Abstract
The U.S. Tox21 program has developed in vitro assays to test large collections of environmental chemicals in a quantitative high-throughput screening (qHTS) format, using triplicate 15-dose titrations to generate over 100 million data points to date. Counter screens are also employed to minimize interferences from non-target specific assay artifacts, such as compound auto fluorescence and cytotoxicity. New data analysis approaches are needed to integrate these data and characterize the activities observed from these assays. Here, we describe a complete analysis pipeline that evaluates these qHTS data for technical quality in terms of signal reproducibility. We integrate signals from repeated assay runs, primary readouts and counter screens to produce a final call on on-target compound activity.
Keywords: HTS, concentration response, in vitro assay, activity profile, Tox21
1. Introduction
The U.S. Tox21 program [1–4], a collaboration among the National Institute of Environmental Health Sciences (NIEHS)/National Toxicology Program (NTP), the U.S. Environmental Protection Agency’s (EPA) National Center for Computational Toxicology (NCCT), the National Institutes of Health (NIH) National Center for Advancing Translational Sciences (NCATS), and the U.S. Food and Drug Administration (FDA), is aimed at developing alternative testing methods that can quickly and efficiently assess the toxic potential of tens of thousands of environmental chemicals. Working toward this goal, the Tox21 program has successfully developed various cell-based assays to serve as in vitro models for toxicity assessment [5–7,4,8–13]. These assays have been miniaturized and validated in a 1536-well plate format at NCATS for quantitative high-throughput screening (qHTS) [14]. These assays are currently being screened against a collection of ~10,000 compounds (Tox21 10K) [15] composed of environmental chemicals and approved drugs as triplicate 15-dose titrations, generating over 100 million data points to date [16,17,8].
The use of qHTS to produce high quality and biologically relevant data is critical in correlation to in vivo activity, low dose extrapolation, and risk assessment. However, as with any technology, these assays are not immune to noise or artifacts that may interfere with the true biological activity. When a signal is observed in an assay, it is important to be able to distinguish a true biological effect from an artifact. In addition to “noise” and experimental variations, common artifacts found with fluorescence or luminescence based reporter assays, such as those employed by Tox21, include compound auto fluorescence [18,19], interference with the assay reporter gene [20], and cytotoxicity [6,9]. Compound auto fluorescence often interferes with agonist mode assays, in which an increase in signal indicates activity. Compound interaction with the assay reporter gene itself could be mistaken for either agonist activity, when the compound activates the reporter gene, or antagonist activity, when the compound inhibits the reporter gene. Luciferase and β-lactamase reporters are commonly used in Tox21 and other HTS assays. Finally, cell-based antagonist mode assays are often confounded with cytotoxicity interference because both cell death and inhibition of the target of interest result in a decrease in assay signal.
To minimize compound or assay technology dependent artifacts, all compounds in the Tox21 10K library are tested for auto fluorescence at wavelengths used for assay readouts and for luciferase activity. In addition, each assay is multiplexed with cell viability measurements to identify cytotoxicity interference. The challenge is then to devise methods to 1) evaluate these qHTS data for technical quality, e.g. signal reproducibility, and 2) integrate signals from repeated assay runs, primary readouts and counter screens to produce a final call on on-target compound activity. Here, we describe a complete qHTS data analysis pipeline developed at NCATS that begins with plate level raw data processing, followed by concentration response curve fitting and classification, data reproducibility evaluation, and assignment of activity outcomes to compounds through integration of data from multiple readouts and counter screens. This approach has been applied to all the qHTS data generated from the Tox21 assay validation runs and 10K screens, and can be adapted to analyze other qHTS data generated in a similar fashion.
2. qHTS data pipeline
2.1. Plate level data processing, curve fitting and classification
During the execution of the screen, quality metrics, such as CV, S/B, and Z-factor [21], are calculated using raw fluorescence or luminescence reads from each plate to monitor gross assay performance. These metrics are recorded for each plate and examined. “Failed plates,” identified by abnormally poor values, are inspected visually and, if necessary, excluded from further data analysis. Upon completion of an assay run, raw plate reads for each titration point are first normalized relative to the positive control compound (100% for agonist mode and −100% for antagonist mode assays) and DMSO-only wells (0%) placed in the first four columns of each plate as follows: % Activity = ((Vcompound − VDMSO)/(Vpos − VDMSO)) × 100, where Vcompound denotes the compound well values, Vpos denotes the median value of the positive control wells, and VDMSO denotes the median values of the DMSO-only wells, and then corrected using compound-free control plates (i.e., DMSO-only plates) at the beginning and end of the compound plate stack to remove background patterns and subtle abnormalities such as tip effects or blotting from cell dispenses [22].
Corrected plate data are pivoted to form concentration–response series, which are subsequently fit to a four-parameter Hill equation [23] yielding concentrations of half-maximal activity (AC50) and maximal response (efficacy) values [24]. Concentration-response curves are designated as Class 1–4 based on efficacy, the number of data points observed above background activity, and the quality of fit [25]. Curve classes are heuristic measures of data confidence. The qHTS curve classification scheme has been later amended to better suit the needs of toxicology research (Table 1) [6]. The most problematic concentration responses are automatically assigned curve class 5 based on considerations like the direction of activity (observing alternately both increases and decreases in signal over a short concentration range) and unusually large signal at low sample concentrations (activity at zero concentration is estimated to be >3SD of control) [6]. Class 5 curves and other cases in which an inconsistency between the highest compound activity and the curve class assigned is identified, such as assigning a compound with a positive response a negative curve class, are manually inspected to correct the curve class, if necessary. Adjustment is normally done by masking or by unmasking data points improperly masked by the automated curve fitting process to adjust the curve fit (Figure 1). To facilitate analysis and activity profiling, each curve class is further combined with an efficacy cutoff and converted to a numerical curve rank such that more potent and efficacious compounds with higher quality curves are assigned a higher rank (Table 2). Curve ranks should be viewed as a numerical measure of compound activity.
Table 1.
Amended qHTS curve classification
| Curve Class | Description | Efficacy | p-value* | Asymptotes | Inflection |
|---|---|---|---|---|---|
| 1.1 | Complete curve | >6SD† | <0.05 | 2 | Yes |
| 1.2 | Complete curve | ≤6SD; >3SD | <0.05 | 2 | Yes |
| 1.3 | Complete curve | >6SD | ≥0.05 | 2 | Yes |
| 1.4 | Complete curve | ≤6SD; >3SD | ≥0.05 | 2 | Yes |
| 2.1 | Incomplete curve | >6SD | <0.05 | 1 | Yes |
| 2.2 | Incomplete curve | ≤6SD; >3SD | <0.05 | 1 | Yes |
| 2.3 | Incomplete curve | >6SD | ≥0.05 | 1 | Yes |
| 2.4 | Incomplete curve | ≤6SD; >3SD | ≥0.05 | 1 | Yes |
| 3 | Single point activity | >3SD | NA | 1 | No |
| 4 | Inactive | ≤3SD | ≥0.05 | 0 | No |
| 5‡ | Inconclusive | NA | NA | NA | NA |
p-value is derived from a F-test that measures the quality of curve fit.
SD is the standard deviation of sample activities at the lowest tested concentration and values of the DMSO control wells.
Class 5 is a special class for samples with activity at zero concentration (zero activity; extrapolated) exceeding 6SD or with zero activity>3SD and the difference between the maximal change in activity observed in the tested concentration range and zero activity is <3SD.
Figure 1.
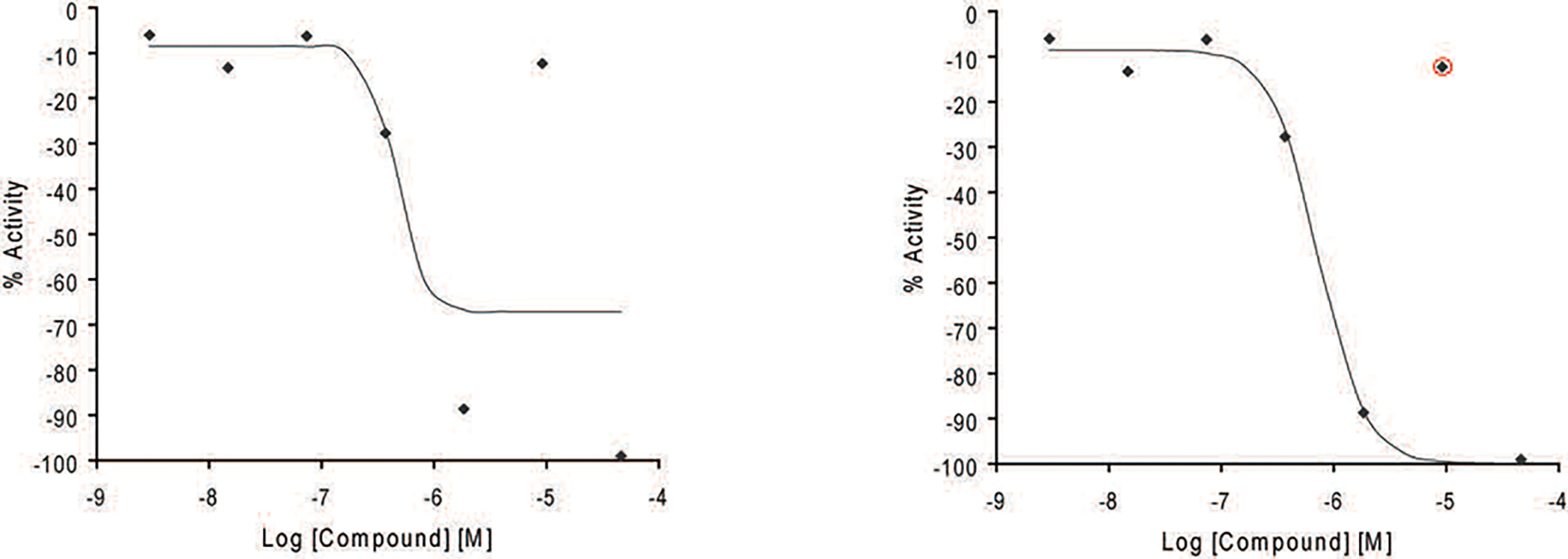
Example of outlier masking. Curves are manually inspected to mask or unmask data points improperly masked by the automated curve fitting process to adjust the curve fit when necessary.
Table 2.
Definition of curve rank as a numeric measure of compound activity
| Curve class | Efficacy | Curve rank | Activity Category |
|---|---|---|---|
| 1.1 | 9 | agonist | |
| 1.2 | >50% | 8 | agonist |
| 2.1 | 7 | agonist | |
| 1.2 | ≤50% | 6 | agonist |
| 2.2 | >50% | 5 | agonist |
| 2.2 | ≤50% | 4 | inconclusive |
| 1.3 | 3 | inconclusive | |
| 1.4 | 3 | inconclusive | |
| 2.3 | 2 | inconclusive | |
| 2.4 | 2 | inconclusive | |
| 3 | 2 | inconclusive | |
| 5 | 1 | inconclusive | |
| 4 | 0 | inactive | |
| −2.3 | −2 | inconclusive | |
| −2.4 | −2 | inconclusive | |
| −3 | −2 | inconclusive | |
| −1.3 | −3 | inconclusive | |
| −1.4 | −3 | inconclusive | |
| −2.2 | ≤50% | −4 | inconclusive |
| −2.2 | >50% | −5 | antagonist |
| −1.2 | ≤50% | −6 | antagonist |
| −2.1 | −7 | antagonist | |
| −1.2 | >50% | −8 | antagonist |
| −1.1 | −9 | antagonist |
2.2. Assay performance measured by reproducibility
After manual curation, the “clean” curve fitting results from the replicate assay runs are assessed for activity reproducibility to determine the final assay performance. Each sample curve is first assigned an activity outcome based on its curve class as follows: inactive (class 4), active agonist/antagonist (class 1.1, 2.1; class 5 due to super potency (AC50<lowest test concentration)), agonist/antagonist (class 1.2, 2.2), inconclusive agonist/antagonist (all other non-5 classes), no call (other cases of class 5). Each activity outcome category (excluding the “no call” category, which is treated as missing data) is then assigned a score: active agonist (3), agonist (2), inconclusive agonist (1), active antagonist (−3), antagonist (−2), inconclusive antagonist (−1), inactive (0). The pair-wise activity outcome score differences for all replicate curves of each sample are then averaged and the % of inactive calls for the sample calculated to determine the final reproducibility call of the sample: active match (average score difference <1.1, %inactive call <25%), inactive match (average score difference <1.1, %inactive call >50%), mismatch (average score difference >2.5), inconclusive (all other cases). Each assay is assigned a performance score as follows: reproducibility score = 2×%active match + %inactive match − %inconclusive − 2×%mismatch.
2.3. Identification of auto fluorescence and cytotoxicity artifacts
In fluorescence based agonist mode assays, auto fluorescent compounds can show the same phenotype as those of agonists. Two approaches are used to identify potential auto fluorescent artifacts to distinguish them from true agonists. One approach is using the auto fluorescence detection counter screen [18,19] measured at the same wavelength as the assay readout (e.g., 460 nm for β-lactamase assays). Any compound with the agonist phenotype in the assay signal channel that also shows activation in the auto fluorescence counter screen with an AC50 difference <3 fold is flagged as a potential auto fluorescent false positive. The second approach is examining the activity of each compound in all the assays screened having the same reporter (e.g. β-lactamase). Compounds with the agonist phenotype in the assay signal channel (e.g., the 460 nm channel of β-lactamase assays) that also had an >4 average curve rank in the signal channel of all the assays with the same reporter are considered promiscuously active in such reporter gene assays and potentially auto fluorescent. The compounds identified by either approach are assigned the “inconclusive agonist (fluorescent)” activity outcome category. The auto fluorescence counter screen data on the Tox21 10K have been made publicly available in PubChem [26] (assay IDs 720678, 720680, 720679, 720681, 720682, 720683, 720687, 720675, 720674, 720685, 720686, 720684). The same methods could be applied to luciferase reporter gene assays to identify compounds that are promiscuous luciferase stabilizers. Luciferase counter screens have also been conducted as an auxiliary approach to identify such compounds in the Tox21 10K library and the data are available in PubChem (assay ID 1224835).
In antagonist mode assays, cytotoxic compounds can show the same inhibitory phenotype as those of antagonists that need to be distinguished from true antagonists and flagged as cytotoxicity-related false positive responses. For this reason, each antagonist mode assay screened for Tox21 is accompanied with a cell viability readout that serves as the counter screen. Any compound with the antagonist phenotype in the assay signal channel that also shows inhibition in the cell viability counter screen with an AC50 difference <3 fold or p>0.05 (t-test) is flagged as a potential cytotoxic false positive. As an alternative to the cell viability counter screen, the control channel in assays with multiple channel readouts (e.g. the 530 nm readout of β-lactamase assays) can be used to identify potential cytotoxic compounds. Either activation or inhibition shown in this channel can be an indication of cytotoxicity [6]. The effectiveness of using the control readout to identify potential cytotoxicity artifacts has been compared with the cell viability counter screen. The two approaches achieve similar specificity in correctly distinguishing true antagonists from cytotoxic artifacts, while filtering with the cell viability counter screen results in better sensitivity compared to the control readout [27].
2.4. Compound activity assignment
Compounds are assigned one of the following activity outcome categories: active agonist, inconclusive agonist (due to poor curve quality), inconclusive agonist (due to auto fluorescence), active antagonist, inconclusive antagonist (due to poor curve quality), inconclusive antagonist (due to cytotoxicity), inconclusive (activity direction cannot be determined), or inactive. The antagonist outcome labels in agonist mode assays are for compounds that show inhibition, which does not necessarily reflect true antagonism but rather might reflect increased cytotoxicity or promiscuous reporter gene inhibition. The agonist outcome labels in antagonist mode assays are for compounds that show activation, which does not necessarily reflect true agonism but rather may reflect compound auto fluorescence or promiscuous reporter gene activation. To generate these assignments, curve ranks from all replicates of a compound are first averaged for each of the assay readouts, and the activity outcome of the compound in the assay readout is assigned based on the compound’s average curve rank and reproducibility call as shown in Table 3. For luminescence assays with a single readout that is run in agonist mode, such as the estrogen receptor alpha luciferase reporter gene assay (BG1-ER-luc) [27], this activity outcome is assigned as the final activity outcome for a compound. For the same assay run in antagonist mode in complex with a cell viability counter screen, an activity outcome is assigned to both the antagonist mode readout and the cell viability readout first, and the final assay activity outcome for a compound is determined according to Table 4(c). For fluorescence based assays with multiple channel readouts (signal, control and ratio), such as the estrogen receptor alpha β-lactamase reporter gene assay (ER-bla) [27] or the mitochondrial membrane potential assay [16,12], the final activity outcome of a compound is determined based on its multi-channel activity as shown in Tables 4(a) and (b). For antagonist mode assays, the cell viability counter screen data are used to flag potential cytotoxic artifacts. For agonist mode assays, potential artifacts produced by compounds that auto fluoresce in the signal channel (e.g., 460 nm readout in the ER-bla assay) are flagged using both the compound auto fluorescence profiling data and the promiscuous compound activity shown in the signal readout of all the assays screened in Tox21 that have the same reporter (e.g. β-lactamase). The complete activity assignment process is illustrated in Figure 2.
Table 3.
Compound single channel activity outcome assignments based on curve rank and reproducibility
| Curve rank | Reproducibility call | Activity outcome |
|---|---|---|
| >−1 and <1 | inactive match | inactive |
| >−1 and <1 | inconclusive | inconclusive |
| >=1 | mismatch | inconclusive agonist |
| >=1 | active match | active agonist |
| >4 | inconclusive | active agonist |
| >=1 and <=4 | inconclusive | inconclusive agonist |
| <=−1 | mismatch | inconclusive antagonist |
| <=−1 | active match | active antagonist |
| <−4 | inconclusive | active antagonist |
| >=−4 and <=−1 | inconclusive | inconclusive antagonist |
Table 4.
Compound activity outcome assignments based on multi-channel assay readouts (a) multi-channel fluorescence agonist mode assay (b) multi-channel fluorescence antagonist mode assay (c) luminescence antagonist mode assay with cytotoxicity counter screen
| (a) | ||||
| *Ratio outcome | Signal channel outcome | Same reporter assay promiscuity | Auto fluorescence outcome | Activity outcome |
| inactive | N/A | N/A | N/A | inactive |
| inconclusive | N/A | N/A | N/A | inconclusive |
| active agonist | agonist | average curve rank ≤4 | inactive or AC50 fluor/AC50 signal ≥3 | active agonist |
| inconclusive agonist | agonist | average curve rank ≤4 | inactive or AC50 fluor/AC50 signal ≥3 | inconclusive agonist |
| agonist | agonist | average curve rank >4 | agonist and AC50 fluor/AC50 signal <3 | inconclusive agonist (fluorescent) |
| active antagonist | antagonist | N/A | N/A | active antagonist |
| inconclusive antagonist | antagonist | N/A | N/A | inconclusive antagonist |
| (b) | ||||
| *Ratio outcome | Signal channel outcome | Cell viability outcome | Other conditions | Activity outcome |
| inactive | N/A | N/A | N/A | inactive |
| inconclusive | N/A | N/A | N/A | inconclusive |
| active agonist | agonist | inactive or agonist | N/A | active agonist |
| active agonist | agonist | antagonist | AC50 viability/AC50 signal ≥3 (p<0.05) | active agonist |
| inconclusive agonist | agonist | N/A | N/A | inconclusive agonist |
| agonist | agonist | antagonist | AC50 viability/AC50 signal <3 or p≥0.05 | inconclusive agonist (cytotoxic) |
| active antagonist | antagonist | inactive or agonist | N/A | active antagonist |
| active antagonist | antagonist | antagonist | AC50 viability/AC50 signal ≥3 (p<0.05) | active antagonist |
| inconclusive antagonist | antagonist | N/A | N/A | inconclusive antagonist |
| antagonist | antagonist | antagonist | AC50 viability/AC50 signal <3 or p≥0.05 | inconclusive antagonist (cytotoxic) |
| (c) | ||||
| Signal channel outcome | Cell viability outcome | Other conditions | Activity outcome | |
| inactive | N/A | N/A | inactive | |
| inconclusive | N/A | N/A | inconclusive | |
| active agonist | inactive or agonist | N/A | active agonist | |
| active agonist | antagonist | AC50 viability/AC50 signal ≥3 (p<0.05) | active agonist | |
| inconclusive agonist | N/A | N/A | inconclusive agonist | |
| agonist | antagonist | AC50 viability/AC50 signal <3 or p≥0.05 | inconclusive agonist (cytotoxic) | |
| active antagonist | inactive or agonist | N/A | active antagonist | |
| active antagonist | antagonist | AC50 viability/AC50 signal ≥3 (p<0.05) | active antagonist | |
| inconclusive antagonist | N/A | N/A | inconclusive antagonist | |
| antagonist | antagonist | AC50 viability/AC50 signal <3 or p≥0.05 | inconclusive antagonist (cytotoxic) | |
Ratio = signal channel/control channel
Abbreviations: AC50 fluor = AC50 in the auto fluorescence assay, AC50 signal = AC50 in the ratio channel
Ratio = signal channel/control channel
Abbreviations: AC50 viability = AC50 in the cell viability assay, AC50 signal = AC50 in the ratio channel
Abbreviations: AC50 viability = AC50 in the cell viability assay, AC50 signal = AC50 in the antagonist mode assay
Figure 2.
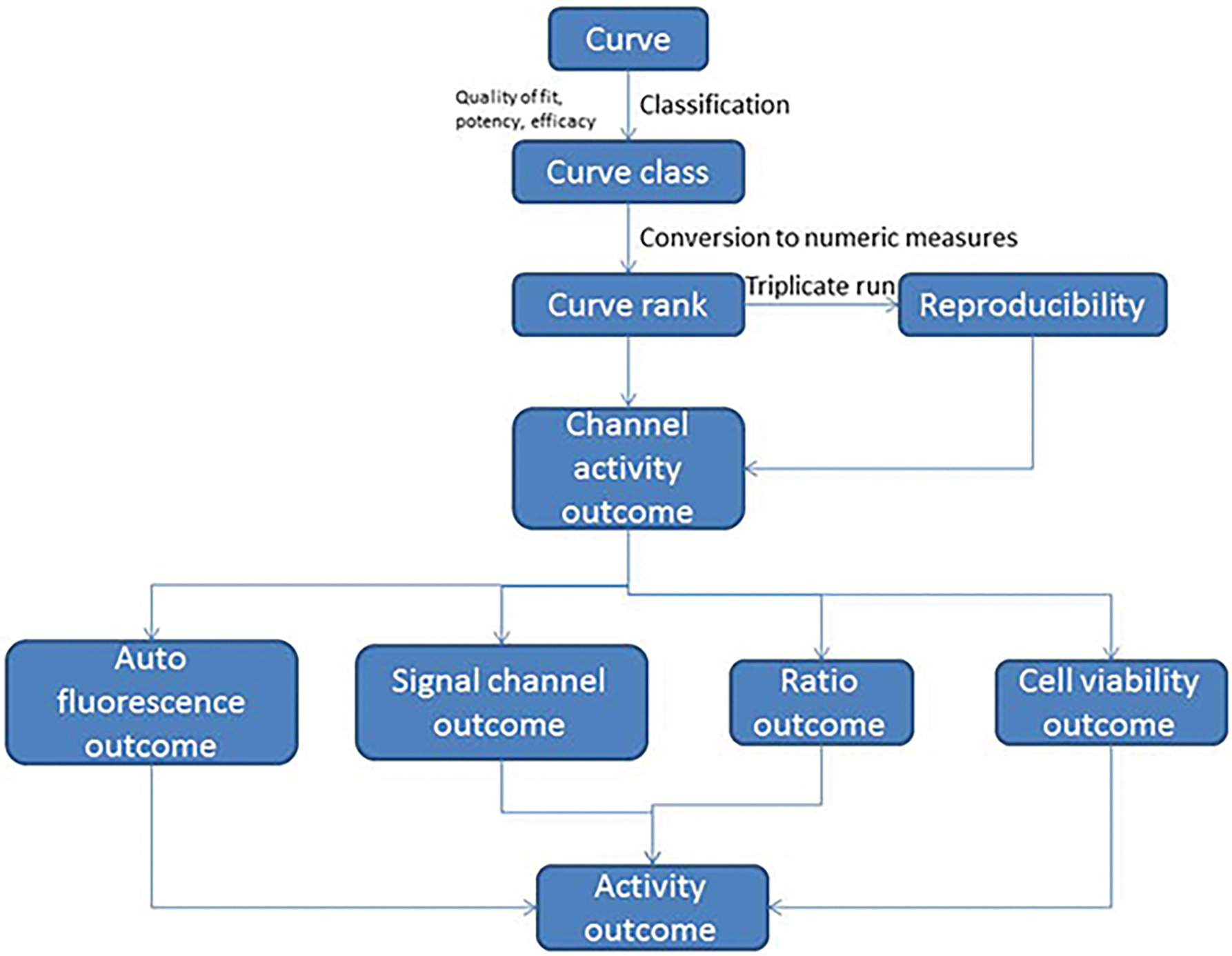
Compound activity assignment process. This flowchart shows the process of assigning a final assay activity outcome to a compound based on its qHTS data from replicate assay runs, multiple readouts and counter screens.
3. Data sharing
As soon as the initial data parsing and assessment at the NCATS are complete, the concentration response data, curve fitting results, the raw plate reads, the assay conditions, and the sample mapping information are shared with the Tox21 partners through a suite of databases and software tools custom built by the NCATS for the Tox21 program (http://tripod.nih.gov/tox/). Within the first six months of data generation, the assay data are only made available to the Tox21 partners through the aforementioned controlled-access site in which the data are further scrutinized for quality and utility. The data are then released to the public domain in a number of public databases including PubChem (http://pubchem.ncbi.nlm.nih.gov/), CEBS (http://tools.niehs.nih.gov/cebs3/ui/), the EPA CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/) and the NCATS Tox21 Data Browser (https://tripod.nih.gov/tox21/assays/). The high quality concentration response data generated on a wide spectrum of pathways and phenotypic toxicity endpoints provide a valuable resource for predictive toxicity modeling. These data can not only serve as in vitro signatures that can be used to predict in vivo toxicity endpoints and to prioritize chemicals for more in depth toxicity testing [28,12] that helps fulfill the goals of the Tox21 program, but also provide rich training data sets for the QSAR (quantitative structure–activity relationship) modeling community to build more robust models [29–33] such as the ones in the recent Tox21 Data Challenge hosted by NCATS in 2014 (https://tripod.nih.gov/tox21/challenge/) [34].
References
- 1.Collins FS, Gray GM, Bucher JR (2008) Toxicology. Transforming environmental health protection. Science 319 (5865):906–907. doi:319/5865/906 [pii] 10.1126/science.1154619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kavlock RJ, Austin CP, Tice RR (2009) Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal 29 (4):485–487; discussion 492–487. doi:RISK1168 [pii] 10.1111/j.1539-6924.2008.01168.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NRC (2007) Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press, Washington, DC [Google Scholar]
- 4.Tice RR, Austin CP, Kavlock RJ, Bucher JR (2013) Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121 (7):756–765. doi: 10.1289/ehp.1205784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attene-Ramos MS, Huang R, Sakamuru S, Witt KL, Beeson GC, Shou L, Schnellmann RG, Beeson CC, Tice RR, Austin CP, Xia M (2013) Systematic study of mitochondrial toxicity of environmental chemicals using quantitative high throughput screening. Chem Res Toxicol 26 (9):1323–1332. doi: 10.1021/tx4001754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang R, Xia M, Cho MH, Sakamuru S, Shinn P, Houck KA, Dix DJ, Judson RS, Witt KL, Kavlock RJ, Tice RR, Austin CP (2011) Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ Health Perspect 119 (8):1142–1148. doi: 10.1289/ehp.1002952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla SJ, Huang R, Austin CP, Xia M (2010) The Future of Toxicity Testing: A Focus on In Vitro Methods Using a Quantitative High Throughput Screening Platform. Drug Discovery Today 15 (23–24):997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang R, Xia M, Sakamuru S, Zhao J, Shahane SA, Attene-Ramos M, Zhao T, Austin CP, Simeonov A (2016) Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat Commun 7:10425. doi: 10.1038/ncomms10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh JH, Huang R, Lin JA, Sedykh A, Zhao J, Tice RR, Paules RS, Xia M, Auerbach SS (2017) Real-time cell toxicity profiling of Tox21 10K compounds reveals cytotoxicity dependent toxicity pathway linkage. PLoS One 12 (5):e0177902. doi: 10.1371/journal.pone.0177902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng CT, Hsieh JH, Zhao J, Huang R, Xia M, Martin N, Gao X, Dixon D, Auerbach SS, Witt KL, Merrick BA (2017) Development of Novel Cell Lines for High-Throughput Screening to Detect Estrogen-Related Receptor Alpha Modulators. SLAS Discov 22 (6):720–731. doi: 10.1177/2472555216689772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witt KL, Hsieh JH, Smith-Roe SL, Xia M, Huang R, Zhao J, Auerbach SS, Hur J, Tice RR (2017) Assessment of the DNA damaging potential of environmental chemicals using a quantitative high-throughput screening approach to measure p53 activation. Environ Mol Mutagen 58 (7):494–507. doi: 10.1002/em.22112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia M, Huang R, Shi Q, Boyd WA, Zhao J, Sun N, Rice JR, Dunlap PE, Hackstadt AJ, Bridge MF, Smith MV, Dai S, Zheng W, Chu PH, Gerhold D, Witt KL, DeVito M, Freedman JH, Austin CP, Houck KA, Thomas RS, Paules RS, Tice RR, Simeonov A (2018) Comprehensive Analyses and Prioritization of Tox21 10K Chemicals Affecting Mitochondrial Function by in-Depth Mechanistic Studies. Environ Health Perspect 126 (7):077010. doi: 10.1289/EHP2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch C, Mackowiak B, Huang R, Li L, Heyward S, Sakamuru S, Wang H, Xia M (2019) Identification of Modulators That Activate the Constitutive Androstane Receptor From the Tox21 10K Compound Library. Toxicol Sci 167 (1):282–292. doi: 10.1093/toxsci/kfy242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attene-Ramos MS, Miller N, Huang R, Michael S, Itkin M, Kavlock RJ, Austin CP, Shinn P, Simeonov A, Tice RR, Xia M (2013) The Tox21 robotic platform for the assessment of environmental chemicals - from vision to reality. Drug Discov Today 18 (15–16):716–723. doi:S1359–6446(13)00161-X [pii] 10.1016/j.drudis.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard AM, Huang R, Waidyanatha S, Shinn P, Collins BJ, Thillainadarajah I, Grulke CM, Williams AJ, Lougee RR, Judson RS, Houck KA, Shobair M, Yang C, Rathman JF, Yasgar A, Fitzpatrick SC, Simeonov A, Thomas RS, Crofton KM, Paules RS, Bucher JR, Austin CP, Kavlock RJ, Tice RR (2021) The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem Res Toxicol 34 (2):189–216. doi: 10.1021/acs.chemrestox.0c00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attene-Ramos MS, Huang R, Michael S, Witt KL, Richard A, Tice RR, Simeonov A, Austin CP, Xia M (2015) Profiling of the Tox21 chemical collection for mitochondrial function to identify compounds that acutely decrease mitochondrial membrane potential. Environ Health Perspect 123 (1):49–56. doi: 10.1289/ehp.1408642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu CW, Zhao J, Huang R, Hsieh JH, Hamm J, Chang X, Houck K, Xia M (2014) Quantitative high-throughput profiling of environmental chemicals and drugs that modulate farnesoid X receptor. Sci Rep:doi: 10.1038/srep06437. doi: 10.1038/srep06437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J (2008) Fluorescence spectroscopic profiling of compound libraries. J Med Chem 51 (8):2363–2371. doi: 10.1021/jm701301m [DOI] [PubMed] [Google Scholar]
- 19.Borrel A, Huang R, Sakamuru S, Xia M, Simeonov A, Mansouri K, Houck KA, Judson RS, Kleinstreuer NC (2020) High-Throughput Screening to Predict Chemical-Assay Interference. Sci Rep 10 (1):3986. doi: 10.1038/s41598-020-60747-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorne N, Auld DS, Inglese J (2010) Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol 14 (3):315–324. doi: 10.1016/j.cbpa.2010.03.020 S1367–5931(10)00046–3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR (1999) A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4 (2):67–73 [DOI] [PubMed] [Google Scholar]
- 22.Southall NT, Jadhav A, Huang R, Nguyen T, Wang Y (2009) Enabling the Large Scale Analysis of Quantitative High Throughput Screening Data. In: Seethala R, Zhang L (eds) Handbook of Drug Screening. 2nd edn. Taylor and Francis, New York, pp 442–463 [Google Scholar]
- 23.Hill AV (1910) The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol (London) 40:4–7 [Google Scholar]
- 24.Wang Y, Jadhav A, Southal N, Huang R, Nguyen DT (2011) A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genomics 4:57–66. doi: 10.2174/1875397301004010057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP (2006) Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A 103 (31):11473–11478. doi:0604348103 [pii] 10.1073/pnas.0604348103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.PubChem (2013) Tox21 phase II data. http://www.ncbi.nlm.nih.gov/pcassay?term=tox21. Accessed 11/16/2013 2013
- 27.Huang R, Sakamuru S, Martin MT, Reif DM, Judson RS, Houck KA, Casey W, Hsieh JH, Shockley KR, Ceger P, Fostel J, Witt KL, Tong W, Rotroff DM, Zhao T, Shinn P, Simeonov A, Dix DJ, Austin CP, Kavlock RJ, Tice RR, Xia M (2014) Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci Rep:doi: 10.1038/srep05664. doi: 10.1038/srep05664 srep05664 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, Dix DJ (2010) In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect 118 (4):485–492. doi: 10.1289/ehp.0901392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, Zakharov A, Worth A, Richard AM, Grulke CM, Trisciuzzi D, Fourches D, Horvath D, Benfenati E, Muratov E, Wedebye EB, Grisoni F, Mangiatordi GF, Incisivo GM, Hong H, Ng HW, Tetko IV, Balabin I, Kancherla J, Shen J, Burton J, Nicklaus M, Cassotti M, Nikolov NG, Nicolotti O, Andersson PL, Zang Q, Politi R, Beger RD, Todeschini R, Huang R, Farag S, Rosenberg SA, Slavov S, Hu X, Judson RS (2016) CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ Health Perspect 124 (7):1023–1033. doi: 10.1289/ehp.1510267 ehp.1510267 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansouri K, Kleinstreuer N, Abdelaziz AM, Alberga D, Alves VM, Andersson PL, Andrade CH, Bai F, Balabin I, Ballabio D, Benfenati E, Bhhatarai B, Boyer S, Chen J, Consonni V, Farag S, Fourches D, Garcia-Sosa AT, Gramatica P, Grisoni F, Grulke CM, Hong H, Horvath D, Hu X, Huang R, Jeliazkova N, Li J, Li X, Liu H, Manganelli S, Mangiatordi GF, Maran U, Marcou G, Martin T, Muratov E, Nguyen DT, Nicolotti O, Nikolov NG, Norinder U, Papa E, Petitjean M, Piir G, Pogodin P, Poroikov V, Qiao X, Richard AM, Roncaglioni A, Ruiz P, Rupakheti C, Sakkiah S, Sangion A, Schramm KW, Selvaraj C, Shah I, Sild S, Sun L, Taboureau O, Tang Y, Tetko IV, Todeschini R, Tong W, Trisciuzzi D, Tropsha A, Van Den Driessche G, Varnek A, Wang Z, Wedebye EB, Williams AJ, Xie H, Zakharov AV, Zheng Z, Judson RS (2020) CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity. Environ Health Perspect 128 (2):27002. doi: 10.1289/EHP5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slavov S, Stoyanova-Slavova I, Li S, Zhao J, Huang R, Xia M, Beger R (2017) Why are most phospholipidosis inducers also hERG blockers? Arch Toxicol 91 (12):3885–3895. doi: 10.1007/s00204-017-1995-9 10.1007/s00204–017-1995–9 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Zakharov AV, Zhao T, Nguyen DT, Peryea T, Sheils T, Yasgar A, Huang R, Southall N, Simeonov A (2019) Novel Consensus Architecture To Improve Performance of Large-Scale Multitask Deep Learning QSAR Models. J Chem Inf Model 59 (11):4613–4624. doi: 10.1021/acs.jcim.9b00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimenko K, Rosenberg SA, Dybdahl M, Wedebye EB, Nikolov NG (2019) QSAR modelling of a large imbalanced aryl hydrocarbon activation dataset by rational and random sampling and screening of 80,086 REACH pre-registered and/or registered substances. PLoS One 14 (3):e0213848. doi: 10.1371/journal.pone.0213848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang R, Xia M, Nguyen D-T, Zhao T, Sakamuru S, Zhao J, Shahane SA, Rossoshek A, Simeonov A (2016) Tox21 Challenge to Build Predictive Models of Nuclear Receptor and Stress Response Pathways as Mediated by Exposure to Environmental Chemicals and Drugs. Frontiers in Environmental Science 3 (85):1–9. doi:doi: 10.3389/fenvs.2015.00085 [DOI] [Google Scholar]


