Abstract
Objective diagnosis and prognosis in major depressive disorder (MDD) remains a challenge due to the absence of biomarkers based on physiological parameters or medical tests. Numerous studies have been conducted to identify functional magnetic resonance imaging‐based biomarkers of depression that either objectively differentiate patients with depression from healthy subjects, predict personalized treatment outcome, or characterize biological subtypes of depression. While there are some findings of consistent functional biomarkers, there is still lack of robust data acquisition and analysis methodology. According to current findings, primarily, the anterior cingulate cortex, prefrontal cortex, and default mode network play a crucial role in MDD. Yet, there are also less consistent results and the involvement of other regions or networks remains ambiguous. We further discuss image acquisition, processing, and analysis limitations that might underlie these inconsistencies. Finally, the current review aims to address and discuss possible remedies and future opportunities that could improve the search for consistent functional imaging biomarkers of depression. Novel acquisition techniques, such as multiband and multiecho imaging, and neural network‐based cleaning approaches can enhance the signal quality in limbic and frontal regions. More comprehensive analyses, such as directed or dynamic functional features or the identification of biological depression subtypes, can improve objective diagnosis or treatment outcome prediction and mitigate the heterogeneity of MDD. Overall, these improvements in functional MRI imaging techniques, processing, and analysis could advance the search for biomarkers and ultimately aid patients with MDD and their treatment course.
Keywords: biomarkers, major depressive disorder, MRI, review
INTRODUCTION
Major depressive disorder (MDD) is a prevalent neuropsychiatric disorder with at least one episode occurring in the lifetime of 15%‐18% of the people worldwide. 1 Various hypotheses have been proposed regarding its underlying pathology that involve abnormal levels of monoamine, cortisol, inflammatory processes, or structural and/or functional abnormalities of the brain. 1 Many researchers have attempted to explore the latter hypothesis by identifying brain imaging biomarkers and biological subtypes (biotypes) of depression. Ultimately, these insights and biomarkers could contribute to a better diagnosis and an improved prediction of depression treatment outcome.
To study the functional hypothesis of MDD, one of the most frequently employed modalities in depression studies is functional magnetic resonance imaging (fMRI), both in task‐based and resting‐state conditions. This technique indirectly measures brain activity and therefore provides a powerful way of examining the underlying aberrant brain mechanisms in psychiatric disorders. 2 fMRI detects changes in blood deoxyhemoglobin concentrations that reflect the oxygen consumption. The Blood‐Oxygen‐Level‐Dependent (BOLD) signal, an indirect measure of the concentration of (de)oxygenated hemoglobin in blood, changes in active brain regions due to hemodynamic responses that occur as a consequence of the oxygen consumption. 2 , 3
MDD studies have used fMRI to identify biomarkers for different purposes, including understanding of affected functioning and connectivity of brain regions or networks and prediction of treatment response. 4 Here, a biomarker is defined as an objectively measurable image feature that is an indicator of the diagnosis—or reflects or predicts the treatment course—of MDD subjects. 5 In order to make a diagnosis of prognosis in MDD, both task‐based fMRI (tb‐fMRI) and resting‐state fMRI (rs‐fMRI)—during which subjects perform dedicated tasks or do not perform any task and lie still in the scanner, respectively—have frequently been employed. Even though a small number of consistent MDD biomarkers has been found, many contradictory findings are still present in the literature. 6
In order to improve the search for functional imaging biomarkers in depression, this review provides an overview of the most consistent findings and pinpoints future opportunities in fMRI studies of MDD. The current limitations and pitfalls that could underlie the discrepant findings are also briefly discussed. The “FUTURE OPPORTUNITIES” section focusses on promising developments and future directions in image acquisition, processing, and analysis approaches. The principal aim of the review is to address and discuss current developments and future directions that could improve the search for consistent functional imaging biomarkers in MDD. We will address the following research questions: what are future opportunities for novel fMRI acquisition, processing, and analysis methods to improve the search for imaging biomarkers of MDD?
FINDINGS AND DEVELOPMENTS
In task‐based fMRI depression studies, subjects perform tasks requiring activation of brain areas or circuits that are presumably affected in MDD, which mostly assess the affective and cognitive domain. Activity in limbic (anterior cingulate cortex [ACC] and amygdala) and frontal cortical areas (dorsolateral prefrontal cortex [dlPFC]) has been shown to be altered in depression during viewing or processing of emotional stimuli—mainly facial expressions. 7 , 8 , 9 , 10 There is some consensus that activity of limbic regions is increased, whereas in frontal regions (involved in numerous cognition processes) the activity is decreased. During cognitive tasks, mostly prefrontal activity increases have been found in depressed subjects despite task performance scores that were similar to healthy controls (HCs), whereas reduced activity is often observed in combination with poor performance. 11 , 12 This suggests that depression is associated with cortical inefficiency and is compensated by enhancement of brain activation in these regions.
In terms of prediction of treatment outcome, less consistent tb‐fMRI biomarkers have been identified so far with one exception: the rostral ACC (rACC). Increased baseline activity of the rACC during viewing of images of negative emotional faces has often been found to be predictive of antidepressant treatment response. 8 , 10 , 13 Yet, there are still many studies that find controversial results, for example, regarding the role of the amygdala. This brain area is assumed to be essential in the altered brain networks of depression as it is associated with fear and other emotional processing functions. Some researchers have found differences in amygdala response during affective tasks, whereas others found contradictory findings or no significant differences at all. 14 , 15 , 16 , 17
rs‐fMRI studies have identified several network functional connectivity (FC) differences between MDD patients and HCs. Networks that have found to be altered include the default mode network (DMN), salience network, central executive network (CEN; also sometimes referred to as frontoparietal network), dorsal attention network, and affective network, almost all of which play a role in affective processing and cognitive control. 18 , 19 , 20 More specifically, hyperconnectivity within the DMN and hypoactivity within the CEN have frequently been observed in reviews and meta‐analyses. Contradictory, a recent mega‐analysis did not find decreases within the CEN and DMN, whereas another reported DMN hypoactivation. 21 , 22
rs‐fMRI has also been studied to identify biomarkers of treatment outcome. Increased FC within areas of the DMN such as the posterior cingulate cortex and medial prefrontal cortex (mPFC) and decreased FC within areas of the cognitive control network such as the insula, ACC, and dlPFC have shown to separate antidepressant treatment‐resistant from treatment‐responsive MDD patients. 17 , 23 , 24 An overview of the most consistent tb‐fMRI and rs‐fMRI biomarkers of objective diagnosis and treatment response in MDD is shown in Figures 1A and 1B, respectively.
FIGURE 1.
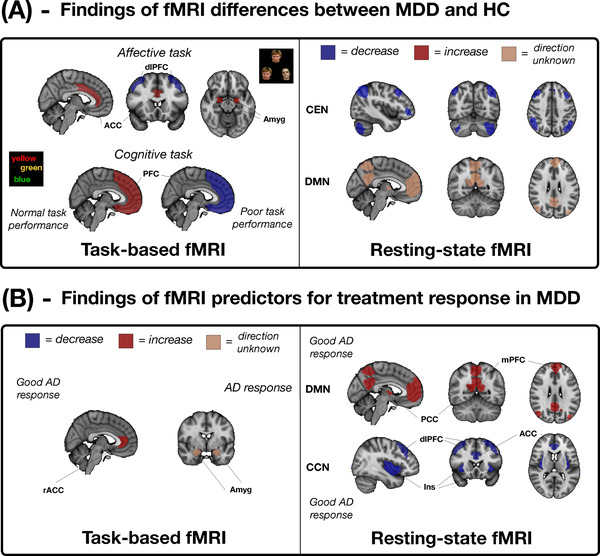
Identified relative consistent functional major depressive disorder biomarkers for task‐based and resting‐state fMRI for (A) objective diagnosis (MDD > HC) and (B) treatment response. ACC, anterior cingulate cortex; AD, antidepressant treatment; Amyg, amygdala; CCN, cognitive control network; CEN, central executive network; dlPFC, dorsolateral PFC; DMN, default mode network; HC, healthy controls; Ins, insula; MDD, major depressive disorder; mPFC, medial PFC; PCC, posterior cingulate cortex; PFC, prefrontal cortex; rACC, rostral ACC
LIMITATIONS AND PITFALLS
Structural and functional MRI studies of MDD have recently been criticized. A crucial factor is the small sample size in case‐control and brain‐behavioral association studies. 25 , 26 Typical neuroimaging sample sizes often lead to statistically underpowered results and low replication rates. Multicohort, within‐participant study designs with multivariate and data‐driven analyses have been suggested as improvements for statistical power. 26 , 27 Furthermore, effect size measures of HC‐MDD group differences are often small despite harmonization of study acquisition and methodology. 27 The previously highlighted consistent biomarkers should therefore be taken with caution, even though they been reported in numerous studies. Importantly, however, fMRI measures show less distributional overlap between groups and increased out‐of‐sample replication rates on average compared to structural MRI and diffusion tensor imaging. 26 , 27 Moreover, improvements in functional image quality and more advanced analyses might enhance reproducibility and the chances of identifying robust MDD biomarkers. 26 , 27 In this review, we first provide further elucidation of the received criticism by discussing the most notorious limitations and pitfalls of fMRI image acquisition, processing, and analysis methods in MDD studies. The topics that will be discussed explain why these approaches may restrain researchers from identifying robust functional biomarkers of depression. Subsequently, we highlight future directions that could mitigate these problems and lead to improved data acquisition and analysis methodology.
Acquisition limitations
Conventional fMRI acquisitions limit the reliability of quantification of functional measures by several factors including susceptibility to noise and artifacts, relatively low temporal and spatial resolutions, and low signal‐to‐noise ratio (SNR). For example, regions such as the orbitofrontal cortex (OFC), cingulate, parahippocampus, amygdala, and striatum have been found to be functionally abnormal in MDD. 28 , 29 These limbic and frontal areas are often affected by signal dropout as their T2* values are dramatically reduced due to tissue‐air boundaries surrounding the nasal cavity, mastoid air cells, and sphenoid sinus. 30 , 31 Tissue‐air boundaries distort the field because they are diamagnetic versus paramagnetic, bending the magnetic fields differently resulting in increased dephasing and signal loss. Additionally, only a few depression studies acquired fMRI scans with a repetition time (TR) below 2 seconds to assess FC between brain regions or networks. 19 , 24 Low temporal resolutions time‐series often suffer from a relatively high amount of noise and decrease the power in statistical tests (less time points) that are conducted to derive functional measures. 32 Typical TRs from a conventional echoplanar imaging acquisition demonstrated lower signal‐noise separation and test‐retest reliability values in FC analyses compared to acquisitions with lower TRs. 33
Conventional cleaning methods
Presumably the most acknowledged disadvantage of fMRI is its susceptibility to contamination from motion, physiological, and scanner‐related (eg, thermal) sources. Respiratory (around 0.3 Hz) and cardiac (1‐2 Hz) signals can alias into the lower frequencies at typical TRs of 2 seconds or higher during acquisition. 34 In addition, arteriole CO2, respiratory volume per time, and heart rate variability could introduce low frequencies into the fMRI spectrum as they slowly fluctuate at around the same frequency as the BOLD signal (<0.1 Hz). 34 Notably, depression is being associated with reduced heart rate variability according to a meta‐analysis. 35 Prevention or cleaning of fMRI data is therefore especially relevant in MDD studies. Model‐based and data‐driven methods exist to limit the amount of aliased noise in the fMRI data.
Of the model‐based denoising methods, the most common is RETROICOR. 36 RETROICOR requires external recordings of cardiac and respiratory signals and models them using low‐order Fourier series taking into account time‐varying phases of physiological signals. The models are then treated as nuisance regressors and removed from the fMRI signal. 36 , 37 Despite high performance, RETROICOR is model based and thus depends on the accuracy of the contribution of the models to the BOLD signals. 38 In addition, monitoring of external signals can be difficult and sensitive to noise induced in the MR setting. 39 The data‐driven method CompCor generates nuisance regressors from principal components of time‐series of voxels with a high standard deviation, in which cardiac and respiratory noise is assumed to be abundant. 40 This assumption is invalid if motion artifacts are dominating. 40 , 41 Moreover, CompCor could introduce additional artifacts into the signal. 39 Other popular data‐driven denoising methods are FMRIB's independent component analysis‐based Xnoiseifier (FIX‐ICA) 42 and ICA‐AROMA 43 that have shown to perform well but have other limitations or restrictions. 44 , 45 FIX‐ICA, a method to classify independent components (ICs) as either noise or neural components, is slow as it extracts over 180 features and depends on the pretraining of the classifier with manually labeled data. 46 , 47 Unlike FIX‐ICA, ICA‐AROMA is fully automated but solely cleans motion artifacts. 44
Limitations of current analysis methods
Among depression studies, the most popular rs‐fMRI analysis methods are seed‐based correlation analysis (SCA) and ICA, which provide insights about the activation synchronicity between multiple brain regions or networks. Other popular methods include regional homogeneity (ReHo) and amplitude of low‐frequency fluctuations (ALFF), which measure local synchronicity and power of low‐frequency oscillations, respectively. 48
Some inconsistent findings have been found in the literature of rs‐fMRI and MDD that perform SCA to assess FC. For example, connectivity between the anterior and posterior DMN is often altered in MDD but both increases and decreases have been reported. Moreover, SCA is sensitive to the seed placement, 20 , 49 which can differ significantly even between adjacent voxels and is study specific or standardized across subjects. Subject‐specific defined seeds could mitigate a part of this problem since individual spatial differences are taken into account. 50
ICA identifies functional resting‐state networks that are statistically independent. Results from ICA in MDD patients have been found to be more consistent than SCA. 20 Consensus exists regarding the clear distinction of the anterior and posterior DMN in MDD, which were found to be spatially independent in depressed patients. Mostly, increases are reported in both parts of the DMN compared to HCs. The advantage of ICA is that this method does not require an a priori assumption and identifies whole brain networks. A disadvantage is that the number of components is rather an arbitrary choice, possibly resulting in subnetwork components or spatially global network components if the input number is chosen too large or small, respectively. 51 The number of components is still variable over different fMRI studies despite attempts to harmonize the number of components. 52 Adding to this, the underlying source of the independent networks is difficult to trace as ICA is also very sensitive toward coherent nonneuronal signals. 20 , 48
ReHo measures the synchronicity of the BOLD signal in a voxel with its neighboring voxels, an indirect measure of local neural activity. Controversial ReHo findings have been found in MDD studies. One meta‐analysis confirmed the presence of increases in ReHo in the left hippocampus and decreases in the OFC, another found decreases in the insula and superior temporal gyrus, and a third meta‐analysis reported a significant increase in the mPFC. 53 , 54 , 55 An explanation for the different findings could be attributed to the frequency dependence of ReHo in MDD patients as alterations in brain regions have been found for different low‐frequency bands that might have distinct physiological meaning. 56 Finally, ReHo is very sensitive to smoothing because of its local properties, whereas the studies of the meta‐analyses applied different Gaussian kernel sizes. 57
ALFF measures the regional brain activity and is calculated as the sum of spectral amplitudes in the range of 0.01‐0.1 Hz after Fourier transforming each voxel's time‐series. Like ReHo, ALFF is similar with regard to that it reflects neural activity but only provides local information. Nowadays, methods such as SCA and ICA are more often applied in neuroimaging studies of depression to discover potential biomarkers such as large‐scale networks alterations and their temporal interactions that may be more predictive of antidepressant treatment outcome. 20 , 48
Task design inconsistencies
A possible reason for contradictory results is the dissimilar task paradigms among depression studies. In MDD studies, affective and cognitive tasks have been most commonly used, while some have implemented reward‐based and self‐referential tasks.
MDD studies that have implemented affective facial recognition tasks in MDD include various emotions such as fear, sadness, happiness, and neutral faces that are processed differently in the brain. Yet, there is still no consensus on which emotions are particularly regulated abnormally in depression patients and which brain region or networks are responsible for the processing of each valence. For example, in one MDD study amygdala increases were only observed after viewing sad faces but not fearful faces, whereas numerous of studies found similar increases toward fearful faces. Furthermore, some researchers use contrasts between the BOLD signal of negative and positive stimuli tasks, while others compare negative or positive valences to neutral or scrambled faces. 7 , 58 , 59 , 60 , 61 , 62
Most of the fMRI experiments in MDD require the participants to respond to stimuli, for example, with an MRI‐compatible keypad. Responses can be categorized in explicit (emotion directed, eg, which emotion was shown) or implicit (not related to emotions, eg, which gender was shown). Next to that, there are studies in which participants are asked to passively view the images. Activation tasks, for example, demanding the participant to press a button, could activate brain areas involved in decision‐making and motor control, which could have a confounding effect on the affective or cognitive aspect of fMRI tasks. 63 It is therefore essential to implement task contrasts by not only including rest but also control blocks that are designed to capture the confounding factors without the activity of interest. 14 , 64 , 65 , 66 , 67 An example is the Hariri task in which not only faces are matched but also shapes. 68
Differences in task difficulty could influence the observed brain activity patterns between studies that assess cognition through executive tasks. In MDD, patterns of hyperactivation in cortical and frontal brain areas are often observed combined with unaffected cognitive performance and reduced cortical activation with poor performance. 12 , 69 , 70 These contradictory findings have prompted suggestions that it is possible that this pattern is only evident in more difficult tasks. 12 Furthermore, some antidepressants might impair cognitive functioning of depressed patients as a result of sedative effects, while others have no or an improved effect on cognition. 71
FUTURE OPPORTUNITIES
This section discusses the future opportunities of fMRI studies in MDD, supported by relevant approaches, findings, and recommendations of novel depression research. It is divided into a subsequent order from first acquiring MRI images, followed by image processing to the subsequent analysis of the images. Some of the described sections are also relevant for fMRI studies or psychiatric disorders in general. An overview of the highlighted future opportunities and their benefits is presented in Table 1.
TABLE 1.
Summary of future directions of fMRI studies to improve the identification of biomarkers in major depressive disorder
| Future opportunities | Benefits | |
|---|---|---|
| Image acquisition |
|
|
| Image processing |
|
|
| Image analysis |
|
|
Note: A summary of future opportunities of functional MRI studies to identify more consistent major depressive disorder biomarkers.
Abbreviations: BOLD, blood‐oxygen‐level dependent; MDD, major depressive disorder; SNR, signal‐to‐noise ratio.
Image acquisition
Higher magnetic field strengths
Higher SNR of fMRI scans can be obtained by imaging at a higher magnetic field strength (B 0). 72 This is because the induced voltage (MR signal) in the receiver coil is proportional to the square of B 0 as it is dependent on the precession rate of the spins and the net magnetization. 72 With increasing B 0, both the precession rate and net magnetization increase linearly. The latter is a result of the larger fraction of protons that flip to the high‐energy state. The noise only increases linearly with B 0 and consequently the SNR increases linearly with B 0. In line with this, an MDD study demonstrated significant increases in temporal SNR (tSNR) of brain scans at 7T. These ultrahigh‐field images enhanced sensitivity of detecting mood‐related neurocircuit disturbances. 73 This is of special interest in psychiatric diseases that are associated with affected subcortical brain regions or networks. Moreover, the BOLD contrast increases as a result of the increase in T2* dephasing from the more prominent local field inhomogeneities. 74 Yet, physiological noise is also more comprised at higher magnetic field strengths. 75 During fMRI imaging of subcortical brain regions, physiological noise and BOLD sensitivity losses due to high iron content and larger distances from the coil are more pronounced. 76 Despite the increased SNR, BOLD contrast, and percent signal change, images acquired at higher magnetic fields suffer from increased B 0‐inhomogeneities, particularly in subcortical and midbrain regions. One way to minimize signal dropout is by using multiecho (ME) imaging, which will be discussed later. Future studies minimizing the B 0‐imhomogeneities, for example, by improvements in shimming, may further improve 7T fMRI imaging. 77 Another way to reduce the effect of susceptibility artifacts is by placing dielectric pads close to the region of interest that increases the magnetic field homogeneity. 78 Next to that, optimization of fast imaging protocols could prevent physiological noise from aliasing into the BOLD frequency range. Such a protocol optimization study for 7T fMRI was recently published in which BOLD sensitivity was optimized for subcortical structures while maintaining sensitivity in cortical areas. 79 Lastly, advancements in receiver coils could diminish the effects of physiological artifacts. 80
Multiband imaging
Preventing aliasing of nonneural physiological signals would reduce the abundance of noise to which fMRI is susceptible to. One way in which this could be achieved is by significantly reducing the TR. Parallel imaging methods such as SENSitivity encoding and GeneRalized Autocalibrating Partial Parallel Acquisition (GRAPPA) reduce scanning time by partially acquiring k‐space data and recovering signals by utilizing properties and spatial sensitivity from multiple receiver coils. 81 This comes at the cost of SNR loss due to undersampled data and amplification of noise from the coils. 82 Additionally, images are often affected by distortions. Other acceleration techniques include compressed sensing and k‐t reconstruction. More recently, multiband (MB) imaging was introduced. 83 MB imaging offers the opportunity to simultaneously acquire slices and thus decrease the total scan time. The SNR penalty of MB imaging is minimal and only reduced by amplified reconstruction noise and lower net magnetization. 84 Lastly, MB imaging can cause slice leakage artifacts, that is, slice aliasing. A specific GRAPPA technique has been developed that optimizes reconstruction of simultaneously acquired slices by minimizing the influence of slice leakage. 85
Some resting‐state and task‐based depression studies have already shown the potential of MB acquisition, yielding TRs of around 1 second. 73 , 86 Recently, an fMRI echoplanar imaging MB sequence with a TR of 350 milliseconds was developed, resulting in, both, an improved SNR ratio compared to conventional fMRI images and more reliable brain networks after only 3 minutes of scanning. 33
ME imaging
In ME sequences, each slice is acquired at multiple echo times (TEs). This results in multiple time‐series of the same run with different signal intensities and contrasts. The number of time‐series is equal to the number of different TEs at which each slice has been acquired. Volumes of the time‐series acquired at smaller TEs contain higher voxel intensities but lower contrast between white and gray matter and cerebral spinal fluid. Vice versa, higher TEs lead to lower signal intensities but higher contrast. By fitting the signal intensity decay over TE, one estimates the so‐called T2* decay, reflecting the magnetic field inhomogeneities. The T2* decay is different for non‐BOLD signals compared to BOLD signals, where the latter is dependent on the TE. 87 By taking this into account, MRI signals can be decomposed into separate ICs using ICA and classified as either BOLD or non‐BOLD. 88 Subsequently, they can be regressed out from the fMRI signal to reduce the amount of non‐BOLD signals.
As the T2* value is estimated per voxel, the T2* variation across regions is taken into account with ME imaging. 89 Moreover, ME imaging decreases the effect of dropout. A recent study demonstrated increases in test‐retest reliability of individual‐specific FC in the subgenual cingulate, basal ganglia, and cerebellum in an ME sequence. 90 FC reliability of 10 minutes of ME fMRI was found to be comparable with 30 minutes of single‐echo fMRI. Moreover, reliability of functional brain networks was enhanced in ME fMRI. The researchers attributed these improvements partly to the decrease in signal dropout and S0‐dependent artifacts such as head motion or MRI hardware instabilities. Finally, the results of this study are highly valuable for the search of neuroimaging biomarkers. As demonstrated, ME imaging increased the FC reliability on an individual level. Individual differences are often neglected in group analysis studies. In terms of individualized treatment outcome prediction, however, this unshared variance could increase accuracy and should be taken into account.
Time‐series acquired at different TEs can be combined to a single time‐series in several ways. One of the most widely applied approaches is to calculate weights based on TE and T2* values and derive its weighted average to obtain an “optimally combined” time‐series representing a balanced combination of signal intensity and contrast. 88 , 91 By fitting a monoexponential function over the signal at each TE, the decay rate, that is, the inverse of T2*, and the S0‐value can be estimated per voxel. TEs closer to the T2* value are weighted more heavily to enhance BOLD contrast, whereas signals from shorter TEs are also included as they contain higher signal intensities. The T2* and S0 values are calculated per voxel over the whole time‐series in this optimal combination method. Recently, another ME combination approach, called T2*FIT‐weighted combination, was proposed that calculates the T2* and S0 values using the same algorithm per volume instead of over the whole‐time series. 92 Despite fitting the monoexponential decay on significantly fewer data points, the tSNR of tb‐fMRI and rs‐fMRI time‐series increased the most compared to other weighted ME combination methods. Interestingly, when the T2* maps were used as time‐series—instead of deriving their weights and calculating the weighted average—the highest effect sizes, functional contrasts, and temporal contrast‐to‐noise ratio in motor and emotion processing tasks were achieved. Signal quality gains of ME fMRI data could be maximized by implementation of an optimal combination scheme in terms of effects on signal loss, BOLD contrast and sensitivity. It is therefore of uttermost importance to evaluate their differences on several functional measures and investigate the opportunities in psychiatric studies in which regions that are prone to susceptibility artifacts and lower SNR are of exceptional interest.
The combination of novel acquisition techniques
As acquiring multiple echoes per slice comes at the cost of imaging time, acceleration techniques such as MB imaging offer a solution by speeding up the process. 93 In addition, increases in spatial resolution, while maintaining a good temporal SNR, can be achieved by using MRI at higher magnetic field strengths. 87 Nevertheless, more dropout of signals is common in 7T as the magnetic field is more inhomogeneous leading to faster signal decay. T2* values are even decreased by around 50% at 7T compared to 3T. 87 ME imaging therefore seems essential in 7T fMRI studies focusing on orbitofrontal or inferior temporal regions. The combination of ME in 7T requires further validation before wide application in the neuroimaging research field.
Few studies in MDD have examined the combination of these novel image acquisitions. A study involving ME and MB imaging at 7T improved tSNR and FC metrics with 200%‐300% compared to the same acquisition at 3T. 73 The researchers showed the existence of mood‐related neurocircuit disturbances in MDD patients and increased power in regions related to MDD such as the ventral tegmental area, a small midbrain area that is prone to poor tSNR. Both findings were not evident in the 3T acquisition protocol. Another study that used ME‐MB fMRI at 7T, including ME ICA denoising, examined rumination in MDD. They found that rumination was related to lower FC within the DMN, in particular the connectivity of the left medial OFC with the right precuneus. 94
Single‐echo fMRI scans may suffer from signal loss in iron‐rich subcortical regions. ME‐MB imaging at 7T increases the tSNR, contrast‐to‐noise ratio, and t‐statistics in the caudate nucleus and putamen. 89 Interestingly, the iron concentration of subcortical regions such as the bilateral putamen and left thalamus has been shown to be altered in MDD and correlated positively with depression severity. 95
Overall, the combination of ME, MB, and 7T imaging seems to complement one another and is warranted in fMRI studies of depression and other neuropsychiatric disorders. Nonetheless, the choice of acquisition is highly dependent on the research question and hardware and software availability. For example, if parallel imaging is not an option, studies focusing on regions that are known to have relatively low T2* values could benefit more from ME imaging and longer acquisition times than increased temporal resolution to limit signal loss and susceptibility‐related artifacts. Likewise, shorter TRs could be favored in studies that focus on obtaining clean functional networks in healthy subjects as small TRs limit the amount of aliasing from nonneural signals. An overview of these acquisition improvements is shown in Figure 2.
FIGURE 2.
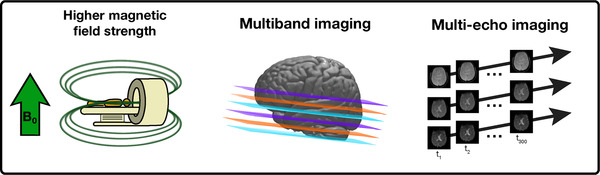
Novel acquisition methods that could increase the identification of robust functional biomarkers. Left: a higher magnetic field strength (B0) increases the signal‐to‐noise ratio (SNR), and blood‐oxygen‐level‐dependent sensitivity of fMRI data. In major depressive disorder (MDD), 7T imaging revealed enhanced sensitivity of detecting mood‐related neurocircuit disturbances. Centre: in multiband imaging, multiple slices are acquired simultaneously (color coded here), allowing an increase in temporal or spatial resolution in the same scanning time. Right: in multiecho imaging, multiple brain volumes are acquired from the same excitation pulse but at different echo times, resulting in different contrast images. These different contrasts increase SNR and prevent signal loss in regions that are prone to susceptibility artifacts, which have frequently been associated with MDD. B 0, main magnetic field strength; tn , time at acquisition of volume n.
Image processing
Neural network‐based cleaning methods
In the case when fast or ME‐MB acquisition protocols are insufficient or not available to minimize the confound from nonneuronal signals, application of additional denoising steps is warranted. Recently, the rs‐fMRI data‐driven denoising methods have entered the new era of neural networks. One of such novel machine learning studies described the development of an automated classifier to assess whether an IC is considered noise or neuronal signal. 47 The classifier is based on three fused convolutional neural networks (CNNs) of which each classifies the IC spatial maps, time‐series, or both simultaneously. Subsequently, majority voting can be applied to allocate either a neuronal or artifact label to each of the ICs. The models performed robust with accuracies over 95%. Spatial maps only showed little improvement compared to the model based on time‐series. 47 In the same category of component classification, researchers developed a deep neural network that was able to achieve over 98% accuracy in classifying ICs from an rs‐fMRI dataset consisting of 394 subjects. Several combinations and different voting schemes demonstrated that spatial information provided the highest performance compared with temporal and spectral features. A voting scheme with a voting weight of 0.5 for the spatial CNN, 0.25 for the temporal CNN, and 0.25 for a spectral CNN resulted in a balanced sensitivity and specificity and a precision of 99.4%. 96 It should still be noted, however, that for both studies the components were rated by trained experts so the ground truth is merely subjective. Moreover, a single IC could contain BOLD and nonneural signals, potentially leading to inaccurate labelling.
Finally, another study introduced a CNN that is able to obtain optimal motion regressors used for nuisance regression in rs‐fMRI. After head motion correction, six estimated motion parameters (translation and rotation in each direction) were used as input to the CNN network consisting of two temporal convolutional layers. Following the second layer, the 12 output regressors were correlated with voxel time‐series of the white matter and cerebral spinal fluid to calculate the loss function and update the model. Lower variance and higher network modularity was found in filtered data of 76 healthy subjects with the 12 CNN regressors compared to the 12 traditional regressors (including first‐order derivative) and FC difference before and after regression was higher for the 12 CNN regressors, suggesting improved motion regression with the CNN. 97
The high‐performance metrics of these studies indicate the promise of data‐driven noise cleaning methods in fMRI. Its main applications for now are the classification of components, particularly spatial maps, and development of optimized motion regressors. In order to implement these cleaning approaches in other clinical studies, the neural networks should be trained on neuropathological fMRI data as noise and artifact patterns might differ in psychiatric disorders compared to HCs. 98 Future optimization studies of motion regressors could focus on more local, for example, voxel‐specific corrections. External cardiac and respiratory signals could be integrated to further enhance cleaning of the fMRI signals. 96 A combination of both, improved acquisitions and novel noise removal methods, would be a promising approach for enhancement of the reliability of detecting altered functional activity or connectivity in deeper located brain regions in psychiatric disorders, including depression.
Image analysis
Causality in brain activation
Most fMRI studies involving MDD patients have implemented undirected FC measures. Yet, it has been demonstrated that directional FC is altered in MDD patients compared to HCs 99 , 100 , 101 , 102 and before and after antidepressant treatment. 103
The most common methods for assessing fMRI causality are Granger causality and dynamic causal modeling (DCM). Granger causality, which is model free, is based on the principle that information, that is, samples, of the past from a network can predict future information of another network. 104 DCM relies on models that link the firing of neural populations to BOLD signals. 57
DCM analyses of tb‐fMRI have highlighted the enhanced effective connectivity from the dorsal ACC to the rostral ACC in the cognitive Stroop task 105 and in another study, direction‐ and valence‐dependent abnormal effective connectivity between the subgenual ACC, amygdala, and ventromedial prefrontal cortex in an implicit emotional face task. 106 Moreover, efficient connectivity from the amygdala to the subgenual ACC was found to be lower in MDD patients following a task involving negative emotion processing. 107 In remitted depressed patients, DCM has revealed bidirectional connectivity alterations within frontotemporal connections and from the fusiform gyrus to the OFC compared to controls while viewing happy and sad faces, respectively. 108 In a recent larger study, decreased effective connectivity from the mPFC to the striatum was correlated with increased depressive symptoms. 109 In short, there is some evidence of the involvement of the ACC, amygdala, and PFC in the directional connectivity in depression; its role in predicting treatment outcome is still largely unknown. It is also important to note that many of the studies implementing these effective connectivity analyses had low sample sizes, while all of them were cross‐sectional studies.
Interestingly, there is one paper that focused on treatment outcome prediction using task‐based directional connectivity features in MDD. Following an emotional face perception task, these features were able to distinguish fast remission from chronic depression trajectories in a 2‐year longitudinal study with an accuracy of 79%. 110 Some of the most contributing features that were found during this classification were the connections from the right amygdala to the right fusiform area for happy faces and many face‐processing and emotion‐sensitive regions for negatively valenced emotions such as the fusiform area, amygdala, and occipital face area. This study shows the potential in predicting individual clinical depression trajectories in the long‐term using tb‐fMRI and could therefore be valuable in future MDD studies with larger sample sizes, potentially in combination with other MRI modalities.
From two rs‐fMRI causality studies, the DMN appears to play a major role in depression and response to antidepressant treatment. 101 , 103 Moreover, changes in causal dependencies in MDD patients compared to HCs were found with or from the (anterior) insula 99 , 100 and from the lateral visual network. 102
Functional neurodynamics
The FC measures that have been discussed so far are static in the sense that each time‐series is analyzed and correlated as an entity, for example, resulting in a single FC value between two pairs of time‐series instead of a value per time point. The interaction between functional brain networks might, however, be more complex than that and could change over time depending on cognitive or emotional states. 111 , 112 The neuroimaging field has recognized this time‐varying nature as reflected by the increase in the number of these neurodynamic fMRI studies, including various MDD studies. A recent study showed increased temporal variability and efficiency and decreased temporal clustering in MDD patients, mostly evident within the DMN and some subcortical and sensorimotor structures. They speculate that these results could indicate more aberrant brain connections and fluctuations, possibly related to common MDD symptoms as rumination and anhedonia. 113 The finding of increased temporal variability within the DMN in MDD patients is in agreement with many studies 114 , 115 , 116 but contradictory to the results of another paper. 117
Of those studies, dynamic FC has frequently been found to be correlated with depression severity scores 113 , 114 , 118 suggesting that this metric could potentially serve as a biomarker of depression classification or treatment outcome. Not long ago, the former notion was supported by two studies that improved MDD classification by including dynamic FC features in their prediction model. 118 , 119 Another classification study found dynamic FC differences between treatment‐resistant MDD, MDD without treatment‐resistance, and healthy controls, 120 thereby demonstrating the value of functional neurodynamics in differentiating between treatment groups. Further research is required to predict depression trajectories and treatment outcomes in an early stage to improve treatment selection.
Similarly, wavelet coherence assesses the time‐dependent activation patterns in functional time‐series. In contradiction to dynamic FC, wavelet coherence calculates the time‐changing coherence, that is, synchronous oscillations, per frequency of nonstationary time‐series. 121 An rs‐fMRI study demonstrated improved sensitivity of detecting group differences in wavelet‐based regularity (stability of recurrent temporal patterns) of activity within the DMN and executive control networks between HCs and patients with mild cognitive impairment. 122 This instability of recurrent temporal patterns could be attributed to disrupted information feedback or cognitive impairment with which MDD is often characterized. In autism spectrum disorder, an accuracy of over 86% was achieved for the classification of patients based on time of in‐phase coherence between the ventral stream network and frontoparietal networks. 123 Both studies indicate the potential of wavelet coherence in psychiatric disorders for classification purposes. To date, no depression fMRI study has implemented wavelet coherence. Nonetheless, in an electroencephalography study a CNN relying on wavelet coherence of the DMN was able to reliably classify MDD patients with over 98%. 124
Concluding, there is a promise in neurodynamic analyses for identifying MDD‐associated biomarkers, but the limited number of studies so far only led to a relatively consistent finding of the instability of the DMN network in MDD. More consistent findings of other potentially affected networks related to MDD are needed to draw reliable conclusions.
Identification of depression biotypes
MDD is a very heterogeneous disease as patients differ in symptoms, trajectory course (chronic, recurrent, single episode), and treatment response. This heterogeneity contributes to the divergent number of potential biomarkers as suggested by researchers. Not long ago, supported by the development of novel machine learning methods, the approach of biotyping gained attention. 125 Biotyping is the identification of subtypes of depression based on differences in biological characteristics (eg, features derived from neuroimaging of the brain) that are associated with specific symptoms. Genetic and neuroimaging biotyping studies are increasingly being conducted, whereas the latter have investigated features mostly derived from rs‐fMRI. These neuroimaging studies found several biotypes, which they claimed to be distinguishable by anxiety, anhedonia, childhood trauma, or depression severity in combination with specific brain activation patterns. 126 , 127 , 128 , 129 An important rationale for conducting these MDD biotyping studies is that a certain treatment may be more beneficial for particular biotypes of depression compared to others.
A study based on rs‐fMRI found two biotypes of depression, differing from each other by high DMN versus high dorsal ACC effective connectivity. 130 The latter subgroup consisted of a high percentage of females (87%), comorbid anxiety diagnoses (43%), and recurrent depression (63%). A second study discriminated biotypes of depression based on FC between the right angular gyrus and areas in the DMN and scores from a child abuse trauma questionnaire. Subjects from two out of the three biotypes scored lower depression severity scores following a 6‐week antidepressant treatment. 129 The authors developed a custom co‐clustering approach that produces so‐called “views” of the datasets, which cluster both subjects and features simultaneously. The most prominent view distinguished depression subjects in three clusters based on differences in degree of FC, child abuse trauma, and depression severity scores at baseline and after 6 weeks. Other views could be prescribed to subjects with fatigue symptoms or to duration of depression episodes, both in combination with abnormal FC patterns between specific regions. This algorithm has the advantage of generating several cluster solutions simultaneously, possibly revealing less dominant clusters that would normally have been ignored.
A third and probably the most prominent biotype research recently discovered four biotypes based on different resting‐state FC and symptomatology patterns. 127 Subjects from biotypes 1 and 2 represented groups with anergia and fatigue symptoms in combination with reduced connectivity in the ACC and OFC. Instead, patients from the biotype group 3 and 4 showed the most severe anhedonic and psychomotor retardation symptoms along with FC increases in the thalamus and frontostriatal networks. Biotype 1 and 4 suffered from severe anxiety symptoms with reduced FC in frontoamygdala networks. Revolutionary, it was found that 83% and 66% of patients with biotypes 1 and 3 responded to transcranial magnetic stimulation of the dorsomedial PFC, respectively, compared to only 25% and 30% of patients from biotypes 2 and 4. The paper also received criticism from researchers that replicated the same methodology on a separate dataset, arguing that overfitting took place and p‐values were inflated because their feature reduction method was not taken into consideration in the statistical analyses. They showed that by leaving out a single subject from the cluster population, clusters were not stable anymore and they were unable to reject the null hypothesis that the distribution was non‐Gaussian. 131
Overall, these studies show the promise of biotyping for prediction of treatment outcome or supporting treatment decision‐making in depression. For example, a clinician might treat a patient with a more invasive method such as electroconvulsive therapy earlier with the support of such biotype models than without. Similar efforts could be made for psychotherapy and pharmacotherapy, the most widely provided treatments for depression. Nonetheless, only a couple of biotyping studies have been conducted so far in MDD and published results should still be taken with caution. One of the potential pitfalls in biotype studies is the high ratio of the number of subjects to variables. 131 For a single subject, often a few dozen symptoms are correlated with multiple thousands of FC values, while most sample sizes are less than 100. Overfitting is therefore likely to occur. Yet, these studies instigate a potential future direction and approach the search for a biomarker from a different angle. Future biotype studies could limit the amount of included symptoms and connections in the analyses by forming a priori hypotheses, thereby increasing statistical power. 26 If a large number of features is included or a large sample size is not realistic, however, identified subtypes should be verified on independent samples (test set) or datasets. This ensures the generalizability and validity of the biotypes. As shown recently, the replicability of multivariate FC correlations with cognitive ability and psychopathology scores increases with an increase in the number of samples and decreases with an increase in the number of features after an optimum of about 20% explained variance has been reached. 26 Examples of methods to test generalizability include cross‐validation, bootstrapping, or Jackknife resampling. An overview of the discussed recent progress in rs‐fMRI analysis methods of MDD is presented in Figure 3.
FIGURE 3.
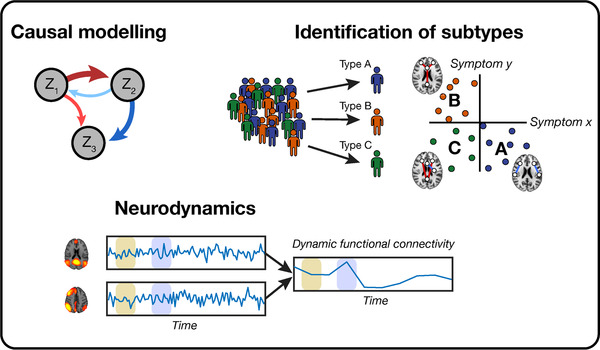
An overview of recent developments in analysis methods of functional MRI studies used in major depressive disorder (MDD). Top left: causal modeling is a method that is directional and compares the functional connectivity (FC) between n regions or networks (Zn ). Below: neurodynamical analyses measure the difference in FC over time. On the bottom left, the FC is calculated in the beige and blue window between two different networks, after which the dynamic FC over time is shown on the bottom right. Top right: from MDD samples, multiple biotypes can be identified (here A, B, and C) in which different connectivity patterns associated with specific symptoms.
CONCLUSIONS
Despite the numerous published fMRI studies in MDD, only a few biomarkers are consistently found to be altered in depression or associated with treatment outcome. In this review, first, the most prominent findings and developments were discussed, followed by an overview of limitations and pitfalls of fMRI studies in depression. Most notably, we found that the most common functional outcome measures might not suffice to represent the MDD patients’ brain and that analytical approaches thereof are not coherent among MDD studies. In this review, an overview and discussion of future directions in functional image acquisition, processing, and analysis in MDD studies was presented. Novel acquisitions techniques, such as ME and MB imaging, as well as machine learning‐based noise reduction methods improve image quality and BOLD sensitivity as demonstrated by several MDD studies. Regarding image analysis, more extensive functional features, such as directional FC or neurodynamics, and the segregation of biological depression subtypes enhance the chances of identifying consistent functional MDD biomarkers. Overall, we see promising developments in fMRI data acquisition, processing, and analysis that potentially could lead to objective diagnosis and prognosis in MDD.
ACKNOWLEDGMENT and DISCLOSURE
We would like to provide our gratitude to Prof. Dr. Bert Aldenkamp for inspiring this study and our team as well as for creating the opportunity for collaboration between the authors of the involved institutions. The authors WH and RL are employees of Philips Research, Eindhoven, the Netherlands and MB is an employee of Philips Healthcare, Best, the Netherlands. The other authors declare no conflict of interest.
Pilmeyer J, Huijbers W, Lamerichs R, Jansen JFA, Breeuwer M, Zinger S. Functional MRI in major depressive disorder: A review of findings, limitations, and future prospects. J Neuroimaging. 2022;32:582–95. 10.1111/jon.13011
Funding informaton
This research is partly funded by allowance from Top Consortia for Knowledge and Innovation Public‐Private Partnerships (TKI‐PPP) under subsidy number TK11812P07 and additional funding from Philips and Eindhoven Engine.
REFERENCES
- 1. Malhi GS, Mann JJ. Depression. Lancet 2018;392:2299‐312. [DOI] [PubMed] [Google Scholar]
- 2. Glover GH. Overview of functional magnetic resonance imaging. Neurosurg Clin N Am 2011;22:133‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barth M, Poser BA. Advances in high‐field BOLD fMRI. Materials 2011;4:1941‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunlop BW, Mayberg HS. Neuroimaging advances for depression. Cerebrum 2017;2017:cer16‐cer17. [PMC free article] [PubMed] [Google Scholar]
- 5. Prescott JW. Quantitative imaging biomarkers: the application of advanced image processing and analysis to clinical and preclinical decision making. J Digit Imaging 2013;26:97‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhuo C, Li G, Lin X, et al. The rise and fall of MRI studies in major depressive disorder. Transl Psychiatry 2019;9:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stuhrmann A, Suslow T, Dannlowski U. Facial emotion processing in major depression: a systematic review of neuroimaging findings. Biol Mood Anxiety Disord 2011;1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hacimusalar Y, Eşel E. Suggested biomarkers for major depressive disorder. Noro Psikiyatr Ars 2018;55:280‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ebneabbasi A, Mahdipour M, Nejati V, et al. Emotion processing and regulation in major depressive disorder: a 7T resting‐state fMRI study. Hum Brain Mapp 2021;42:797‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lener MS, Iosifescu DV. In pursuit of neuroimaging biomarkers to guide treatment selection in major depressive disorder: a review of the literature. Ann N Y Acad Sci 2015;1344:50‐65. [DOI] [PubMed] [Google Scholar]
- 11. Ebmeier K, Rose E, Steele D. Cognitive impairment and fMRI in major depression. Neurotox Res 2006;10:87‐92. [DOI] [PubMed] [Google Scholar]
- 12. Thomas EJ, Elliott R. Brain imaging correlates of cognitive impairment in depression. Front Hum Neurosci 2009;3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palmer SM, Crewther SG, Carey LM, et al. A meta‐analysis of changes in brain activity in clinical depression. Front Hum Neurosci 2015;8:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Canli T, Cooney R, Goldin P, et al. Amygdala reactivity to emotional faces predicts improvement in major depression. Neuroreport 2005;16:1267‐70. [DOI] [PubMed] [Google Scholar]
- 15. Siegle GJ, Carter CS, Thase ME. Use of fMRI to predict recovery from unipolar depression with cognitive behavior therapy. Am J Psychiatry 2006;163:735‐8. [DOI] [PubMed] [Google Scholar]
- 16. Williams LM, Korgaonkar MS, Song YC, et al. Amygdala reactivity to emotional faces in the prediction of general and medication‐specific responses to antidepressant treatment in the randomized iSPOT‐D trial. Neuropsychopharmacology 2015;40:2398‐408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perlman K, Benrimoh D, Israel S, et al. A systematic meta‐review of predictors of antidepressant treatment outcome in major depressive disorder. J Affect Disord 2019;243:503‐15. [DOI] [PubMed] [Google Scholar]
- 18. Dai L, Zhou H, Xu X, et al. Brain structural and functional changes in patients with major depressive disorder: a literature review. PeerJ 2019;7:e8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaiser RH, Andrews‐Hanna JR, Wager TD, et al. Large‐scale network dysfunction in major depressive disorder: a meta‐analysis of resting‐state functional connectivity. JAMA Psychiatry 2015;72:603‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mulders PC, van Eijndhoven PF, Schene AH, et al. Resting‐state functional connectivity in major depressive disorder: a review. Neurosci Biobehav Rev 2015;56:330‐44. [DOI] [PubMed] [Google Scholar]
- 21. Javaheripour N, Li M, Chand T, et al. Altered resting‐state functional connectome in major depressive disorder: a mega‐analysis from the PsyMRI consortium. Transl Psychiatry 2021;11:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan C‐G, Chen X, Li L, et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc Natl Acad Sci USA 2019;116:9078‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexopoulos GS, Hoptman MJ, Kanellopoulos D, et al. Functional connectivity in the cognitive control network and the default mode network in late‐life depression. J Affect Disord 2012;139:56‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dichter GS, Gibbs D, Smoski MJ. A systematic review of relations between resting‐state functional‐MRI and treatment response in major depressive disorder. J Affect Disord 2015;172:8‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kennis M, Gerritsen L, van Dalen M, et al. Prospective biomarkers of major depressive disorder: a systematic review and meta‐analysis. Mol Psychiatry 2020;25:321‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marek S, Tervo‐Clemmens B, Calabro FJ, et al. Reproducible brain‐wide association studies require thousands of individuals. Nature 2022;603:654‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winter NR, Leenings R, Ernsting J, et al. More alike than different: quantifying deviations of brain structure and function in major depressive disorder across neuroimaging modalities. ArXiv 2021. 10.48550/arXiv.2112.10730. Accessed 20 Dec 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drevets WC. Orbitofrontal cortex function and structure in depression. Ann N Y Acad Sci 2007;1121:499‐527. [DOI] [PubMed] [Google Scholar]
- 29. Gabbay V, Ely BA, Li Q, et al. Striatum‐based circuitry of adolescent depression and anhedonia. J Am Acad Child Adolesc Psychiatry 2013;52:628‐41.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kirilina E, Lutti A, Poser BA, et al. The quest for the best: the impact of different EPI sequences on the sensitivity of random effect fMRI group analyses. Neuroimage 2016;126:49‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cordes D, Turski PA, Sorenson JA. Compensation of susceptibility‐induced signal loss in echo‐planar imaging for functional applications. Magn Reson Imaging 2000;18:1055‐68. [DOI] [PubMed] [Google Scholar]
- 32. Demetriou L, Kowalczyk OS, Tyson G, et al. A comprehensive evaluation of increasing temporal resolution with multiband‐accelerated protocols and effects on statistical outcome measures in fMRI. Neuroimage 2018;176:404‐16. [DOI] [PubMed] [Google Scholar]
- 33. Jahanian H, Holdsworth S, Christen T, et al. Advantages of short repetition time resting‐state functional MRI enabled by simultaneous multi‐slice imaging. J Neurosci Methods 2019;311:122‐32. [DOI] [PubMed] [Google Scholar]
- 34. Pais‐Roldán P, Biswal B, Scheffler K, et al. Identifying respiration‐related aliasing artifacts in the rodent resting‐state fMRI. Front Neurosci 2018;12:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kemp AH, Quintana DS, Gray MA, et al. Impact of depression and antidepressant treatment on heart rate variability: a review and meta‐analysis. Biol Psychiatry 2010;67:1067‐74. [DOI] [PubMed] [Google Scholar]
- 36. Glover GH, Li T‐Q, Ress D. Image‐based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med 2000;44:162‐7. [DOI] [PubMed] [Google Scholar]
- 37. Caballero‐Gaudes C, Reynolds RC. Methods for cleaning the BOLD fMRI signal. Neuroimage 2017;154:128‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kay K, Rokem A, Winawer J, et al. GLMdenoise: a fast, automated technique for denoising task‐based fMRI data. Front Neurosci 2013;7:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Agrawal U, Brown EN, Lewis LD. Model‐based physiological noise removal in fast fMRI. Neuroimage 2020;205:116231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Behzadi Y, Restom K, Liau J, et al. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage 2007;37:90‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soltysik DA, Thomasson D, Rajan S, et al. Improving the use of principal component analysis to reduce physiological noise and motion artifacts to increase the sensitivity of task‐based fMRI. J Neurosci Methods 2015;241:18‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salimi‐Khorshidi G, Douaud G, Beckmann CF, et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage 2014;90:449‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pruim RHR, Mennes M, van Rooij D, et al. ICA‐AROMA: a robust ICA‐based strategy for removing motion artifacts from fMRI data. Neuroimage 2015;112:267‐77. [DOI] [PubMed] [Google Scholar]
- 44. Pruim RHR, Mennes M, Buitelaar JK, et al. Evaluation of ICA‐AROMA and alternative strategies for motion artifact removal in resting state fMRI. Neuroimage 2015;112:278‐87. [DOI] [PubMed] [Google Scholar]
- 45. Carone D, Licenik R, Suri S, et al. Impact of automated ICA‐based denoising of fMRI data in acute stroke patients. Neuroimage Clin 2017;16:23‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dipasquale O, Sethi A, Laganà MM, et al. Comparing resting state fMRI de‐noising approaches using multi‐ and single‐echo acquisitions. PLoS ONE 2017;12:e0173289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kam T‐E, Wen X, Jin B, et al. A deep learning framework for noise component detection from resting‐state functional MRI. Med Image Comput Comput Assist Interv 2019;11766:754‐62. [Google Scholar]
- 48. Lv H, Wang Z, Tong E, et al. Resting‐state functional MRI: everything that nonexperts have always wanted to know. AJNR Am J Neuroradiol 2018;39:1390‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mueller S, Wang D, Fox MD, et al. Individual variability in functional connectivity architecture of the human brain. Neuron 2013;77:586‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sohn WS, Yoo K, Lee Y‐B, et al. Influence of ROI selection on resting state functional connectivity: an individualized approach for resting state fMRI analysis. Front Neurosci 2015;9:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Majeed W, Avison MJ. Robust data driven model order estimation for independent component analysis of fMRI data with low contrast to noise. PLoS ONE 2014;9:e94943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y‐O, Adali T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp 2007;28:1251‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Z‐Q, Du M‐Y, Zhao Y‐J, et al. Voxel‐wise meta‐analyses of brain blood flow and local synchrony abnormalities in medication‐free patients with major depressive disorder. J Psychiatry Neurosci 2015;40:401‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hao H, Chen C, Mao W, et al. Aberrant brain regional homogeneity in first‐episode drug‐naïve patients with major depressive disorder: a voxel‐wise meta‐analysis. J Affect Disord 2019;245:63‐71. [DOI] [PubMed] [Google Scholar]
- 55. Iwabuchi SJ, Krishnadas R, Li C, et al. Localized connectivity in depression: a meta‐analysis of resting state functional imaging studies. Neurosci Biobehav Rev 2015;51:77‐86. [DOI] [PubMed] [Google Scholar]
- 56. Xue S, Wang X, Wang W, et al. Frequency‐dependent alterations in regional homogeneity in major depression. Behav Brain Res 2016;306:13‐9. [DOI] [PubMed] [Google Scholar]
- 57. Bijsterbosch J, Smith SM, Beckmann CF. An introduction to resting state fMRI functional connectivity. Oxford: Oxford University Press; 2017. [Google Scholar]
- 58. Arnone D, McKie S, Elliott R, et al. Increased amygdala responses to sad but not fearful faces in major depression: relation to mood state and pharmacological treatment. Am J Psychiatry 2012;169:841‐50. [DOI] [PubMed] [Google Scholar]
- 59. Hamilton JP, Etkin A, Furman DJ, et al. Functional neuroimaging of major depressive disorder: a meta‐analysis and new integration of base line activation and neural response data. Am J Psychiatry 2012;169:693‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nielen MMA, Heslenfeld DJ, Heinen K, et al. Distinct brain systems underlie the processing of valence and arousal of affective pictures. Brain Cogn 2009;71:387‐96. [DOI] [PubMed] [Google Scholar]
- 61. Ramani R. Functional MRI: basic principles and emerging clinical applications for anesthesiology and the neurological sciences. Oxford: Oxford University Press; 2019. [Google Scholar]
- 62. Yang Y, Zhong N, Imamura K, et al. Task and resting‐state fMRI reveal altered salience responses to positive stimuli in patients with major depressive disorder. PLoS ONE 2016;11:e0155092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Warbrick T, Reske M, Shah NJ. Transferring cognitive tasks between brain imaging modalities: implications for task design and results interpretation in fMRI studies. J Vis Ex 2014;91:e51793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Godlewska BR, Browning M, Norbury R, et al. Predicting treatment response in depression: the role of anterior cingulate cortex. Int J Neuropsychopharmacol 2018;21:988‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hall LMJ, Klimes‐Dougan B, Hunt RH, et al. An fMRI study of emotional face processing in adolescent major depression. J Affect Disord 2014;168:44‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hilland E, Landrø NI, Harmer CJ, et al. Attentional bias modification is associated with fMRI response towards negative stimuli in residual depression: a randomized controlled trial. J Psychiatry Neurosci 2020;45:22‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Victor TA, Furey ML, Fromm SJ, et al. Relationship of emotional processing to masked faces in the amygdala to mood state and treatment in major depressive disorder. Arch Gen Psychiatry 2010;67:1128‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hariri AR, Tessitore A, Mattay VS, et al. The amygdala response to emotional stimuli: a comparison of faces and scenes. Neuroimage 2002;17:317‐23. [DOI] [PubMed] [Google Scholar]
- 69. Fitzgerald PB, Srithiran A, Benitez J, et al. An fMRI study of prefrontal brain activation during multiple tasks in patients with major depressive disorder. Hum Brain Mapp 2008;29:490‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rose EJ, Simonotto E, Ebmeier KP. Limbic over‐activity in depression during preserved performance on the n‐back task. Neuroimage 2006;29:203‐15. [DOI] [PubMed] [Google Scholar]
- 71. Amado‐Boccara I, Gougoulis N, Poirier Littré MF, et al. Effects of antidepressants on cognitive functions: a review. Neurosci Biobehav Rev 1995;19:479‐93. [DOI] [PubMed] [Google Scholar]
- 72. Collins CM. Electromagnetics in magnetic resonance imaging: physical principles, related applications, and ongoing developments. San Rafael, CA: Morgan & Claypool Publishers; 2016. [Google Scholar]
- 73. Morris LS, Kundu P, Costi S, et al. Ultra‐high field MRI reveals mood‐related circuit disturbances in depression: a comparison between 3‐Tesla and 7‐Tesla. Transl Psychiatry 2019;9:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van der Zwaag W, Francis S, Head K, et al. fMRI at 1.5, 3 and 7 T: characterising BOLD signal changes. Neuroimage 2009;47:1425‐34. [DOI] [PubMed] [Google Scholar]
- 75. Krüger G, Glover GH. Physiological noise in oxygenation‐sensitive magnetic resonance imaging. Magn Reson Med 2001;46:631‐7. [DOI] [PubMed] [Google Scholar]
- 76. Marquis R, Muller S, Lorio S, et al. Spatial resolution and imaging encoding fMRI settings for optimal cortical and subcortical motor somatotopy in the human brain. Front Neurosci 2019;13:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stockmann JP, Wald LL. In vivo B0 field shimming methods for MRI at 7T. Neuroimage 2018;168:71‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang QX, Mao W, Wang J, et al. Manipulation of image intensity distribution at 7.0 T: passive RF shimming and focusing with dielectric materials. J Magn Reson Imaging 2006;24:197‐202. [DOI] [PubMed] [Google Scholar]
- 79. Miletić S, Bazin P‐L, Weiskopf N, et al. fMRI protocol optimization for simultaneously studying small subcortical and cortical areas at 7 T. Neuroimage 2020;219:116992. [DOI] [PubMed] [Google Scholar]
- 80. Havsteen I, Ohlhues A, Madsen KH, et al. Are movement artifacts in magnetic resonance imaging a real problem?—A narrative review. Front Neurol 2017;8:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hamilton J, Franson D, Seiberlich N. Recent advances in parallel imaging for MRI. Prog Nucl Magn Reson Spectrosc 2017;101:71‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Blaimer M, Choli M, Jakob PM, et al. Multiband phase constrained parallel MRI. Magn Reson Med 2013;69:974‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nunes RG, Hajnal JV, Golay X, et al. Simultaneous slice excitation and reconstruction for single shot EPI. Presented at the 14th Annual Meeting ISMRM, Seattle; 2006:293.
- 84. Barth M, Breuer F, Koopmans PJ, et al. Simultaneous multislice (SMS) imaging techniques. Magn Reson Med 2016;75:63‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cauley SF, Polimeni JR, Bhat H, et al. Inter‐slice leakage artifact reduction technique for simultaneous multi‐slice acquisitions. Magn Reson Med 2014;72:93‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bijsterbosch JD, Ansari TL, Smith S, et al. Stratification of MDD and GAD patients by resting state brain connectivity predicts cognitive bias. Neuroimage Clin 2018;19:425‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kundu P, Voon V, Balchandani P, et al. Multi‐echo fMRI: a review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage 2017;154:59‐80. [DOI] [PubMed] [Google Scholar]
- 88. Kundu P, Inati SJ, Evans JW, et al. Differentiating BOLD and non‐BOLD signals in fMRI time series using multi‐echo EPI. Neuroimage 2012;60:1759‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Puckett AM, Bollmann S, Poser BA, et al. Using multi‐echo simultaneous multi‐slice (SMS) EPI to improve functional MRI of the subcortical nuclei of the basal ganglia at ultra‐high field (7T). Neuroimage 2018;172:886‐95. [DOI] [PubMed] [Google Scholar]
- 90. Lynch CJ, Power JD, Scult MA, et al. Rapid precision functional mapping of individuals using multi‐echo fMRI. Cell Rep 2020;33:108540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Posse S, Wiese S, Gembris D, et al. Enhancement of BOLD‐contrast sensitivity by single‐shot multi‐echo functional MR imaging. Magn Reson Med 1999;42:87‐97. [DOI] [PubMed] [Google Scholar]
- 92. Heunis S, Breeuwer M, Caballero‐Gaudes C, et al. The effects of multi‐echo fMRI combination and rapid T2*‐mapping on offline and real‐time BOLD sensitivity. Neuroimage 2021;238:118244. [DOI] [PubMed] [Google Scholar]
- 93. Gonzalez‐Castillo J, Panwar P, Buchanan LC, et al. Evaluation of multi‐echo ICA denoising for task based fMRI studies: block designs, rapid event‐related designs, and cardiac‐gated fMRI. Neuroimage 2016;141:452‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jacob Y, Morris LS, Huang K‐H, et al. Neural correlates of rumination in major depressive disorder: a brain network analysis. Neuroimage Clin 2020;25:102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yao S, Zhong Y, Xu Y, et al. Quantitative susceptibility mapping reveals an association between brain iron load and depression severity. Front Hum Neurosci 2017;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Theodoropoulos C, Chatzichristos C, Van Huffel S. Automatic artifact removal of resting‐state fMRI with deep neural networks. ArXiv 2020. 10.48550/arXiv.2011.12113. Accessed 18 Feb 2022. [DOI] [Google Scholar]
- 97. Yang Z, Zhuang X, Sreenivasan K, et al. Robust motion regression of resting‐state data using a convolutional neural network model. Front Neurosci 2019;13:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Makowski C, Lepage M, Evans AC. Head motion: the dirty little secret of neuroimaging in psychiatry. J Psychiatry Neurosci 2019;44:62‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guo W, Liu F, Zhang Z, et al. Unidirectionally affected causal connectivity of cortico‐limbic‐cerebellar circuit by structural deficits in drug‐naive major depressive disorder. J Affect Disord 2015;172:410‐6. [DOI] [PubMed] [Google Scholar]
- 100. Kandilarova S, Stoyanov D, Kostianev S, et al. Altered resting state effective connectivity of anterior insula in depression. Front Psychiatry 2018;9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li G, Liu Y, Zheng Y, et al. Large‐scale dynamic causal modeling of major depressive disorder based on resting‐state functional magnetic resonance imaging. Hum Brain Mapp 2020;41:865‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Luo L, Wu H, Xu J, et al. Abnormal large‐scale resting‐state functional networks in drug‐free major depressive disorder. Brain Imaging Behav 2020;15:96‐106. [DOI] [PubMed] [Google Scholar]
- 103. Li L, Li B, Bai Y, et al. Abnormal resting state effective connectivity within the default mode network in major depressive disorder: a spectral dynamic causal modeling study. Brain Behav 2017;7:e00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Seth AK, Barrett AB, Barnett L. Granger causality analysis in neuroscience and neuroimaging. J Neurosci 2015;35:3293‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Schlosser RGM, Wagner G, Koch K, et al. Fronto‐cingulate effective connectivity in major depression: a study with fMRI and dynamic causal modeling. Neuroimage 2008;43:645‐55. [DOI] [PubMed] [Google Scholar]
- 106. de Almeida JRC, Kronhaus DM, Sibille EL, et al. Abnormal left‐sided orbitomedial prefrontal cortical‐amygdala connectivity during happy and fear face processing: a potential neural mechanism of female MDD. Front Psychiatry 2011;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Musgrove DR, Eberly LE, Klimes‐Dougan B, et al. Impaired bottom‐up effective connectivity between amygdala and subgenual anterior cingulate cortex in unmedicated adolescents with major depression: results from a dynamic causal modeling analysis. Brain Connect 2015;5:608‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Goulden N, McKie S, Thomas EJ, et al. Reversed frontotemporal connectivity during emotional face processing in remitted depression. Biol Psychiatry 2012;72:604‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rupprechter S, Romaniuk L, Series P, et al. Blunted medial prefrontal cortico‐limbic reward‐related effective connectivity and depression. Brain 2020;143:1946‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Frässle S, Marquand AF, Schmaal L, et al. Predicting individual clinical trajectories of depression with generative embedding. Neuroimage Clin 2020;26:102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chang C, Glover GH. Time–frequency dynamics of resting‐state brain connectivity measured with fMRI. Neuroimage. 2010;50:81‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Preti MG, Bolton TA, Van De Ville D. The dynamic functional connectome: state‐of‐the‐art and perspectives. Neuroimage 2017;160:41‐54. [DOI] [PubMed] [Google Scholar]
- 113. Long Y, Cao H, Yan C, et al. Altered resting‐state dynamic functional brain networks in major depressive disorder: findings from the REST‐meta‐MDD consortium. Neuroimage Clin 2020;26:102163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kaiser RH, Whitfield‐Gabrieli S, Dillon DG, et al. Dynamic resting‐state functional connectivity in major depression. Neuropsychopharmacology 2016;41:1822‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wei M, Qin J, Yan R, et al. Association of resting‐state network dysfunction with their dynamics of inter‐network interactions in depression. J Affect Disord 2015;174:527‐34. [DOI] [PubMed] [Google Scholar]
- 116. Wise T, Marwood L, Perkins AM, et al. Instability of default mode network connectivity in major depression: a two‐sample confirmation study. Transl Psychiatry 2017;7:e1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Demirtas M, Tornador C, Falcon C, et al. Dynamic functional connectivity reveals altered variability in functional connectivity among patients with major depressive disorder. Hum Brain Mapp 2016;37:2918‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zheng Y, Chen X, Li D, et al. Treatment‐naïve first episode depression classification based on high‐order brain functional network. J Affect Disord 2019;256:33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yan B, Xu X, Liu M, et al. Quantitative identification of major depression based on resting‐state dynamic functional connectivity: a machine learning approach. Front Neurosci 2020;14:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Byun HY, Lu JJ, Mayberg HS, et al. Classification of resting state fMRI datasets using dynamic network clusters. In: Workshops at the Twenty‐Eighth AAAI Conference on Artificial Intelligence, Québec City; 2014:2‐6.
- 121. Müller K, Lohmann G, Neumann J, et al. Investigating the wavelet coherence phase of the BOLD signal. J Magn Reson Imaging 2004;20:145‐52. [DOI] [PubMed] [Google Scholar]
- 122. Smith RX, Jann K, Ances B, et al. Wavelet‐based regularity analysis reveals recurrent spatiotemporal behavior in resting‐state fMRI. Hum Brain Mapp 2015;36:3603‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bernas A, Aldenkamp AP, Zinger S. Wavelet coherence‐based classifier: a resting‐state functional MRI study on neurodynamics in adolescents with high‐functioning autism. Comput Methods Programs Biomed 2018;154:143‐51. [DOI] [PubMed] [Google Scholar]
- 124. Khan DM, Masroor K, Jailani MFM, et al. Development of wavelet coherence EEG as a biomarker for diagnosis of major depressive disorder. IEEE Sens J 2022;22:4315‐25. [Google Scholar]
- 125. Wager TD, Woo C‐W. Imaging biomarkers and biotypes for depression. Nat Med 2017;23:16‐7. [DOI] [PubMed] [Google Scholar]
- 126. Beijers L, Wardenaar KJ, van Loo HM, et al. Data‐driven biological subtypes of depression: systematic review of biological approaches to depression subtyping. Mol Psychiatry 2019;24:888‐900. [DOI] [PubMed] [Google Scholar]
- 127. Drysdale AT, Grosenick L, Downar J, et al. Resting‐state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 2017;23:28‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Feder S, Sundermann B, Wersching H, et al. Sample heterogeneity in unipolar depression as assessed by functional connectivity analyses is dominated by general disease effects. J Affect Disord 2017;222:79‐87. [DOI] [PubMed] [Google Scholar]
- 129. Tokuda T, Yoshimoto J, Shimizu Y, et al. Identification of depression subtypes and relevant brain regions using a data‐driven approach. Sci Rep 2018;8:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Price RB, Gates K, Kraynak TE, et al. Data‐driven subgroups in depression derived from directed functional connectivity paths at rest. Neuropsychopharmacology 2017;42:2623‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Dinga R, Schmaal L, Penninx BWJH, et al. Evaluating the evidence for biotypes of depression: methodological replication and extension of Drysdale et al. (2017). Neuroimage Clin 2019;22:101796. [DOI] [PMC free article] [PubMed] [Google Scholar]


