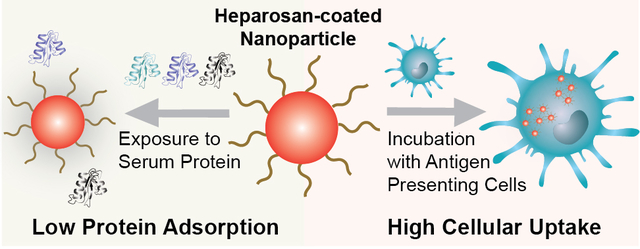
Abstract
Nanoparticle modification with poly(ethylene glycol) (PEG) is a widely used surface engineering strategy in nanomedicine. However, since the artificial PEG polymer may adversely impact nanomedicine safety and efficacy, alternative surface modifications are needed. Here, we explored the ‘self’ polysaccharide heparosan (HEP) to prepare colloidally stable HEP-coated nanoparticles, including gold and silver nanoparticles and liposomes. We found that the HEP-coating reduced the nanoparticle protein corona formation as efficiently as PEG coatings upon serum incubation. Liquid chromatography-mass spectrometry revealed the protein corona profiles. Heparosan-coated nanoparticles exhibited up to 230-fold higher uptake in certain innate immune cells, but not in other tested cell types, than PEGylated nanoparticles. No noticeable cytotoxicity was observed. Serum proteins did not mediate the high cell uptake of HEP-coated nanoparticles. Our work suggests that HEP polymers may be an effective surface modification technology for nanomedicines to safely and efficiently target certain innate immune cells.
Keywords: Nanoparticles, liposome, cellular uptake, heparosan, protein corona, surface engineering, PEGylation, nanomedicine, drug delivery
Graphical Abstract
Introduction
Nanoparticles provide flexible platforms for the development of drug delivery technologies, disease diagnostics, and vaccines1–6. Yet, upon exposure to physiological fluids, proteins adsorb onto the nanoparticle surface to form a layer termed the protein corona7,8. This protein corona can alter the biological fate and immunogenicity of nanoparticles7–10. For example, certain proteins can undergo configurational changes upon adsorption to nanoparticle surfaces, potentially resulting in nanoparticle aggregation or the presentation of novel antigenic sites11,12. To address this challenge, nanoparticle surface modifications with synthetic polymers are commonly used in nanomedicine to enhance colloidal stability and reduce the non-specific protein adsorption13–17.
While the FDA has approved the clinical use of nanoparticles with polymer coatings, such as poly(ethylene glycol) (PEG) and dextran, these coating agents have been reported in some cases to impact nanomedicine safety and efficacy adversely18–20. These reports have raised growing clinical concern about anti-PEG immunogenicity, which may be amplified by the widespread use of PEG in cosmetics, health care products, and over-the-counter medications21,22. Anti-PEG antibodies can bind to PEGylated nanoparticles, which may induce undesired immune responses, including premature clearance of nanomedicines, allergic reactions, and anaphylaxis21,23–29. There is a need to investigate alternative nanoparticle surface modifications that can address the shortcomings of PEGylated nanomedicines30.
Here, we explored the polysaccharide heparosan (HEP) as a biocompatible nanoparticle surface modification agent. The HEP polysaccharide is a biosynthetic precursor in the anticoagulant polysaccharides pathway in animals31,32. In a previous report, no immunogenicity was observed when HEP was used as a surface coating agent for drug delivery liposomes in vivo33. While some reported studies used HEP-coated liposomes and micelles33–36, we demonstrate here the broad applicability of HEP surface modifications for various inorganic and organic nanomaterials (i.e. gold nanoparticles (AuNPs), silver nanoparticles (AgNPs), and synthetic liposomes). Using AuNPs as a model system, we systematically characterized HEP-based surface modifications and quantitatively assessed the associated biological interactions.
First, we adopted two different methods, salt aging and pH reduction, to efficiently functionalize the negatively charged HEP on the surface of AuNPs of various sizes. Then, AuNPs with various HEP surface densities were exposed to serum-containing media, and we compared the protein corona characteristics to PEGylated nanoparticles. Next, we performed label-free liquid chromatography tandem mass spectrometry (LC-MS/MS) to study the protein corona profiles of HEP- or PEG- functionalized AuNPs. We quantified the cytotoxicity, cytokine release profiles, and the cellular uptake of these HEP- or PEG-coated AuNPs upon exposure to various cell types. Additionally, we assessed the cellular uptake profiles of HEP- or PEG-coated AgNPs and liposomes. Our results indicate that HEP polymers may be an effective surface modification technology for nanomedicines.
Results and Discussion
We demonstrated in previous studies that orthopyridyl disulfide (OPSS) is an effective linker to bind OPSS-modified polymers to citrate-coated gold nanoparticles (AuNPs)16,37. Therefore, we covalently attached an OPSS group via an amide bond to a modified HEP polysaccharide chain containing an amine at the reducing-end terminus to form OPSS-HEP. Figure S1 depicts the chemical structure of OPSS-HEP. The qualitative characterization of the successful OPSS conjugation to HEP is shown in Figure S2. We selected 10-kDa OPSS-PEG as a comparative control for the 13-kDa OPSS-HEP to match the molecular weights of both polymers.
To modify colloidally dispersed citrate-coated AuNPs with negatively charged HEP polymers (Figure 1A), we established two different surface modification strategies: (I) an increase in ionic strength to 0.7 M through the step-wise addition of a saline solution (salt aging method, Figure 1B), or (II) a single-step pH reduction to pH 3 by addition of an aqueous hydrochloric acid solution (pH reduction method, Figure 1C). We applied these two methods to increase surface coating effectiveness by reducing the electrostatic repulsion between individual HEP polymers38,39.
Figure 1: Schematic of gold nanoparticle (AuNP) surface modification with heparosan (HEP) polymers.
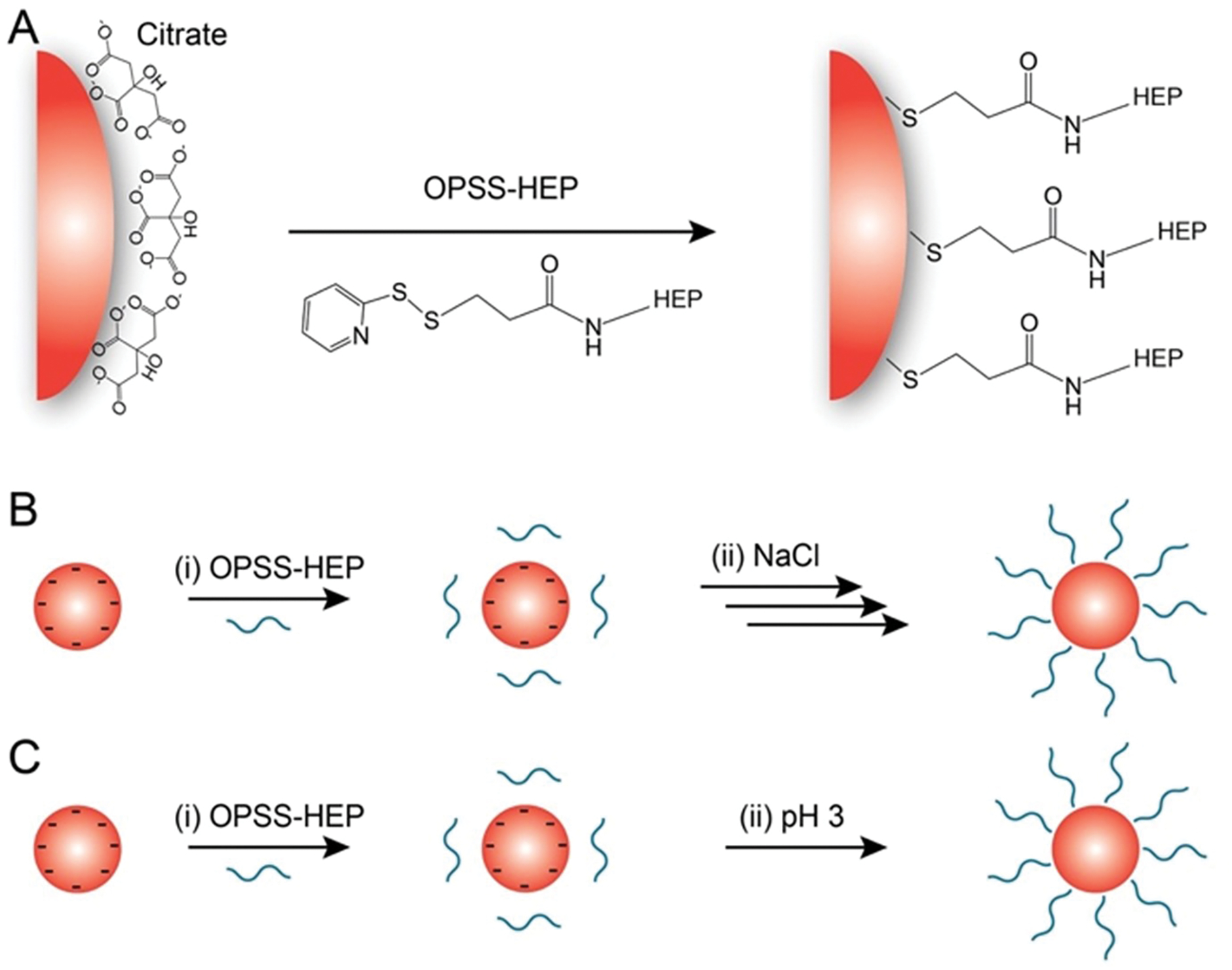
(A) General surface attachment strategy of OPSS-terminated HEP (OPSS-HEP). (B) Salt aging method: (i) OPSS-HEP is mixed with colloidally dispersed citrate-coated AuNPs. (ii) The ionic strength of the dispersion is then increased by the step-wise addition of a NaCl solution (denoted with multiple arrows). (C) pH method. (i) OPSS-HEP is mixed with colloidally dispersed citrate-coated AuNPs. (ii) The pH of the colloidal dispersion is subsequently decreased to ~pH 3 by the one-step addition of a hydrochloric acid solution.
To establish the feasibility of the salt aging and pH methods for the attachment of OPSS-HEP onto AuNPs, we used 15-nm AuNPs as a model nanoparticle system. These AuNPs can be synthesized reproducibly with high yield (typically >80%) and narrow size distribution (<10% deviation)37. As shown in Figure 2A, simply mixing AuNPs with OPSS-HEP did not result in substantial increases in AuNPs hydrodynamic diameter, measured by dynamic light scattering (DLS), which is likely due to the electrostatic repulsion between individual negatively charged HEP polymers. In contrast, both salt aging and pH methods increased the AuNPs hydrodynamic diameters similarly up to ~49 nm as a function of the amount of OPSS-HEP added per nanoparticle surface area in a coating reaction (Fig. 2B). Saturation of the surface was indicated by a plateau when maximal hydrodynamic size was achieved. These DLS results were supported qualitatively by agarose gel electrophoresis experiments, where the migration of the nanoparticles was reduced with an increase in size and HEP surface coverage (Fig. S3). Transmission electron microscopy (TEM) of negative-stained nanoparticles indicated the presence of a dense surface coating layer around HEP-modified AuNPs and revealed an average increase in nanoparticle size of ~25 nm (Fig. 2C–E). This size increase is smaller than the hydrodynamic size increase observed with DLS, most likely due to a partial collapse of the polysaccharide structure during the sample dehydration process required for TEM imaging.
Figure 2: Characterization of heparosan (HEP) surface modification using 15-nm AuNPs.
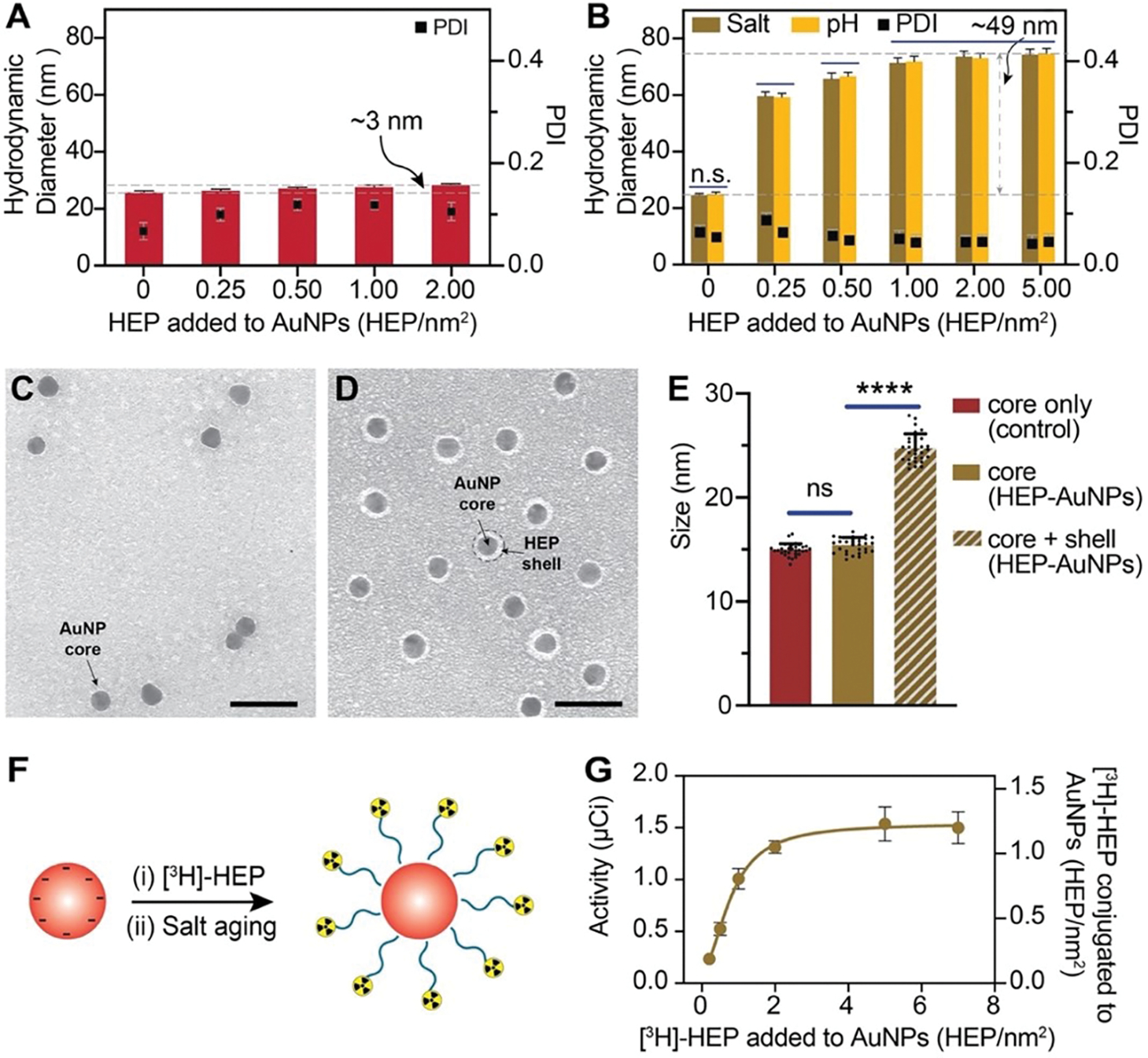
(A) Dynamic light scattering (DLS) was used to measure the hydrodynamic diameter of 15-nm AuNPs after simply mixing with various amounts of HEP per nm2 of nanoparticle surface area with vortexing (without changes in salt concentration (salt aging) or pH reduction). The increase in hydrodynamic diameter of only ~3 nm suggests that HEP did not efficiently conjugate AuNPs. Bars indicate mean ± SD (n=3). (B) DLS results of 15-nm AuNPs mixed with various amounts of HEP per nm2 nanoparticle surface area and addition of saline (salt aging) or subsequent decrease in pH (pH reduction method). The increase in hydrodynamic diameter of ~49 nm suggests efficient HEP conjugation. Bars indicate mean ± SD (n=3). Statistical tests were performed by two-way ANOVA; n.s. indicates no statistically significant differences. (C-D) Representative TEM micrograph of 15-nm citrate-coated AuNPs with a diameter of 14.9 ± 0.6 nm (C) and HEP-AuNPs with a diameter of 24.7 ± 1.4 nm (D). Citrate-coated AuNPs (C) and HEP-AuNPs (D) were stained with 2% uranyl acetate. The light grey halo around the dark AuNP core corresponds to the coating or shell of the surface conjugated HEP. Scale bar indicates 50 nm. (E) Size analysis of 15 nm citrate- and HEP-AuNPs by TEM imaging. The core only is the core size of citrate-AuNPs of panel C (control; red bar). The core of HEP-AuNPs of panel D is represented by a brown bar. The diameter of the core and shell of HEP-AuNPs of panel D is represented by a slanted lined brown bar. Bars indicate mean ± SD. Statistical tests were performed by one-way ANOVA (p<0.0001 (****); n.s. indicates no statistically significant differences). (F) Schematic of AuNP surface modification with radiolabeled HEP. (G) Radiochemical assessment of HEP coating density: Liquid scintillation analysis was used to measure the 3H radioactivity in comparison to coating density (the addition of [3H]-HEP per nm2) conjugated to 15-nm AuNPs.
To determine the HEP coating efficiency on AuNPs, we prepared tritium [3H] radiolabeled OPSS-HEP polymers and used liquid scintillation counting measurements to quantify the amount of HEP conjugated to AuNPs (Fig. 2F, 2G). As shown in Fig. 2G, the maximum achievable HEP surface coating density was ~1.1 HEP/nm2. In addition, we observed that the colloidal stability of HEP-conjugated AuNPs did not change noticeably for various storage conditions (Fig. S4). Collectively, our data confirmed that both salt aging and pH methods resulted in effective and stable HEP surface coating of 15-nm AuNPs.
Next, we expanded this surface modification strategy to larger nanoparticles to demonstrate the generalizability of our approach. As shown in Figures S5–S8, we used both surface modification methods to successfully coat 55-nm and 100-nm AuNPs with OPSS-HEP, resulting in similar overall increases in hydrodynamic diameter (~49 nm) as observed with 15-nm AuNPs. These results indicate that both surface modification strategies were functional and consistent across a wide range of nanoparticle sizes. Additionally, we found that the long-term colloidal stability of AuNPs coated with low HEP surface density (<0.1 HEP/nm2) could be increased to over one year when using the pH method without citrate (Fig. S8). In summary, both surface modification strategies allowed the successful coating of HEP polymers onto various AuNPs systems. It is worth mentioning that these surface coating strategies could be used as effective general approaches to modify nanoparticles with negatively charged polymers, which is in line with reports by Hurst et al. and Xu et al. for DNA coatings38,39.
The hydrophilicity of PEG has been reported to reduce protein adsorption through repulsion between the PEGylated nanoparticle surface and serum proteins40,41. HEP polymers exhibit a high number of hydroxyl and amide groups ([−4-N-acetylglucosamine-α1,4-glucuronic acid-β1-]n) that render the polymer overall hydrophilic. This hydrophilicity and the overall negative charge of the polysaccharide may reduce HEP polymer-protein interactions. Based on this rationale, we then hypothesized that HEP would not only enhance the colloidal stability but further reduce the serum protein adsorption onto the nanoparticle surface. To evaluate heparosan’s ability to reduce protein adsorption, we exposed HEP-modified AuNPs coated with various surface densities to 100% fetal bovine serum (FBS; Fig. 3A) as a model serum7,42,43. We qualitatively assessed the serum protein adsorption before and after FBS incubation on 15-, 55-, or 100-nm HEP-AuNPs with two different methods: (i) by DLS via changes in hydrodynamic diameter (Fig. S9), and (ii) by agarose gel shift experiments via changes in nanoparticle electrophoretic mobility (Fig. S10). After the FBS exposure, nanoparticles without HEP surface modification exhibited a substantial and consistent DLS size increase of ~26 nm (Fig. 3B) and an overall reduced electrophoretic mobility (Fig. S10). We did not observe significant differences in hydrodynamic diameter for AuNPs modified with >0.5 HEP/nm2 before and after FBS incubation (Fig. 3C and Fig. S9). To demonstrate the broad applicability of this HEP surface modification strategy, we additionally synthesized HEP-coated AgNPs and liposomes44–46. We observed no significant changes in hydrodynamic diameter for HEP-coated AgNPs and liposomes before and after FBS incubation indicating minimal interactions between the nanoparticle surfaces and serum proteins (Fig. S11). Our findings suggest that the HEP surface modification strategy effectively minimizes serum protein adsorption onto the surfaces of various nanoparticles, i.e. AuNPs, AgNPs, and liposomes. We corroborated these findings qualitatively with sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) of isolated proteins from AuNPs surfaces (Fig. 3E–F; S12). Using a quantitative bicinchoninic acid (BCA) assay, we confirmed that the observed nanoparticle surface protein adsorption correlated inversely with increasing surface density of the HEP coating (Fig. 3D). This ability to reduce the protein adsorption of HEP-coated AuNPs was similar for AuNPs that were surface-modified with OPSS-PEG. We used PEG as a control surface modification due to PEG’s widespread use in nanomedicine6,47,48. We summarized the physicochemical characterization results of PEG-modified AuNPs in Figure S13. Overall, our findings confirmed that the HEP surface modification effectively reduced protein adsorption onto nanoparticles, and this effect was more pronounced with increasing HEP surface coating densities similar to PEGylated nanoparticles.
Figure 3: Nanoparticle surface engineering with heparosan reduces protein corona formation.
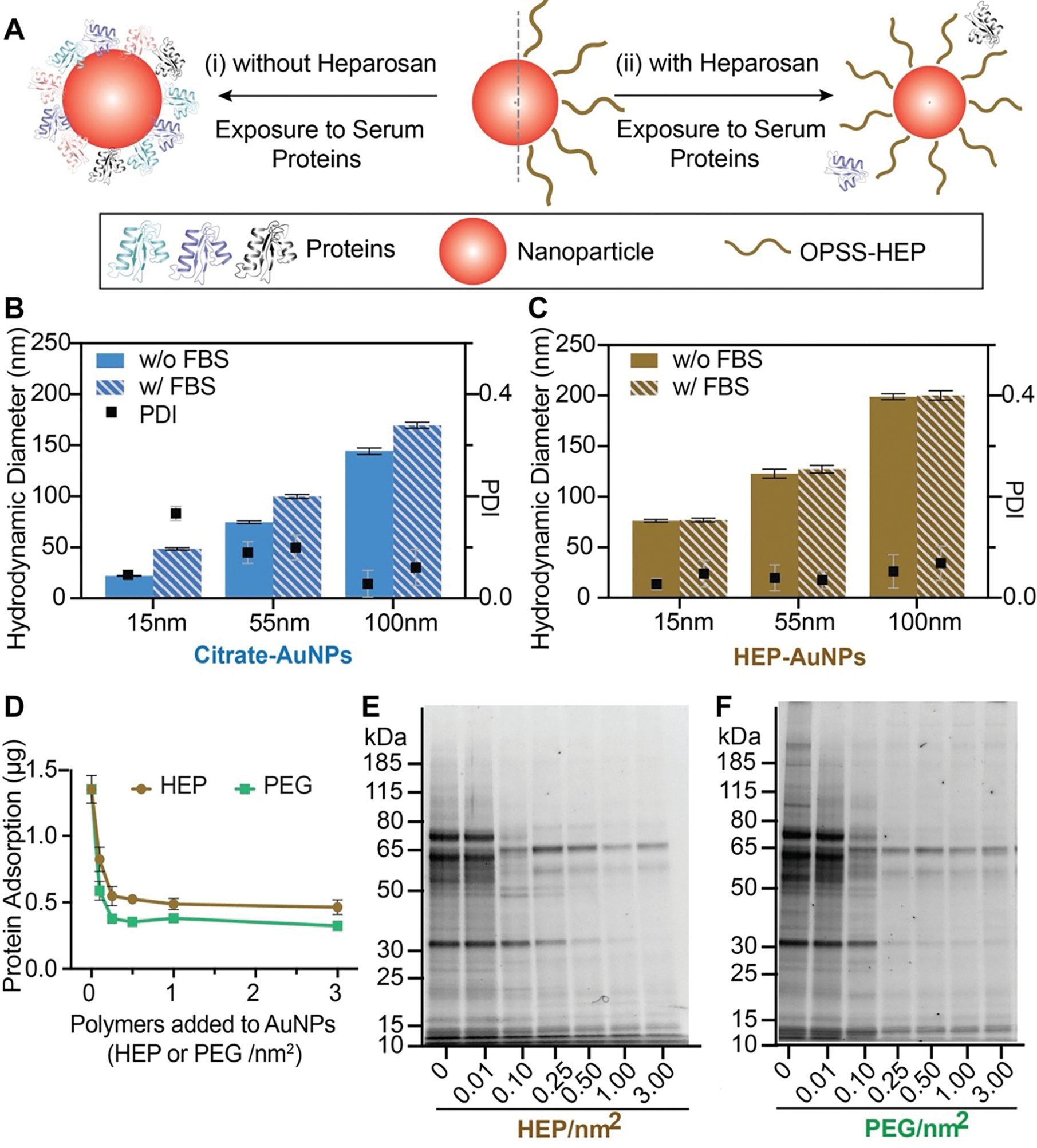
(A) Schematic representation of nanoparticle protein corona formation with and without HEP coating. (B-C) Dynamic light scattering (DLS) was used to compare the hydrodynamic diameter differences before and after FBS incubation (slanted lined bars stand for incubation with FBS) of citrate-coated (B; 0 HEP/nm2) and HEP-coated (panel C)15-, 55-, or 100-nm AuNPs. Bar graphs indicate mean ± SD (n=3). Statistical tests were performed by two-way ANOVA (p<0.0001 (****)). (D) Quantitative BCA protein assay results for HEP- or PEG-coated 55-nm AuNPs created by increasing amounts of polymer added to coating reactions. Results are presented as mean ± SD (n=3). (E-F) SDS-PAGE showing the qualitative biomolecular composition of the adsorbed FBS protein layer on 55-nm AuNPs with various surface HEP (panel E) or PEG densities (panel F). The coating densities represent the added amount of polymers in a coating reaction per nanoparticle surface area.
Next, we used label-free LC-MS/MS to characterize the adsorbed proteins isolated from the nanoparticle surfaces (Fig. 4). Table S1 summarizes the complete list of protein names, molecular weights, and known biological activities of the 16 detected protein species identified on the HEP- and PEG-coated AuNPs. The HEP and PEG surface coatings shared 12 proteins (Fig. 4E). Average spectral counts for each identified protein from HEP-AuNPs and PEG-AuNPs are reported in Tables S2 and S3, respectively. The spectral counts varied with both the densities and types of surface coating, as summarized in the corresponding heat maps (Fig. 4A, C). We performed hierarchy clustering to organize proteins into groups based on the correlation of relative abundances (Fig. 4B, D). We observed that the identified proteins presented distinct preferential surface adsorption as a function of the nanoparticle surface coating types and densities. We summarized the similarities and differences between the five protein cluster groups in a Venn diagram (Fig. 4F). Our proteomic analysis showed that changes in nanoparticle surface coating affect the types and quantities of surface-adsorbed proteins.
Figure 4: Proteomic analysis of nanoparticle protein corona by LC-MS/MS.
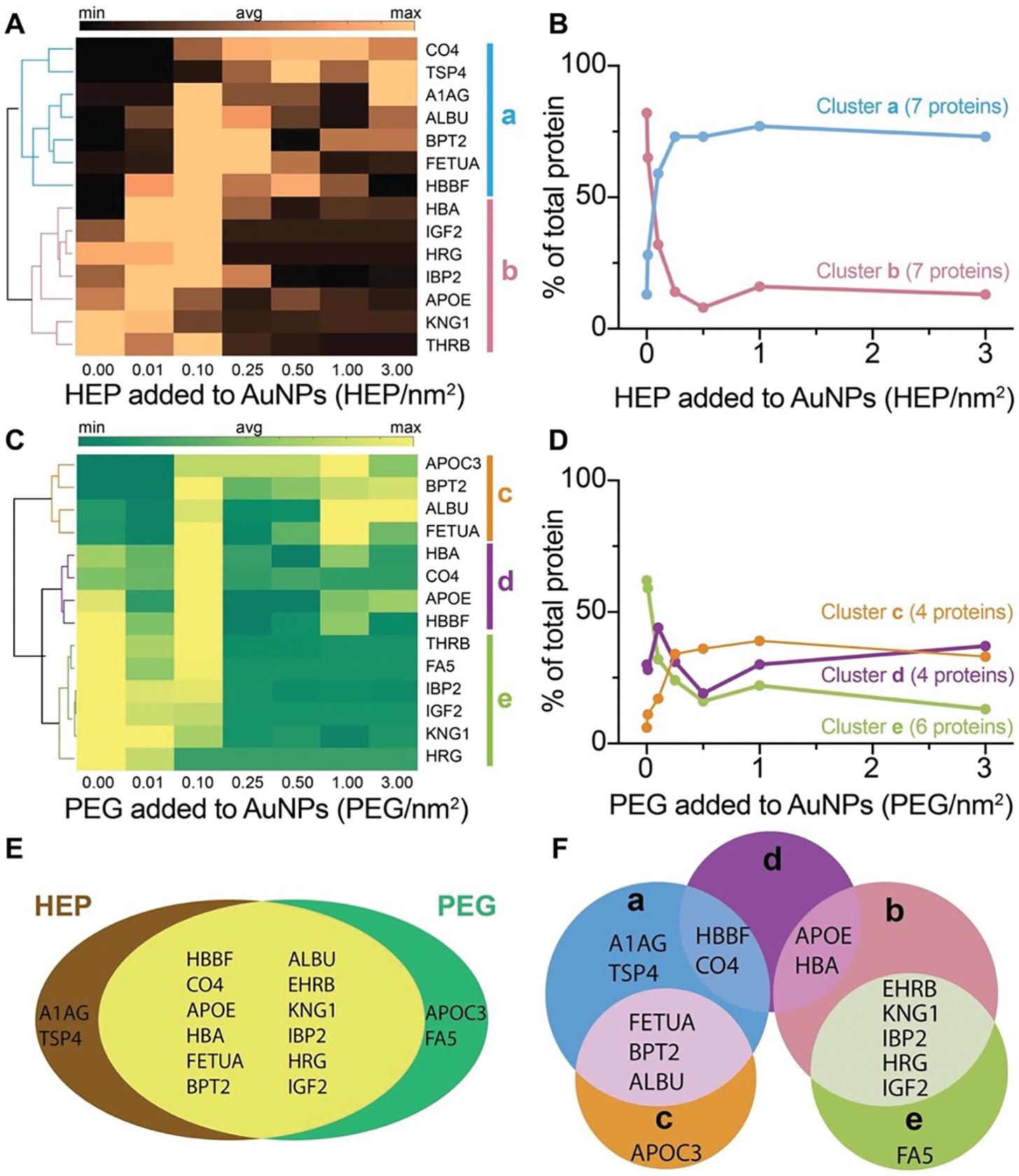
(A), (C) Heat maps and clustergrams of the most abundant proteins isolated from HEP- (panel A) or PEG-coated (panel C) AuNPs. The dendrogram on the left side of the heat map shows the hierarchical relationship between proteins in each row across the densities. According to the correlation in the dendrogram, proteins were clustered into groups a, b, c, d, and e (represented by colored bars on the right side of the heat maps and colored lines in the dendrograms). The average amount of each protein was calculated from three independent experimental replicates. The scale bars on the top of the heat maps stand for the relative abundance of proteins (min, avg, and max represent the minimum, average and maximum amount of proteins in the heat map, respectively). Only proteins with relative abundance larger than 0.25% were included. (B), (D) The number of proteins in a cluster group was reported. The data points represent proteins amount of the same cluster groups over different densities. The connections of data points show the trend of protein amount change in each cluster group along with different densities. (E-F) The proportional Venn diagrams of protein corona isolated from HEP- or PEG-coated 55-nm AuNPs and the clustered groups. The intersection of proteins from HEP- with PEG-coated AuNPs (panel E). Proteins from the clustered groups a, b, c, d, and e were intersected (panel F). The cluster groups a, b, c, d, e are defined in panels A and C.
Considering the potential impact of the nanoparticle protein corona on biological function and toxicity, we then compared the cytotoxicity, hemolysis potential, and cytokine release profiles of HEP- or PEG-modified AuNPs (Fig. 5A–B, S14). On average, we added about five polymers/nm2 to modify AuNPs with HEP or PEG. We evaluated the cytotoxicity for various nanoparticle doses, nanoparticle sizes, and incubation periods in different cell types (Fig. S15A–C). We did not observe any noticeable cytotoxicity at the highest nanoparticle dose tested. In addition, we did not detect any pronounced hemoglobin release upon incubation of human red blood cells with HEP- or PEG-modified nanoparticles (Fig. 5B, S15D). We analyzed the cytokine release levels in supernatants of RAW264.7 macrophages after 24 h of incubation with either citrate-, HEP-, or PEG-modified AuNPs (Fig. S14). In comparison to the untreated cell control, no significant changes were observed in this panel of over three dozen cytokines, interleukins, or factors known to be involved in stress and inflammatory reactions. These results highlight the biocompatibility of HEP coatings and warrant the future investigation of HEP for safe and effective nanomedicine coatings.
Figure 5: Cytotoxicity, hemolysis, and cell uptake of HEP- and PEG-modified 55-nm AuNPs.
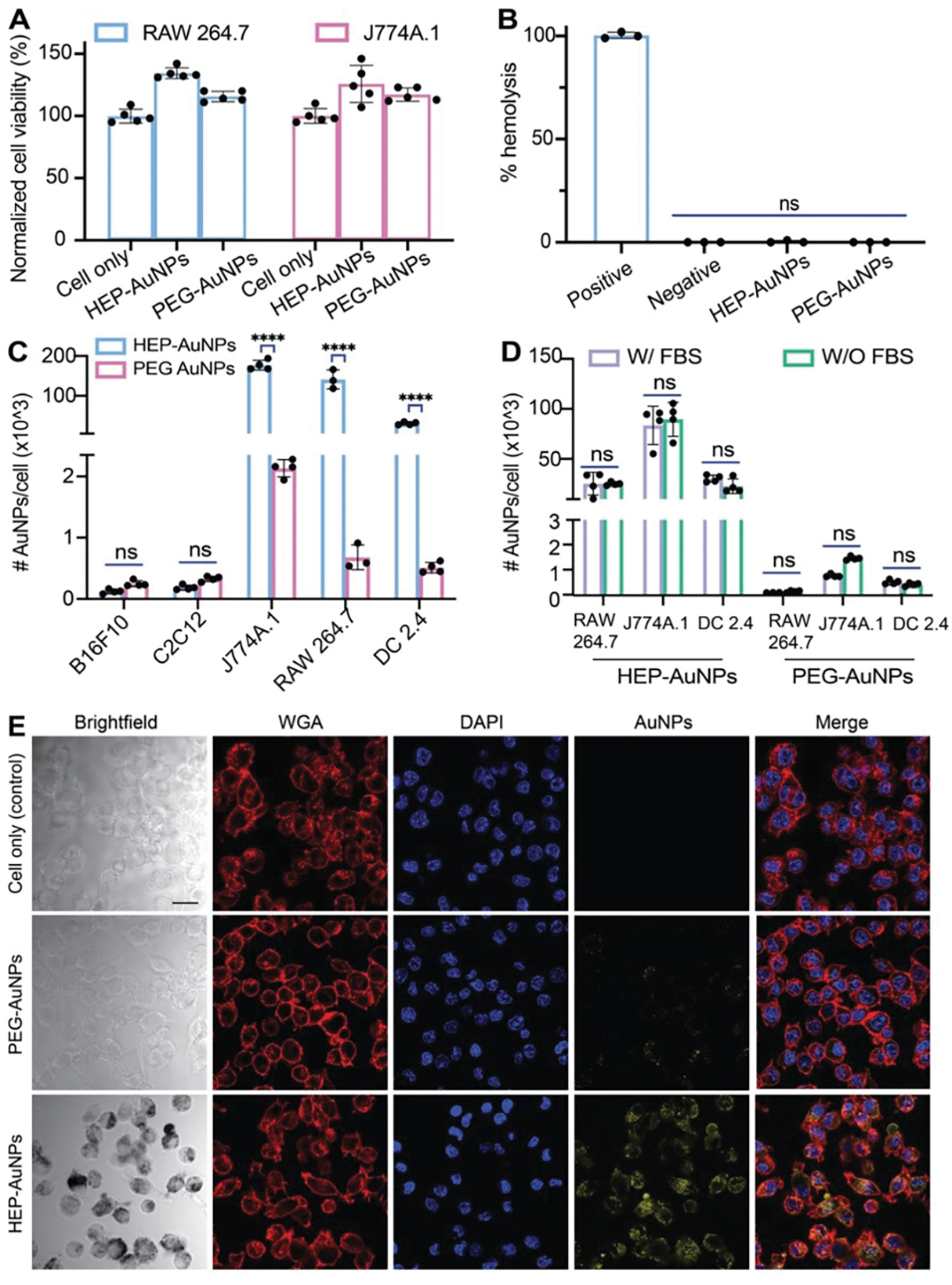
(A) Cell viability test of RAW 264. 7 or J774A macrophages treated with 1-nM HEP-AuNPs for 48 h with control groups (cells with PEG- AuNPs or without AuNPs) by XTT assay. Bar graphs indicate mean ± SD (n=5). (B) Hemolysis assay of 55-nm nanoparticles (1-nM HEP- or PEG-AuNPs final). 1x PBS or 1% Triton-X 100 were used as negative and positive controls, respectively. Bar graphs indicate mean ± SD (n=3). (C) Cell uptake assays: HEP-AuNPs or control PEG-AuNPs were incubated with B16F10 murine melanoma, C2C12 murine muscle cells, J774A.1 murine macrophages, RAW 264.7 murine macrophages, or DC2.4 murine dendritic cells. ICP-MS was performed to quantify cell uptake of nanoparticles. About 70x, 230x, and 45x more HEP-AuNPs were internalized than PEG-AuNPs in J774A.1 macrophage, RAW 264.7 macrophage, and DC 2.4 dendric cells, respectively. Bar graphs indicate mean ± SD (n=3–4). (D) The effect of FBS (protein corona) on cellular uptake of HEP-AuNPs with control (PEG-AuNPs) when incubated with J774A.1 murine macrophages, RAW 264.7 murine macrophages, and DC2.4 murine dendritic cells. ICP-MS was performed to quantify nanoparticles cell uptake. No significant difference was observed with FBS-treatment as in panel C. Bar graphs indicate mean ± SD (n=3–4). (E) Confocal laser scanning microscopy images of HEP- and PEG-coated AuNPs incubated with DC2.4 dendritic cells for 3 h. The added coating density of HEP-AuNPs and PEG-AuNPs in this figure was ~5 polymers/nm2. The scale bar indicates 20 𝜇m.
Since the protein corona molecular composition is critical in governing the nanoparticles’ biological fate and cellular interactions, we wondered about potential differences in cell uptake efficiencies between HEP- and PEG-modified nanoparticles. We incubated HEP-AuNPs or PEG-AuNPs with various healthy and cancerous cell lines, including J774A.1 macrophages, RAW264.7 macrophages, DC2.4 dendritic cells, HUVEC human endothelial cells, B16f10 melanoma cells, and C2C12 muscle cells. Our nanoparticle-cell incubation experiments revealed high associations of HEP-AuNPs with certain cell types of the innate immune system (Fig. S16). To prove that the observed nanoparticle-cell interactions were due to the intracellular uptake of nanoparticles, we conducted confocal laser scanning microscopy (CLSM) to visualize AuNPs in a label-free manner via light scattering49,50. Figure 5E shows intracellular CLSM image sections, which confirmed the substantial uptake of HEP-coated AuNPs into these cells (Fig. S17).
To further confirm the intracellular localization of AuNPs, we performed a gold etching experiment by exposing cells to KI/I2 etchant, a highly effective etchant of gold. Our rationale for the etching experiment was that any externally located AuNPs, for example, AuNPs attached to the cell membrane, will dissolve during this etching treatment51,52. As shown in Figure S18, quantitative inductively coupled plasma mass spectrometry (ICP-MS) of cells exposed to nanoparticles revealed no notable changes in the extent of nanoparticle cell uptake after the etchant treatment. These results suggest that most AuNPs were located inside the cells in line with our CLSM images. To visualize the subcellular distribution of AuNPs, we performed transmission electron microscopy (TEM) of cells incubated with AuNPs. The TEM micrographs shown in Figure S19 reveal the localization of AuNPs in intracellular vesicles. We additionally confirmed the intracellular uptake of HEP-coated AgNPs and liposomes with CLSM (Fig. S20).
We then used ICP-MS to quantify nanoparticle cellular uptake in various cell types. Compared with 55-nm PEG-AuNP, we observed up to 230-fold higher 55-nm HEP-AuNPs uptake in immune cells, including J774A.1 and RAW264.7 macrophages, as well as in DC2.4 dendritic cells (Fig. 5C). For B16f10 melanoma cells, C2C12 muscle cells, murine 4T1 breast cancer cells, and human endothelial cells (HUVEC), the uptake results of HEP-AuNPs were similar to those obtained for PEG-AuNPs. (Fig. 5C, Fig. S18, S21).
Since our previous LC-MS/MS studies revealed that HEP- and PEG-AuNPs exhibited different protein corona profiles, we wondered about the role of the protein corona in driving nanoparticle cell uptake. We performed cellular uptake experiments with and without FBS in the cell media (Fig. 5D and Fig. S22). Our results indicate that heparosan’s intrinsic properties rather than the protein corona determine the observed cellular uptake efficiencies.
To investigate whether the nanoparticle size mediated the high cellular uptake of HEP-AuNPs, we incubated RAW264.7 macrophages with 15-nm AuNPs. Interestingly, we observed a 21-fold higher cell uptake for 15-nm HEP-AuNPs than 15-nm PEG-AuNPs (Fig. S18). This finding confirms that relatively high cell uptake can be achieved even with small HEP-modified nanoparticles and further suggests that HEP has a specific role in driving cellular interactions. Further mechanistic studies are needed to determine how HEP-based surface modifications facilitate the internalization of nanoparticles in cells.
In summary, our data suggest that HEP-AuNPs exhibit no apparent cytotoxicity or hemolysis, and very interestingly, display relatively high cellular uptake by specific innate immune cells. There is no correlation between this high cellular uptake and the protein corona (as shown by controlled FBS incubation experiments), thus suggesting that the uptake behavior is intrinsic to the HEP polysaccharide. The high cellular uptake of HEP-coated nanoparticles may increase the effectiveness of macrophage and antigen-presenting cell targeting deliveries and therapies compared to the existing PEG-coatings. The important translational impacts for such improved behavior may include higher potency of HEP-modified nanomedicines yielding drug sparing (i.e. less active ingredient is needed and thus cost savings and more doses per manufacturing batch become available) and reduced potential side effects due to lowered bioburden. In future studies, we will investigate the mechanisms involved in the observed cellular uptake behavior of HEP-modified nanoparticles.
Conclusions
We demonstrated that the salt aging and pH reduction methods effectively coated negatively charged HEP onto various sized AuNPs. Similar to PEG-based modifications, surface engineering of AuNPs with HEP reduced protein adsorption as a function of HEP surface density. While HEP coatings exhibited a comparable ability to reduce protein adsorption as PEG, HEP substantially enhanced cellular uptake in certain antigen-presenting cells, but not in other tested cell types from various lineages. While we observed some differences in serum protein corona profiles between HEP-AuNPs and PEG-AuNPs, we found that the high nanoparticle cell uptake was not affected by serum proteins. We further demonstrated our HEP-coating strategy’s broad applicability for various inorganic and organic nanomaterials. In future studies, we will investigate the mechanisms for the enhanced cellular uptake of HEP-coated nanoparticles. This research will guide the translation of HEP-based nanoparticle surface engineering to enable nanomedicine-based immunotherapies, such as vaccines and CAR-T therapies, that safely and efficiently target immune cells.
Supplementary Material
Acknowledgments
The authors thank Ms. S. Butterfield for assistance with experiments and Dr. P. Larson for assistance with TEM. The Irvine and Chen labs kindly provided C2C12, B16F10, and DC 2.4 cells. This research was supported in part by the University of Oklahoma (OU) IBEST-OUHSC Seed Grant for Interdisciplinary Research, the Oklahoma Center for the Advancement of Science and Technology OCAST (HR20–106), the NSF CAREER (2048130), the University of Oklahoma Faculty Investment Program, and the Oklahoma Tobacco Settlement Endowment Trust (TSET) awarded to the University of Oklahoma - Stephenson Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Oklahoma Tobacco Settlement Endowment Trust.
Footnotes
Supporting Information
This material is available free of charge via the internet at http://pubs.acs.org.
The supporting information includes supplementary figures S1–S22, supplementary tables S1–S3, and the methods and materials.
References
- (1).Sang W; Zhang Z; Dai Y; Chen X Recent Advances in Nanomaterial-Based Synergistic Combination Cancer Immunotherapy. Chem Soc Rev 2019, 48 (14), 3771–3810. 10.1039/c8cs00896e. [DOI] [PubMed] [Google Scholar]
- (2).Suk JS; Xu Q; Kim N; Hanes J; Ensign LM PEGylation as a Strategy for Improving Nanoparticle-Based Drug and Gene Delivery. Adv. Drug Deliv. Rev. 2016, 99 (Pt A), 28–51. 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Salata O Applications of Nanoparticles in Biology and Medicine. Journal of Nanobiotechnology 2004, 2 (1), 3. 10.1186/1477-3155-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WCW Analysis of Nanoparticle Delivery to Tumours. Nature Reviews Materials 2016, 1 (5), 1–12. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- (5).Sindhwani S; Syed AM; Ngai J; Kingston BR; Maiorino L; Rothschild J; MacMillan P; Zhang Y; Rajesh NU; Hoang T; Wu JLY; Wilhelm S; Zilman A; Gadde S; Sulaiman A; Ouyang B; Lin Z; Wang L; Egeblad M; Chan WCW The Entry of Nanoparticles into Solid Tumours. Nature Materials 2020, 1–10. 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- (6).Narum SM; Le T; Le DP; Lee JC; Donahue ND; Yang W; Wilhelm S Chapter 4 - Passive Targeting in Nanomedicine: Fundamental Concepts, Body Interactions, and Clinical Potential. In Nanoparticles for Biomedical Applications; Chung EJ, Leon L, Rinaldi C, Eds.; Micro and Nano Technologies; Elsevier, 2020; pp 37–53. 10.1016/B978-0-12-816662-8.00004-7. [DOI] [Google Scholar]
- (7).Walkey CD; Chan WCW Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem Soc Rev 2012, 41 (7), 2780–2799. 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- (8).Cai R; Chen C The Crown and the Scepter: Roles of the Protein Corona in Nanomedicine. Adv. Mater. Weinheim 2019, 31 (45), e1805740. 10.1002/adma.201805740. [DOI] [PubMed] [Google Scholar]
- (9).Aggarwal P; Hall JB; McLeland CB; Dobrovolskaia MA; McNeil SE Nanoparticle Interaction with Plasma Proteins as It Relates to Particle Biodistribution, Biocompatibility and Therapeutic Efficacy. Adv Drug Deliv Rev 2009, 61 (6), 428–437. 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Monopoli MP; Aberg C; Salvati A; Dawson KA Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat Nanotechnol 2012, 7 (12), 779–786. 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- (11).Dominguez-Medina S; Kisley L; Tauzin LJ; Hoggard A; Shuang B; Indrasekara DS, A. S.; Chen S; Wang L-Y; Derry PJ; Liopo A; Zubarev ER; Landes CF; Link S Adsorption and Unfolding of a Single Protein Triggers Nanoparticle Aggregation. ACS Nano 2016, 10 (2), 2103–2112. 10.1021/acsnano.5b06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cedervall T; Lynch I; Lindman S; Berggård T; Thulin E; Nilsson H; Dawson KA; Linse S Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (7), 2050–2055. 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Veronese FM; Pasut G PEGylation, Successful Approach to Drug Delivery. Drug Discovery Today 2005, 10 (21), 1451–1458. 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- (14).Larson TA; Joshi PP; Sokolov K Preventing Protein Adsorption and Macrophage Uptake of Gold Nanoparticles via a Hydrophobic Shield. ACS Nano 2012, 6 (10), 9182–9190. 10.1021/nn3035155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fang RH; Kroll AV; Gao W; Zhang L Cell Membrane Coating Nanotechnology. Adv. Mater. Weinheim 2018, 30 (23), e1706759. 10.1002/adma.201706759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee JC; Donahue ND; Mao AS; Karim A; Komarneni M; Thomas EE; Francek ER; Yang W; Wilhelm S Exploring Maleimide-Based Nanoparticle Surface Engineering to Control Cellular Interactions. ACS Appl. Nano Mater. 2020. 10.1021/acsanm.9b02541. [DOI] [Google Scholar]
- (17).Wilhelm S; Hirsch T; Patterson WM; Scheucher E; Mayr T; Wolfbeis OS Multicolor Upconversion Nanoparticles for Protein Conjugation. Theranostics 2013, 3 (4), 239–248. 10.7150/thno.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chen L; Hong W; Ren W; Xu T; Qian Z; He Z Recent Progress in Targeted Delivery Vectors Based on Biomimetic Nanoparticles. Sig Transduct Target Ther 2021, 6 (1), 1–25. 10.1038/s41392-021-00631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Banda NK; Mehta G; Chao Y; Wang G; Inturi S; Fossati-Jimack L; Botto M; Wu L; Moghimi SM; Simberg D Mechanisms of Complement Activation by Dextran-Coated Superparamagnetic Iron Oxide (SPIO) Nanoworms in Mouse versus Human Serum. Particle and Fibre Toxicology 2014, 11 (1), 64. 10.1186/s12989-014-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yang W; Wang L; Mettenbrink EM; DeAngelis PL; Wilhelm S Nanoparticle Toxicology. Annu. Rev. Pharmacol. Toxicol. 2021, 61 (1), 269–289. 10.1146/annurev-pharmtox-032320-110338. [DOI] [PubMed] [Google Scholar]
- (21).Zhang P; Sun F; Liu S; Jiang S Anti-PEG Antibodies in the Clinic: Current Issues and beyond PEGylation. Journal of Controlled Release 2016, 244, 184–193. 10.1016/j.jconrel.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Fang J-L; Beland FA; Tang Y; Roffler SR Flow Cytometry Analysis of Anti-Polyethylene Glycol Antibodies in Human Plasma. Toxicology Reports 2021, 8, 148–154. 10.1016/j.toxrep.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kozma GT; Mészáros T; Vashegyi I; Fülöp T; Örfi E; Dézsi L; Rosivall L; Bavli Y; Urbanics R; Mollnes TE; Barenholz Y; Szebeni J Pseudo-Anaphylaxis to Polyethylene Glycol (PEG)-Coated Liposomes: Roles of Anti-PEG IgM and Complement Activation in a Porcine Model of Human Infusion Reactions. ACS Nano 2019, 13 (8), 9315–9324. 10.1021/acsnano.9b03942. [DOI] [PubMed] [Google Scholar]
- (24).Povsic TJ; Lawrence MG; Lincoff AM; Mehran R; Rusconi CP; Zelenkofske SL; Huang Z; Sailstad J; Armstrong PW; Steg PG; Bode C; Becker RC; Alexander JH; Adkinson NF; Levinson AI Pre-Existing Anti-PEG Antibodies Are Associated with Severe Immediate Allergic Reactions to Pegnivacogin, a PEGylated Aptamer. Journal of Allergy and Clinical Immunology 2016, 138 (6), 1712–1715. 10.1016/j.jaci.2016.04.058. [DOI] [PubMed] [Google Scholar]
- (25).Sellaturay P; Nasser S; Islam S; Gurugama P; Ewan PW Polyethylene Glycol (PEG) Is a Cause of Anaphylaxis to the Pfizer/BioNTech MRNA COVID-19 Vaccine. Clinical & Experimental Allergy 2021, 51 (6), 861–863. 10.1111/cea.13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Troelnikov A; Perkins G; Yuson C; Ahamdie A; Balouch S; Hurtado PR; Hissaria P Basophil Reactivity to BNT162b2 Is Mediated by PEGylated Lipid Nanoparticles in Patients with PEG Allergy. Journal of Allergy and Clinical Immunology 2021, 148 (1), 91–95. 10.1016/j.jaci.2021.04.032. [DOI] [PubMed] [Google Scholar]
- (27).Shimabukuro TT; Cole M; Su JR Reports of Anaphylaxis After Receipt of MRNA COVID-19 Vaccines in the US—December 14, 2020-January 18, 2021. JAMA 2021, 325 (11), 1101–1102. 10.1001/jama.2021.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sellaturay P; Nasser S; Ewan P Polyethylene Glycol–Induced Systemic Allergic Reactions (Anaphylaxis). The Journal of Allergy and Clinical Immunology: In Practice 2021, 9 (2), 670–675. 10.1016/j.jaip.2020.09.029. [DOI] [PubMed] [Google Scholar]
- (29).Bigini P; Gobbi M; Bonati M; Clavenna A; Zucchetti M; Garattini S; Pasut G The Role and Impact of Polyethylene Glycol on Anaphylactic Reactions to COVID-19 Nano-Vaccines. Nat. Nanotechnol. 2021, 1–3. 10.1038/s41565-021-01001-3. [DOI] [PubMed] [Google Scholar]
- (30).Nogueira SS; Schlegel A; Maxeiner K; Weber B; Barz M; Schroer MA; Blanchet CE; Svergun DI; Ramishetti S; Peer D; Langguth P; Sahin U; Haas H Polysarcosine-Functionalized Lipid Nanoparticles for Therapeutic MRNA Delivery. ACS Appl. Nano Mater. 2020, 3 (11), 10634–10645. 10.1021/acsanm.0c01834. [DOI] [Google Scholar]
- (31).Rippe M; Stefanello TF; Kaplum V; Britta EA; Garcia FP; Poirot R; Companhoni MVP; Nakamura CV; Szarpak-Jankowska A; Auzély-Velty R Heparosan as a Potential Alternative to Hyaluronic Acid for the Design of Biopolymer-Based Nanovectors for Anticancer Therapy. Biomater. Sci. 2019, 7 (7), 2850–2860. 10.1039/C9BM00443B. [DOI] [PubMed] [Google Scholar]
- (32).Wang Z; Dordick JS; Linhardt RJ Escherichia Coli K5 Heparosan Fermentation and Improvement by Genetic Engineering. Bioeng Bugs 2011, 2 (1), 63–67. 10.4161/bbug.2.1.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lane RS; Haller FM; Chavaroche AAE; Almond A; DeAngelis PL Heparosan-Coated Liposomes for Drug Delivery. Glycobiology 2017, 27 (11), 1062–1074. 10.1093/glycob/cwx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Qiu L; Shan X; Long M; Ahmed KS; Zhao L; Mao J; Zhang H; Sun C; You C; Lv G; Chen J Elucidation of Cellular Uptake and Intracellular Trafficking of Heparosan Polysaccharide-Based Micelles in Various Cancer Cells. International Journal of Biological Macromolecules 2019, 130, 755–764. 10.1016/j.ijbiomac.2019.02.133. [DOI] [PubMed] [Google Scholar]
- (35).Chen J-X; Zhang M; Liu W; Lu G-Z; Chen J-H Construction of Serum Resistant Micelles Based on Heparosan for Targeted Cancer Therapy. Carbohydrate Polymers 2014, 110, 135–141. 10.1016/j.carbpol.2014.03.084. [DOI] [PubMed] [Google Scholar]
- (36).Chen J-X; Liu W; Zhang M; Chen J-H Heparosan Based Negatively Charged Nanocarrier for Rapid Intracellular Drug Delivery. International Journal of Pharmaceutics 2014, 473 (1), 493–500. 10.1016/j.ijpharm.2014.07.045. [DOI] [PubMed] [Google Scholar]
- (37).Dai Q; Wilhelm S; Ding D; Syed AM; Sindhwani S; Zhang Y; Chen YY; MacMillan P; Chan WCW Quantifying the Ligand-Coated Nanoparticle Delivery to Cancer Cells in Solid Tumors. ACS Nano 2018, 12 (8), 8423–8435. 10.1021/acsnano.8b03900. [DOI] [PubMed] [Google Scholar]
- (38).Hurst SJ; Lytton-Jean AKR; Mirkin CA Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal. Chem. 2006, 78 (24), 8313–8318. 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Xu Q; Lou X; Wang L; Ding X; Yu H; Xiao Y Rapid, Surfactant-Free, and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA under Physiological PH and Its Application in Molecular Beacon-Based Biosensor. ACS Appl. Mater. Interfaces 2016, 8 (40), 27298–27304. 10.1021/acsami.6b08350. [DOI] [PubMed] [Google Scholar]
- (40).Schöttler S; Becker G; Winzen S; Steinbach T; Mohr K; Landfester K; Mailänder V; Wurm FR Protein Adsorption Is Required for Stealth Effect of Poly(Ethylene Glycol)- and Poly(Phosphoester)-Coated Nanocarriers. Nature Nanotechnology 2016, 11 (4), 372–377. 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- (41).Bernhard C; Bauer KN; Bonn M; Wurm FR; Gonella G Interfacial Conformation of Hydrophilic Polyphosphoesters Affects Blood Protein Adsorption. ACS Appl. Mater. Interfaces 2019, 11 (1), 1624–1629. 10.1021/acsami.8b17146. [DOI] [PubMed] [Google Scholar]
- (42).Walkey CD; Olsen JB; Guo H; Emili A; Chan WCW Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J. Am. Chem. Soc. 2012, 134 (4), 2139–2147. 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- (43).Xu Q; Ensign LM; Boylan NJ; Schön A; Gong X; Yang J-C; Lamb NW; Cai S; Yu T; Freire E; Hanes J Impact of Surface Polyethylene Glycol (PEG) Density on Biodegradable Nanoparticle Transport in Mucus Ex Vivo and Distribution in Vivo. ACS Nano 2015, 9 (9), 9217–9227. 10.1021/acsnano.5b03876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Donahue ND; Kanapilly S; Stephan C; Marlin MC; Francek ER; Haddad M; Guthridge J; Wilhelm S Quantifying Chemical Composition and Reaction Kinetics of Individual Colloidally Dispersed Nanoparticles. Nano Lett. 2022, 22 (1), 294–301. 10.1021/acs.nanolett.1c03752. [DOI] [PubMed] [Google Scholar]
- (45).Syed AM; MacMillan P; Ngai J; Wilhelm S; Sindhwani S; Kingston BR; Wu JLY; Llano-Suárez P; Lin ZP; Ouyang B; Kahiel Z; Gadde S; Chan WCW Liposome Imaging in Optically Cleared Tissues. Nano Lett. 2020, 20 (2), 1362–1369. 10.1021/acs.nanolett.9b04853. [DOI] [PubMed] [Google Scholar]
- (46).Hossen MN; Wang L; Chinthalapally HR; Robertson JD; Fung K-M; Wilhelm S; Bieniasz M; Bhattacharya R; Mukherjee P Switching the Intracellular Pathway and Enhancing the Therapeutic Efficacy of Small Interfering RNA by Auroliposome. Science Advances 2020. 10.1126/sciadv.aba5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lazarovits J; Chen YY; Song F; Ngo W; Tavares AJ; Zhang Y-N; Audet J; Tang B; Lin Q; Tleugabulova MC; Wilhelm S; Krieger JR; Mallevaey T; Chan WCW Synthesis of Patient-Specific Nanomaterials. Nano Lett. 2019, 19 (1), 116–123. 10.1021/acs.nanolett.8b03434. [DOI] [PubMed] [Google Scholar]
- (48).Sheth V; Wang L; Bhattacharya R; Mukherjee P; Wilhelm S Strategies for Delivering Nanoparticles across Tumor Blood Vessels. Advanced Functional Materials 2021, 31 (8), 2007363. 10.1002/adfm.202007363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wang F; Chen B; Yan B; Yin Y; Hu L; Liang Y; Song M; Jiang G Scattered Light Imaging Enables Real-Time Monitoring of Label-Free Nanoparticles and Fluorescent Biomolecules in Live Cells. J. Am. Chem. Soc. 2019, 141 (36), 14043–14047. 10.1021/jacs.9b05894. [DOI] [PubMed] [Google Scholar]
- (50).Syed AM; Sindhwani S; Wilhelm S; Kingston BR; Lee DSW; Gommerman JL; Chan WCW Three-Dimensional Imaging of Transparent Tissues via Metal Nanoparticle Labeling. J. Am. Chem. Soc. 2017, 139 (29), 9961–9971. 10.1021/jacs.7b04022. [DOI] [PubMed] [Google Scholar]
- (51).Zhang Y; Wu JLY; Lazarovits J; Chan WCW An Analysis of the Binding Function and Structural Organization of the Protein Corona. J. Am. Chem. Soc. 2020, 142 (19), 8827–8836. 10.1021/jacs.0c01853. [DOI] [PubMed] [Google Scholar]
- (52).Albanese A; Chan WCW Effect of Gold Nanoparticle Aggregation on Cell Uptake and Toxicity. ACS Nano 2011, 5 (7), 5478–5489. 10.1021/nn2007496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


