Abstract
Chromatin structure, transcription, DNA replication, and repair are regulated via locus-specific incorporation of histone variants and posttranslational modifications that guide effector chromatin-binding proteins. Here we report unbiased, quantitative interactomes for the replication-coupled (H3.1) and replication-independent (H3.3) histone H3 variants based on BioID proximity labeling, which allows interactions in intact, living cells to be detected. Along with a significant proportion of previously reported interactions detected by affinity purification followed by mass spectrometry, three quarters of the 608 histone-associated proteins that we identified are new, uncharacterized histone associations. The data reveal important biological nuances not captured by traditional biochemical means. For example, we found that the chromatin assembly factor-1 histone chaperone not only deposits the replication-coupled H3.1 histone variant during S-phase but also associates with H3.3 throughout the cell cycle in vivo. We also identified other variant-specific associations, such as with transcription factors, chromatin regulators, and with the mitotic machinery. Our proximity-based analysis is thus a rich resource that extends the H3 interactome and reveals new sets of variant-specific associations.
Keywords: H3.3, H3.1, BioID, CAF-1, histone chaperone, mitosis
Abbreviations: AP-MS, affinity purification followed by mass spectrometry; ASF1, anti-silencing factor 1; ATRX, alpha-thalassemia mental retardation syndrome X-linked; CAF-1, chromatin assembly factor 1; ChIP, chromatin immunoprecipitation; DAXX, death domain-associated protein 6; EdU, 5-ethynyl-2′-deoxyuridine; HIRA, histone cell cycle regulator; MNase, Micrococcal nuclease; PLA, proximity ligation assay; PPI, protein–protein interaction; SAINT, Significance Analysis of INTeractome; TF, transcription factor
Graphical Abstract
Highlights
-
•
BioID interactomes of histone variants H3.1 and H3.3.
-
•
Novel interactors and pathways were revealed.
-
•
H3, in particular H3.1, interacts with mitotic complexes.
-
•
H3.3 associates with transcription-related proteins, notably MYC interactors.
-
•
The chromatin assembly factor 1 chaperone interacts with both H3.1 and H3.3 in vivo.
In Brief
Proximity-dependent (BioID) interactome analysis of the histone H3 variants H3.1 and H3.3 identified 608 interaction partners and expanded the characterized H3 interaction landscape. Among the most significant findings were that the chaperone chromatin assembly factor 1, previously thought to be H3.1-specific, interacts with H3.3 throughout the cell cycle. Preferential binding was identified between H3.1 and components of the mitotic machinery and between H3.3 and transcription factors, notably MYC interactors.
Eukaryotic cells package genomic DNA into chromatin by first assembling nucleosomes. These repetitive structures are formed by wrapping DNA around histone octamers comprised of two copies each of histones H2A, H2B, H3, and H4 (1). Mammalian cells express a large number of histone H3 variants, including H3.1 and H3.3 that play important regulatory roles in somatic cells. Canonical H3.1 is expressed mainly in S-phase and considered replication-coupled, while H3.3 expression occurs throughout the cell cycle independently of replication (2). They differ by just five amino acids, yet their usage and genomic localization varies broadly. H3.1 is deposited genome wide with relative uniformity, while H3.3 is preferentially incorporated in actively transcribed regions, telomeres, and pericentric heterochromatin (3).
Early attention to differences between H3 interaction partners focussed on the histone chaperones that deposit them into chromatin. Biochemical purifications from nuclear lysates revealed that H3.1 binds the chromatin assembly factor 1 (CAF-1) complex, which deposits H3-H4 tetramers behind replication forks and during DNA damage repair (4, 5, 6, 7). In vitro, CAF-1 was shown to preferentially incorporate H3.1 in histone deposition assays using Xenopus egg extracts and plasmid DNA (4). In contrast, H3.3 was shown to be deposited by histone cell cycle regulator (HIRA) and death domain-associated protein 6/alpha-thalassemia mental retardation syndrome X-linked (DAXX/ATRX) (3, 4, 8, 9, 10, 11, 12). Histone deposition by HIRA predominates over gene promoters and transcribed regions, while DAXX/ATRX deposits H3.3 at telomeres and pericentric heterochromatin (3, 8, 12). In contrast, other H3 chaperones are not seemingly selective between H3.1 and H3.3 (4, 13). A prime example is ASF1 (ASF1A/B; anti-silencing factor 1), which facilitates nuclear import of newly synthesized H3-H4 dimers and their transfer to HIRA/CAF-1 (14, 15, 16). Although these data suggest that CAF-1 is H3.1-specific, some species, such as yeasts including S. pombe and S. cerevisiae, encode only an H3.3 variant H3, that is nevertheless deposited by CAF-1 (13), implying that CAF-1 may exhibit flexibility in vivo with respect to H3 selectivity.
For much of the data generated thus far on histone interaction partners, including by affinity purification followed by mass spectrometry (AP-MS), the first step was to extract nuclear proteins using conditions including high salt and/or detergent, which are needed to solubilize histones from chromatin. However, biochemically labile, or transient, protein–protein interactions (PPIs), such as those with an enzyme, can be missed by these approaches, suggesting that additional interactors could have been missed among the H3 PPIs identified to date (4, 8, 17, 18, 19, 20). In contrast, methods such as proximity biotinylation (BioID) assay PPIs in their native environment by fusing the protein of interest with a biotin ligase from E. coli (BirA-R118G or BirA∗) (21) which, in the presence of biotin, biotinylates free ε-amino groups on interacting proteins within approximately 10 nm (22). BioID has previously been employed to profile PPIs occurring in hard-to-extract locations such as the nuclear lamina (21, 23) and centrosome (24, 25), as well as chromatin- and transcription-associated proteins (18, 26, 27).
Here, we used BioID to compare the interactomes of histone variants H3.3 and H3.1, finding hundreds of PPIs not previously characterized by AP-MS. These carry out important biological functions and include many DNA repair and replication proteins, chromatin modifiers, and remodelers. We uncovered variant-specific interactions, such as between H3.1 and components of the mitotic machinery or H3.3 and a large number of transcription factors (TFs), broadening the range of histone variant-specific biological functions. Finally, we revealed unexpected flexibility in the CAF-1 histone deposition pathway in living cells, finding that it interacts with H3.3 throughout the cell cycle.
Experimental Procedures
Plasmids and Reagents
Lentiviral plasmids to express H3.1 and H3.3 were generated by cloning HIST1H3B (primers: ACCTCCATAGAAGATTCTAGAGCCACCATGGCTCGTACTAAACAGACAG and GTCGTCCTTGTAGTCGGATCCCGCTCTTTCTCCGCGAATGC) and H3F3A (primers: ACCTCCATAGAAGATTCTAGAGCCACCATGGCTCGTACAAAGCAGACTG and GTCGTCCTTGTAGTCGGATCCAGCACGTTCTCCGCGTATGC) cDNA between the XbaI/BamHI sites of a pCDH-CMV-MCS-EF1α-copGFP (SystemBioscience) that was previously modified to encode a FLAG/HA tag between the BamHI/NotI sites. pcDNA5-FLAG-BirA∗ plasmids to create Flp-In T-REx HEK293 cells were generated as previously described (24). Bacterial histone expression vectors were created by cloning cDNA into pET3a (Novagen). CAF-1 p150 (CHAF1A, primers: TAAGCAGTCGACATGGGCTACTACCATCACCATCACCATCACGATTACGACATGCTGGAGGAGCTGGAGTGCG and TGCTTAAAGCTTTCAGGATGCACCCAGTGGGCTC) and p60 (CHAF1B, primers: TAAGCAGCTAGCATGAAAGTCATCACTTGTGAAATAGCCTGGC and TGCTTAGCATGCTCAAGCACTCTTTTCGAATTGCGGATGACTCCAAGAGCCAGCACTGCGCTTTTCGAACTGCGGGTGAGACCAGCCGGTCATAGGGTCCAGACTTTCCGTGCC) cDNA was cloned from a HEK293T cDNA library and introduced into pFastBacDual (Thermo), introducing a 5′ 6xHis tag on p150 and a 3′ Strep II tag on p60. All plasmids were sequenced before use.
Tetracycline (Sigma; 10,000× stock solution; 1 mg/ml) was resuspended in 70% ethanol. Biotin (BioShop and Sigma) was prepared as a 1000× stock solution (50 mM) by dissolving 250 mg in 2 ml 30% v/v NH4OH on ice and neutralized by slowly adding 18 ml HCl (1 N), before sterile filtration through a 0.22 μm filter and storage at 4 °C.
Cell Lines
HeLa-Fucci(CA2) cells (female) were a kind gift from Atsushi Miyawaki (28). Flp-In T-REx HEK293 (female) were purchased from Thermo Fisher. HEK293T (female) cells were purchased from ATCC. Flp-In T-REx HEK293, HEK293T and HeLa-Fucci(CA2) cells were cultured in Dulbecco's modified Eagle's medium (Gibco), supplemented with 10% FBS (Wisent) and 1% penstrep (Invitrogen), at 37 °C in 5% CO2. All cell lines were routinely tested for mycoplasma.
Flp-In T-REx HEK293 cells were established as previously described by hygromycin selection following transfection (24).
Plasmids were packaged into lentivirus by cotransfection into HEK293T cells with psPAX2 (Addgene#12260) and pMD2.G (Addgene#12259) using Lipofectamine 2000 (Invitrogen). Media were changed the next day. The supernatant was collected after 30 h, precipitated overnight with Lenti-X concentrator (Clontech), and resuspended in Optimem (Gibco). HEK293T cells were subsequently transduced for 24 h with lentivirus.
Proximity Ligation Assay
Cells grown on coverslips to approximately 80% confluence were washed in PBS, fixed in 4% PFA diluted in PBS for 10 min at room temperature, and permeabilized in 0.2% Triton X-100 for 10 min at room temperature. Proximity ligation assays were carried out using Duolink In Situ Red Mouse/Rabbit kit (Sigma), according to the manufacturers’ instructions. Samples were imaged on a Leica Spinning Disk confocal microscope using a 40× lens. Images were analyzed with ImageJ. Antibodies used are in supplemental Table S2.
Western Blotting
Samples were boiled in 2× SDS-PAGE loading buffer (78.0 mM Tris [pH 6.8], 4% SDS, 20% glycerol, 100 mM DTT, 0.2% bromophenol blue) and loaded on SDS-PAGE gels (fixed % between 10 and 15% acrylamide [37.5:1], Bio-Rad, or Novex Tris-Glycine 4–20% precast gels [Invitrogen]). Proteins were transferred onto PVDF (Immobilon), which were blocked in TBS-T containing 5% nonfat milk or BSA before primary antibodies were added overnight at 4 °C. After three TBS-T washes, membranes were incubated with HRP-conjugated secondary antibodies (Bio-Rad, 1:10,000) and developed with ECL reagents (Pierce). Antibodies used are in supplemental Table S2.
Cell Fractionation and Micrococcal Nuclease Digestion
Cell pellets were separated into cytoplasmic plus nuclear soluble and insoluble fractions (29). Cells were suspended on ice for 10 min in nuclear isolation buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1× protease inhibitor cocktail [PIC, Sigma]). NP-40 substitute was added to 0.2% for 5 min, and nuclei collected at 600g, 5 min. Cytoplasmic supernatant was removed, and nuclei lysed in nuclear extraction buffer (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 25% glycerol, 1× PIC) on ice for 10 min before centrifugation at 10,000g, 10 min.
Salt fractionation was performed by suspending cell pellets on ice in lysis buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 0.2% Triton X-100, 1× PIC) containing the appropriate NaCl concentration. Lysate was incubated at 4 °C with rotation, 1 h, before centrifugation at 16,000g, 20 min.
Micrococcal nuclease (MNase) digests were performed on cells suspended in digestion buffer (50 mM Tris [pH 8.0], 1 mM CaCl2, 0.2% Triton X-100) at a concentration of 10 million/ml. Lysate was prewarmed for 1 min at 37 °C, then 0.5 μl MNase (M0247S; NEB) added and incubated at 37 °C. Sixty microliter samples were taken at the indicated times, and the reactions stopped by adding EDTA (pH 8.0) to 8 mM on ice. NaCl and SDS were added to 1.2 M and 1%, respectively, and DNA precipitated after phenol–chloroform extraction. DNA was resolved on 1.5% agarose gels.
Chromatin Immunoprecipitation
Cells were grown in 10 cm plates to 80% confluence, fixed for 10 min, room temperature, in 1% PFA, quenched with 125 mM glycine for 10 min, and collected in ice-cold PBS. Pellets were suspended in 500 μl lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS, 0.5 mM PMSF, 1× PIC) and sonicated for 30 cycles (30 s on/off) in a cooled Bioruptor (Diagenode) to an average fragment size of 250 bp. Fifty microliter chromatin was diluted 9× in dilution buffer (16.7 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.2 mM EDTA [pH 8.0], 0.01% SDS, 1.1% Triton X-100) and incubated overnight with either 10 μl prewashed FLAG-M2 magnetic beads (Sigma) or 5 μg antibodies plus 10 μl prewashed Dynabeads (Invitrogen). The next day, beads were moved to a new tube, washed twice each for 10 min with low salt buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA [pH 8.0], 0.1% SDS, 1% Triton X-100), high-salt buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 2 mM EDTA [pH 8.0], 0.1% SDS, 1% Triton X-100), lithium chloride buffer (0.25 M LiCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% NP-40, and 1% sodium deoxycholate), moved to a second new tube, and washed twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Chromatin was eluted at 62 °C in 100 μl fresh chromatin immunoprecipitation (ChIP) elution buffer (100 mM NaHCO3, 1% SDS) for 10 min with shaking before Proteinase K (20 μg; Thermo) was added in supplementary buffer (300 mM NaCl, 4 mM Tris-HCl, 10 mM EDTA, pH 8.0), and the mixture shaken at 62 °C for a further 2 h. DNA was purified with NucleoSpin columns (Clontech). ACTB enrichment was determined by qPCR (primers: GCAGAAGGAGCTCTTGGAGG and AGGGCAGTTGCTCTGAAGTC) using iTAQ SYBR Green master mix (Bio-Rad) and a StepOne Plus machine (ABI Biosystems) to assess the β-actin promoter.
BioID
BioID experiments were performed by seeding Flp-In T-REx HEK293 cells to be ∼60% confluent the next day. Cells were induced with 1 μg/ml sterile-filtered tetracycline in the presence of 50 μM sterile-filtered biotin for 24 h, washed, and scraped in ice-cold PBS and cell pellets snap frozen.
For streptavidin affinity purifications, five 15 cm plates of cells were prepared per sample. Cells were collected and pelleted (2000 rpm, 3 min), the pellet was washed twice with PBS, and dried pellets were snap frozen. Cell pellets were resuspended in 10 ml of lysis buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, 1:500 protease inhibitor cocktail (Sigma), 1:1000 benzonase nuclease (Novagen)] and incubated on an end-over-end rotator at 4 °C for 1 h, briefly sonicated to disrupt any visible aggregates, then centrifuged at 45,000g for 30 min at 4 °C. After retaining an input fraction, the remaining cleared supernatants were transferred to a fresh 15 ml conical tube. 30 μl of packed, preequilibrated streptavidin sepharose beads (GE) were added, and the mixture incubated for 3 h at 4 °C with end-over-end rotation. Beads were pelleted by centrifugation at 2000 rpm, 2 min, and transferred with 1 ml of lysis buffer to a fresh Eppendorf tube. Beads were washed once with 1 ml lysis buffer and twice with 1 ml of 50 mM ammonium bicarbonate (pH 8.3). Beads were transferred in ammonium bicarbonate to a fresh centrifuge tube and washed two more times with 1 ml ammonium bicarbonate buffer. Tryptic digestion was performed by incubating the beads with 1 μg MS-grade TPCK trypsin (Promega) dissolved in 200 μl of 50 mM ammonium bicarbonate (pH 8.3) overnight at 37 °C. The following morning, 0.5 μg MS-grade TPCK trypsin was added, and beads were incubated two additional hours at 37 °C. Beads were pelleted by centrifugation at 2000g, 2 min, and the supernatant was transferred to a fresh Eppendorf tube. Beads were washed twice with 150 μl of 50 mM ammonium bicarbonate, and these washes were pooled with the first eluate. The sample was lyophilized and resuspended in buffer A (0.1% formic acid). 1/5th of the sample was analyzed per MS run.
For Western blotting experiments, two 15 cm plates of cells were lysed in 1 ml RIPA buffer and subjected to one freeze-thaw cycle. Benzonase (250 U; Novagen) was added, and samples rotated at 4 °C for 1 h before brief sonication on ice and clarification at 16,000g, 20 min at 4 °C. After retaining an input fraction, the remaining lysate was incubated for 3 h with 60 μl of a 50% streptavidin-conjugated agarose bead (Sigma) slurry, previously equilibrated in RIPA buffer. Beads were moved to a fresh tube and washed in three changes of ice-cold RIPA buffer before elution. For Western blotting, beads were boiled for 20 min in 2× SDS lysis buffer supplemented with 3 mM biotin, and the supernatant transferred to a new tube while still hot.
Mass Spectrometry
Mass spectrometry was performed and analyzed as previously described (24). Briefly, high-performance liquid chromatography was conducted using a 2 cm precolumn (Acclaim PepMap 50 mm × 100 μm inner diameter) and 50 cm analytical column (Acclaim PepMap, 500 mm × 75 μm diameter; C18; 2 μm; 100 Å, Thermo Fisher Scientific), running a 120 min reversed-phase buffer gradient at 225 nl/min on a Proxeon EASY-nLC 1000 pump in-line with a Thermo Q-Exactive HF quadrupole-Orbitrap mass spectrometer. A parent ion scan was performed using a resolving power of 60,000, then up to the 20 most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation) using higher energy collision-induced dissociation fragmentation. Dynamic exclusion was activated such that MS/MS of the same m/z (within a range of 10 ppm; exclusion list size = 500) detected twice within 5 s were excluded from analysis for 15 s.
For protein identification, Thermo.RAW files were converted to the mzXML format using Proteowizard (30), then searched using X!Tandem Jackhammer TPP (2013.06.15.1 - LabKey, Insilicos, ISB) (31) and COMET (32) against the human Human RefSeq Version 45 database (containing 36,113 entries). Data were analyzed using the transproteomic pipeline via the ProHits software suite (v3.3) (33, 34, 35). Search parameters specified a parent ion mass tolerance of 10 ppm, and an MS/MS fragment ion tolerance of 0.4 Da, with up to two missed cleavages allowed for trypsin. Variable modifications of +16@M and W, +32@M and W, +42@N-terminus, and +1@N and Q were allowed. Proteins identified with an iProphet cut-off of 0.9 (corresponding to ≤1% FDR), and at least two unique peptides were analyzed with SAINT Express v.3.3.1. Ten control runs (from cells expressing the FlagBirA∗ epitope tag) were collapsed to the two highest spectral counts for each prey and compared to the two biological and two technical replicates of H3.1 and H3.3 BioID. High confidence interactors were defined as those with Bayesian false discovery rate ≤0.01. Raw mass-spec data are available for download from https://massive.ucsd.edu/, accession number MSV000087736.
Protein Purification
Spodoptera frugiperda Sf9 insect cells were cultured in suspension in I-Max media (Wisent) at 27 °C at 110 rpm. 2 μg of bacmid DNA was transfected using Effectene Transfection Reagent (QIAGEN). The cells were incubated with the transfection mix for 6 h, with gentle agitation every 30 to 60 min at 27 °C. The cells were then placed in fresh media and incubated for 7 to 10 days at 27 °C. Supernatant was collected by centrifugation at 1000g for 5 min at 4 °C and filtered with a 0.2 μm syringe filter.
For large-scale expression of CAF-1, 1 l of Sf9 insect cell shaking culture at a concentration of 1 to 2 × 106 cells/ml was infected with 10 ml of a P3 baculovirus preparation, incubated for 72 h at 27 °C shaking at 110 rpm. Cells were harvested by centrifugation at 1000g for 10 min at 4 °C. The pellets were washed with 10 ml of cold 1× PBS, flash frozen, and stored at −80 °C. The pellets were resuspended in five pellet volumes lysis buffer (20 mM Tris [pH 8.0], 350 mM NaCl, 1 mM EDTA) as described (36). The cells were lysed through a freeze-thaw cycle on dry ice and at 37 °C, and then sonicated for three cycles (30 s on, 30 s off with gentle mixing by inversion in between) at an amplitude of 40 on a Misonix S-4000 Sonicator. The lysate was then centrifuged at 25,000g for 1 h at 4 °C to remove the insoluble material. The supernatant was used for the affinity purification of CAF-1 with the StrepTrap HP column (Cytiva). The lysis buffer was used for the washes, and the protein was eluted with 2.5 mM of desthiobiotin in lysis buffer. Elution fractions were dialyzed against BC100 with 20% glycerol at 4 °C to remove desthiobiotin.
pET3a plasmids were transformed into BL21(DE3) E.coli cells. Colonies were expanded in LB overnight and a 1 l culture inoculated, grown to A600 = 0.4, and induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside for 4 h at 37 °C. Cultures were harvested by centrifugation at 3000g, and the pellets stored at −80 °C. Recombinant histones were purified as described previously, with minor changes (37). Bacterial cell pellets expressing recombinant (untagged) histone proteins were dounce homogenized 10 to 15 times in 10× cell pellet volume of Histone Wash Buffer (50 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, supplemented with 5 mM PMSF) with a tight pestle. To lyse the cells, the suspension was freeze-thawed three times by incubating on dry ice and at 37 °C. To fully lyse the cells, samples were then treated with 10 μl/ml of lysozyme (VWR) and incubated for 2 h on ice, with occasional gentle mixing. The lysate was Dounce homogenized again, 10 to 15 times with tight pestle, and centrifuged for 20 min at 4 °C at 23,000g. Recombinant histones expressed in bacteria aggregate in inclusion bodies. The pellet that contained the inclusion bodies was washed by resuspending in 100 ml histone wash buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA) supplemented with 1% Triton X-100, Dounce homogenized 10 to 15 times with tight pestle and centrifuged at 25,000g at 4 °C for 10 min. The process was repeated, and the samples left to incubate in wash buffer at 4 °C overnight with gentle rotation. The inclusion bodies were recentrifuged the next day, and the pellet washed twice more with 100 ml histone wash buffer with 1% Triton X-100 and twice again with histone wash buffer alone by repeating the Dounce homogenization and centrifugation. The final sample pellet was homogenized in 25 ml of Protein unfolding buffer (7 M guanidinium HCl, 20 mM Tris [pH 7.5]) supplemented fresh with 10 mM of DTT and gently stirred at 4 °C overnight. The samples were centrifuged at 25,000g for 30 min at 20 °C, and the supernatant which contained the purified histone proteins was collected and stored at −80 °C.
Recombinant H3.1 and H3.3 were mixed in equimolar amounts with recombinant Xenopus H2A, H2B, and H4 proteins. The mixture was dialyzed against Refolding Buffer (10 mM Tris, pH 7.5, 2 M NaCl, 20% glycerol, 1 mM EDTA, 5 mM β-mercaptoethanol) at 4 °C, with at least three changes of the buffer and one dialysis step performed overnight. Insoluble material was removed by centrifugation at 25,000g for 30 min at 4 °C. The supernatant contained the folded histones.
In Vitro Pulldowns
MagStrep Type 3 XT beads (IBA Lifesciences) were preblocked by incubating in binding buffer (50 mM Tris pH 7.6, 350 mM KCl, 2 mM EDTA, 10% glycerol) + 1% BSA at 4 °C overnight. Twenty picomoles each of histone octamers and CAF-1 were premixed in 500 μl of the binding buffer for 1 h at 4 °C, after which 2 μl of a 5% preblocked bead slurry was added, and mixtures incubated at 4 °C with gentle rotation overnight. Beads were immobilized on a magnetic rack and washed 3× with binding buffer containing either 350 mM or 1 M KCl. A final wash was done in BC100 to remove excess salts, and reactions analyzed by Western blot. Band intensities were determined using Image J.
Chromatin Immunoprecipitation With High-Throughput Sequencing Analysis
Raw reads for FLAG/HA-H3.1 and FLAG/HA-H3.3 chromatin immunoprecipitation with high-throughput sequencing (ChIP-Seq) data plus corresponding input controls from HeLa cells (European Nucleotide Archive PRJEB27519) (38) were quality trimmed with Trimmomatic-v0.36 (39) and aligned to human genome build GRCh37-v75 using bowtie2 (40). Duplicate reads were marked with PicardTools-v2.5.0. Bigwig files accounting for H3 sequencing depth and input were generated with deepTools (41). MYC ChIP-Seq from HeLa cells was from ENCODE (ENCFF609BPN, ENCFF950LQM). MYC, H3.1 and H3.3 density was profiled in 10 kb windows centred on MYC binding sites. Regions were ranked by MYC density.
To compare H3.3 (ENCODE ENCFF735XMI, ENCFF509WEL, ENCFF484HZI, ENCFF276HPR, ENCFF718YSW, ENCFF549AYZ, ENCFF421FUZ, ENCFF052DKK, ENCFF864SBW, ENCFF376NEQ, ENCFF926ZHI, ENCFF509ONF, ENCFF595UXF, ENCFF516NJV) and CAF-1 p60 (Gene Expression Omnibus GSE120063) (42) ChIP-Seq colocalization, CAF-1 p60 peaks were first merged before being overlapped calculated with 14 H3.3 datasets. Significance was calculated by randomly shuffling each of the H3.3 and CAF-1 peak sets 10,000 times and repeating the overlaps with true peaks from the other factor to create a null distribution. Overlap significance was then calculated against a T-distribution. Genome-wide distribution of peaks was determined with HOMER (43).
Bioinformatics
Enrichment of CORUM protein complexes (44), Reactome pathways, and Gene Ontology biological processes were tested using g:profiler (45). Terms with adjusted p-value <0.05 were retained, and networks constructed with EnrichmentMap in Cytoscape v3.5.1 (46). Node size and color reflects term significance, and edge weight reflects the similarity between proteins contributing to each node. Previously annotated H3 interactors were extracted from BioGRID v3.4 and STRING v10.5 (47, 48), pooling the results of searches against all human and mouse histone H3 variant genes with those from previous AP-MS studies (4, 8, 17, 18, 19, 20). For networks of H3.3- and H3.1-specific interactors, node size reflects peptide count. Known PPIs were retrieved from STRING, and edge weight reflects interaction confidence.
Experimental Design and Statistical Rationale
BioID datasets were analyzed using the significance analysis of interactome method (49), retaining high-confidence interactors with Bayesian false discovery rate ≤0.01. Experiments were conducted in biological and technical duplicate, using a background of ten independent runs using parental Flp-In T-REx HEK293 cells to control for nonspecific biotinylation events. Proteins differentially bound by H3.1 and H3.3 were identified as those with absolute log2-fold change >1.5 and adjusted p-value <0.05. Proximity ligation assay (PLA) experiments were carried out in at least two biological replicates. Unless otherwise stated, all p-values were calculated by two-tailed t-tests, not assuming equal variance between samples. Analysis was conducted with GraphPad-v8.3 or R-v4.0.4.
Results
Biotin Ligase-Labeled Histones Are Appropriately Localized
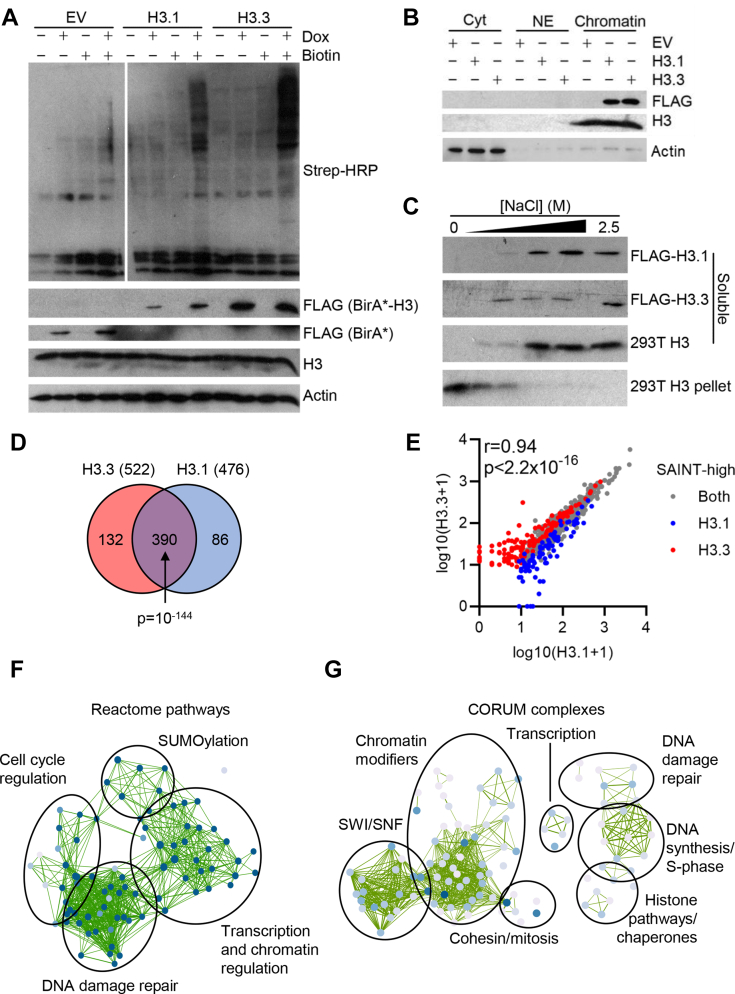
Isogenic Flp-In T-REx HEK293 cells were established to express the FLAG-tagged BirA∗ biotin ligase either alone or fused with H3.1 or H3.3, from which a 24-h induction period led to a moderate chimeric protein expression (Fig. 1A). Addition of biotin to the culture media resulted in robust biotinylation over background levels, or those arising from induced control cells. Cell fractionation experiments showed a similar distribution to endogenous H3, with almost all the protein located in the chromatin fraction (Fig. 1B). To further ascertain that the FLAG-BirA∗-H3 was properly incorporated into chromatin, histones were extracted from chromatin with increasing NaCl concentrations. Initial experiments using HEK293T cells comparing bulk H3 that solubilized in each fraction or that remained insoluble in the pellet revealed that at least 1 M NaCl was required for efficient extraction (Fig. 1C). FLAG-BirA∗-H3.1 had a similar profile to bulk H3, suggesting that the nucleosomes were not destabilized by the presence of the BirA∗ biotin ligase. Depending on context, H3.3-containing nucleosomes can have reduced stability relative to those containing H3.1 (50). Consistent with this, FLAG-BirA∗-H3.3 was extracted from chromatin at slightly lower salt concentrations compared with its FLAG-BirA∗-H3.1 counterpart. Next, we wished to test whether incorporation of BirA∗-tagged histones alters chromatin by digesting cell pellets with MNase, which preferentially digests internucleosomal DNA. The MNase digest showed that global chromatin accessibility was unaffected by the incorporation of the FLAG-BirA∗-H3 chimeric proteins into DNA and that, overall, FLAG-BirA∗-H3 does not grossly perturb nucleosome patterns at the whole nuclear level (supplemental Fig. S1A). Induction of FLAG-BirA∗-histones did not induce DNA damage (supplemental Fig. S1B). In keeping with its preferential incorporation at transcribed regions, ChIP followed by quantitative real-time PCR showed that FLAG-BirA∗-H3.3 had greater enrichment than FLAG-BirA∗-H3.1 at the promoter of a housekeeping gene (ACTB; supplemental Fig. S1C). Taken together, these data confirm that the FLAG-BirA∗ fusion is properly incorporated into chromatin.
Fig. 1.
The BioID landscape of H3.A, Flp-In T-REx HEK293 BirA∗-fusion cells were induced or not with doxycycline (1 μg/ml) or biotin (50 μM) for 24 h before being harvested and subjected to Western blot analysis. B, Flp-In T-REx HEK293 BirA∗-fusion cells were induced with tetracycline (1 μg/ml) for 24 h and fractionated into cytoplasmic (Cyt), soluble nuclear (NE), and insoluble chromatin fractions. C, BirA∗-fusion or HEK293T cells were extracted in increasing concentrations of NaCl (0, 0.5, 1, 1.5, 2.5 M) and soluble or insoluble proteins analyzed by Western blotting. D, Venn diagram of H3 interactors scored as high-confidence interactors by SAINT analysis. p-value: hypergeometric test. E, correlation of peptide counts from all proteins recovered. Gray proteins were SAINT-scored for both H3.1 and H3.3; blue proteins for just H3.1, and red proteins for just H3.3. r: Pearson correlation. p-value: t test. F, network of Reactome pathways significantly enriched among combined H3.1 and H3.3 BioID hits. Node size and color reflects term significance and edge weight the similarity between proteins contributing to each node. G, network of CORUM protein complexes significantly enriched among combined H3.1 and H3.3 BioID hits. Node size and color reflects term significance and edge weight the similarity between proteins contributing to each node. SAINT, significance analysis of interactome.
The BioID Interactomes of H3.1 and H3.3
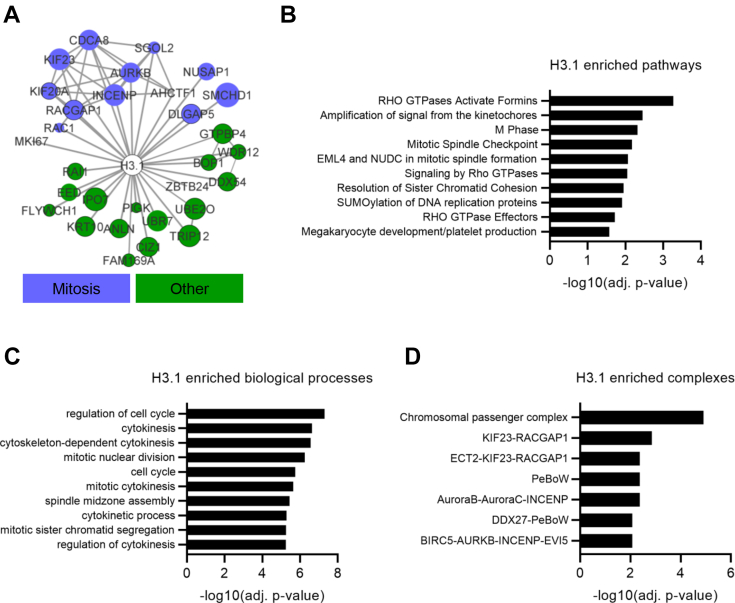
To delineate the proximal interaction landscape of H3.1 and H3.3, we performed BioID experiments twice with two technical replicates each. Cells were induced for 24 h in the presence of tetracycline and biotin. Purification of biotinylated interactors on streptavidin beads allows for rigorous washes to remove nonspecific proteins. A pool of ten runs using control cells expressing just BirA∗ provided a stringent background control, and material recovered from BioID pulldowns was analyzed by mass spectrometry. Significance analysis of interactome analysis (49) identified 608 high-confidence proximal associations for H3.1 and/or H3.3. Of these, 132 proteins were specific to H3.3 and 86 to H3.1, with the remaining proteins representing a significant but expected large number of shared associations (p = 10−144; Fig. 1D and supplemental Table S1). The peptide counts of proteins interacting with each H3 variant were highly correlated, including for many proteins scored as high-confidence interactors of just one variant (r = 0.94, p < 2.2 × 10−16; Fig. 1E). Analysis of the overall H3 BioID interactome of 608 proteins showed that the most significant (adjusted p-value < 0.05). Reactome pathways and CORUM protein complexes enriched with H3 were related to transcription, chromatin, or DNA-damage repair (Fig. 1, E and F and supplemental Fig. S1, D and E). Cell cycle regulation was also enriched, including not only DNA synthesis/S-phase protein complexes but also several mitotic complexes including cohesin, the chromosomal passenger complex, and the MIS12 complex (Fig. 1, E and F and supplemental Fig. S1, D and E).
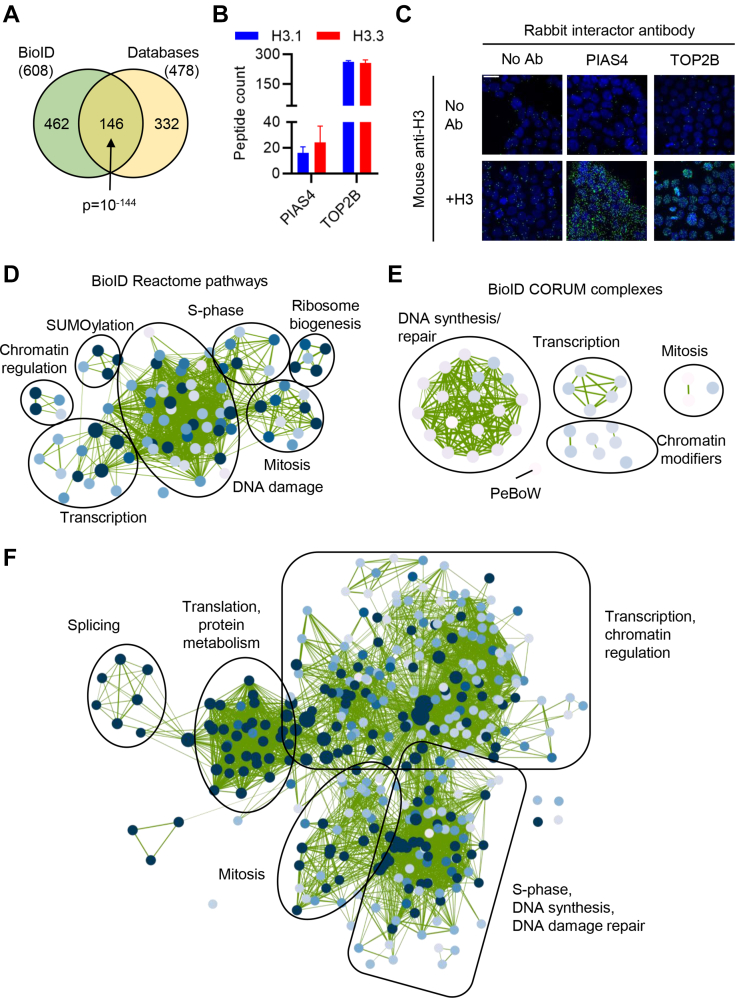
BioID Reveals Novel Histone Interactors
Of the 608 total interactors that we identified, 462 were not previously identified in the STRING or BioGRID databases or through AP-MS studies (4, 8, 17, 18, 19, 20). However, 44% of the previously characterized interacting proteins were identified (p = 10−144, hypergeometric test), indicating that BioID can extend the H3 interactome (Fig. 2A). We confirmed that these were bona fide interactors, by validating our findings with an orthogonal method, PLAs. In PLA, primary antibodies (from different species) raised against two putative interacting proteins are bound to a sample. If the two antibodies bind associating proteins, a template for a rolling circle reaction is generated, allowing for visualization of individual interactions as foci within cells (51). To show that PLA can detect H3 interactions that are of high and low abundance, we first performed PLA on PIAS4 and TOP2B, which had very different peptide counts (Fig. 2B) that were similar for both H3.1 and H3.3. PLA using an endogenous H3 antibody in HEK293T cells showed a clear increase in foci when combined with PIAS4 or TOP2B antibodies (Fig. 2C). Analysis of the significant biological pathways and protein complexes associated with the proteins uniquely identified by BioID showed strong enrichment of proteins involved in transcription (including numerous components of the basal TF complexes TFIIIC, TFIIE, and TFIIF as well as specific TFs such as MGA, HOXD13, and GATAD1), chromatin regulation (for example histone modifiers including NSD1/3, KDM2A, KDM4B, and PHF2 and remodeler complex components including BAZ2A, ARID2, and MTA3) and the DNA damage response (Fig. 2, D and E and supplemental Fig. S2, A and B). The mitotic enrichment seen in the overall BioID H3 interactome was also present in the BioID-unique pathways, suggesting an underappreciated association between H3 and the mitotic machinery. Also enriched was the PeBoW complex (comprised of Pescadillo 1 [PES1], block of proliferation1 [BOP1] and WDR12), which is essential for cell cycle progression and also functions in 60S ribosome biogenesis in the nucleolus (52, 53), consistent with enrichment of cell cycle–related proteins.
Fig. 2.
BioID extends the H3 interaction landscape.A, Venn diagram comparing proteins identified by BioID and H3 interactors curated by the STRING/BioGRID databases. p-value: hypergeometric test. B, peptide counts from BioID (n = 4). Bars show mean ± standard deviation. C, representative PLA images between H3 and the indicated interactors in HEK293T cells. Scale bar: 24 μm. D, network of Reactome pathways enriched among 462 H3-interacting proteins uniquely identified by BioID. Node size and color reflects term significance and edge weight the similarity between proteins contributing to each node. E, network of CORUM protein complexes enriched among 462 H3-interacting proteins uniquely identified by BioID. Node size and color reflects term significance and edge weight the similarity between proteins contributing to each node. F, combined interacting proteins identified here, in Lambert et al. or Liu et al., or annotated in the STRING or BioGRID databases, were used as g:profiler input for Reactome pathways and CORUM complexes. Node size and color reflects term significance and edge weight the similarity between proteins contributing to each node. PLA, proximity ligation assay.
The majority of the H3 interactors found in either the STRING/BioGRID databases or AP/MS datasets, and that were not identified by BioID, are associated with the ribosome or spliceosome as well as some transcription, chromatin, and cell cycle proteins (supplemental Fig. S2, C–F). Finally, we combined all the H3 interactome datasets, revealing a densely interconnected network of related proteins and processes that is dominated by transcription and chromatin regulation and DNA synthesis (replication and repair) (Fig. 2F). Notably, as well as the expected enrichment of DNA replication proteins, BioID further identified a substantial enrichment of mitotic proteins, expanding the number of known cell cycle–specific histone associations.
Regulation of Transcription and Chromatin by H3.3-Specific Interactors
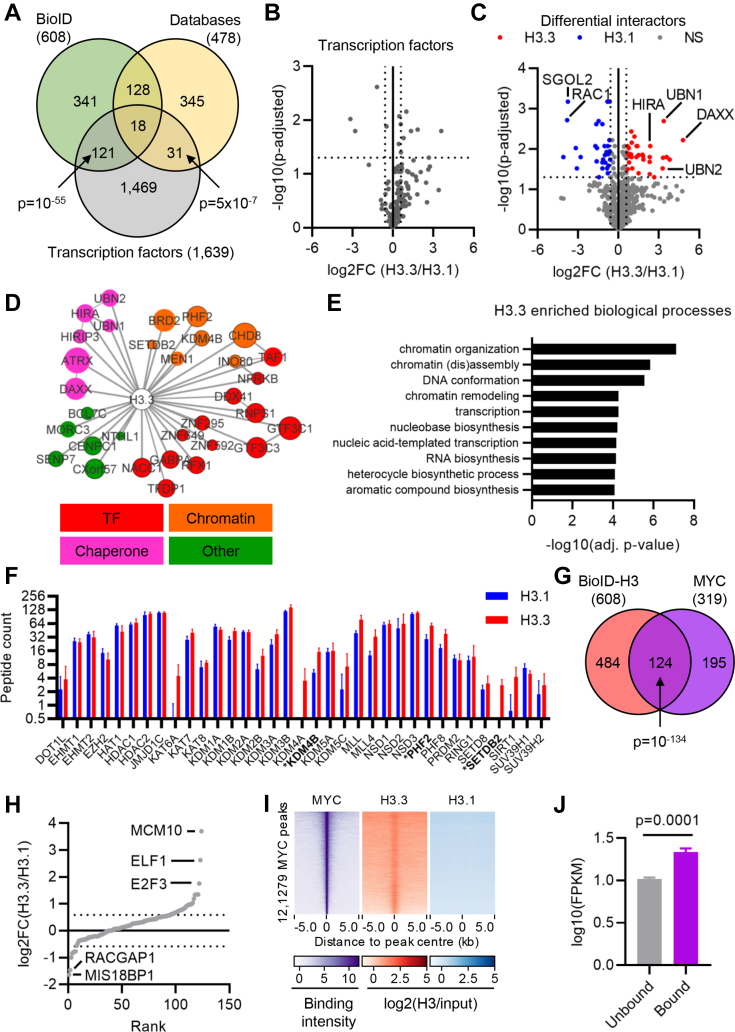
We anticipated that many H3 interactors identified by BioID but not captured through affinity purification methods could be either tightly bound to chromatin or biochemically labile. We therefore expected to identify proteins such as TFs or chromatin modifiers that are frequently missed in AP-MS approaches. To test this hypothesis, we compared our BioID H3 interactors and those found in the STRING and BioGRID databases with known human TFs (54). BioID identified 139 TFs, of which only 18 were previously identified as H3 interactors. Although the previously described interactors included 49 TFs (p = 5 × 10−7), BioID recovered 2.8× more TFs than previous methods (p = 10−56; Fig. 3A), including both basal and specific TFs. Consistent with a preferential role of H3.3 in active chromatin, TFs were more enriched with H3.3 than H3.1 (Fig. 3B).
Fig. 3.
The H3.3-specific interactome converges on transcriptional regulators.A, Venn diagram comparing BioID and STRING/BioGRID H3 interactors with human transcription factors (TFs). p-values: hypergeometric test. B, volcano plot comparing TF association with H3.3 and H3.1. C, volcano plot comparing differential interaction between H3.3 and H3.1, highlighting those passing a cut-off of absolute log2FC >1.5 and adjusted p-value <0.05 in blue (H3.1) and red (H3.3). D, network of H3.3-specific interactors colored by to function (red, TFs; orange, chromatin; pink, histone chaperone; green, other). Node size reflects peptide count. Edges between nodes denote protein–protein interactions between the proteins with edge weight denoting confidence. E, biological processes enriched among H3.3-specific interactors. F, peptide counts from BioID for histone modifiers (n = 4). Bars show mean ± standard deviation. G, Venn diagram comparing BioID H3 interactors with MYC-interacting proteins. H, MYC interacting proteins ranked by log2(H3.3/H3.1). Dotted lines mark log2FC = 1.5 and −1.5. I, ChIP-Seq enrichment of MYC, H3.3, and H3.1 in regions ±5 kb around the centre of MYC binding sites in HeLa cells. J, mean expression of genes bound or not by MYC in HeLa-S3 cells (n = 4). ChIP-Seq, chromatin immunoprecipitation with high-throughput sequencing.
To more fully elucidate potential differences in H3.1 and H3.3 function that could be explained by their respective interaction partners, we next identified the proteins that were differentially bound by H3.3 and H3.1. At cut-offs of absolute log2-fold change of 1.5 and adjusted p-value <0.05, 32 proteins were H3.3 specific and 30 were H3.1 specific (Figs. 3, C and D and 4A, supplemental Table S1). Many of the H3.3-specific interactors have primary functions in either chromatin regulation or transcription, including both general TFs that are part of the basal transcription machinery as well as specific TFs (Fig. 3, D and E). Overall, the BioID approach is advantageous in the identification of new, additional variant-specific protein associations otherwise missed by AP-MS.
Fig. 4.
The H3.1-specific interactome regulates mitotic progression.A, network of H3.1-specific interactors colored by to function (blue, mitosis; green, other). Node size reflects peptide count. Edges between nodes denote protein–protein interactions between the proteins with edge weight denoting confidence. B, Reactome pathways enriched among H3.1-specific interactors. C, biological processes enriched among H3.1-specific interactors. D, CORUM protein complexes enriched among H3.1-specific interactors.
Our data included some interesting observations pertaining to histone modifying enzymes. Overall, we identified 35 proteins with direct histone-modifying activity (Fig. 3F). Of these, the H3K9 methylase SETDB2 and H3K9 demethylases KDM4B/JMJD2B and PHF2 showed a significantly increased association with H3.3. In addition, the H3K9 demethylase KDM4A/JMJD2A interacted exclusively with H3.3 (log2FC = 3.9, unadjusted p = 0.05, supplemental Table S1). Increased binding to both methylases and demethylases of H3K9 could reflect the dual enrichment of H3.3 in actively transcribed regions and constitutive heterochromatin, which have low and high levels of H3K9 methylation, respectively. Finally, MEN1 (menin), a key component of the mixed-lineage leukamia (MLL) complex that deposits H3K4me3 in active regions of the genome (55) and the MLL complex catalytic components MLL (KMT2A; log2FC(H3.3/H3.1) = 0.99, unadjusted p = 0.009) and MLL4 (KMT2D; log2FC(H3.3/H3.1) = 1.34, unadjusted p = 0.03) were also enriched with H3.3.
Interestingly, although MYC itself was not captured, many H3 interactors in our dataset appeared to be MYC-related. To formally test this, we compared H3 BioID interactors with the MYC interactome (56), finding a highly significant overlap (p = 10−134; Fig. 3G). Proteins interacting with both MYC and H3 were more enriched with H3.3 (Fig. 3H). Intriguingly, MCM10, which is important for DNA replication initiation and fork progression (57, 58), was the most H3.3-enriched MYC interactor and not detected in any H3.1 BioID replicate (log2FC(H3.3/H3.1) = 3.7, unadjusted p = 0.059; Fig. 3H and supplemental Table S1). To further examine the bias in histone variants, we next integrated ChIP-Seq data for MYC (ENCODE) plus H3.3 and H3.1 (38). While H3.3 was strongly enriched around the centre of MYC ChIP-Seq binding sites, there was no H3.1 enrichment over background at these locations (Fig. 3I). In keeping with a functional association between H3.3-containing chromatin and MYC, MYC-bound genes had a significantly greater expression than MYC-unbound genes (p = 0.0001; Fig. 3J).
Enrichment of Mitotic But Not S-Phase Proteins Among H3.1 Interacting Proteins
In contrast to the chaperones and transcription/chromatin proteins that were H3.3 specific, 43% (13/30) of H3.1-specific proteins have mitotic-related functions (Fig. 4A), notably multiple members of the chromosome passenger complex, the master regulator of mitosis, and cohesin (Fig. 4, B–D) (59). SGOL2 (Shugoshin 2) and RAC1 were among the most differentially enriched proteins with H3.1 (Fig. 3D). SGOL2 is a mitotic protein that protects centromeric cohesin from eviction during prophase (60, 61), and while RAC1 is largely a cytoplasmic protein, it is nuclear in G2 and promotes cell division (62). Overall, the enrichment of mitotic functions in interactors identified by BioID but not AP-MS (Fig. 2, D and E) suggests that this is an aspect of the H3 interactome under-represented by traditional methods.
Expression of the genes encoding H3.1 is restricted to S-phase, and accordingly, newly synthesized H3.1 is abundantly incorporated into chromatin during S-phase, while H3.3 is expressed and incorporated throughout the cell cycle at basal levels (13). Although biotin labeling took place over the whole cell cycle, it therefore was notable that, despite the enrichment of mitotic proteins among H3.1 interactors, S-phase interactors were not preferentially enriched in the H3.1 interactome.
Unexpected CAF-1 Flexibility Towards H3.3 in Vivo
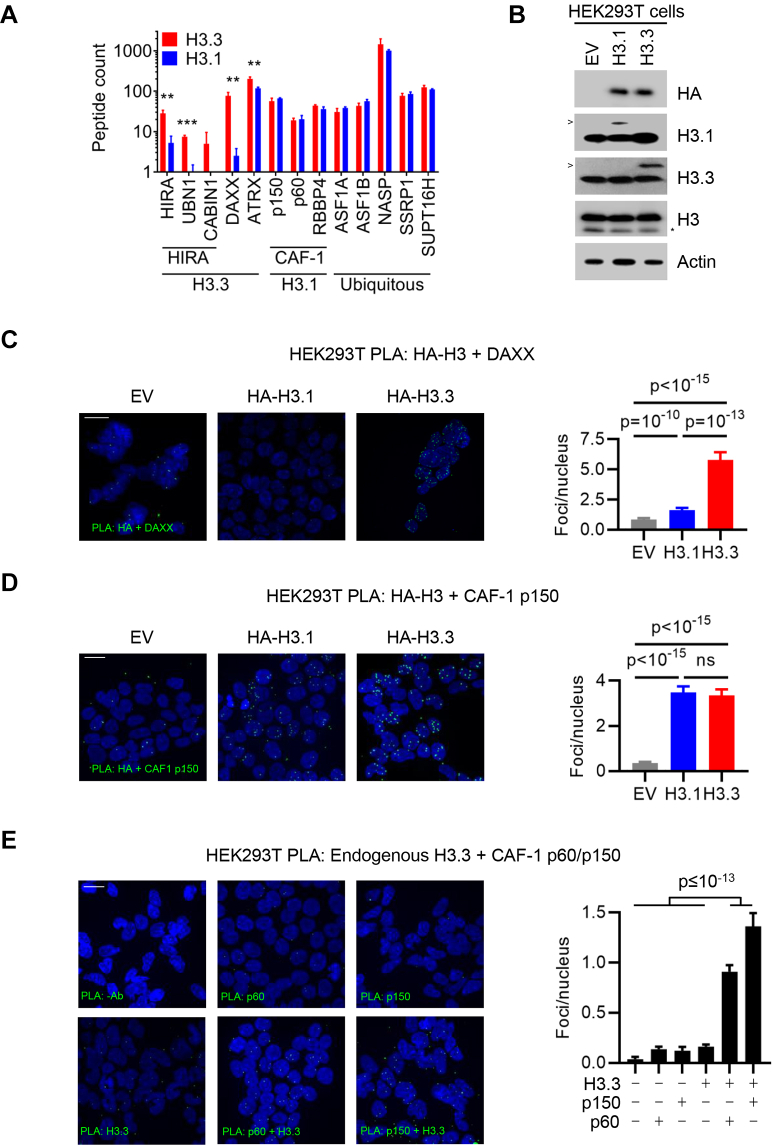
The HIRA (HIRA, UBN1, and CABIN1) and DAXX/ATRX complexes have previously been shown to be H3.3 specific (3, 4, 8, 9, 10, 11, 12, 63). All were significantly more associated with H3.3 than H3.1, consistent with previous reports (Figs. 3C and 5A), indicating that BioID recapitulated known histone-variant specific interactions. This was especially striking when looking at the histone binding subunits (UBN1, log2FC = 3.37; and DAXX, log2FC = 4.81) of the H3.3 histone chaperone complexes.
Fig. 5.
CAF-1 and H3.3 interact in vivo.A, peptide counts from BioID for indicated histone chaperones (n = 4). Bars show mean ± standard deviation. p-values: t-test. ∗∗: p < 0.01. ∗∗∗: p < 0.0001. B, Western blot of HEK293T cells transduced with control, H3.1 or H3.3 lentiviruses. >: ectopic H3.3 or H3.1 detected with variant-specific antibodies. ∗: clipped H3. C, proximity ligation assay (PLA) between HA-H3 and DAXX in HEK293T cells from (B). Scale bar: 20 μm. Bars show mean ± 95% CI of n = 73 (EV), 150 (H3.1), 95 (H3.3) cells. p-values: ANOVA (Welch with Games-Howell’s multiple comparisons test). D, PLA between HA-H3 and CAF-1 p150 in HEK293T cells in HEK293T cells from (B). Scale bar: 20 μm. Quantification shows mean ± 95% CI of n = 446 (EV), 387 (H3.1), 408 (H3.3) cells from two independent experiments. p-values: ANOVA (Welch with Games-Howell’s multiple comparisons test). E, PLA between endogenous H3.3 and p60 or p150 in parental HEK293T cells. Scale bar: 20 μm. Quantification shows mean ± 95% CI of n = 276 (no antibody), 1050 (p60), 327 (p150), 1356 (H3.3), 1029 (H3.3+p60), 328 (H3.3+p150) cells from 2 to 3 independent experiments. p-values: ANOVA (Welch with Games-Howell’s multiple comparisons test). CAF-1, chromatin assembly factor 1; DAXX, death domain–associated protein 6; HIRA, histone cell cycle regulator.
In contrast to HIRA and DAXX/ATRX, ASF1 and sNASP are not known to discriminate between H3 variants, although the tNASP isoform has been reported to preferentially incorporate H3.3 (64). These, along with both components of the FACT complex (SUPT16H, SSRP1) that is involved in nucleosome reorganization were similarly enriched in the H3.1 and H3.3 BioID interactomes (Fig. 5A). Unexpectedly, CAF-1, which deposits the replication-coupled H3.1 histone variant on replicating DNA (4), had no difference in its association between H3.1 and H3.3. This held true for either the main subunits (p150/CHAF1A and p60/CHAF1B) or the accessory protein RBBP4 (Fig. 5A). To confirm this, we first carried out streptavidin pulldowns from biotin-induced Flp-In T-REx HEK293 BirA∗-H3 cells. Subsequent western analysis confirmed a clear bias toward HIRA in the H3.3 proximal associations; while ASF1B, but also p150 and p60, showed similar levels of enrichment with both H3.1 and H3.3 (supplemental Fig. S3A).
To confirm that the H3.3–CAF-1 interaction was not due to the presence of the BirA∗ ligase, we expressed FLAG-HA epitope-tagged H3.1 and H3.3 at similar levels in HEK293T cells (Fig. 5B). We then carried out PLAs between HA-H3 and either CAF-1 p150 or DAXX. For DAXX, we observed the expected preferential binding to H3.3: there was 1.9-fold enrichment over empty vector control cells with HA-H3.1 (p = 2 × 10−10) and a further 3.5-fold increase with HA-H3.3 (p = 10−13 versus HA-H3.1; Fig. 5C). This confirmed the ability of PLA to quantitatively detect differential protein interactions. For CAF-1 p150, both HA-H3.1 and HA-H3.3 had significant enrichment in interaction foci compared to empty vector controls (p < 10−15 for both), but there was little difference in the number of foci per nucleus between HA-H3.1 and HA-H3.3 cells (Fig. 5D). Next, we confirmed the interaction between endogenous H3.3 and both p60 and p150 subunits of CAF-1. To do this, we used an H3.3-specific antibody (Fig. 5B) and, as a further control, an independent CAF-1 p150 antibody. In parental HEK293T cells, with both the p60 and p150 subunits of CAF-1, we detected substantial interaction compared to single/no-antibody controls (p < 0.0001; Fig. 5E). These data indicate that the interaction we observe is not a function of H3.3 overexpression or of BirA∗-fusion but rather a bona fide underappreciated interaction between H3.3 and CAF-1.
The specificity of CAF-1 and HIRA toward H3.1 and H3.3, respectively, was initially defined from nuclear lysates prepared with a high-salt concentration (4). We therefore tested the stability of the interaction of H3.1 and H3.3 with CAF-1 using in vitro binding assays between CAF-1 and nucleosomes assembled with either H3.1 or H3.3. When binding reactions were washed in high salt, H3.1 nucleosomes were readily bound by CAF-1, while H3.3 binding was barely detected, consistent with data from AP-MS studies. In contrast, however, with lower salt (350 mM KCl, which is close to physiological condition and thus replicates the salt concentrations of intact nuclei in living cells), as well as an increase in the total amount of H3 bound by CAF-1, there was no significant difference between the amount of each variant bound (Fig. 6, A and B).
Fig. 6.
H3.3 interacts with CAF-1 throughout the cell cycle.A, recombinant Strep-tagged CAF-1 was incubated with nucleosomes containing H3.1 or H3.3. Reactions were washed with 350 mM or 1 M KCl. Gels are representative of n = 3 replicates. B, quantification of in vitro CAF-1 H3 binding reactions from (F) (n = 3). p-values: t test. C, representative PLA images in HeLa-Fucci(CA2) cells between H3.3 and p150 or p60. Scale bar: 20 μm. D, quantification of PLA signal from C. Bars show mean ± 95% CI of n = 140 (p60), 148 (p150), 126 (H3.3), 119 (H3.3+p60), 122 (H3.3+p150). p-values: ANOVA (Welch with Games-Howell’s multiple comparisons test). Results are representative of two biological replicate experiments. E, mean ± 95% CI of PLA signal from cells in cell cycle phases from H3.3+p60 (G1, n = 28; S, n = 35; G2, n = 49) and H3.3+p150 (G1, n = 42; S, n = 39; G2, n = 35) in C. Results are representative of two biological replicate experiments. F, representative PLA images in HeLa-Fucci(CA2) cells between endogenous H3.1 and CAF-1 p150. Scale bar: 20 μm. G, quantification of PLA signal between H3.1 and CAF-1 p150 from F. Bars show mean ± 95% CI of n = 105 (no primary), 95 (p150), 114 (H3.1), 100 (H3.1+p150) cells. p-value: ANOVA with Tukey’s multiple comparisons test. See also supplemental Fig. S4D. Results are representative of two biological replicate experiments. H, mean ±95% CI of PLA signal between H3.1 and CAF-1 p150 from cells in either G1 (n = 50) or S/G2 (n = 49) cell cycle phases. p-value: t test. See also supplemental Fig. S4D. Results are representative of two biological replicate experiments. CAF-1, chromatin assembly factor 1; PLA, proximity ligation assay.
Given that CAF-1 deposits H3 into newly synthesized DNA during both DNA replication and repair, we labeled cells with 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analog that can be incorporated in newly synthesized DNA. After labeling, we performed PLA between p150 and either H3.3 or bulk H3 (supplemental Fig. S3B). In cycling cells, H3.3 represents a limited fraction of H3 in the cell (2), and accordingly, there were more foci with H3 than with H3.3 (supplemental Fig. S3C). For both H3 and H3.3, the majority of interaction foci were in EdU-positive cells with no significant difference between the two (supplemental Fig. S3D). However, some of the foci still lay outside of strongly EdU-positive nuclei, suggesting that CAF-1 and H3 can interact outside of the context of DNA replication.
To better test the cell cycle distribution of the H3.3–CAF-1 interaction, we next used HeLa-Fucci(CA2) cells expressing the fluorescent cell-cycle sensors mVenus-hGem and mCherry-hCdt1 (28). In this system, G1-phase cells are red, and S-phase cells are green, while G2 and mitotic cells are both red and green and can be distinguished by the nuclear counterstain. In PLAs between H3.3 and p150 or p60, there was a significant interaction between H3.3 and CAF-1 subunits (p < 10−15; Fig. 6, C and D). Importantly, HeLa cells are the principal cell type used in studies identifying CAF-1 as an H3.1-specific interactor. Given that our BioID and PLA assays were so far conducted in HEK293-derivative cell lines, this result confirms that the in vivo interaction between CAF-1 and H3.3 is not HEK293 specific. Importantly, when we examined the cell cycle distribution of HeLa-Fucci(CA2) cells containing p60/p150–H3.3 interaction foci, we found that there was no significant difference in the abundance of either CAF-1 subunit with H3.3 in G1, S, and G2 phases (2-way ANOVA; Fig. 6, C and E and supplemental Fig. S3E).
Next, we used an H3.1-specific antibody (Fig. 5B) for PLA in HeLa-Fucci(CA2) cells between endogenous H3.1 and CAF-1 p150. Staining with both antibodies gave significant enrichment over controls (p < 10−15; Fig. 6, F and G), with ∼15× more interaction foci per nucleus between H3.1 and CAF-1 p150 than between H3.3 and CAF-1 p150 (Fig. 6, D and G). This is expected given the greater amount of H3.1 present in chromatin in comparison to that of H3.3 in cycling cells (2). The cell cycle distribution of the H3.1-CAF-1 p150 interaction was, as expected, increased in S/G2-phase nuclei (p = 10−10; Fig. 6H). Collectively, this indicates that H3.3 interacts with CAF-1 throughout the cell cycle, while the H3.1–CAF-1 interaction is highly abundant during DNA replication.
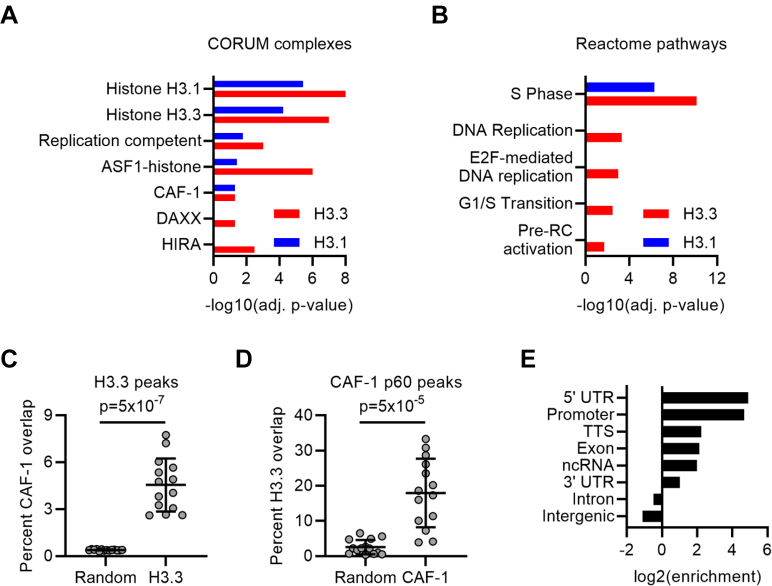
While H3.1–CAF-1 interactions predominate in S-phase, H3.3 did, however, also make other S-phase–specific protein associations. When we considered all the proteins identified by BioID for each variant, we found that DNA replication-specific gene ontology terms were nevertheless enriched with H3.3. Specifically, members of the replication competent complex (RFC1,2,4,5; POLA1,2) were enriched with H3.3 along with proteins involved in the G1/S transition and initiation of DNA replication (Fig. 7, A and B).
Fig. 7.
S-phase enrichment of H3.3 interactors.A, enrichment of indicated CORUM complexes among H3.1 and H3.3 interacting proteins. B, enrichment of indicated Reactome complexes among H3.1 and H3.3 interacting proteins. C, percent H3.3 ChIP-Seq peaks overlapping with CAF-1 p60 peaks compared with randomly permuted CAF-1 peaks. p-value: hypergeometric test. D, percent CAF-1 p60 ChIP-Seq peaks overlapping with H3.3 peaks compared with randomly permuted H3.3 peaks. p-value: hypergeometric test. E, log2 enrichment of different genomic locations bound by both CAF-1 p60 and H3.3 was calculated with HOMER. ASF1, anti-silencing factor 1; CAF-1, chromatin assembly factor 1; ChIP-Seq, chromatin immunoprecipitation with high-throughput sequencing; DAXX, death domain–associated protein 6; HIRA, histone cell cycle regulator.
Collectively, these data support an in vivo interaction between H3.3 and CAF-1 at replication forks. Finally, to further examine whether CAF-1 and H3.3 may functionally interact, we examined CAF-1 p60 (42) and H3.3 (ENCODE) ChIP-Seq data (cell lines: n = 3, CAF-1; n = 14, H3.3). Despite differences between cell lines, significant genome-wide colocalization was observed between H3.3 and CAF-1, with 18 ± 5.6% (95% confidence interval, CI) of CAF-1 p60 peaks co-occupied by H3.3 (p = 5 × 10−7, hypergeometric test; Fig. 7C), while 4.5 ± 1% (95% CI) of H3.3 peaks co-occupied by CAF-1 p60 (p = 5 × 10−5, hypergeometric test; Fig. 7D). Overall, 6042 out of 9787 CAF-1 p60 peaks overlapped with H3.3 peaks (p = 0, hypergeometric test), with these peaks highly enriched in promoter regions (Fig. 7E).
Collectively, our data demonstrate that BioID is able to capture nuances to PPIs that traditional AP-MS approaches fail to identify and that, in vivo, CAF-1 is able to associate with not only H3.1, but also with H3.3.
Discussion
In this study, we report the BioID interactomes of H3.1 and H3.3, uncovering numerous novel, distinct interactions, and providing an important new resource for the scientific community. BioID is a complementary method to the traditional method of AP-MS. In AP-MS, antibodies against the protein of interest are used to purify both it and its interactors. For chromatin-related proteins, this is most usually done starting with nuclear lysates that are typically prepared under high-salt conditions (29). These harsh methods can lead to disruption of PPIs and interactions being missed, as complexes need to be maintained throughout the entire purification. In other words, although an invaluable technique, AP-MS can lead to an underappreciation of PPIs as well as a need to carefully control for nonspecific antibody-binding events. In contrast, the BioID approach uses in vivo biotinylation to biotinylate proteins that are directly adjacent to the protein of interest in living cells. These proteins do not need to be maintained in complexes and can be stringently purified on streptavidin, thus circumventing any biases that may be introduced by buffer composition during nuclear lysis. As well as identifying stable interactors, BioID is also able to successfully identify proteins that have labile and/or transient interactions with the protein of interest. Because BioID is a proximity-based method that can label proteins within a narrow radius of the fused biotin ligase, distal members of protein complexes, or proteins buried within complexes, are less likely to be recovered by this method. Therefore, BioID serves as an experimental approach that can work in tandem with other existing methodologies to better characterize protein interactomes. In this work, we used BioID to uncover many chromatin modifiers and TFs, which would not necessarily be expected to stably associate with histones for long time periods, many of which were not captured by previous high-throughput interaction studies. We were thereby able to offer new biological insight to previously described histone interactions.
As an additional quality control step in our analysis, we compared the H3.1 BioID data with previous datasets (18, 26) and confirmed a highly significant overlap between our prey proteins and those identified before (supplemental Fig. S4). Our BirA∗ construct was an N-terminal fusion (24), Lambert et al. (18) employed a C-terminal fusion, while Liu et al. (26) used both C- and N-terminal fusions but included an additional StrepIII tag between the BirA∗ and H3. While proteins not identified in each study could be attributable to spatial differences between the biotin ligase locations or experimental approach, including data analysis methods, the prey proteins identified by all three are nevertheless highly related (p ≤ 10−43, hypergeometric test), in further confirmation of the quality of our dataset.
Amongst the many interesting novel associations, our analysis yielded three interesting findings: namely, that BioID revealed enrichment of mitotic proteins with H3, in particular H3.1, that H3.3 is associated with MYC, and that CAF-1 interacts with both H3.1 and H3.3 in vivo.
The interactors identified by BioID, in particular the H3.1-specific hits, were highly enriched in mitotic proteins, in particular mitotic spindle processing and the chromosome passenger complex (59, 65). SGOL2 (Shugoshin 2) and RAC1 were among the most differentially enriched proteins with H3.1. SGOL2 is a mitotic protein that protects centromeric cohesin from eviction during prophase (60, 61), and while RAC1 is largely a cytoplasmic protein, it is nuclear in G2 and promotes cell division (62). There is evidence that H3 has important functions in mitosis. It has been shown to physically interact with shugoshin/SGOL1 through residues K42, G44, and T45, which are conserved between H3.1 and H3.3 (66). The H3-shugoshin interaction is important for its function in preventing cohesin from dissociating from centromeres after prophase, thus playing an essential role in the proper separation of chromosomes (60, 61, 67). The H3-shugoshin interaction is integral to the spindle assembly checkpoint and maintaining spindle tension during mitosis (68). Here we found that although SGOL1 had 1.3-fold greater binding to H3.1 than H3.3, SGOL2, which has similar functions to SGOL1, was 13-fold more enriched with H3.1, suggesting there may be nuanced differences between the H3-binding functions of different shugoshin proteins. Finally, biochemical studies have shown that old H3 has higher mitotic phosphorylation levels than new H3 that was incorporated during the previous S-phase (69). Notably, phosphorylation of H3.1-S28, but not H3.3-S28, is enriched among these marks.
Mitotic proteins would perhaps only be expected to be associated with histones during mitosis, which is short compared with the rest of the cell cycle, thus making it difficult to detect large amounts of them by conventional AP-MS from cycling cells. BioID will tag each of these proteins as the cell passes through mitosis, however. Given that cells for BioID were induced for at least 1 cell cycle, this allowed their later recovery from all the cells used rather than just the small fraction of mitotic cells at the point of harvesting. A number of these proteins, particularly spindle and cytokinesis proteins, were enriched with H3.1 compared to H3.3. This perhaps results from H3.1 being incorporated in the centromeric attachment points for these complexes during S-phase and then there being no cause for its replacement by H3.3 during G2.
Given the different genomic locations in which these variants are incorporated and their reported functions to date, their interactomes were very similar, consistent with the overall findings from AP-MS (4). While the studies carried out to date have been in cycling cells, H3 variant usage varies throughout development, with H3.3 gradually replacing H3.1/2 as the dominant variant during differentiation. In future, it will be important to explore how the interactome of H3 variants varies during differentiation.
Secondly, as well as directly identifying H3-associated TFs, we found strong enrichment of MYC-associated proteins with H3 that was more pronounced with H3.3. MYC-bound gene promoters are enriched with H3.3 but not H3.1 and are expressed at higher levels than unbound genes, consistent with the role of MYC in driving transcription and the association of H3.3 with transcriptionally active chromatin. Overall, this indicates the ability of BioID to probe interaction networks deeper than immediate bait–prey interactions.
The chaperones that deposit H3.1 and H3.3 have previously been characterized through AP-MS, with CAF-1 being shown to deposit H3.1 during DNA replication and repair (4), and HIRA and DAXX/ATRX depositing H3.3 into active chromatin and heterochromatin, while ASF1, NASP, and FACT do not discriminate between the two variants. We found HIRA and DAXX/ATRX to be enriched with H3.3, while ASF1, NASP, and FACT interacted similarly with each variant, confirming these previous findings. Our BioID, PLA, and in vitro data, however, unexpectedly revealed flexibility in the CAF-1 histone deposition pathway, a nuance that carries important biological implications.
The H3.3-specificity of binding to HIRA and DAXX/ATRX by BioID and PLA, which recapitulates data from immunoprecipitation (4, 12), argues strongly that the detection of an in vivo association between CAF-1 and H3.3 was enabled by the emergence of recently developed proximity-labeling tools (70).
Our study shows that while the H3.1–CAF-1 interaction predominates in mammalian cells, there is also a nonnegligeable association between CAF-1 and H3.3. This is an important nuance for which there are precedents. Prior H3.3 immunoprecipitation studies detected little biochemically stable association with CAF-1. However, there was clear CAF-1 coprecipitation with H3.3 when H3.3-specific chaperones are depleted (8, 12). This further highlights the flexibility in the CAF-1 deposition pathway. H3.1 is highly expressed from multiple genes in S-phase, while H3.3 is expressed at basal levels from two genes throughout the cell cycle. Consistent with this and prior studies on CAF-1, we found that the cell cycle distribution of the H3.1–CAF-1 interaction is highly enriched in S/G2-phase nuclei. We now provide evidence for basal H3.3–CAF-1 interactions that are independent of the cell cycle. We therefore propose that the relative histone levels primarily modulate the differences seen in CAF-1 binding patterns.
The H3 chaperone interactions were initially characterized on the basis of purifications from high-salt nuclear extracts and in vitro chromatin assembly assays (4, 29). While there is an apparent discrepancy with our BioID and PLA data, our data confirms that the H3.1–CAF-1 interaction largely predominates. However, the interaction with H3.3 requires further investigation. There are different reasons that could explain why little CAF-1 was previously detected in H3.3 immunoprecipitates, notably the salt concentrations used Experimental Procedures (71). To address this, we performed in vitro binding assays between CAF-1 p60 and H3.1 and H3.3 with reactions washed at different salt concentrations. When binding assays were performed under lower salt concentrations, there was no difference between binding of the histone variants to CAF-1. Overall, our data suggests that, although its binding to H3.1 is more abundant—owing to its higher expression level in cells—and stable, CAF-1 has far more flexibility in vivo than initially thought and can associate with both H3.1 and H3.3 in mammalian cells.
These observations were pivotal to our understanding of H3.1 and H3.3 histone deposition, and our data fully agree with the CAF-1 and H3.1 interaction predominating in cycling mammalian cells. Because BioID labels interacting proteins in real time, we now report interactions between CAF-1 and H3.3 that occur throughout the cell cycle. In agreement with our findings, Drosophila H3.3 is deposited in both replication-coupled and replication-independent manners (72). Consistent with this, integration of ChIP-Seq, Repli-Seq and nascent RNA-Seq datasets showed that H3.3 is enriched at early replicating origins (38). Although early origins tended to be located in more transcriptionally active regions, perhaps due to their more relaxed epigenetic state, the H3.3-enrichment in these regions could not be explained only on the basis of transcriptional activity. Furthermore, H3.3 is required for replication origin firing and its loss leads to defects in DNA replication (73). We found that the H3.3 interactome was enriched for proteins associated with the G1/S transition and DNA replication. In addition, we found that H3.3 preferentially interacted with MCM10. MCM10 functions in DNA replication initiation and fork elongation by stimulating and stabilizing the CDC45-MCM2-7-GINS (CMG) helicase (57, 58).
It is also possible that the H3.3–CAF-1 interaction plays a role in the DNA damage response. H3.3 is required for replication fork progression through UV-damaged regions (74). Furthermore, H3.1 has been shown to be deposited by CAF-1 outside of S-phase in UV-irradiated cells as part of the nucleotide excision repair pathway (75). These authors also reported observing H3.3 deposition at sites of UV-irradiation, suggesting a similar mechanism could be operating.
Collectively, these data support a model whereby CAF-1 can deposit H3.3-containing nucleosomes in vivo, but where the variant-specific levels also vary with available histone pools. There are examples of CAF-1-mediated H3.3 deposition that support this model. First, H3.3-containing nucleosomes were deposited into SV40 minichromosomes assembled in the presence of HEK293 histones and CAF-1 biochemically purified from human cells (76, 77). In these experiments, the amount of histone variants incorporated onto the DNA template was proposed to reflect their relative amounts in the histone preparations. More recently, CAF-1 depletion in Epstein-Barr virus–positive host cells was also shown to cause a loss of both H3.1 and H3.3 in the Epstein-Barr virus genome (78). Furthermore, by integrating CAF-1 (42) and H3.3 (ENCODE) ChIP-Seq datasets, we found a sizeable degree of signal colocalization between CAF-1 and H3.3.
Certain species, such as S. pombe and S. cerevisiae, express a single H3 variant whose sequence, while divergent from humans, matches H3.3 at the critical ‘gatekeeper’ amino acids (A87, I89, G90 in H3.3; S87, V89, M90 in H3.1). These organisms nevertheless use CAF-1 to deposit histones during DNA replication (13), implying that CAF-1 must be able to assemble H3.3-containing nucleosomes. Indeed, while H3.3 occurs near-universally throughout eukaryotes, H3.2 evolved in early branching animals and H3.1 is mammalian-specific (79, 80). While plants have both H3.1 and H3.3 variants, they have evolved independently (81, 82).
In summary, in this work we quantitatively characterized the interactomes of H3 variants H3.1 and H3.3 using BioID to uncover novel aspects of their in vivo interaction landscapes that were previously determined by affinity purifications and in vitro assays. BioID is highly complementary to traditional techniques for difficult-to-access proteins such as those in chromatin locations. Our data speak to the importance of combining existing and new methodologies together with comprehensive analysis of all available datasets. In particular, this allowed a complete snapshot of the in vivo histone interaction landscape, revealing a network of transcription and chromatin regulators along with protein complexes that function throughout all phases of the cell cycle.
Data Availability
Raw mass-spec data are available for download from https://massive.ucsd.edu/, accession number MSV000087736. All ChIP-Seq and RNA-Seq data are publicly available as outlined in Experimental Procedures. Other data are contained within the manuscript.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank the Hawkins lab for their insight during this work.
Funding and additional information
C. H. was funded by Canadian Cancer Society Research Institute (702296 and 706160), Canadian Institutes of Health Research (PJT-156407), ChadTough Foundation, DIPG Collaborative, Meagan’s Walk. E. I. C. was funded by the Canadian Institutes of Health Research (PJT-159683) and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-05559).
Author contributions
R. S., S. P., B. R., and C. H. conceptualization; R. S., B. R., E. I. C., and C. H. methodology; R. S. and É. C. formal analysis; R. S., S. M., É. C., H. Y. Y., S. P., and S. M. M. investigation; R. S. visualization; E. I. C. resources; R. S. and C. H. writing-original draft; B. R., E. I. C., and C. H. supervision; E. I. C. and C. H. funding acquisition.
Supplemental Data
References
- 1.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Wu R.S., Tsai S., Bonner W.M. Patterns of histone variant synthesis can distinguish go from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 5.Shibahara K., Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 6.Gaillard P.H.L., Martini E.M., Kaufman P.D., Stillman B., Moustacchi E., Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 7.Hoek M., Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsässer S.J., Huang H., Lewis P.W., Chin J.W., Allis C.D., Patel D.J. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricketts M.D., Frederick B., Hoff H., Tang Y., Schultz D.C., Singh Rai T., et al. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 2015;6:7711. doi: 10.1038/ncomms8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoelper D., Huang H., Jain A.Y., Patel D.J., Lewis P.W. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 2017;8:1193. doi: 10.1038/s41467-017-01206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis P.W., Elsaesser S.J., Noh K.M., Stadler S.C., Allis C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grover P., Asa J.S., Campos E.I. H3-H4 histone chaperone pathways. Annu. Rev. Genet. 2018;52:109–130. doi: 10.1146/annurev-genet-120417-031547. [DOI] [PubMed] [Google Scholar]
- 14.Green E.M., Antczak A.J., Bailey A.O., Franco A.A., Wu K.J., Yates J.R., 3rd, et al. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munakata T., Adachi N., Yokoyama N., Kuzuhara T., Horikoshi M. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- 16.Tyler J.K., Adams C.R., Chen S.R., Kobayashi R., Kamakaka R.T., Kadonaga J.T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 17.Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., 3rd, Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 18.Lambert J.P., Tucholska M., Go C., Knight J.D., Gingras A.C. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos E.I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W.H., et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos E.I., Smits A.H., Kang Y.H., Landry S., Escobar T.M., Nayak S., et al. Analysis of the histone H3.1 interactome: a suitable chaperone for the right event. Mol. Cell. 2015;60:697–709. doi: 10.1016/j.molcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D.I., Jensen S.C., Roux K.J. Identifying protein-protein associations at the nuclear envelope with BioID. Methods Mol. Biol. 2016;1411:133–146. doi: 10.1007/978-1-4939-3530-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birendra K.C., May D.G., Benson B.V., Kim D.I., Shivega W.G., Ali M.H., et al. VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol. Biol. Cell. 2017;28:2241–2250. doi: 10.1091/mbc.E17-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta G.D., Coyaud E., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comartin D., Gupta G.D., Fussner E., Coyaud E., Hasegan M., Archinti M., et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Liu X., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Öhman T., et al. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018;9:1188. doi: 10.1038/s41467-018-03523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dingar D., Kalkat M., Chan P.K., Srikumar T., Bailey S.D., Tu W.B., et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J. Proteomics. 2015;118:95–111. doi: 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 28.Sakaue-Sawano A., Yo M., Komatsu N., Hiratsuka T., Kogure T., Hoshida T., et al. Genetically encoded tools for optical dissection of the mammalian cell cycle. Mol. Cell. 2017;68:626–640.e5. doi: 10.1016/j.molcel.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig R., Beavis R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 32.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 33.Hubbard S.J., Jones A.R. Humana; New York, NY: 2010. Proteome Bioinformatics. [DOI] [PubMed] [Google Scholar]
- 34.Deutsch E.W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., et al. A guided tour of the trans-proteomic pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z.Y., et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 2010;28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W.H., Roemer S.C., Port A.M., Churchill M.E. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 2012;40:11229–11239. doi: 10.1093/nar/gks906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loyola A., Reinberg D. Histone deposition and chromatin assembly by RSF. Methods. 2003;31:96–103. doi: 10.1016/s1046-2023(03)00093-8. [DOI] [PubMed] [Google Scholar]
- 38.Clément C., Orsi G.A., Gatto A., Boyarchuk E., Forest A., Hajj B., et al. High-resolution visualization of H3 variants during replication reveals their controlled recycling. Nat. Commun. 2018;9:3181. doi: 10.1038/s41467-018-05697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volk A., Liang K., Suraneni P., Li X., Zhao J., Bulic M., et al. A CHAF1B-dependent molecular switch in hematopoiesis and leukemia pathogenesis. Cancer Cell. 2018;34:707–723.e7. doi: 10.1016/j.ccell.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47:D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatr-Aryamontri A., Breitkreutz B.J., Heinicke S., Boucher L., Winter A., Stark C., et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alam M.S. Proximity ligation assay (PLA) Curr. Protoc. Immunol. 2018;123 doi: 10.1002/cpim.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., et al. J. Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., et al. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 2007;27:3682–3694. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., et al. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 55.Hughes C.M., Rozenblatt-Rosen O., Milne T.A., Copeland T.D., Levine S.S., Lee J.C., et al. Menin associates with a trithorax family histone methyltransferase complex and with the Hoxc8 locus. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 56.Kalkat M., Resetca D., Lourenco C., Chan P.K., Wei Y., Shiah Y.J., et al. MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell. 2018;72:836–848.e7. doi: 10.1016/j.molcel.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 57.Lõoke M., Maloney M.F., Bell S.P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Im J.S., Ki S.H., Farina A., Jung D.S., Hurwitz J., Lee J.K. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carmena M., Wheelock M., Funabiki H., Earnshaw W.C. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krishnan S., Smits A.H., Vermeulen M., Reinberg D. Phospho-H1 decorates the inter-chromatid axis and is evicted along with shugoshin by SET during mitosis. Mol. Cell. 2017;67:579–593.e6. doi: 10.1016/j.molcel.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 62.Michaelson D., Abidi W., Guardavaccaro D., Zhou M., Ahearn I., Pagano M., et al. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J. Cell Biol. 2008;181:485–496. doi: 10.1083/jcb.200801047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawatsubashi S., Murata T., Lim J., Fujiki R., Ito S., Suzuki E., et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kato D., Osakabe A., Tachiwana H., Tanaka H., Kurumizaka H. Human tNASP promotes in vitro nucleosome assembly with histone H3.3. Biochemistry. 2015;54:1171–1179. doi: 10.1021/bi501307g. [DOI] [PubMed] [Google Scholar]
- 65.Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. The spindle assembly checkpoint. Curr. Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Luo J., Xu X., Hall H., Hyland E.M., Boeke J.D., Hazbun T., et al. Histone H3 exerts a key function in mitotic checkpoint control. Mol. Cell. Biol. 2010;30:537–549. doi: 10.1128/MCB.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ng T.M., Lenstra T.L., Duggan N., Jiang S., Ceto S., Holstege F.C., et al. Kinetochore function and chromosome segregation rely on critical residues in histones H3 and H4 in budding yeast. Genetics. 2013;195:795–807. doi: 10.1534/genetics.113.152082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo J., Deng X., Buehl C., Xu X., Kuo M.H. Identification of tension sensing motif of histone H3 in Saccharomyces cerevisiae and its regulation by histone modifying enzymes. Genetics. 2016;204:1029–1043. doi: 10.1534/genetics.116.192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin S., Yuan Z.F., Han Y., Marchione D.M., Garcia B.A. Preferential phosphorylation on old histones during early mitosis in human cells. J. Biol. Chem. 2016;291:15342–15357. doi: 10.1074/jbc.M116.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scott W.A., Campos E.I. Interactions with histone H3 & tools to study them. Front. Cell Dev. Biol. 2020;8:701. doi: 10.3389/fcell.2020.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dick D.A. The distribution of sodium, potassium and chloride in the nucleus and cytoplasm of Bufo bufo oocytes measured by electron microprobe analysis. J. Physiol. 1978;284:37–53. doi: 10.1113/jphysiol.1978.sp012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 73.Strobino M., Wenda J.M., Padayachy L., Steiner F.A. Loss of histone H3.3 results in DNA replication defects and altered origin dynamics in C. elegans. Genome Res. 2020;30:1740–1751. doi: 10.1101/gr.260794.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frey A., Listovsky T., Guilbaud G., Sarkies P., Sale J.E. Histone H3.3 is required to maintain replication fork progression after UV damage. Curr. Biol. 2014;24:2195–2201. doi: 10.1016/j.cub.2014.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Polo S.E., Roche D., Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 76.Smith S., Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 77.Smith S., Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y., Jiang C., Trudeau S.J., Narita Y., Zhao B., Teng M., et al. Histone loaders CAF1 and HIRA restrict Epstein-Barr virus B-cell lytic reactivation. mBio. 2020;11 doi: 10.1128/mBio.01063-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Postberg J., Forcob S., Chang W.J., Lipps H.J. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol. Biol. 2010;10:259. doi: 10.1186/1471-2148-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malik H.S., Henikoff S. Phylogenomics of the nucleosome. Nat. Struct. Mol. Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 81.Wells D., Hoffman D., Kedes L. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 1987;15:2871–2889. doi: 10.1093/nar/15.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ingouff M., Berger F. Histone3 variants in plants. Chromosoma. 2010;119:27–33. doi: 10.1007/s00412-009-0237-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass-spec data are available for download from https://massive.ucsd.edu/, accession number MSV000087736. All ChIP-Seq and RNA-Seq data are publicly available as outlined in Experimental Procedures. Other data are contained within the manuscript.