Abstract
Formation of DNA adducts following conversion of dichloromethane by bacterial dichloromethane dehalogenase/glutathione S-transferase was demonstrated. Adducts included dichloromethane carbon and glutathione sulfur atoms. A reaction with DNA occurred preferentially at guanine bases. Increased DNA degradation in a polA mutant of Methylobacterium dichloromethanicum DM4 grown with dichloromethane confirmed the genotoxicity associated with dichloromethane degradation, suggesting an important role of DNA repair in the metabolism of halogenated, DNA-alkylating compounds by bacteria.
Glutathione S-transferases (GSTs) are ubiquitous and versatile enzymes promoting the degradation or the inactivation of electrophilic compounds by their conjugation to glutathione (GSH) (1, 18). Compared to eukaryotic representatives of this enzyme family, bacterial GSTs are more diverse in sequence and often appear to catalyze specific reactions in the catabolism of compounds used as carbon sources for growth by the host bacteria (17, 18). For example, dichloromethane dehalogenase/GST (20) allows growth of methylotrophic bacteria with dichloromethane (DCM), a solvent and widespread environmental contaminant (13), as the sole carbon and energy source (19). Similar enzymes have been characterized in mammals in the past decade (12). DCM dehalogenases represent a special type of GSTs in that the GSH cofactor is not incorporated into the reaction product, but is regenerated after the reaction CH2Cl2 + GSH + H2O → CH2O + 2 HCl + GSH.
The genotoxicity of DCM conversion by GSTs has been well documented in both bacterial (7, 15) and mammalian (5, 8) systems (reviewed in references 12 and 19), but its molecular basis has not yet been fully elucidated. Mechanistic considerations and indirect experimental evidence (e.g., see reference 10) have suggested that S-chloromethylglutathione, the presumed short-lived intermediate in the reaction catalyzed by DCM dehalogenase/GST, may be involved in this process by reacting with DNA. Transient formation of a compound with an 19F-NMR (nuclear magnetic resonance) signal compatible with S-fluoromethylglutathione was observed when DCM dehalogenase from Methylophilus sp. strain DM11 was incubated with GSH and chlorofluoromethane (3). The lesser reactive fluorinated homolog of S-chloromethylglutathione was hydrolyzed with a half-life of 5.8 min at room temperature in D2O (4). Chemically synthesized S-chloromethylglutathione was shown to react with deoxyguanosine in vitro (4), and the product of this reaction was identified as S-[1-N2-deoxyguanosinylmethyl]glutathione (4, 15). Direct evidence for enzymatic GST-catalyzed conversion of DCM resulting in DNA modification, however, has not yet been obtained. The present report documents the formation of GST-dependent DNA adduct formation as a result of the degradation of DCM by bacterial DCM dehalogenases.
DNA adduct formation by GST-mediated conversion of DCM.
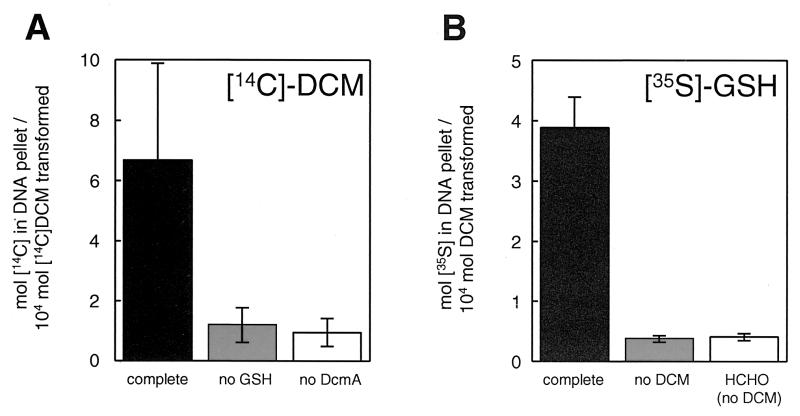
Purified DCM dehalogenase from Methylophilus sp. strain DM11 overexpressed in Escherichia coli (21) was incubated with 50 mM [14C]DCM (Sigma) (Fig. 1A) and 1 mM GSH in the presence of calf thymus DNA (Sigma). Radioactivity incorporation into DNA recovered after alcohol precipitation demonstrated the formation of DNA adducts. Recovery of radiolabel in the DNA fraction increased with the amount of DCM dehalogenase added (data not shown), and both DCM dehalogenase and GSH were required for this process to occur. The same experiment, but performed with [35S]GSH (Moravek Biochemicals, Brea, Calif.) and unlabeled DCM, also led to incorporation of radioactive label into DNA (Fig. 1B). No significant 35S labeling of DNA was observed in the absence of DCM. Thus, the carbon atom of DCM and at least the sulfur atom of GSH were incorporated into DNA following DCM dehalogenase-mediated turnover of DCM, a finding compatible with S-chloromethylglutathione being the causative agent of DNA adduct formation. Furthermore, incubation of formaldehyde in excess of that produced by DCM dehalogenation in the presence of radiolabeled GSH (Fig. 1B) did not result in DNA labeling. In other words, no DNA adduct formation was observed with the product of the DCM dehalogenase reaction or its GSH conjugate, which in the presence of GSH is the predominant form of formaldehyde in solution (16). This is in agreement with previous evidence that formaldehyde does not play a major role in DCM genotoxicity (see reference 19 for review and discussion). For instance, the spectrum of mutations induced by formaldehyde and by DCM are clearly different (5, 8). Moreover, a polA mutant of the DCM-degrading strain Methylobacterium dichloromethanicum DM4 lacking DNA polymerase I, whose role in DNA repair is well known (6), was unable to grow with DCM as the sole carbon source but still grew with formaldehyde (11). The DNA adducts detected here, however, could not be further characterized because of the low yield in which they were produced relative to the extent of DCM conversion (∼0.05%) (Fig. 1), despite the use of a large excess of DNA (approximately 1 adduct was formed per 200,000 bp). Nevertheless, this observation further substantiates the idea that a reactive, short-lived compound such as S-chloromethylglutathione is responsible for DNA adduct formation.
FIG. 1.
DNA adduct formation following DCM conversion by DCM dehalogenase/GST. Purified DCM dehalogenase (DcmA) (60 μg, 1 to 2 μmol/min/mg) was incubated with calf thymus DNA (0.5 mg/ml), 1 mM GSH and 50 mM DCM in 0.2 ml of 0.1 M phosphate buffer (pH 8.0) at 30°C for an hour. Experiments were performed at least twice with either [14C]DCM (0.58 mCi/mmol) (A) or [35S]GSH (20 Ci/mmol) (B). Reactions were stopped by addition of 0.3 M sodium acetate, and the DNA was precipitated by addition of 0.6 volumes of isopropanol and centrifugation (15,000 rpm, 1 h, 4°C). The DNA pellet was reprecipitated twice, washed with 70% ethanol, redissolved in 0.1 M phosphate (pH 8.0). The DNA was quantified spectrophotometrically at 260 nm, and DNA-associated radioactivity was measured by scintillation spectrometry (Beckman LS5801) and expressed as the percentage of DCM conversion during the experiment. Solid bars, all reaction components added (complete); grey bars, no GSH (A) or DCM (B) added; open bars, no DCM dehalogenase added (A) or no DCM added, but incubated with 50 mM formaldehyde (HCHO) (B).
Base specificity of DNA adduct formation and stability of DNA adducts.
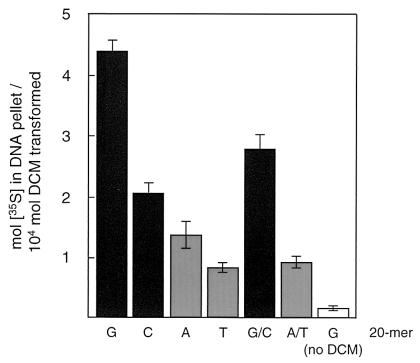
In keeping with previous suggestions of a higher reactivity of guanine and cytosine bases with GST-activated DCM (4, 5, 15), incorporation of radiolabel into synthetic 20-mer homo-oligonucleotides (Microsynth, Balgach, Switzerland) as DNA substrates followed the order G20 > C20 > A20 > T20 (Fig. 2). Also, single-stranded oligonucleotides were better substrates than double-stranded complementary oligonucleotides (Fig. 2). The stability of the formed DNA adducts was estimated by incubating aqueous solutions of 35S-labeled DNA preparations recovered after DCM conversion by DCM dehalogenase/GST for various times before alcohol precipitation. Interestingly, heterogeneity of the DNA adducts was apparent from the multiphasic behavior of the decrease in DNA-associated radioactivity (data not shown). The half-lives (in hours) of the initial major phase of DNA adduct decay (G20, 1.7 ± 0.3; C20, 2.4 ± 0.4; A20, 2.0 ± 0.2; T20, 3.2 ± 0.5; G20/C20, 3.3 ± 0.5; A/T, 5.7 ± 0.7; calf thymus DNA, 1.6 ± 0.2) were of the same order of magnitude as those determined earlier for model single-base adducts obtained by chemical synthesis (4, 15). In contrast, the rate and magnitude of the minor second, slower phase of DNA adduct degradation was similar (half-life, 50 to 100 h) for all DNA preparations.
FIG. 2.
Base specificity of DNA adduct formation. Synthetic homo-oligonucleotides (20-mers; 0.1 mg) were incubated with DCM dehalogenase and [35S]GSH and processed and analyzed exactly as described in the legend to Fig. 1. Double-stranded DNA was obtained by denaturation of pairs of complementary oligonucleotides at 100°C for 5 min followed by incubation at 50°C for 30 min, and the resulting preparation was checked by acridine orange staining.
DNA damage associated with DCM conversion in vivo.
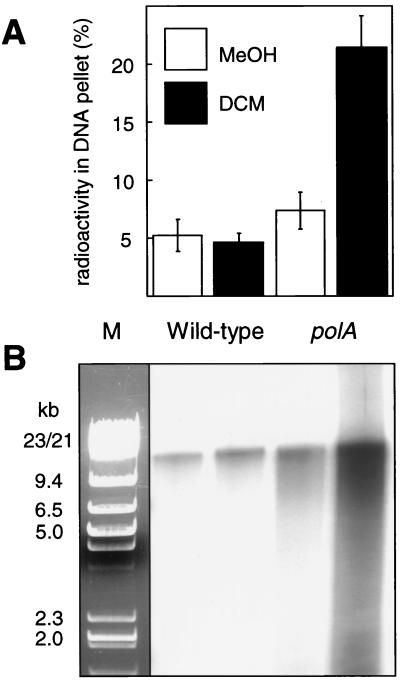
The data presented above suggested that methylotrophic bacteria are confronted with severe problems of genotoxicity while growing with DCM by virtue of DCM dehalogenase. To explore the extent of DNA damage caused by GST-mediated DCM turnover in Methylobacterium in vivo, Methylobacterium dichloromethanicum DM4 and its polA mutant (11), which was expected to be impaired in the repair of DNA lesions occurring upon GST-mediated DNA adduct formation, were grown with 40 mM methanol or with a mixture of 40 mM methanol and 10 mM DCM as carbon sources, after which total DNA was isolated from the cultures (Fig. 3). Both strains expressed DCM dehalogenase at similar levels during growth in the presence of DCM as judged by the chloride released into the medium (5 and 4.2 mM chloride for the wild-type and the polA mutant strains, respectively) (data not shown). Repair of damaged DNA typically involves the formation of DNA strand breaks at the site of DNA lesions (9, 22). Therefore, equal amounts of isolated total DNA were treated with terminal deoxyribonucleotide transferase (Roche Diagnostics) and [α-32P]dATP (Moravek Biochemicals). DNA was recovered by alcohol precipitation, and the radioactivity in the DNA pellet was quantified (Fig. 3A). In addition, gel-filtered samples of 3′-labeled total DNA were separated by agarose gel electrophoresis and autoradiographed (Fig. 3B). Increased labeling, indicating a more frequent occurrence of DNA strand breaks (14), was apparent in DNA from the polA mutant grown in the presence of DCM.
FIG. 3.
Labeling of DNA 3′ ends with terminal deoxynucleoside transferase. Cultures of M. dichloromethanicum DM4 wild type and its polA mutant were grown in mineral minimal medium (11) with 40 mM methanol (MeOH) with or without addition of 10 mM DCM for 12 h, and total DNA was isolated by a standard cetyltetramethylammonium bromide-based method (2). DNA samples (5 μg) were treated with terminal deoxyribonucleotide transferase (15 U) and 30 mCi of [α-32P]dATP (3,000 Ci/mmol) in 0.1 ml of 0.2 M cacodylate–25 mM Tris buffer (pH 6.6) at 37°C for 1 h. DNA was precipitated as described in the legend to Fig. 1 and purified by gel filtration with disposable NAP-10 columns (Amersham Pharmacia Biotech). (A) Terminal transferase-treated DNA samples recovered by precipitation were resuspended in 40 μl of 0.1 M Tris-HC1 buffer (pH 8.0), and the radioactivity associated with the DNA was quantified by scintillation spectrometry (Beckman LS5801) and expressed as the percentage of the total radiolabel used. (B) Corresponding autoradiograph of [α32P]dAMP-labeled samples of total DNA (2.5 μg of DNA/lane) size separated by agarose gel electrophoresis. M, marker (1:1 mixture of HindIII- and EcoRI/HindIII-digested lambda DNA) (Fermentas).
Thus, both in vitro and in vivo data reported here provide direct evidence for DNA damage following DCM dehalogenase/GST-mediated conversion of DCM. As an aside, these experiments also provided new evidence that the formaldehyde product of the enzymatic dehalogenation reaction is most likely not involved in this process. To conclude, it appears increasingly clear that efficient mechanisms to cope with the genotoxicity of products arising from the metabolism of DCM are an asset for bacteria growing with this compound by means of a GST-dependent pathway. This raises the question as to whether methylotrophic bacteria growing with DCM as the sole carbon source are more resistant to the toxic effects of DCM than other bacteria (19). Indeed, preliminary experiments suggest that Methylobacterium extorquens AM1, the genome of which is currently being sequenced, is unable to grow with DCM as the sole carbon source when provided with a plasmid-expressed DCM dehalogenase/GST (our unpublished data). A search for accessory genes and proteins that allow methylotrophic bacteria to grow with DCM may reveal new perspectives on the degradation of halogenated compounds by bacteria.
Acknowledgments
We gratefully acknowledge Thomas Leisinger for discussions, encouragement, and support.
This research was supported by the Swiss National Research Foundation (grant 3100-50602.97 to S.V.).
ADDENDUM IN PROOF
A characterization of the adducts formed by the reaction of chemically synthesized S-(1-acetoxymethyl)glutathione (a mimick for the glutathione conjugate of dichloromethane) with nucleosides and DNA was reported after this paper was submitted (G. A. Marsch et al., Chem. Res. Toxicol. 14:600–608, 2001).
REFERENCES
- 1.Armstrong R N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 2001. [Google Scholar]
- 3.Blocki F A, Logan M S P, Baoli C, Wackett L P. Reaction of rat liver glutathione S-transferases and bacterial dichloromethane dehalogenase with dihalomethanes. J Biol Chem. 1994;269:8826–8830. [PubMed] [Google Scholar]
- 4.Dechert S. Untersuchungen zum Wirkmechanismus der Mutagenität and Tumorigenität von Dichlormethan and seinen Metaboliten. Ph.D. thesis. Würzburg, Germany: Universität Würzburg; 1995. [Google Scholar]
- 5.DeMarini D M, Shelton M L, Warren S H, Ross T M, Shim J-Y, Richard A M, Pegram R A. Glutathione S-transferase-mediated induction of GC-AT transitions by halomethanes in Salmonella. Environ Mol Mutagen. 1997;30:440–447. [PubMed] [Google Scholar]
- 6.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 7.Gisi D, Leisinger T, Vuilleumier S. Enzyme-mediated dichloromethane toxicity and mutagenicity of bacterial and mammalian dichloromethane-active glutathione S-transferases. Arch Toxicol. 1999;73:71–79. doi: 10.1007/s002040050589. [DOI] [PubMed] [Google Scholar]
- 8.Graves R J, Trueman P, Jones S, Green T. DNA sequence analysis of methylene chloride-induced HPRT mutations in Chinese hamster ovary cells: comparison with the mutation spectrum obtained for 1,2-dibromoethane and formaldehyde. Mutagenesis. 1996;11:229–233. doi: 10.1093/mutage/11.3.229. [DOI] [PubMed] [Google Scholar]
- 9.Grossman L, Lin C-I, Ahn C. Nucleotide excision repair in Escherichia coli. In: Nickoloff J A, Hoekstra M F, editors. DNA repair in prokaryotes and lower eukaryotes. Vol. 1. Totowa, N.J: Humana Press; 1998. pp. 11–27. [Google Scholar]
- 10.Hashmi M, Dechert S, Dekant W, Anders M W. Bioactivation of [13C]dichloromethane in mouse, rat, and human liver cytosol: 13C nuclear magnetic resonance spectroscopic studies. Chem Res Toxicol. 1994;7:291–296. doi: 10.1021/tx00039a004. [DOI] [PubMed] [Google Scholar]
- 11.Kayser M F, Stumpp M T, Vuilleumier S. DNA polymerase I is essential for growth of Methylobacterium dichloromethanicum DM4 with dichloromethane. J Bacteriol. 2000;182:5433–5439. doi: 10.1128/jb.182.19.5433-5439.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landi S. Mammalian class theta GST and differential susceptibility to carcinogens: a review. Mutat Res. 2000;463:247–283. doi: 10.1016/s1383-5742(00)00050-8. [DOI] [PubMed] [Google Scholar]
- 13.Mackay D, Shiu W Y, Ma K C. Volatile organic chemicals. Vol. 3. Boca Raton, Fla: Lewis Publishers; 1993. pp. 400–406. [Google Scholar]
- 14.Rohwer F, Azam F. Detection of DNA damage in prokaryotes by terminal deoxyribonucleotide transferase-mediated dUTP nick end labeling. Appl Environ Microbiol. 2000;66:1001–1006. doi: 10.1128/aem.66.3.1001-1006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thier R, Taylor J B, Pemble S E, Humphreys W G, Persmark M, Ketterer R, Guengerich F P. Expression of mammalian glutathione S-transferase 5-5 in Salmonella typhimurium TA1535 leads to base-pair mutations upon exposure to dihalomethanes. Proc Natl Acad Sci USA. 1993;90:8576–8580. doi: 10.1073/pnas.90.18.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uotila L, Koivusalo M. Glutathione-dependent oxidoreductases: formaldehyde dehydrogenase. In: Dolphin D, Avramovic O, Poulson R, editors. Glutathione chemical, biochemical, and medical aspects. A. New York, N.Y: John Wiley & Sons; 1989. pp. 517–551. [Google Scholar]
- 17.Vuilleumier S. Bacterial dichloromethane dehalogenases and the detoxification of xenobiotics: dehalogenation through glutathione conjugation and beyond. ACS Symp Ser. 2001;777:240–252. [Google Scholar]
- 18.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vuilleumier, S. Coping with a halogenated one-carbon diet: aerobic dichloromethane-mineralising bacteria. In S. Agathos and W. Reineke (ed.), Biotechnology for the environment. Focus on biotechnology series, vol. 3, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 20.Vuilleumier S, Ivos N, Dean M, Leisinger T. Sequence variation in dichloromethane dehalogenases/glutathione S-transferases. Microbiology. 2001;147:611–616. doi: 10.1099/00221287-147-3-611. [DOI] [PubMed] [Google Scholar]
- 21.Vuilleumier S, Leisinger T. Protein engineering studies of dichloromethane dehalogenase/glutathione S-transferase from Methylophilus sp. strain DM11. Ser 12 but not Tyr6 is required for enzyme activity. Eur J Biochem. 1996;239:410–417. doi: 10.1111/j.1432-1033.1996.0410u.x. [DOI] [PubMed] [Google Scholar]
- 22.Wilson D M, III, Engelward B P, Samson L. Prokaryotic base excision repair. In: Nickoloff J A, Hoekstra M F, editors. DNA repair in prokaryotes and lower eukaryotes. Vol. 1. Totowa, N.J: Humana Press; 1998. pp. 29–64. [Google Scholar]