoheterotrophic plants. The network analysis indicated that this pattern occurs more often than expected by chance. Based on the highest fungal overlap between each mycoheterotrophic species with different subsets of highly connected autotrophic species, this pattern appears to be driven by a further preference of mycoheterotrophs for fungi that are well linked to specific mutualistic plants. These autotrophic plants are the ultimate sources of the carbon that mycoheterotrophic plants take up from the fungi shared between autotrophs and mycoheterotrophs. Therefore, we suggest that the observed pattern reflects a strategy in which the maintenance of antagonistic interactions is maximised by targeting well linked fungi, thereby minimising the risk of carbon supply shortages.
Fungal preferences of mycoheterotrophic plants
We found that plant species identity had a significant influence on the fungal community composition, regardless of the plant type, which indicates that these communities are nonrandom subsets of the local fungal taxon pool. This supports previous evidence that co‐occurring plant species showed differences in selectivity towards available AM fungi (Davison et al., 2011). Mycoheterotrophic plant species are known to select particular groups of fungi, often a narrower range than the surrounding autotrophic plants (Bidartondo et al., 2002; Gomes et al., 2017a,b). Here we observed that five co‐occurring mycoheterotrophic plant species collectively associate with approximately half of the available fungal taxa (Fig. 4). Antagonistic interactions can therefore be supported by a relatively wide array of AM taxa, as shown previously (Merckx et al., 2012; Gomes et al., 2017b; Sheldrake et al., 2017). Although fungi from three different fungal families (Glomeraceae, Acaulosporaceae and Gigasporaceae) were detected in the roots of mycoheterotrophic plants, a clear preference for Glomeraceae taxa, and Rhizophagus irregularis relatives in particular, was observed. The taxa of this clade were the most frequently encountered in the roots of the autotrophic plants as well (Fig. 4). Rhizophagus contains some of the most globally widespread and common AM fungi (Kivlin et al., 2011; Moora et al., 2011; Davison et al., 2015; Gomes et al., 2018) although this finding is mostly derived from studies in temperate areas. Our results indicate that the tropical rainforest offers no exception to this pattern. Glomeraceae are usually not only the most dominant clade in natural AM communities, often accounting for c. 70% of all species (Montesinos‐Navarro et al., 2012), but they also have been found consistently to include the most generalist AM fungi in other network studies (Montesinos‐Navarro et al., 2012; Chagnon et al., 2015; Chen et al., 2017). The ability to interact with many autotrophic plant species can be a potential reason for why mycoheterotrophic plants generally target Glomeraceae fungi (Merckx et al., 2012; Renny et al., 2017). Ecological theory predicts that generalist species tend to have large distribution ranges (Brown, 1984) and, consequently, are less vulnerable to (local) extinction than specialised species (Schleuning et al., 2016). Therefore, associations with generalist fungi may be advantageous for the evolutionary persistence of mycoheterotrophs. Furthermore, associations with multiple autotrophic plant partners may increase fungal resilience to disturbance, while mediating temporal fluctuations in carbon flow and interaction dynamics (Bennett et al., 2013). Therefore, this would guarantee a continuous carbon supply to the entire network without the pronounced negative effects, even in the presence of antagonists. In addition, in the context of mycorrhizal fungi, which can be linked to different plant species simultaneously (Montesinos‐Navarro et al., 2012), generalist fungi are therefore likely to be more reliable carbon sources for mycoheterotrophs. An alternative and perhaps not mutually exclusive explanation for why mycoheterotrophic plants preferentially target well connected fungi may be that these fungi are less effective in detecting and excluding nonphotosynthetic plant partners (Bruns et al., 2002; Egger & Hibbett, 2004; Bidartondo, 2005; Walder & Van Der Heijden, 2015). In contrast with our local‐scale study, Perez‐Lamarque et al. (2020), who performed a global‐scale study on AM mycoheterotrophic plants, reported that these plants tended to interact with (globally) specialised fungi. Although their results may have been influenced by the limited availability of global data – data of less than c. 0.2% of all autotrophic AM plants were available – it is possible that the most‐connected fungi in our study are less well connected in other habitats. In this case, the pattern of global‐scale reciprocal specialisation between mycoheterotrophs and AM fungi might be influenced by the specific local environmental conditions under which mycoheterotrophy occurs, such as low soil fertility (Gomes et al., 2019b).
Fungal links between mutualistic and antagonistic networks
We detected that pairs of fungi that interact with similar sets of autotrophic plants share links with overlapping mycoheterotrophic plants. Therefore, there is a high level of interaction symmetry between mutualistic and antagonistic mycorrhizal networks. Also, we measured a significant influence of the fungal phylogenetic relationships on both the mutualistic and antagonistic interactions, showing that closely related fungi interact with similar autotrophic and mycoheterotrophic plants respectively. Because biotic interactions are mediated by functional traits, and most functional traits are evolutionarily conserved, a shared evolutionary history of fungi can serve as a proxy for functional similarity (Chagnon et al., 2015). We therefore hypothesise that both mutualistic and antagonistic interactions are shaped partially by evolutionary conserved functional traits of the fungi. In this case, the apparent preference for members of the Glomeraceae family may indicate a higher reliance on ruderal AM fungi (Chagnon et al., 2013) for both autotrophic and mycoheterotrophic plants. Moreover, multiple clades within this family seem to be preferentially associated with mycoheterotrophic plants, which could reflect more fine discrimination of traits that we cannot discern with the current knowledge on AM fungal strategies. In addition, the network analysis indicated that pairs of fungi shared a mycoheterotrophic and an autotrophic plant more often than expected by chance. This analysis solely indicates that the diamond‐shaped module is overrepresented in the empirical network (Fig. 2), without reference to species degree or species identity. However, our results support the idea that the observed pattern is driven by the tendency of mycoheterotrophic plants to target fungi that are well linked to autotrophic plants (Fig. 1b). The autotrophic plants with the highest fungal overlap in relation to the mycoheterotrophic plants are among those with the highest ranked degree and phylogenetic species variability from the pool of detected autotrophic plant species (Fig. 3). Moreover, fungi with the highest number of interactions in the mutualistic network are also among the best connected fungi in the antagonistic network (Fig. 4). Our findings therefore reveal that mycoheterotrophic plants preferentially associate with fungi that are simultaneously linked to a wide range of autotrophic plants. Targeting well connected fungi in the mutualistic network could be a strategy for mycoheterotrophic plants to increase their resistant and resilient facing perturbations. Although many mycoheterotrophic plants share a large number of fungi with Aspidosperma sp., which has the highest normalised degree among the autotrophic plants, mycoheterotrophic plant species also indirectly associate with nonoverlapping sets of autotrophic plants, as indicated by their divergent positions in the plant–plant interaction network (Fig. 2b). Therefore, building on the hypothesis that mycoheterotrophic plants maximise their coexistence by increasing the phylogenetic diversity of the AM fungi with which they associate as the overlap among co‐occurring species increases (Gomes et al., 2017b), the present study suggests that the differential preference of mycoheterotrophic species for connections with nonoverlapping autotrophic species may contribute to competition avoidance among mycoheterotrophic plants.
Potential sampling biases
Considering that rainforests are species‐rich ecosystems (ter Steege et al., 2013), it is likely that, despite our efforts, the sampling of the belowground diversity of AM fungi, and their plant partners remained incomplete, in part because both plant identity and fungal communities of only 35% of autotrophic plants samples could be obtained. Therefore, not all autotrophic plant species present at the sites were included in the network and the reported patterns must be interpreted with caution. However, the large fungal overlap between the mutualistic and antagonistic networks suggested that it is unlikely that exclusive fungal connections of mycoheterotrophs to nonrepresented autotrophic plants are prevalent.
Furthermore, the representation of roots from mycoheterotrophic and autotrophic plants was necessarily imbalanced, as whole and partial root systems, respectively, were collected. This probably has an impact on the completeness of the fungal communities of the autotrophic plant species, and made the use of read abundances to estimate interaction strengths impossible. Moreover, the choice of primer set can introduce biases in the discovery of fungal diversity (Lekberg et al., 2018). We used the fITS7/ITS4 primer pair to characterise the fungal communities associated with the plants in our study, and found that mycoheterotrophic plants were associated primarily with fungi in the Glomeraceae family, which agrees with previous studies that used the SSU region (Merckx et al., 2012; Renny et al., 2017). Glomeraceae fungi have also been revealed to predominate in roots of autotrophic plant species (Davison et al., 2015).
Importantly, previous studies have highlighted the importance of sampling intensity (i.e. the number of possible interactions per node, which directly impacts the normalised degree per species) on network metrics (Blüthgen et al., 2007; Dormann et al., 2009). As the plant species in our study are represented by different numbers of samples, we assessed carefully any potential impacts of sampling bias on the results of the network analysis using multiple strategies. First, we randomised the mycoheterotrophic and autotrophic matrices to build the null models separately as the ratio of autotrophic and mycoheterotrophic species was only 4 : 1. Second, we calculated the number of modules over 100 rarefied matrices (which greatly reduced the number of discovered fungi for the mycoheterotrophic plants) and null models showed the presence of a network motif in 100% of the cases. Third, as the number of samples per species was also unequal across plant species, we repeated the module search procedure with random resampling of three individual samples per species and while discarding species for which fewer than three samples were available (even though this led to an unrealistic proportion of mycoheterotrophic to autotrophic plant species). This approach showed that the overrepresentation of the module in the network with all fungi was not influenced by unequal sampling across plant species. For the network with overlapping fungi, the empirical overrepresentation of the diamond‐shaped module was potentially influenced by unequal sampling, therefore we also verified the consistency of this result across multiple rarefactions depths by the use of multiple rarefied matrices at each depth (Gotelli & Colwell, 2001).
While we acknowledge that the completeness of sampling in our study is not ideal – a typical challenge for any study on mycorrhizal diversity – we also considered several factors that allowed us to separate the statistical patterns in our data from the influence of sampling effort, both in terms of the plant species detected in the sampled roots, and in their potentially incomplete number of fungal associations. Whether our results are potentially influenced by spatial patterns in the distribution of autotrophs, mycoheterotrophs and fungi remains to be determined.
Conclusions and future perspectives
To our knowledge, our study is the first to assess how mycoheterotrophic plant species are embedded in mutualistic mycorrhizal networks. We found that mycoheterotrophic plants as a group interacted with a subset of the available fungal partners, and generally targeted fungi that were well connected to autotrophic plants. Although mycoheterotrophic species show overlap in their fungal associations, we found that they were indirectly linked to different sets of autotrophic plants, suggesting a potential mechanism to avoid competition by preferentially relying on different carbon sources (Gomes et al., 2017b). The phylogenetic relationships between the fungi, probably a proxy for fungal traits, had a significant influence on these nonrandom tripartite interactions. Therefore, we concluded that the persistence of mycoheterotrophs in AM networks is dependent on particular well connected ‘keystone’ mycorrhizal fungi, which provide the mycoheterotrophs with carbon from a wide range of plants. Our observations that fungi connected mutualistic and antagonistic networks in a nonrandom fashion and that well connect fungal nodes in AM networks were more prone to be targeted by mycoheterotrophs, are similar to those of Sauve et al. (2016) for a plant–pollinator–herbivore network when considering binary interactions. Further research is needed to assess whether this is a general feature of interactions within species‐rich communities, also when taking interaction strength into account. Our study emphasises the raising of awareness of considering multiple interaction types simultaneously (e.g. antagonistic and mutualistic) to deepen our understanding of complex biodiversity patterns (Losapio et al., 2021).
In contrast with ectomycorrhizal symbiosis, for which it has been known for decades that several plant species are able to combine photosynthesis and carbon uptake from fungi, in a strategy termed ‘partial mycoheterotrophy’ (Selosse & Roy, 2009), only recently this mode of life has been suggested to be widespread within the AM symbiosis. Giesemann et al. (2021) have shown that many photosynthetic understory plants are potentially able to take up carbon from associated AM fungi. Future work will enlighten us on whether these partially mycoheterotrophic plants rely on similar sets of fungi and rely on similar interaction patterns within the mycorrhizal network as the fully mycoheterotrophic plants in in the present study.
Author contributions
All authors designed the study; SIFG and VSFTM collected the materials; SIFG performed the laboratory work and data analysis with input from MAF, JB and VSFTM authors. SIFG, MAF, JB and VSFTM wrote the manuscript.
Supporting information
Fig. S1 Accumulation curves considering the cumulative number of reads and the cumulative number of samples for autotrophic and mycoheterotrophic plants.
Fig. S2 Venn diagrams representing variation explained by plant species identity, plant type and subplot.
Fig. S3 Phylogenetic signal analysis repeated on multiple rarefaction depths.
Fig. S4 Motif analysis repeated on multiple rarefaction depths.
Methods S1 Effect of plant identity, plant type and subplot on the structure of fungal communities at the plant individual level.
Methods S2 Plant root identification.
Table S1 Plant identity of autotrophic and mycoheterotrophic plants from this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This research was supported by a Veni grant from the Netherlands Organisation of Scientific Research (NWO) to VSFTM (863.11.018) and field work was funded by the Royal Academy of Arts and Sciences of the Netherlands (KNAW) Ecology fund, grant J1606/Eco/G437 to SIFG. JB is supported by the Swiss National Science Foundation (grant no. 31003A‐169671 3). The authors thank the field assistance of Mélanie Roy, Vincent, Mathieu Gerard, the molecular laboratory assistance of Elza Duijm, and Hans ter Steege for verification of plant species occurrence in French Guiana. The authors also thank the valuable comments of five anonymous reviewers and editor Marc‐André Selosse that greatly improved the quality of the manuscript.
Data availability
The raw data that support the findings of this study are openly available in the Short Read Archive under project no. PRJNA846290.
References
- Bascompte J, Jordano P. 2007. Plant‐animal mutualistic networks: the architecture of biodiversity. Annual Review of Ecology, Evolution, and Systematics 38: 567–593. [Google Scholar]
- Bascompte J, Jordano P. 2014. Mutualistic networks. Princeton, NJ, USA: Princeton University Press. [Google Scholar]
- Bascompte J, Jordano P, Melián CJ, Olesen JM. 2003. The nested assembly of plant–animal mutualistic networks. Proceedings of the National Academy of Sciences, USA 100: 9383–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascompte J, Melián CJ. 2005. Simple trophic modules for complex food webs. Ecology 86: 2868–2873. [Google Scholar]
- Bennett AE, Daniell TJ, Öpik M, Davison J, Moora M, Zobel M, Selosse MA, Evans D. 2013. Arbuscular mycorrhizal fungal networks vary throughout the growing season and between successional stages. PLoS ONE 8: e83241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. 2009. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecology Letters 12: 13–21. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI. 2005. The evolutionary ecology of myco‐heterotrophy. New Phytologist 167: 335–352. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, Duckett JG. 2011. The dawn of symbiosis between plants and fungi. Biology Letters 7: 574–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Redecker D, Hijri I, Wiemken A, Bruns TD, Domínguez L, Sérsic A, Leake JR, Read DJ. 2002. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature 419: 389–392. [DOI] [PubMed] [Google Scholar]
- Blüthgen N, Menzel F, Hovestadt T, Fiala B, Blüthgen N. 2007. Specialization, constraints, and conflicting interests in mutualistic networks. Current Biology 17: 341–346. [DOI] [PubMed] [Google Scholar]
- Brown JH. 1984. On the relationship between abundance and distribution of species. The American Naturalist 124: 255–279. [Google Scholar]
- Bruns TD, Bidartondo MI, Taylor DL. 2002. Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integrative and Comparative Biology 42: 352–359. [DOI] [PubMed] [Google Scholar]
- Chagnon PL, Bradley RL, Klironomos JN. 2015. Trait‐based partner selection drives mycorrhizal network assembly. Oikos 124: 1609–1616. [Google Scholar]
- Chagnon PL, Bradley RL, Maherali H, Klironomos JN. 2013. A tait‐based framework to understand life history of mycorrhizal fungi. Trends in Plant Science 18: 484–491. [DOI] [PubMed] [Google Scholar]
- Chen L, Zheng Y, Gao C, Mi XC, Ma KP, Wubet T, Guo LD. 2017. Phylogenetic relatedness explains highly interconnected and nested symbiotic networks of woody plants and arbuscular mycorrhizal fungi in a Chinese subtropical forest. Molecular Ecology 26: 2563–2575. [DOI] [PubMed] [Google Scholar]
- Davison J, Moora M, Öpik M, Adholeya A, Ainsaar L, Bâ A, Burla S, Diedhiou AG, Hiiesalu I, Jairus T et al. 2015. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349: 970–973. [DOI] [PubMed] [Google Scholar]
- Davison J, Öpik M, Daniell TJ, Moora M, Zobel M. 2011. Arbuscular mycorrhizal fungal communities in plant roots are not random assemblages. FEMS Microbiology Ecology 78: 103–115. [DOI] [PubMed] [Google Scholar]
- Dormann CF, Fruend J, Gruber B. 2009. Indices, graphs and null models: analysing bipartite ecological networks. The Open Ecology Journal 2: 7–24. [Google Scholar]
- Dormann CF, Gruber B, Fruend J. 2018. Introducing the bipartite package: analysing ecological networks. R News 8: 8–11. [Google Scholar]
- Edgar RC. 2010. Search and clustering hundreds of times faster than Blast . Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Egger KN, Hibbett DS. 2004. The evolutionary implications of exploitation in mycorrhizas. Canadian Journal of Botany 82: 1110–1121. [Google Scholar]
- Flynn JM, Brown EA, Chain FJJ, Macisaac HJ, Cristescu ME. 2015. Toward accurate molecular identification of species in complex environmental samples: testing the performance of sequence filtering and clustering methods. Ecology and Evolution 5: 2252–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine C, Guimarães PR, Kéfi S, Loeuille N, Memmott J, van der Putten WH, van Veen FJF, Thébault E. 2011. The ecological and evolutionary implications of merging different types of networks. Ecology Letters 14: 1170–1181. [DOI] [PubMed] [Google Scholar]
- Fortuna MA, Bascompte J. 2006. Habitat loss and the structure of plant‐animal mutualistic networks. Ecology Letters 9: 281–286. [DOI] [PubMed] [Google Scholar]
- Funk V, Hollowell T, Berry P, Kelloff C, Alexander SN. 2007. Checklist of the plants of the Guiana Shield (Venezuela: Amazonas, Bolivar, Delta Amacuro; Guyana, Surinam, French Guiana). Contributions from the United States National Herbarium 55: 1–584. [Google Scholar]
- Giesemann P, Rasmussen HN, Gebauer G. 2021. Partial mycoheterotrophy is common among chlorophyllous plants with Paris‐type arbuscular mycorrhiza. Annals of Botany 127: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes SIF, Aguirre‐Gutiérrez J, Bidartondo MI, Merckx VSFT. 2017a. Arbuscular mycorrhizal interactions of mycoheterotrophic Thismia are more specialized than in autotrophic plants. New Phytologist 213: 1418–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes SIF, van Bodegom PM, Merckx VSFT, Soudzilovskaia NA. 2019a. Global distribution patterns of mycoheterotrophy. Global Ecology and Biogeography 28: 1133–1145. [Google Scholar]
- Gomes SIF, van Bodegom PM, Merckx VSFT, Soudzilovskaia NA. 2019b. Environmental drivers for cheaters of arbuscular mycorrhizal symbiosis in tropical rainforests. New Phytologist 223: 1575–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes SIF, Merckx VSFT, Hynson NA. 2018. Biological invasions increase the richness of arbuscular mycorrhizal fungi from a Hawaiian subtropical ecosystem. Biological Invasions 20: 2421–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes SIF, Merckx VSFT, Saavedra S. 2017b. Fungal‐host diversity among mycoheterotrophic plants increases proportionally to their fungal‐host overlap. Ecology and Evolution 7: 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotelli NJ, Colwell RK. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecology Letters 4: 379–391. [Google Scholar]
- Helmus MR, Bland TJ, Williams CK, Ives AR. 2007. Phylogenetic measures of biodiversity. The American Naturalist 169: E68–E83. [DOI] [PubMed] [Google Scholar]
- Ihrmark K, Bödeker ITM, Cruz‐Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandström‐Durling M, Clemmensen KE et al. 2012. New primers to amplify the fungal ITS2 region – evaluation by 454‐sequencing of artificial and natural communities. FEMS Microbiology Ecology 82: 666–677. [DOI] [PubMed] [Google Scholar]
- Imhof S, Massicotte HB, Melville LH, Peterson RL. 2013. Subterranean morphology and mycorrhizal structures. In: Merckx VSFT, ed. Mycoheterotrophy: the biology of plants living on fungi. New York, NY, USA: Springer, 157–214. [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A et al. 2011. Reciprocal rewards stabilize in the mycorrhizal symbiosis. Science 333: 880–882. [DOI] [PubMed] [Google Scholar]
- Kivlin SN, Hawkes CV, Treseder KK. 2011. Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biology and Biochemistry 43: 2294–2303. [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson‐Palme J, Callaghan TM et al. 2013. Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A. 2012. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytologist 193: 970–984. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Molecular Biology and Evolution 34: 1812–1819. [DOI] [PubMed] [Google Scholar]
- Leake JR. 1994. The biology of myco‐heterotrophic (‘saprophytic’) plants. New Phytologist 127: 171–216. [DOI] [PubMed] [Google Scholar]
- Lee EH, Eo JK, Ka KH, Eom AH. 2013. Diversity of arbuscular mycorrhizal fungi and their roles in ecosystems. Mycobiology 41: 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekberg Y, Gibbons SM, Rosendahl S. 2014. Will different OTU delineation methods change interpretation of arbuscular mycorrhizal fungal community patterns? New Phytologist 202: 1101–1104. [DOI] [PubMed] [Google Scholar]
- Lekberg Y, Vasar M, Bullington LS, Sepp SK, Antunes PM, Bunn R, Larkin BG, Öpik M. 2018. More bang for the buck? Can arbuscular mycorrhizal fungal communities be characterized adequately alongside other fungi using general fungal primers? New Phytologist 220: 971–976. [DOI] [PubMed] [Google Scholar]
- Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J et al. 2013. Fungal community analysis by high‐throughput sequencing of amplified markers – a user's guide. New Phytologist 199: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losapio G, Schöb C, Staniczenko PPA, Carrara F, Palamara GM, de Moraes CM, Mescher MC, Brooker RW, Butterfield BJ, Callaway RM et al. 2021. Network motifs involving both competition and facilitation predict biodiversity in alpine plant communities. Proceedings of the National Academy of Sciences, USA 118: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín González AM, Dalsgaard B, Olesen JM. 2010. Centrality measures and the importance of generalist species in pollination networks. Ecological Complexity 7: 36–43. [Google Scholar]
- McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Computational Biology 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melián CJ, Bascompte J, Jordano P, Krivan V. 2009. Diversity in a complex ecological network with two interaction types. Oikos 118: 122–130. [Google Scholar]
- Merckx V, Bidartondo MI. 2008. Breakdown and delayed cospeciation in the arbuscular mycorrhizal mutualism. Proceedings of the Royal Society B: Biological Sciences 275: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx VSFT. 2013. Mycoheterotrophy: the biology of plants living on fungi. New York, NY, USA: Springer. [Google Scholar]
- Merckx VSFT, Janssens SB, Hynson NA, Specht CD, Bruns TD, Smets EF. 2012. Mycoheterotrophic interactions are not limited to a narrow phylogenetic range of arbuscular mycorrhizal fungi. Molecular Ecology 21: 1524–1532. [DOI] [PubMed] [Google Scholar]
- Merckx VSFT, Mennes CB, Peay KG, Geml J. 2013. Evolution and diversification. In: Merckx VSFT, ed. Mycoheterotrophy: the biology of plants living on fungi. New York, NY, USA: Springer, 215–244. [Google Scholar]
- Mills LS, Doak DF. 1993. The keystone‐species concept in ecology and conservation. Bioscience 43: 219–224. [Google Scholar]
- Milo R, Shen‐Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298: 824–827. [DOI] [PubMed] [Google Scholar]
- Montesinos‐Navarro A, Segarra‐Moragues JG, Valiente‐Banuet A, Verdú M. 2012. The network structure of plant‐arbuscular mycorrhizal fungi. New Phytologist 194: 536–547. [DOI] [PubMed] [Google Scholar]
- Montesinos‐Navarro A, Segarra‐Moragues JG, Valiente‐Banuet A, Verdu M. 2015. Evidence for phylogenetic correlation of plant‐AMF assemblages? Annals of Botany 115: 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moora M, Berger S, Davison J, Öpik M, Bommarco R, Bruelheide H, Kühn I, Kunin WE, Metsis M, Rortais A et al. 2011. Alien plants associate with widespread generalist arbuscular mycorrhizal fungal taxa: evidence from a continental‐scale study using massively parallel 454 sequencing. Journal of Biogeography 38: 1305–1317. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2015. vegan: community ecology package. R package v.2.3‐1 . [WWW document] URL http://CRAN.R‐project.org/package=vegan [accessed 25 October 2021].
- Pellmyr O, Leebens‐Mack J. 1999. Forty million years of mutualism: evidence for eocene origin of the yucca‐yucca moth association. Proceedings of the National Academy of Sciences, USA 96: 9178–9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Lamarque B, Selosse MA, Öpik M, Morlon H, Martos F. 2020. Cheating in arbuscular mycorrhizal mutualism: a network and phylogenetic analysis of mycoheterotrophy. New Phytologist 226: 1822–1835. [DOI] [PubMed] [Google Scholar]
- Renny M, Acosta MC, Cofré N, Domínguez LS, Bidartondo MI, Sérsic AN. 2017. Genetic diversity patterns of arbuscular mycorrhizal fungi associated with the mycoheterotroph Arachnitis uniflora Phil. (Corsiaceae). Annals of Botany 119: 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra S, Rohr RP, Dakos V, Bascompte J. 2013. Estimating the tolerance of species to the effects of global environmental change. Nature Communications 4: 2350. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Simms EL. 2006. Pathways to mutualism breakdown. Trends in Ecology and Evolution 21: 585–592. [DOI] [PubMed] [Google Scholar]
- Sauve AMC, Fontaine C, Thébault E. 2013. Structure‐stability relationships in networks combining mutualistic and antagonistic interactions. Oikos 123: 378–384. [Google Scholar]
- Sauve AMC, Thébault E, Pocock MJO, Fontaine C. 2016. How plants connect pollination and herbivory networks and their contribution to community stability. Ecology 97: 908–917. [DOI] [PubMed] [Google Scholar]
- Schleuning M, Fründ J, Schweiger O, Welk E, Albrecht J, Albrecht M, Beil M, Benadi G, Blüthgen N, Bruelheide H et al. 2016. Ecological networks are more sensitive to plant than to animal extinction under climate change. Nature Communications 7: 13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse MA, Rousset F. 2011. The plant‐fungal marketplace. Science 333: 828–829. [DOI] [PubMed] [Google Scholar]
- Selosse MA, Roy M. 2009. Green plants that feed on fungi: facts and questions about mixotrophy. Trends in Plant Science 14: 64–70. [DOI] [PubMed] [Google Scholar]
- Sepp S, Davison J, Jairus T, Vasar M, Moora M, Zobel M, Öpik M. 2019. Non‐random association patterns in a plant–mycorrhizal fungal network reveal host–symbiont specificity. Molecular Ecology 28: 365–378. [DOI] [PubMed] [Google Scholar]
- Sheldrake M, Rosenstock NP, Revillini D, Olsson PA, Wright SJ, Turner BL. 2017. A phosphorus threshold for mycoheterotrophic plants in tropical forests. Proceedings of the Royal Society B: Biological Sciences 284: 20162093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis. Cambridge, MA, USA; San Diego, CA, USA: Academic Press. [Google Scholar]
- Southworth D, He X‐H, Swenson W, Bledsoe CS, Horwath WR. 2005. Application of network theory to potential mycorrhizal networks. Mycorrhiza 15: 589–595. [DOI] [PubMed] [Google Scholar]
- Stouffer DB, Camacho J, Jiang W, Amaral LAN. 2007. Evidence for the existence of a robust pattern of prey selection in food webs. Proceedings of the Royal Society B: Biological Sciences 274: 1931–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strullu‐Derrien C, Selosse M‐A, Kenrick P, Martin FM. 2018. The origin and evolution of mycorrhizal symbioses: from palaeomycology to phylogenomics. New Phytologist 172: 589–519. [DOI] [PubMed] [Google Scholar]
- Ter Steege H, Pitman NCA, Sabatier D, Baraloto C, Salomão RP, Guevara JE, Phillips OL, Castilho CV, Magnusson WE, Molino JF et al. 2013. Hyperdominance in the Amazonian tree flora. Science 342: 1243092. [DOI] [PubMed] [Google Scholar]
- Thompson JN. 2005. The geographic mosaic of coevolution. Chicago, IL, USA: University of Chicago Press. [Google Scholar]
- Toju H, Guimaraes PR, Olesen JM, Thompson JN. 2015. Below‐ground plant‐fungus network topology is not congruent with above‐ground plant‐animal network topology. Science Advances 1: e1500291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder F, Van Der Heijden MGA. 2015. Regulation of resource exchange in the arbuscular mycorrhizal symbiosis. Nature Plants 1: 15159. [DOI] [PubMed] [Google Scholar]
- Waterman RJ, Klooster MR, Hentrich H, Bidartondo MI. 2013. Species interactions of mycoheterotrophic plants: specialization and its potential consequences. In: Merckx VSFT, ed. Mycoheterotrophy: the biology of plants living on fungi. New York, NY, USA: Springer, 267–296. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols: a guide to methods and applications. San Diego, CA, USA: Academic Press, 315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Accumulation curves considering the cumulative number of reads and the cumulative number of samples for autotrophic and mycoheterotrophic plants.
Fig. S2 Venn diagrams representing variation explained by plant species identity, plant type and subplot.
Fig. S3 Phylogenetic signal analysis repeated on multiple rarefaction depths.
Fig. S4 Motif analysis repeated on multiple rarefaction depths.
Methods S1 Effect of plant identity, plant type and subplot on the structure of fungal communities at the plant individual level.
Methods S2 Plant root identification.
Table S1 Plant identity of autotrophic and mycoheterotrophic plants from this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The raw data that support the findings of this study are openly available in the Short Read Archive under project no. PRJNA846290.


