Abstract
In the respirofermentative yeast Kluyveromyces lactis, only a single genetic locus encodes glucose transporters that can support fermentative growth. This locus is polymorphic in wild-type isolates carrying either KHT1 and KHT2, two tandemly arranged HXT-like genes, or RAG1, a low-affinity transporter gene that arose by recombination between KHT1 and KHT2. Here we show that KHT1 is a glucose-induced gene encoding a low-affinity transporter very similar to Rag1p. Kht2p has a lower Km (3.7 mM) and a more complex regulation. Transcription is high in the absence of glucose, further induced by low glucose concentrations, and repressed at higher glucose concentrations. The response of KHT1 and KHT2 gene regulation to high but not to low concentrations of glucose depends on glucose transport. The function of either Kht1p or Kht2p is sufficient to mediate the characteristic response to high glucose, which is impaired in a kht1 kht2 deletion mutant. Thus, the KHT genes are subject to mutual feedback regulation. Moreover, glucose repression of the endogenous β-galactosidase (LAC4) promoter and glucose induction of pyruvate decarboxylase were abolished in the kht1 kht2 mutant. These phenotypes could be partially restored by HXT gene family members from Saccharomyces cerevisiae. The results indicate that the specific responses to high but not to low glucose concentrations require a high rate of glucose uptake.
Most organisms have evolved sophisticated regulatory strategies to adapt their metabolism to the availability of nutrients. Substrate uptake is a first key function that is regulated. Signals that control substrate uptake depend on the nature and concentration of available nutrients and the nutritional state of the cell. Since substrate uptake feeds back on the nutritional state, a regulatory circuit exists, the components of which are only beginning to be understood even in such intensely studied pathways as glycolysis in Saccharomyces cerevisiae (see reference 20 for a recent review).
S. cerevisiae cells are apparently able to sense the extracellular glucose concentration and transmit the signal over the membrane into the cytoplasm (20, 32). Two types of receptors have been proposed: hexose transporter-like receptors, which are involved in controlling hexose transporter (HXT) gene expression (30), and a G protein-coupled receptor that is required for the activation of the protein kinase A signaling pathway by cyclic AMP (22, 32, 36).
Intracellularly, glucose-phosphorylating enzymes play an important role in glucose regulation. Whether these enzymes exert their influence on glucose regulation through their metabolic activity or whether they function as intracellular signaling molecules is still a controversial issue (13, 15, 19, 33).
A signaling function has clearly been established for the galactose-phosphorylating enzyme of the yeast Kluyveromyces lactis. This galactokinase (KlGal1p) is required to activate transcription of lactose and galactose metabolic genes (26). Upon binding of its substrates galactose and ATP, KlGal1p can interact with the KlGal80 protein, an inhibitor of the transcription activator KlGal4p (an ortholog of S. cerevisiae Gal4p) (45). KlGal1p-KlGal80p interaction relieves KlGal4p inhibition and does not require galactokinase enzymatic activity. The KlGAL80 gene is also under control of KlGal4p, and KlGAL80 induction counteracts KlGal4p activation (44). Thus, the dynamics of induction depends crucially on the dynamics of KlGal80p inactivation. This, in turn, depends on the rate of lactose and galactose uptake, since the signaling molecule is intracellular galactose.
The induction process can be impaired in the presence of glucose if the concentration of KlGal4p is below a critical threshold (34, 43). Transcription of the LAC4-LAC12 genes, encoding β-galactosidase and lactose permease, respectively, and controlled by KlGal4p from a large bidirectional promoter (9, 17, 35), is particularly sensitive to glucose.
By screening for reduced β-galactosidase expression in glucose-lactose medium, mutations in glucose transporters were obtained that reduced the inhibitory effect of glucose (38).
By complementation of these mutants, two new K. lactis hexose transporter genes were isolated, KHT1 and KHT2 (38). These genes are closely linked and tandemly transcribed. Mutations in any of these genes slightly reduced the repression by glucose of lactose induction, whereas in the kht1 kht2 double mutant, the glucose effect was completely abolished. Not only the inducible LAC/GAL regulon, but also glucose repression of lactate dehydrogenase and malate dehydrogenase, was affected in the kht1 kht2 mutant.
KHT1 and KHT2 map to the same chromosomal locus as the low-affinity glucose transporter gene RAG1 described earlier (11). The sequence of RAG1 is almost identical to that of KHT1, except for the 3′ end (encoding K-A-M-L in RAG1 and K-R-F in KHT1), which is identical to the 3′ end of KHT2, indicating that RAG1 arose by recombination between KHT1 and KHT2 (38).
The presence of KHT1 and KHT2 correlated with a higher sensitivity to glucose repression found in only a few K. lactis strains (7). A natural isolate that was entirely insensitive to glucose repression was shown to carry a defective rag1 allele (18). These findings suggested that glucose repression in K. lactis depends on particular glucose transporters.
The KHT1/RAG1-KHT2 gene cluster is the only genetic locus in the respirofermentative yeast K. lactis that contains glucose permeases relevant for fermentative metabolism.
Another hexose transporter gene, HGT1, encoding a high-affinity transporter has a transport capacity too low to suppress the so-called “Rag−” phenotype of rag1 mutants (Rag+ = resistant against antimycin A on glucose) (18). A rag1 hgt1 double mutant like the kht1 kht2 mutant is still able to grow on glucose as a carbon source, indicating the presence of more still unidentified glucose transporters (5). Here we extend the studies on glucose transport in K. lactis by characterizing the Kht1p and Kht2p transport kinetics and by analyzing regulation of the KHT1 and KHT2 genes. We show that transporter gene regulation is influenced by transport activity and that Kht1p and Kht2p mutually control each other, depending on glucose availability. In the absence of both transporters, the high glucose response is impaired, supporting the view that the rate of sugar transport is a crucial parameter in intracellular glucose signaling.
MATERIALS AND METHODS
Strains and culture conditions.
The yeast strains used in this study are listed in Table 1. Strains JA6/CM57, JA6/CM58, DT12R/57, and DT12R/58 arose from ectopic integration of BamHI-linearized plasmids pBM3157 and pBM3158, respectively, into the genomes of strains JA6/DL4R (lac4) and JA6/DT12R (kht1 kht2). JA6/CM57 and JA6/CM58 carry single integrations, as shown by Southern blot hybridization, whereas DT12R/57 and DT12R/58 carry multiple copies. Cells were grown in batch culture at 30°C in synthetic minimal medium containing (per liter) 6.7 g of yeast nitrogen base (YNB) without amino acids (Difco) supplemented with required amino acids and bases. Carbon sources were routinely added at 2% (wt/vol) for glucose and galactose and 3% (wt/vol) for glycerol. Solid media were prepared by adding Bacto agar (Difco) to a final concentration of 2% (wt/vol). The β-galactosidase activity of K. lactis clones was checked on solid medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [40 μg/ml]).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant characteristics | Parent strain(s) | Source or reference |
|---|---|---|---|
| K. lactis | |||
| JA6 | α ade1-600 adeT-600 trp1-11 ura3-12 | SD11.U2 × W600B | 9 |
| JA6/DL4R | lac4::ura3 | JA6/DL4 (lac4::URA3) fluoroorotic acid-resistant derivative | 17 |
| JA6/CM57 | Chromosomal integration of KHT1-′lacZ fusion | JA6 (integration of pBM3157) | This work |
| JA6/CM58 | Chromosomal integration of KHT2-′lacZ fusion | JA6 (integration of pBM3158) | This work |
| JA6/DT12R | kht1::ura3::kht2 | JA6/DT12 (kht1::URA3::kht2) fluoroorotic acid resistant derivative | 38 |
| DT12R/57 | Chromosomal insertion of KHT1-′lacZ fusion | JA6/DT12R integration of pBM3157 | This work |
| DT12R/58 | Chromosomal insertion of KHT2-′lacZ fusion | JA6/DT12R integration of pBM3158 | This work |
| ST105 | RAG1 | AWJ-8c × MG1/2 | 38 |
| ST106 | rag1::URA3 | ST105 | 38 |
| S. cerevisiae EBY.VW4000 | Δhxt1-17 gal2 stl1 agt1 mph2-3 | CEN.PK2-1C | 40 |
Yeast transformation.
Competent cells of K. lactis were routinely prepared according to the method of Klebe et al. (21) and stored at −70°C (14). Transformation with linearized plasmid DNA for chromosomal integration was performed by the lithium acetate method (1).
DNA manipulation, preparation of yeast RNA, and Northern blot analysis.
The plasmids used are listed in Table 2. pBM3157 and pBM3158 are derivatives of plasmid YEp357R (28) carrying the promoter sequences of KHT1 and KHT2, respectively, as BamHI-EcoRI fragments fused to ′lacZ. Both plasmids were kindly supplied by S. Özcan (Lexington, Ky.). The isolation of total DNA from K. lactis was done as described earlier (7). All DNA techniques were performed according to standard procedures (1, 25).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| pBM3157 | YEp357 with KHT1 promoter (+3 to −1110) fused to ′lacZ | S. Özcan, unpublished data |
| pBM3158 | YEp357 with KHT2 promoter (+3 to −1642) fused to ′lacZ | S. Özcan, unpublished data |
| pKATUC4 | KICEN2-ARS1-KARS12-ScTRP1-ScURA3-vector | 44 |
| pJW10-7 | pUC12 with RAG1 gene | 38 |
| pJW10-23 | PUC12 with KHT1-KHT2 | 38 |
| pY10-7 | KATUC4 cassette in pJW10-7 | 37 |
| pY10-23 | KATUC4 cassette in pJW10-23 (KHT1-KHT2) | 37 |
| pVSH1 | pKATUC42 with HXT1 | This work |
| pVGAL2 | pKATUC42 with GAL2 | This work |
| p33-KHT1 | YCplac33 with KHT1 | 16; this work |
| p33-KHT2 | YCplac33 with KHT2 | 16; this work |
Details available on request.
Total RNA was isolated from K. lactis cells grown to the exponential phase by a hot phenol method. For Northern blot analysis, total RNA was fractionated by electrophoresis in 1.3% agarose–formaldehyde gels. Separated RNA was transferred to nylon membranes (Qiabrane; Qiagen) by capillary blotting with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (25). Prehybridization and hybridization were performed in high-sodium dodecyl sulfate (SDS) buffer (Church buffer) containing 7% (wt/vol) SDS, 50% (vol/vol) formamide, 5× SSC, 2% (wt/vol) Boehringer blocking reagent, 50 mM sodium phosphate (pH 7.0), and 0.1% (wt/vol) N-lauroylsarcosine. Incubation was overnight at 53°C. The following washes were performed: (i) 2× SSC–0.1% SDS at room temperature (twice, 15 min per wash) and (ii) 0.5× SSC–0.1% SDS at 68°C (twice, 15 min per wash). Membranes were exposed to X-ray film at −70°C. Labeling of DNA probes was done by random-primed DNA synthesis with [α-32P]dATP by using the Hexa labeling kit (MBI Fermentas).
β-Galactosidase assays.
β-Galactosidase activity (K. lactis Lac4p) was determined in crude extracts at 30°C as described previously (43). The activity of LacZ fusion proteins was measured in crude extracts at 37°C in LacZ buffer (27). In JA6/DT12-derived strains containing lacZ fusion genes, LAC4-derived background β-galactosidase activity was subtracted from the total activity by including parent strain DT12R in each experiment. The background never exceeded 15% of the lacZ activity and was not influenced by the glucose concentration. Protein concentrations were determined according to the method of Lowry (24) with bovine serum albumin as a standard.
Glucose determination.
Glucose concentrations in the culture supernatants were determined by using the glucose oxidase/peroxidase assay (3).
Glucose transport measurements.
Transport of [14C]glucose was determined in 5-s glucose-uptake assays as described previously (31).
RESULTS AND DISCUSSION
KHT1 and KHT2 encode functional glucose transporters with low and intermediate affinity, respectively.
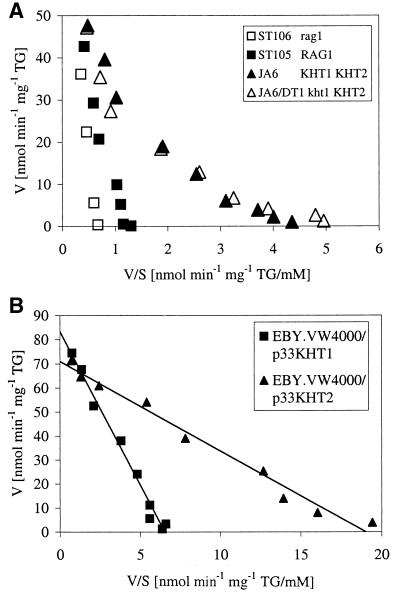
The fact that KHT1 and KHT2 are both able to complement the Rag− phenotype of a K. lactis rag1 mutant indicated that they encode functional glucose transporters (38). To further characterize the gene products, we first compared the kinetics of glucose uptake in a wild-type strain carrying both genes to that of a strain containing only RAG1. Glucose transport kinetics was determined in 5-s-uptake measurements (Fig. 1A). For strain JA6 (KHT1 KHT2), a biphasic curve with a high-affinity branch (Km ≈ 4.5 mM) was obtained. In strain ST105 (RAG1), the high-affinity branch was missing. Exact quantitation of Km was difficult in this strain due to high background at the high substrate concentrations; a rough estimate of 80 mM may not be significantly different from the Km of 20 to 50 mM described before for RAG1 (39).
FIG. 1.
(A) Transport of [14C]glucose in K. lactis strains. Glucose transport rates in different K. lactis strains were determined by 5-s-uptake measurements of [14C]glucose (31). Results are displayed in Eadie-Hofstee plots as uptake rates per milligram of dry weight (TG). The strains differed in the genetic background (squares versus triangles) and the presence (solid symbols) or absence (open symbols) of the low-affinity transporter gene KHT1 or RAG1.Solid triangles, JA6 (KHT1 KHT2); open triangles, JA6 (kht1 KHT2); solid squares, ST105 (RAG1); open squares, ST106 (rag1) (38). (B) Transport of [14C]glucose mediated by Kht1p and Kht2p in the glucose-uptake-deficient S. cerevisiae mutant EBY.VW4000. The strain was transformed with centromeric vectors carrying KHT1 (squares) and KHT2 (triangles), respectively.
Deletion of RAG1 in the latter strain further reduced glucose uptake, but the mutant was still able to grow on glucose at about the same rate (data not shown). Therefore, to determine the kinetic parameters of KHT1- and KHT2-mediated glucose uptake in the absence of any interfering transporters, we transferred these genes individually into the glucose-negative S. cerevisiae strain EBY.VW4000. In this strain, all 17 HXT genes, the galactose permease gene GAL2, and three members of the maltose permease family had been deleted. EBY.VW4000 is unable to grow on glucose medium, and no glucose consumption is detectable (40). Transformation with centromeric vectors carrying the K. lactis gene KHT1 or KHT2 restored growth on glucose, confirming that both KHT gene products mediate glucose uptake. Apparently, no K. lactis-specific factors are required for functional expression of these genes in S. cerevisiae.
Uptake measurements gave linear Eadie-Hofstee plots (Fig. 1B) with a Km for Kht1p of 13 mM and a Km for Kht2p of 3.7 mM. The Km of Kht1p, as determined in S. cerevisae, was somewhat lower than that of Rag1p (39); however, given the general difficulties in determining low-affinity uptake, we doubt that this difference reflects significant differences between Rag1p and Kht1p (38). Kht2p has an affinity for the glucose intermediate between the low-affinity transporter Rag1p and the high-affinity transporter Hgt1p (Km = 1 mM) described before (5, 39).
Expression of KHT1 and KHT2 is differentially regulated by glucose.
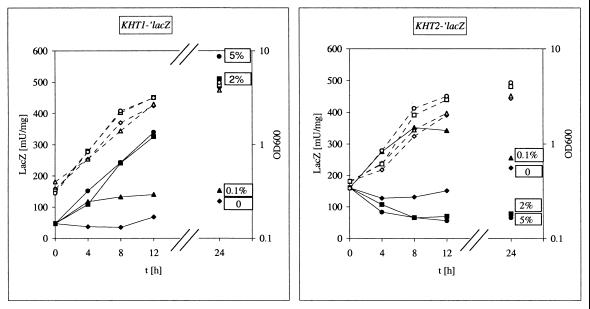
To study the regulation of the KHT1 and KHT2 genes, KHT1 and KHT2 promoter-driven lacZ reporter gene expression was analyzed. The fusion genes were integrated into the chromosome of K. lactis strain JA6/DL4R (KHT1 KHT2 lac4) mutated for the endogenous β-galactosidase gene, and reporter gene expression was measured in cells shifted from glycerol to different concentrations of glucose (Fig. 2). For strain JA6/CM57 carrying the KHT1-′lacZ fusion, β-galactosidase activity was low in glycerol and induced by glucose. Enzyme activity increased with the external glucose concentration up to a concentration of about 2% (100 mM) (Fig. 2) (data not shown) and proceeded slowly with about a twofold increase in 4 h.
FIG. 2.
Regulation by glucose of KHT1 and KHT2 promoter activity. Transformants of K. lactis strain JA6/DL4R (KHT1 KHT2 lac4) carrying single chromosomal insertions of KHT1-′lacZ and KHT2-′lacZ gene fusions, respectively, were grown to the exponential phase in YNB medium with 3% glycerol, collected by centrifugation, and resuspended at time 0 (t0) in the same glycerol medium containing different amounts of glucose as additional carbon sources (no glucose ♦, 0.1% glucose ▴ 2% glucose ▪, and 5% glucose ●). During batch cultivation at 30°C, samples were taken at the indicated time points for determination of lacZ activity (solid symbols) and cell density (open symbols with dotted lines) OD600, optical density at 600 nm.
In contrast, the regulation of KHT2 is more complex. The KHT2-′lacZ fusion (strain JA6/CM58) was expressed at a higher basal level in glycerol and induced about twofold by 0.1% (5 mM) glucose, whereas high glucose (2%) resulted in a weak repression of promoter activity.
The induction by glucose of KHT1 is apparently identical to that of RAG1, a fact that is not surprising given the sequence identity between KHT1 and RAG1 in the 5′-upstream region and most of the coding region. The difference in the 3′ ends between these genes has no obvious influence on regulation of promoter activity.
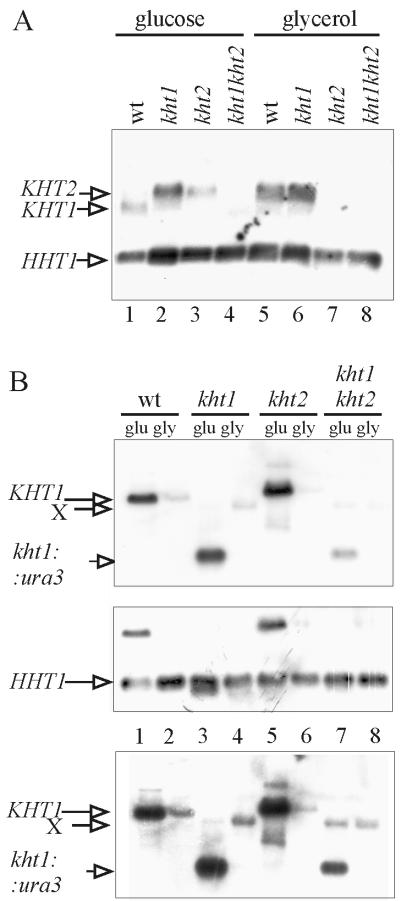
Northern blot analysis with a probe encompassing KHT1 and KHT2 showed that in logarithmic cells, reporter gene expression roughly reflected transcript levels (Fig. 3A). In wild-type cells grown in 2% glucose medium, the shorter KHT1-specific transcript dominated (lane 1), whereas in glycerol, the longer KHT2-specific signal was more prominent (lane 5), confirming a higher level of expression of this gene in the absence of glucose. The ratio between KHT1 and KHT2 mRNAs from glucose-grown cells varied from experiment to experiment, depending on the time in glucose medium and the density of the culture (data not shown). In most experiments, a very low KHT2 transcript level was observed in cells grown in high-glucose medium for extended periods (Fig. 3A, lane 1).
FIG. 3.
Northern blot analysis of KHT1- and KHT2-specific transcription in wild-type (WT) and kht mutant cells. K. lactis strain JA6 and congenic kht disruption mutants were grown to the exponential phase in YNB minimal medium containing 2% glucose (glu) (lanes 1 to 4) or 3% glycerol (gly) (lanes 5 to 8), and poly(A)+ RNA was isolated. Five micrograms of poly(A)+ RNA samples was fractionated by electrophoresis on a 1.3% agarose gel in the presence of formaldehyde. Plasmid pJW10–23 carrying the entire KHT-KHT2 locus (38) (A) and a small KHT1 5′-specific probe (HpaI-XhoI fragment of pJW10-7 [38] [B]) were used as probes. An HHT1 (histone H3) probe was added as a loading control. Note that in panel A, a weak kht1::ura3 fusion transcript overlaps with the HHT1 signal in lanes 2 and 7. In panel B, the two probes were applied sequentially (top and middle panels). A longer exposure of the top panel is shown at the bottom to visualize the weak cross-hybridizing transcript X.
The regulation of KHT2 resembles that of the S. cerevisiae HXT6/7 genes (6, 23). Interestingly, in a Clustal analysis, HXT6 and HXT7, which are nearly identical, turned out to be the closest relatives to KHT2 among the HXT genes. Like the KHT1-KHT2 locus in K. lactis (38), the HXT6-HXT7 locus seems to be recombinogenic, and some strains contain an HXT6/7 chimeric gene (10, 23). In the S. cerevisiae genome, HXT3 and HXT6/7 form a gene cluster similar to KHT1-KHT2, with respect to regulation and properties of the gene products. In a strain retaining only HXT3, HXT6, and HXT7, no growth deficiencies on glucose, fructose, and mannose could be observed (40). We speculate that this gene cluster evolved before the separation of K. lactis and S. cerevisiae species and represents another example of synteny between S. cerevisiae and K. lactis genomes as found at many loci (2, 41).
The kht1 kht2 double mutation eliminates the high glucose response of the KHT1 and KHT2 genes.
To examine the dependence of gene regulation on the structural integrity of the KHT transporter genes, the kht1, kht2, and kht1 kht2 mutants described previously (38) were analyzed in parallel to the wild type (Fig. 3). Interpretation of the banding pattern in Northern blots is complicated by the fact that the kht2 mutant had been generated by introducing the RAG1 gene into the kht1 kht2 deletion instead of KHT1. Since RAG1 differs at the very 3′ end from KHT1, it gives rise to an mRNA that is slightly longer than in the parent strain and migrates at the same position as the KHT2 transcript (Fig. 3A, lane 3). With a short 5′ probe, the KHT1 and RAG1-specific signals were identified (Fig. 3B). KHT1 promoter activity could be monitored even in kht1 mutants, since a small kht1::URA3 fusion was formed. This fusion transcript migrated close to the HHT1 transcript used as a loading control (Fig. 3A); therefore, the two probes were applied sequentially to the same filter (Fig. 3B, top and middle panels).
The intensity of the KHT1- and kht1::URA3 band was stronger on glucose (2%) than on glycerol in all four strains examined (Fig. 3B, lanes 1, 3, 5, and 7). However, in the kht1 kht2 double mutant, the induced level was reduced compared to those of the wild type and single mutants, indicating that at least one of the KHT genes is required for full induction of KHT1 and that the two genes can substitute for one another.
KHT2 transcription could not be analyzed in the same way, since no KHT2-specific transcript was formed in kht2 mutants. (Note that the signal in Fig. 3A, lane 3, is RAG1.)
Therefore, the KHT-′lacZ fusions were integrated into the genome of the kht1 kht2 strain JA6/DT12R, and the influence of the mutations on KHT1 and KHT2 promoter activities was determined by reporter gene assays (Table 3). For the KHT1 promoter, no influence of the kht1 kht2 mutations was detectable in low glucose (0.1%). The wild type and mutant had roughly the same ratio of β-galactosidase activity in glycerol versus glucose (2.7- and 3.3-fold, respectively). However, in 2% glucose, induction in the double mutant was weaker than in the wild type (3.3-fold versus 5.1-fold, respectively), in agreement with the results obtained by Northern analysis (Fig. 3).
TABLE 3.
Influence of hexose transporters on glucose induction and repression
| Genetic background | Gene on plasmid | Glucose induction at glucose concna:
|
|||||
|---|---|---|---|---|---|---|---|
|
KHT1′-lacZb
|
KHT2′-lacZb
|
LAC4b,e (2%) | PDCce (2%) | ||||
| 0.1%d | 2%d | 0.1%d | 2%d | ||||
| Wild-type | Vector | 2.7 | 5.1 | 2.1 | 0.4 | 0.2 | 5.2 |
| kht1 kht2 | Vector | 3.3 | 3.3 | 4.8 | 5.4 | 0.9 | 1.0 |
| kht1 kht2 | KHT1 KHT2 | NDf | 6.2 | ND | 1.0 | ND | ND |
| kht1 kht2 | RAG1 | ND | 4.6 | ND | 1.7 | 0.4 | 3.4 |
| kht1 kht2 | ScHXT1 | ND | 4.3 | ND | 3.3 | 0.6 | 1.0 |
| kht1 kht2 | GAL2 | ND | ND | ND | ND | 0.3 | 5.8 |
Fold- induction relative to glycerol (3%) cultures, mean of at least three independent experiments, deviations <15%.
β-galactosidase activity.
Pyruvate decarboxylase activity.
Glucose (0.1 or 2%) was added to 3% glycerol medium.
In gal80 mutant background.
ND, not determined.
For the KHT2 promoter, the influence of the mutations was even more striking. Whereas in the wild type, a modest induction by low glucose (2.1-fold) and a slight repression (0.4-fold induction) in high glucose were observed, induction was high (4.8-fold) in low glucose and even higher (5.4-fold) in high glucose. These results suggested that Khtp transporter activity was not required for induction but for repression of KHT2 gene expression at a high glucose concentration.
In the double mutant, another transcript of unknown origin, labeled “X” in Fig. 3B, was also released from glucose repression (compare lanes 3 and 4 to lanes 7 and 8 in Fig. 3B, bottom panel).
Taken together, the results indicate that in the absence of both KHT transporters, the specific high-glucose response of the KHT1 and KHT2 promoters does not occur, whereas the responses to low glucose were not affected by the mutations.
Several transporter genes restore the high-glucose response in kht1 kht2 double mutants.
To examine whether complementation of the glucose uptake deficiency was sufficient to restore the high-glucose response in the double mutant, transporter genes were introduced on centromeric plasmids into the kht1 kht2 strains carrying the integrated KHT1-lacZ and KHT2-lacZ fusions, respectively. (Table 3). In transformants with the KHT1-KHT2 tandem genes (plasmid pY10–23), the wild-type regulation of the KHT1 and KHT2 promoters was completely (KHT1-′lacZ) or almost completely (KHT2-′lacZ) restored. Partial induction of the KHT1 promoter and partial repression of the KHT2 promoter by glucose were observed with RAG1 (with plasmid pY10–7), and a weaker effect was seen with the S. cerevisiae HXT1 gene (on pVSH1), encoding a low-affinity hexose transporter.
We also assayed the ability of the S. cerevisiae GAL2 gene to complement the glucose uptake deficiency of the kht1 kht2 mutant strain. The galactose permease encoded by GAL2 had been shown to be able to mediate glucose uptake (23, 31), but the regulation of GAL2 (induction by galactose) suggested that it is not a component of a glucose signaling pathway. To overcome the requirement of galactose for the Gal4p-regulated GAL2 gene, the KlGAL80 gene was disrupted in the kht1 kht2 mutant background. The resulting triple mutant was Rag− like the parent, but a centromeric GAL2 plasmid clearly improved growth on 2% glucose plus antimycin A, an effect not observed when the KlGAL80 gene was functional (data not shown). Thus, Gal2p was able to function as a glucose transporter in K. lactis.
The triple mutant was used to analyze regulation of the LAC4 promoter (Table 3). The KHT1 KHT2 Klgal80 strain showed fivefold-lower LAC4-encoded β-galactosidase activity on glucose compared to glycerol (13,500 mU/mg of protein). This implies that glucose not only inhibits galactose induction of the LAC/GAL genes, but also represses the induced promoter to some extent. This form of glucose repression was abolished in the kht1 kht2 Klgal80 triple mutant. Again, repression could be restored by transformation with the RAG1- as well as with the GAL2-containing centromeric plasmid; ScHXT1 gave a weaker effect.
We also analyzed glucose induction of the glycolytic enzyme pyruvate decarboxylase encoded by the unique glucose-inducible gene, KlPDC1 (4, 12). In glycerol-grown cells, pyruvate decarboxylase activities varied between 60 and 260 mU/mg of protein. A fivefold induction by glucose observed in the Klgal80 mutant was completely abolished in the kht1 kht2 Klgal80 mutant and could be fully or partially restored by GAL2 and RAG1, respectively. No effect was observed with S. cerevisiae HXT1 in this case.
Thus, the kht1 kht2 mutation had a broad influence on glucose-regulated gene expression in general and was not restricted to the inducible LAC/GAL regulon (38). Formally, the possibility exists that the KHT gene products have regulatory activity in addition to their transport function, like the S. cerevisiae Snf3p and Rgt2p proteins, which serve as membrane-bound high- and low-affinity glucose receptors, respectively (30).
However, there is no indication that Kht1p and Kht2p are such glucose sensors. (i) Based on sequence similarity (65% identity), KHT1 and KHT2 clearly belong to the HXT gene family, with much weaker resemblance to SNF3 and RGT2 (25% identity), and the cytoplasmic tail characteristic of the latter gene products is not found in Kht1p and Kht2p. (ii) Both KHT1 and KHT2 encode functional glucose transporters, and the regulatory phenotype displayed by the kht1 kht2 double mutant could be restored by S. cerevisiae hexose transporter genes such as HXT1 and GAL2. (iii) There is a correlation between glucose uptake capacity and glucose signaling. The Hxt1p transporter, which has a very low affinity for glucose and requires high glucose concentrations for induction (29), had a weaker effect on glucose repression and induction than RAG1 and GAL2. The correlation between glucose uptake and the glucose response of gene expression is consistent with recent results from S. cerevisiae showing a correlation between relief from glucose repression and the decrease in glucose transport capacity. Even under high-glucose conditions, an increase in SUC2 expression was detected when cells displayed reduced glucose uptake (31, 42).
We conclude that Kht1p and Kht2p influence regulation through their transport function and that glucose sensing occurs intracellularly.
An intracellular sensing mechanism for glucose does not exclude an additional extracellular sensor. The RAG4 gene conferring a Rag− phenotype when mutated encodes an Snf3/Rgt2-related protein with a long C-terminal cytoplasmic tail containing the conserved sequence motif (3a). It is a regulator of RAG1 and may mediate the low glucose response of KHT1/RAG1, which we have shown here to be independent of KHT1 and KHT2. A combination of extracellular and intracellular sensing mechanisms would ensure that the high-glucose response is only triggered if the cell is able to metabolize the available glucose.
We propose that in K. lactis glucose repression correlates with fermentative metabolism. Both forms of regulation require the transport capacity of the cell to exceed a threshold level. This level cannot be reached in the kht1 kht2 or rag1 mutant. The presence of either KHT1/RAG1 or KHT2 alone is sufficient to overcome the threshold. By a self-sustaining process caused by further induction of KHT1/RAG1 and other fermentative genes (like the pyruvate decarboxylase gene KlPDC1) and by favoring the utilization of glucose over other carbon sources through glucose repression, the metabolism is shifted towards fermentation. The presence of both genes KHT1 and KHT2 favors this metabolic shift due to the high basal expression of KHT2 resulting in a higher uptake capacity in noninduced cells. Since KHT2 itself is subject to glucose repression, the transporter with the higher Km, Kht2p, is replaced by the one with the lower Km if there is a high and constant supply of glucose. The slow and concentration-dependent induction of Kht1p/Rag1p enables the cells to monitor the availability of glucose over time; its low-affinity ensures a concentration-dependent rate of transport at subsaturating glucose levels.
In S. cerevisiae, the abundance of hexose transporter genes and their specific regulation indicate that this yeast tries to sustain the highest possible glucose transport activity, such that the glycolytic flux is largely determined by the extracellular glucose concentration. In contrast, the respirofermentative yeast K. lactis does not fully exploit its glucose uptake capacity during oxidative growth. Sugar uptake may therefore be primarily controlled by the availability of oxygen and by the cellular demand rather than by the extracellular supply (8).
ACKNOWLEDGMENTS
This work was supported by EU grant BIO4-CT96–0003 to K.D.B.
We are grateful to S. Özcan for providing plasmids and unpublished data, to M.Wésolowski-Louvel for communicating results prior to publication, to A. Kruckeberg for stimulating discussion, and to P. Kuger for excellent technical support.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- 2.Bao W G, Fukuhara H. The ubiquitin-encoding genes of Kluyveromyces lactis. Yeast. 2000;16:343–351. doi: 10.1002/1097-0061(20000315)16:4<343::AID-YEA534>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Bergmeyer H U. Methoden der enzymatischen Analyse. Weinheim, Germany: Verlag Chemie; 1974. [Google Scholar]
- 3a.Betina S, Goffrini P, Ferrero I, Wesolowski-Louvel M. RAG4 gene encodes a glucose sensor in Kluyveromyces lactis. Genetics. 2001;158:541–548. doi: 10.1093/genetics/158.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi M M, Tizzani L, Destruelle M, Frontali L, Wésolowski-Louvel M. The ‘petite-negative’ yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol Microbiol. 1996;19:27–36. doi: 10.1046/j.1365-2958.1996.346875.x. [DOI] [PubMed] [Google Scholar]
- 5.Billard P, Ménart S, Blaisonneau J, Bolotin-Fukuhara M, Fukuhara H, Wésolowski-Louvel M. Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J Bacteriol. 1996;178:5860–5866. doi: 10.1128/jb.178.20.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boles E, Hollenberg C P. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 7.Breunig K D. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet. 1989;216:422–427. doi: 10.1007/BF00334386. [DOI] [PubMed] [Google Scholar]
- 8.Breunig K D, Bolotin-Fukuhara M, Bianchi M M, Bourgarel D, Falcone C, Ferrero I, Frontali L, Goffrini P, Krijger J J, Mazzoni C, Milkowski C, Steensma H Y, Wesolowski-Louvel M, Zeeman A M. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb Technol. 2000;26:771–780. doi: 10.1016/s0141-0229(00)00170-8. [DOI] [PubMed] [Google Scholar]
- 9.Breunig K D, Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987;7:4400–4406. doi: 10.1128/mcb.7.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown C J, Todd K M, Rosenzweig R F. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 11.Chen X J, Wésolowski-Louvel M, Fukuhara H. Glucose transport in the yeast Kluyveromyces lactis. II. Transcriptional regulation of the glucose transporter gene RAG1. Mol Gen Genet. 1992;233:97–105. doi: 10.1007/BF00587566. [DOI] [PubMed] [Google Scholar]
- 12.Destruelle M, Menghini R, Frontali L, Bianchi M M. Regulation of the expression of the Kluyveromyces lactis PDC1 gene: carbon source-responsive elements and autoregulation. Yeast. 1999;15:361–370. doi: 10.1002/(SICI)1097-0061(19990330)15:5<361::AID-YEA378>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.De Winde J H, Crauwels M, Hohmann S, Thevelein J M, Winderickx J. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur J Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- 14.Dohmen R J, Strasser A W M, Höner C B, Hollenberg C P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- 15.Entian K-D. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in catabolite repression in yeast. Mol Gen Genet. 1980;178:633–637. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 17.Gödecke A, Zachariae W, Arvanitidis A, Breunig K D. Coregulation of the Kluyveromyces lactis lactose permease and β-galactosidase genes is achieved by interaction of multiple LAC9 binding sites in a 2.6 kbp divergent promoter. Nucleic Acids Res. 1991;19:5351–5358. doi: 10.1093/nar/19.19.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goffrini P, Algeri A A, Donnini C, Wésolowski-Louvel M, Ferrero I. RAG1 and RAG2: nuclear genes involved in the dependence/independence on mitochondrial respiratory function for growth on sugars. Yeast. 1989;5:99–106. doi: 10.1002/yea.320050205. [DOI] [PubMed] [Google Scholar]
- 19.Hohmann S, Winderickx J, De Winde J H, Valckx D, Cobbaert P, Luyten K, de Meirsman C, Ramos J, Thevelein J M. Novel alleles of yeast hexokinase PII with distinct effects on catalytic activity and catabolite repression of SUC2. Microbiology. 1999;145:703–714. doi: 10.1099/13500872-145-3-703. [DOI] [PubMed] [Google Scholar]
- 20.Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 21.Klebe R J, Harris J V, Smart Z D, Douglas M G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983;25:333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- 22.Kraakman L, Lemaire K, Ma P, Teunissen A W, Donaton M C, Van Dijck P, Winderickx J, De Winde J H, Thevelein J M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 23.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 26.Meyer J, Walker-Jonah A, Hollenberg C P. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5454–5461. doi: 10.1128/mcb.11.11.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 28.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 29.Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Özcan S, Dover J, Rosenwald A G, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem. 1997;245:324–333. doi: 10.1111/j.1432-1033.1997.00324.x. [DOI] [PubMed] [Google Scholar]
- 32.Rolland F, De Winde J H, Lemaire K, Boles E, Thevelein J M, Winderickx J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol Microbiol. 2000;38:348–358. doi: 10.1046/j.1365-2958.2000.02125.x. [DOI] [PubMed] [Google Scholar]
- 33.Rose M, Albig W, Entian K-D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991;199:511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- 34.Sheetz R M, Dickson R C. Mutations affecting synthesis of β-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980;95:877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sreekrishna K, Dickson R C. Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc Natl Acad Sci USA. 1985;82:7909–7913. doi: 10.1073/pnas.82.23.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Versele M, De Winde J H, Thevelein J M. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weirich J. Ph.D. thesis. Isolierung und Charakterisierung des Glukose Transporters RAG1 aus Kluyveromyces lactis und Untersuchungen zu dessen Rolle bei der Glukoserepression der β-Galaktosidase. Düsseldorft, Germany: Heinrich-Heine-Universität Düsseldorf; 1992. [Google Scholar]
- 38.Weirich J, Goffrini P, Kuger P, Ferrero I, Breunig K D. Influence of mutations in hexose-transporter genes on glucose repression in Kluyveromyces lactis. Eur J Biochem. 1997;249:248–257. doi: 10.1111/j.1432-1033.1997.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- 39.Wesolowski-Louvel M, Goffrini P, Ferrero I, Fukuhara H. Glucose transport in the yeast Kluyveromyces lactis. I. Properties of an inducible low-affinity glucose transporter gene. Mol Gen Genet. 1992;233:89–96. doi: 10.1007/BF00587565. [DOI] [PubMed] [Google Scholar]
- 40.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg C P, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 41.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 42.Ye L, Kruckeberg A L, Berden J A, Van Dam K. Growth and glucose repression are controlled by glucose transport in Saccharomyces cerevisiae cells containing only one glucose transporter. J Bacteriol. 1999;181:4673–4675. doi: 10.1128/jb.181.15.4673-4675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zachariae W, Kuger P, Breunig K D. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9 (KlGAL4) Nucleic Acids Res. 1993;21:69–77. doi: 10.1093/nar/21.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zenke F, Zachariae W, Lunkes A, Breunig K D. Gal80 proteins of Kluyveromyces lactis and Saccharomyces cerevisiae are highly conserved but contribute differently to glucose repression of the galactose regulon. Mol Cell Biol. 1993;13:7566–7576. doi: 10.1128/mcb.13.12.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zenke F T, Engels R, Vollenbroich V, Meyer J, Hollenberg C P, Breunig K D. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science. 1996;272:1662–1665. doi: 10.1126/science.272.5268.1662. [DOI] [PubMed] [Google Scholar]