Abstract
Spider brain and central nervous system (CNS) have remained unexplored, due in part to the difficulty of observing these organs, usually only possible through histological preparations. Recently, internal anatomy studies have been supplemented by the inclusion of X‐ ray micro‐CT. Unmineralized tissue such as the body of invertebrates requires a staining process to enhance tissue X‐ray absorption and improve contrast during observation. Many current protocols are based on iodine staining requiring critical point drying (CPD) of the sample for optimal contrast. This process induces shrinking of the soft tissue generating artifacts in the morphology, volume, and even position of internal structures. Phosphotungstic acid (PTA) is an alternative staining agent recently used in marine invertebrate and plant studies. Here, we used several specimens of the common spider Araneus diadematus to visualize the spider brain and compare both contrast‐enhancing ethanol‐based solutions. We assessed a gradient of staining times, observed and tested the repercussions of CPD, and examined the use of vacuum to accelerate PTA diffusion. We show that PTA provides the best contrast on micro‐CT scans in ethanol eliminating the need for CPD, and offering more realistic in situ visualizations of the internal organs. In combination with different scanning settings, PTA allowed observation of internal organs like the CNS, digestive system, muscles, and finer structures like the retina, visual nerves, and optic neuropiles. This fast and less invasive method could facilitate the proper documentation of the internal anatomy in the context of evolutionary, developmental and functional studies.
Keywords: arachnida, arthropoda, cerebrum, imaging, nervous system, neuroanatomy, tissue, X‐rays
1. INTRODUCTION
Spider brain and central nervous system (CNS) have remained relatively unexplored due in part to the difficulty of observing these organs, usually only possible through histological preparations (Saint‐Remy 1890; Hanstrom 1921, 1923; Babu & Barth 1984; Strausfeld et al., 1993; Schmid & Becherer 1999; Hill 2006; Long 2016, 2021). Recently, studies of internal anatomy have been supplemented by the inclusion of X‐ ray micro‐CT scanning in spiders (Stafstrom et al., 2017; Steinhoff et al., 2018, 2017, 2020; Lin et al., 2021), as well as other arthropods (Sombke et al., 2019, 2015; Castejón et al., 2018; Alba‐Alejandre et al., 2019; Alba‐Alejandre et al., 2018; Alba‐Tercedor et al., 2021). These invertebrates have a relatively soft unmineralized body that requires a staining process to enhance X‐ray absorption of these tissues and improve contrast during observation. Due in part to their soft bodies, spiders are one of those terrestrial arthropod groups where specimens are preserved in ethanol (Ubick et al., 2017). Dozens of staining agents with different properties, toxicity, and limitations can be found in literature (Keklikoglou et al., 2019; Metscher 2009; Faulwetter et al., 2012). Two of them, iodine and phosphotungstic acid (PTA), are soluble in ethanol and therefore suitable for staining ethanol preserved specimens (Keklikoglou et al., 2019). Iodine staining is commonly used as a contrast‐enhancement agent because of its short staining times and rapid penetration in tissues. Nevertheless its optimal contrast is acquired after drying the sample either by critical point drying (CPD) (Stafstrom et al., 2017; Steinhoff et al., 2017; Sombke et al., 2015) or chemical drying using hexamethyldisilazane (Alba‐Alejandre et al., 2019; Alba‐Alejandre et al., 2018; Alba‐Alejandre et al., 2020; Alba‐Tercedor et al., 2021). The drying process can induce shrinkage of soft internal structures such as muscles, nervous, and digestive tissues (Sombke et al., 2015). This method has proven its value as a tool for making quick observations and measurements of the neuroarchitecture of spiders being used in developmental and descriptive anatomical studies (Steinhoff et al., 2017; Stafstrom, Michalik & Hebets 2017; Steinhoff et al., 2018, 2020). PTA is known to give good overall contrast of structures binding strongly to proteins (e.g., connective tissue and muscles) but having very slow tissue penetration and a tendency to dissolve calcified structures (Keklikoglou et al., 2019). This agent has been used as a contrast enhancer in studies covering a wide range of organisms including orchids (Dirks‐Mulder et al., 2019; Pramanik et al., 2020), marine invertebrates (Faulwetter et al., 2012; Sakurai & Ikeda 2019), insects (Swart et al., 2016), and even mammal embryos (Lesciotto et al., 2020; Metscher, 2009).
Spider micro‐CT scanning studies have not explored the applications of PTA staining; nevertheless, the commonly used iodine method has been pivotal for studies on the anatomy of the male genitalia (Lipke et al., 2015; Dederichs et al., 2019; Rivera‐Quiroz et al., 2021), as well as the description and analysis of the spider CNS (Steinhoff et al., 2017; Stafstrom et al., 2017; Steinhoff et al., 2018, 2020). Spiders are one of the most successful arthropod taxa. They are generalist predators that have developed a myriad of adaptations to almost every terrestrial habitat. These adaptations can be observed in many organs, including, of course, the brain. Only a few studies (Saint‐Remy, 1890; Hanstrom, 1923; Long, 2021) have documented and compared the neuroanatomy of different groups of spiders and only one has related it to the hunting strategies and daily peak of activity (e.g., diurnal/nocturnal) (Long, 2021). Therefore, these arachnids offer a promising model to study the selective pressures posed by different environments, and the adaptation of their nervous systems to these changes. The implementation of quick, powerful, and nondestructive documentation techniques such as the one explored here can help expedite these comparisons and generate deeper knowledge about the evolution of the brain and nervous system.
Here, we used freshly collected specimens of the spider species Araneus diadematus to explore the utilization of PTA and iodine as contrast‐enhancing agents for the observation of nervous tissue in spiders, focusing on the CNS and the visual processing portion of the brain.
2. MATERIAL AND METHODS
Adult females of A. diadematus were collected at the Singelpark, Leiden, Netherlands in October 2020. Spiders were directly fixed in 70% Et‐OH after which, they had their legs and palps removed, and were dehydrated in a graded ethanol series (70–80–85–90–96–99.9%), for about 30 min per step. Spiders were stained with two contrast‐enhancement solutions: 1% iodine‐70% Et‐OH, and 1% PTA–70% Et‐OH (Table 1). Samples were placed inside a 5 ml Eppendorf tube filled with the contrast enhancing solution and left on a rotator for a period of time ranging from 1 to 25 days; staining time per specimen can be found in Table 1. After staining, all spiders were washed several times with 96% Et‐OH and transferred into 1.5 ml Eppendorf tubes filled with 96% Et‐OH for scanning. Specimens were held in place using cotton wool. Micro‐CT scanning was performed with a Zeiss X‐radia 520 Versa; details of each scan can be found in Table 1. Six iodine‐stained specimens were critical point dried in a Leica EM‐CPD 300 using the following program: slow CO2 admittance with 2 min delay, 18 exchange cycles, and slow reheating to medium temperature (36°C) with a slow gas discharge. These dried specimens were then fixed on insect pins using gel glue (Pattex “100% repair gel”; Henkel) to be rescanned.
TABLE 1.
Details of the contrast enhancing and x‐ray scanning parameters tested per specimen
| Specimen code | Part | Staining | Staining | Diffusion | Scan medium | Current (mA) | Exposure times (s) | Optical magnification | Pixel size(μm) | Voltage (kV) | Binning | Scan time |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD_01 | Brain | PTA | 7d | CV | EtOH | 88 | 4 | 4 | 1.69 | 80 | Bin 1 | 4h49m |
| AD_01 | Ceph | PTA | 7d | CV | EtOH | 87 | 1 | 0.4 | 6.6 | 80 | Bin 2 | 1h34m |
| AD_02 | Ceph | PTA | 1d | PD | EtOH | 74 | 15 | 0.4 | 7.57 | 40 | Bin 2 | 7h47m |
| AD_02 | Ceph | PTA | 5d | PD | EtOH | 80 | 6 | 0.4 | 6.55 | 50 | Bin 2 | 3h48m |
| AD_02 | Ceph | PTA | 10d | PD | EtOH | 80 | 10 | 0.4 | ee | 50 | Bin 2 | 5h36m |
| AD_02 | Ceph | PTA | 25d | PD | EtOH | 79 | 8 | 0.4 | 8.26 | 50 | Bin 2 | 4h59m |
| AD_02 a | Ceph | PTA | 25d | PD | EtOH | 86 | 6.5 | 0.4 | 4.13 | 70 | Bin 1 | 8h19m |
| AD_02 | Eyes | PTA | 25d | PD | EtOH | 88 | 13 | 20 | 0.85 | 80 | Bin 2 | 6h53m |
| AD_02 a | Brain | PTA | 25d | PD | EtOH | 88 | 8 | 4 | 1.27 | 80 | Bin 1 | 9h27m |
| AD_03 | Ceph | PTA | 4d | IV | EtOH | 80 | 11 | 0.4 | 6.99 | 50 | Bin 2 | 5h57m |
| AD_03 | Ceph | PTA | 10d | IV | EtOH | 81 | 11 | 0.4 | 6.94 | 50 | Bin 2 | 6h3m |
| AD_03 | Brain | PTA | 10d | IV | EtOH | 8 | 4 | 2.53 | 80 | Bin 1 | 9h27m | |
| AD_04 | Ceph | PTA | 1d | CV | EtOH | 88 | 5 | 0.4 | 3.55 | 80 | Bin 2 | 3h35m |
| AD_04 | Ceph | PTA | 5d | CV | EtOH | 87 | 1 | 0.4 | 8.02 | 80 | Bin2 | 1h31m |
| AD_04 | Brain | PTA | 5d | CV | EtOH | 88 | 8 | 4 | 1.69 | 80 | Bin 1 | 9h50m |
| AD_05 | Ceph | PTA | 2d | CV | EtOH | 8 | 0.4 | 6 | 80 | Bin 2 | 4h32m | |
| AD_05 | Brain | PTA | 2d | CV | EtOH | 6 | 4 | 2.53 | 80 | Bin 1 | 7h38m | |
| AD_06 | Ceph | PTA | 5d | CV | EtOH | 87 | 1 | 0.4 | 8.02 | 80 | Bin2 | 1h31m |
| AD_06 | Brain | PTA | 5d | CV | EtOH | 87 | 7 | 4 | 1.69 | 80 | Bin 1 | 8h34m |
| AD_07 | Ceph | IOD | 1d | PD | EtOH | 88 | 0.7 | 0.4 | 8.02 | 80 | Bin 2 | 1h19m |
| AD_07 | Brain | IOD | 1d | PD | EtOH | 88 | 2.5 | 4 | 1.5 | 80 | Bin 1 | 4h27m |
| AD_07 | Brain | IOD/CPD | Dried | – | Air | 87 | 6 | 4 | 1.41 | 80 | Bin 1 | 7h38m |
| AD_07 | Ceph | IOD/CPD | Dried | – | Air | 88 | 4 | 4 | 5.77 | 80 | Bin 2 | 2h33m |
| AD_08 | Ceph | IOD | 1d | PD | EtOH | 74 | 8 | 0.4 | 6.55 | 40 | Bin 2 | 4h32m |
| AD_08 | Ceph | IOD | 1d | PD | EtOH | 80 | 12 | 4 | 5.4 | 50 | Bin 2 | 6h22 |
| AD_08 | Brain | IOD/CPD | Dried | – | Air | 90 | 3 | 4 | 1.5 | 80 | Bin 1 | 4h54m |
| AD_08 | Ceph | IOD/CPD | Dried | – | Air | 88 | 2 | 4 | 5.4 | 80 | Bin 2 | 1h38m |
| AD_09 | Ceph | IOD | 2d | PD | EtOH | 88 | 0.6 | 0.4 | 8.02 | 80 | Bin 2 | 1h16m |
| AD_09 | Brain | IOD | 2d | PD | EtOH | 87 | 2.5 | 4 | 1.6 | 80 | Bin 1 | 4h27m |
| AD_09 | Brain | IOD/CPD | Dried | – | Air | 75 | 5 | 4 | 1.99 | 80 | Bin 1 | 8h6m |
| AD_09 | Ceph | IOD/CPD | Dried | – | Air | 74 | 1.5 | 0.4 | 8.31 | 80 | Bin 2 | 1h10m |
| AD_10 | Ceph | IOD | 4d | PD | EtOH | 88 | 1 | 0.4 | 8.22 | 80 | Bin 2 | 1h27m |
| AD_10 | Brain | IOD | 4d | PD | EtOH | 87 | 2 | 4 | 1.93 | 80 | Bin 1 | 4h27m |
| AD_10 | Brain | IOD/CPD | Dried | – | Air | 87 | 3 | 4 | 1.69 | 80 | Bin 1 | 4h51m |
| AD_10 | Ceph | IOD/CPD | Dried | – | Air | 88 | 6 | 4 | 5.94 | 80 | Bin 2 | 3h29m |
| AD_11 | Ceph | IOD | 8d | PD | EtOH | 88 | 1 | 0.4 | 8.22 | 80 | Bin 2 | 1h27m |
| AD_11 | Brain | IOD | 8d | PD | EtOH | 88 | 2 | 4 | 1.93 | 80 | Bin 1 | 4h11m |
| AD_11 | Brain | IOD/CPD | Dried | – | Air | 75 | 4 | 4 | 1.93 | 80 | Bin 1 | 5h48m |
| AD_11 | Ceph | IOD/CPD | Dried | – | Air | 75 | 1 | 0.4 | 6.81 | 80 | Bin 2 | 1h10m |
| AD_12 | Ceph | IOD | 2d | PD | EtOH | 87 | 0.7 | 0.4 | 8.02 | 80 | Bin 2 | 1h31m |
| AD_12 | Brain | IOD | 2d | PD | EtOH | 88 | 2 | 4 | 1.93 | 80 | Bin 1 | 4h11m |
| AD_12 | Brain | IOD/CPD | Dried | – | Air | 71 | 6.5 | 4 | 2.27 | 70 | Bin 1 | 8h17m |
| AD_12 | Ceph | IOD/CPD | Dried | – | Air | 72 | 1 | 0.4 | 7.37 | 70 | Bin 2 | 1h40m |
| AD_25 | Brain | PTA | 25d | PD | EtOH | 87 | 4 | 4 | 2.05 | 80 | Bin 1 | 5h50m |
| AD_25 | Ceph | PTA | 25d | PD | EtOH | 88 | 1 | 0.4 | 8.04 | 80 | Bin 2 | 1h11m |
| AD_26 | Brain | PTA | 25d | PD | EtOH | 88 | 4 | 4 | 2.05 | 80 | Bin 1 | 5h50m |
| AD_26 | Ceph | PTA | 25d | PD | EtOH | 87 | 1 | 0.4 | 8.04 | 80 | Bin 2 | 1h11m |
Scanning parameters used. Diffusion column indicates how the staining process was done PD, passive diffusion; CV, continuous vacuum; IV, intermittent vacuum. Some samples were tested more than once under different parameters and staining times. Samples AD_07–AD_12 were scanned before and after CPD. Specimen codes used here correspond to the ones indicated in Table 2, Figures 1, 2, 3, 4, 5, 6, and Supplementary Material.
Indicates our preferred parameters.
For the vacuum‐assisted staining with PTA, we used a medium Kartell plastic vacuum exicator connected to a VWR‐ VP820 vacuum gas pump. Samples were placed inside a 1.5 ml Eppendorf tube filled with the PTA solution, which was placed open at the center of the exicator. The pump was active for a period of 3 min to create a vacuum. For the intermittent vacuum test the samples were left inside for 10 min. After this, air was slowly let in until the vacuum was broken. The Eppendorf tube with the sample was then transferred to a rotator and left there (closed to avoid evaporation) for a period of 24 h after which, the process was repeated. The continuous vacuum test samples received a similar treatment only that in this case they remained inside the exicator the whole time and the vacuum was left for 24‐h periods; PTA solution was topped up when needed.
Scans were visualized using Avizo 2020.2 (Thermo Fisher Scientific, USA). Individual cross sections were obtained using Ortho slices and Slices adjusting histograms individually; volumes were constructed applying the Volume Rendering and adjusting the rendering settings to standard instead of physical with other settings retaining default values; again, histograms were adjusted individually. Reconstruction of the 3D surfaces of the optic nerves and nervous structures was done in the segmentation function; labeling the optic nerves, neuropiles and optic tracks in separate label fields. Surfaces were generated with default settings with smoothing extent and transparency adjusted individually. Labeling of the brain structures was done by topological correspondence to the neuroanatomical descriptions of A. diadematus and Argiope lobata (as Epeira in the original publication) (Saint‐Remy, 1890), Argiope trifasciata (Long, 2016; 2021), and other araneomorph spiders (Hill, 2006; Stafstrom et al., 2017; Steinhoff et al., 2020).
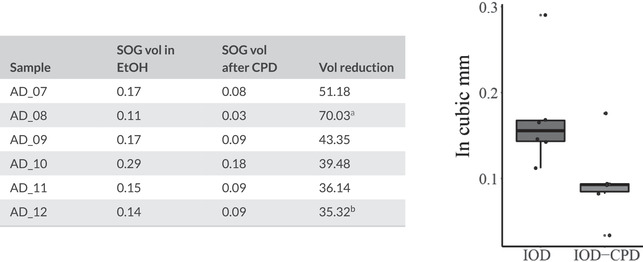
For the evaluation of the impact of CPD, we segmented the supraesophageal ganglion of each specimen twice. The first scan was done with the specimen submerged in 96% EtOH and the second scan done after CPD. We compared the raw volumes to estimate percentage of volume reduction (Table 2) and built boxplots for the SOG volume under each treatment. Afterward, we tested our groups using a Shapiro–Wilk normality test, and used a Welch two sample t‐tests to observe if the differences in volumes before and after CPD were statistically significant. Since our data almost deviated from a normal distribution, we also analyzed it using a nonparametric Mann–Whitney U test. Significance threshold was defined as p = .05. All the analyses and boxplots were done in RStudio (Wickham et al., 2021; Wickham, 2016).
TABLE 2.
Volumes of the supraesophageal ganglion before and after critical point drying
|
|
All the specimens show a considerable reduction of the brain volume after CPD.
Indicates the most affected sample.
bIndicates the best‐preserved sample, even in this case, more than one third of the original brain volume was lost. Boxplot shows the significant differences in volume between samples in EtOH (left) and after drying (right).
3. RESULTS
3.1. Iodine versus PTA
We tested the potential of PTA by comparing different staining times and assessing the penetration in the tissues, visualization of tissues, and overall image quality (Figures 1, 2, 3). We used iodine 1 day submersion as a base for comparing the agent penetration in tissue (Figure 1) and extended the staining times from there. Iodine achieves its best contrast after just 24 h and extending the staining time did not improve but actually hampered the contrast by overstaining tissue and making it impossible to distinguish between different tissues and structures after four days. By comparison, tissue penetration of PTA was extremely slow with only the coxae being stained after 24 h. Samples need to be stained for significantly longer periods with the CNS being stained after 10 days and optimal contrast of the cephalic region being obtained after 25 days. Even after these long staining times, tissues were not visibly overstained as opposed to iodine enhancement.
FIGURE 1.
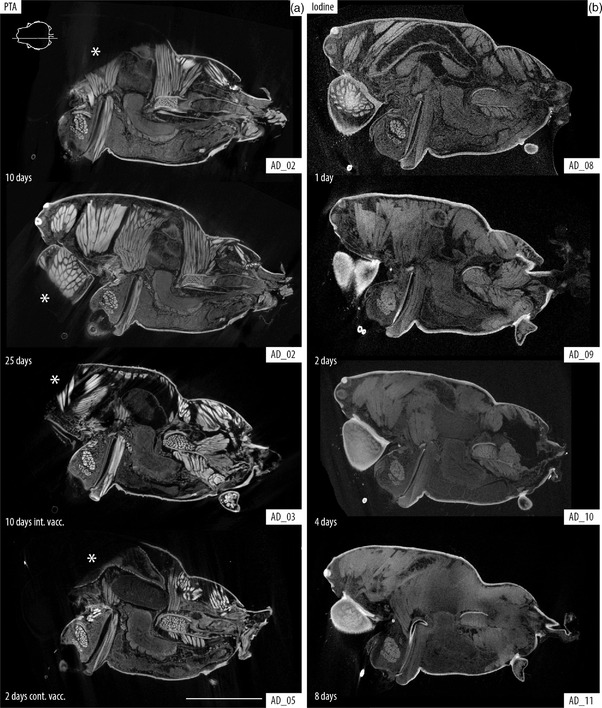
Comparison between PTA and iodine tissue staining. Sagittal slices of the cephalothorax of Araneus diadematus, location indicated with line drawing inset showing cephalothorax in dorsal view. (a) PTA staining after 10 and 25 days of passive diffusion compared with 10 days intermittent and 2 days continuous vacuum. (b) Iodine staining after 1, 2, 4, and 8 days of passive diffusion. Note that although PTA offers a better definition of the structures, there is evident understaining in the cephalic regions and chelicerae (*). Iodine offers better overall staining after 1 day (granted with less definition than PTA); longer times lead to overstaining of the tissues producing a blurry and dull image that lacks definition to discern internal organs. Scale bar: 2 mm
FIGURE 2.
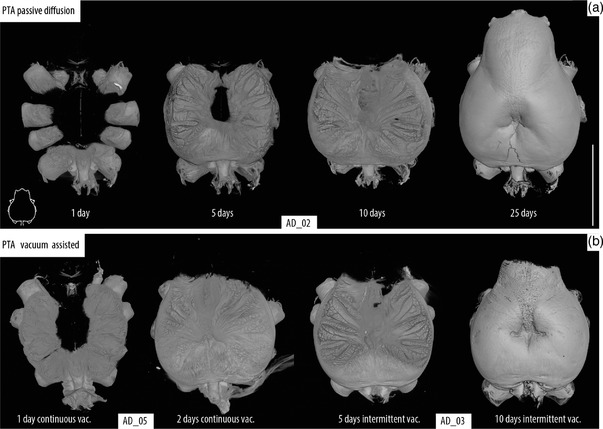
Time series of PTA staining. Volume rendering of the cephalothorax showing the penetration of the PTA solution, location indicated with line drawing inset showing cephalothorax in dorsal view. (a) Passive diffusion. (b) Vacuum assisted diffusion. Brain and CNS staining achieved only after 10 days passively while a similar stage was reached after only 2 days in continuous and 5 days of intermittent vacuum. Scale bar: 3 mm
FIGURE 3.
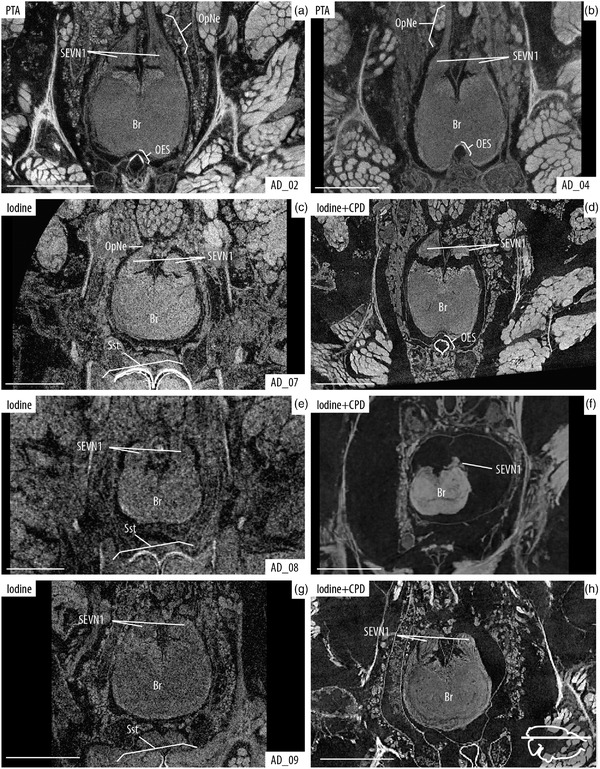
Visual comparison of the brain after PTA and iodine staining, and iodine + CPD. Axial slices of the brain, location indicated with line drawing inset showing cephalothorax in dorsal view. (a and b) PTA staining after 25 days of passive diffusion and 5 days continuous vacuum, respectively. (c–h) Comparisons of iodine‐stained samples before (left) and after CPD (right). Details of staining times and scanning parameters can be found in Table 1. Note the higher contrast and resolution of the structures in PTA (vs. iodine in EtOH), and the shrinkage of tissue after CPD. Br, brain; OpNe, optic nerves; OES, esophagus; SSt, sucking stomach and SEVN1, visual neuropiles 1 (lamina) of the secondary eye pathway. Scale bars: 0.5 mm
To mitigate the time‐consuming process of PTA staining, we tested the implementation of vacuum assistance during the staining process. The two approaches used here, intermittent and continuous vacuum, greatly reduced staining times by about 50 and 80%, respectively (Figure 2). A better overall quality of the internal tissue visualization can be observed in the passively diffused sample versus the vacuum assisted ones (Figure 1); yet, these differences are minimal when zooming in and focusing only on the brain (Figure 3(a)). Additional views of each parameter variation can be found in the Supplementary Materials. Contrast similar to that achieved using PTA was obtained when the iodine sample was dried; although the image contrast is better and noise is greatly reduced, these improvements come at the cost of remarkable impact on tissue shape, position, and volume (Table 2 and Figure 3). We found an important reduction on the brain volume throughout our six dried samples with an average loss of 45.92 ± 13.15%. The most impacted sample lost 70% of its brain volume, while the best preserved lost 35%. Both of our groups were normally distributed according to our Shapiro–Wilk test (SOG volume in EtOH p = .053; SOG volume after CPD p = .18). The Welch two sample t‐test showed that the there is a statistically significant difference (t (9.20) = 2.4073, p = .038) in the volume of this organ after the drying process (Figure 3). Similarly, our Mann–Whitney test showed that difference in the brain volume before and after CPD is significant (p = .041).
3.2. Neuroarchitecture identification and 3D reconstruction
Several organs and tissues could be observed and identified when specimens were stained with PTA (Figures 4 and 5), and detail in our high magnification scans was even sufficient to identify and make 3D reconstruction of several brain neuropiles and substructures (Figures 5 and 6). Also, components of the eyes (e.g., lens, retina, muscles, and optic nerves) could be easily observed (Figures 5(d) and 5(e)). Further comparisons of the detail obtained by the 25‐days passive diffusion versus the 2‐day continuous vacuum show that slight overstaining of the brain tissue does occur during the passive diffusion process with some structures being more defined and more clearly delineated in the vacuum assisted sample (Figures 6(a) and 6(b)).
FIGURE 4.
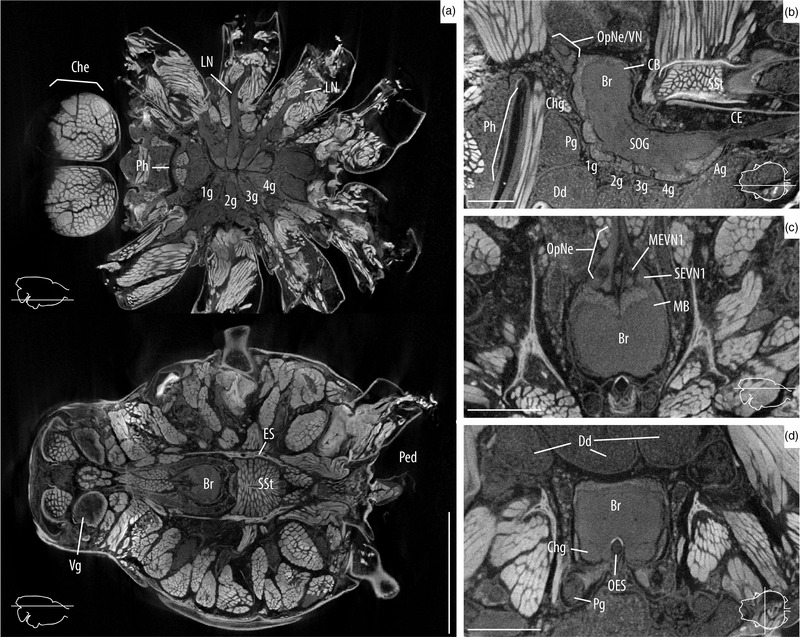
Overview of the internal anatomy of Araneus diadematus. Images shown here were taken from the specimen AD‐02 after 25 days in PTA. Slice locations are indicated with line drawing insets. (a) Axial slices of the whole cephalothorax showing the walking leg ganglia and nerves (top), and the brain and sucking stomach (bottom). (b) Sagittal slice of the CNS showing details of the supra‐ and subesophageal ganglia. (c) Axial slice showing the optic nerves, optic neuropiles, and the brain. (d) Coronal slice of the supraesophageal ganglia surrounded by digestive diverticula and muscles. 1g, walking leg I ganglion; 2g, walking leg II ganglion; 3g, walking leg III ganglion; 4g, walking leg IV ganglion; Ag, abdomen ganglion; Br, brain; CB, central body (arcuate body); CE, cauda equina; Che, chelicerae; Chg, cheliceral ganglion; Dd, digestive diverticula; ES, endosternite; LN, leg nerve; OES, esophagus; OpNe, optic nerves; Ped, pedicel; Pg, pedipalp ganglion; Ph, pharinx; SOG, subesophageal ganglion; SSt, sucking stomach; and Vg, venom gland. Scale bars: a = 2 mm; b–d = 0.5 mm.
FIGURE 5.
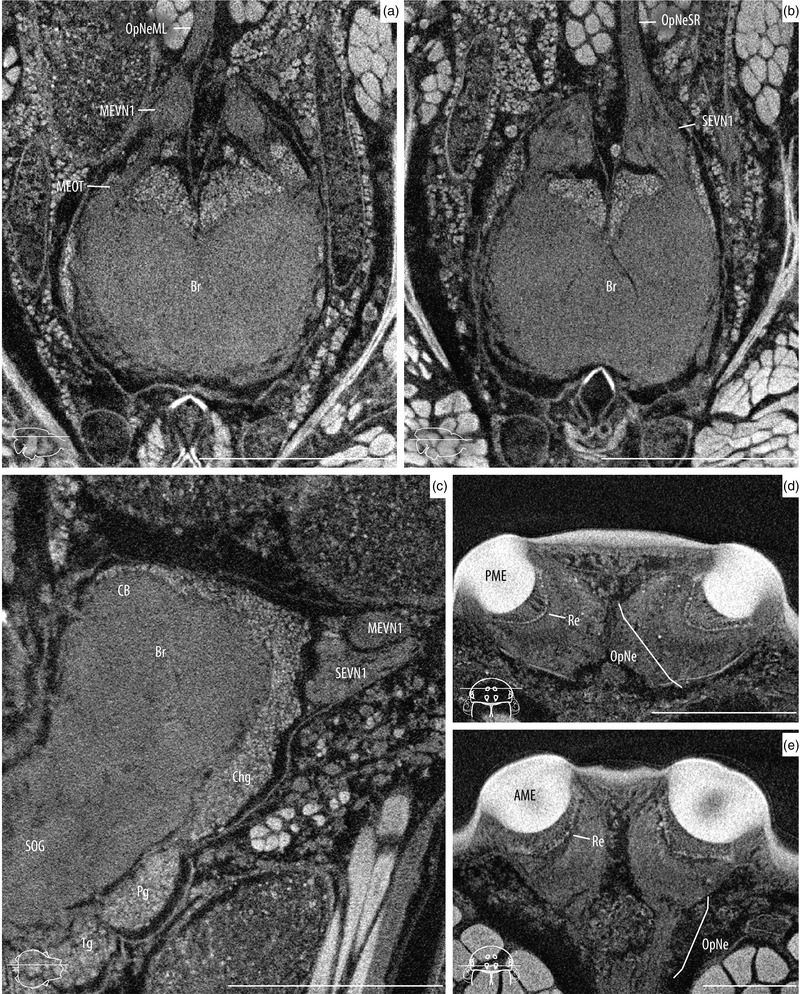
High magnification scans of the supraesophageal ganglia and the eyes. Images shown here were taken from the specimen AD‐02 after 25 days in PTA. Slice locations are indicated with line drawing insets. (a) Axial slice of the brain showing the main eye neuropiles and central body. (b) Same view, showing the secondary eye neuropiles and mushroom bodies. (c) Sagittal slice showing the brain, optic neuropiles, cheliceral ganglia, and anterior portion of the SOG. (d) Axial slice of the posterior median eyes showing the retina and optic nerve. (e) Same for anterior median eyes. 1g, walking leg I ganglion; AME, anterior median eyes; Br, brain; CB, central body (arcuate body); Chg, cheliceral ganglion; MEVN1, visual neuropiles 1 (lamina) of the main eye pathway; MEOT, main optic track; OES, esophagus; OpNe, optic nerves; OpNeML, optic nerves of the left main eye; OpNeSR, optic nerves of the right secondary eyes; Pg, pedipalp ganglion; PME, posterior median eyes; Re, retina; SEVN1, visual neuropiles 1 (lamina) of the secondary eye pathway; and SOG, subesophageal ganglion. Scale bars: a–c = 0.5 mm; d, e = 0.2 mm.
FIGURE 6.
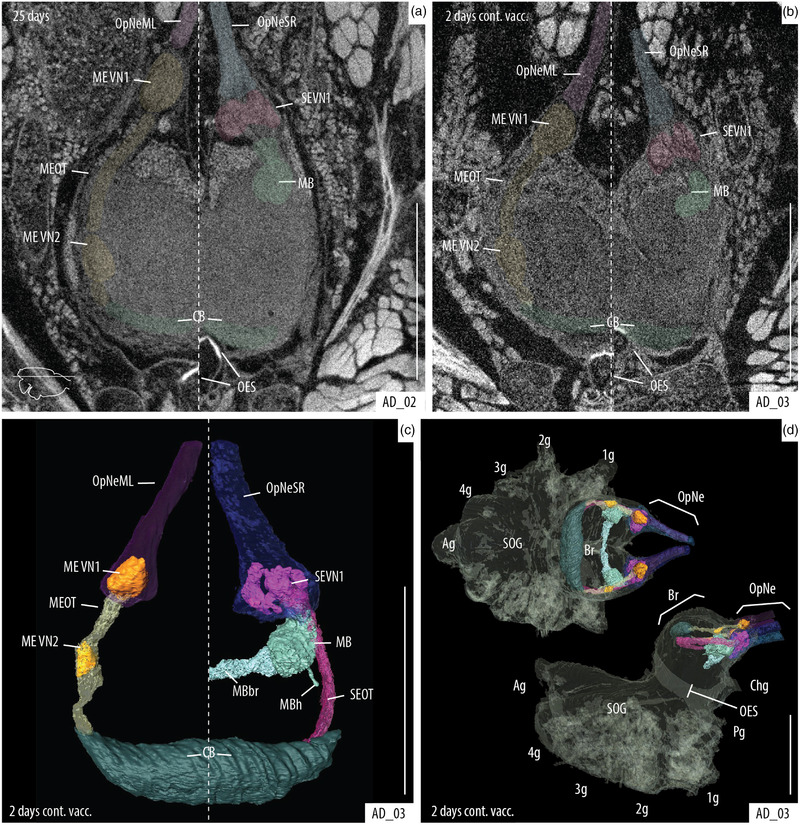
Identification of structures of the main and secondary visual systems within the supraesophageal ganglia. Axial slices of the brain showing the visual ganglia tracks: left side of each picture shows the main eye track; right side shows the secondary eye track. (a) Passive diffusion. (b) Vacuum assisted diffusion. (c, d) 3D reconstruction of the visual pathways. (d) Reconstruction of the visual pathways and whole CNS; dorsal view (top) and lateral view (bottom). Passive diffusion of PTA and 2‐day continuous vacuum are better for the visualization of the optic neuropiles. 1g, walking leg I ganglion; 2g, walking leg II ganglion; 3g, walking leg III ganglion; 4g, walking leg IV ganglion; Ag, abdomen ganglion; Br, brain; CB, central body (arcuate body); Chg, cheliceral ganglion; MB, mushroom body (corpora pedunculata); MBb, mushroom body bridge; MBh, mushroom body heft; MEVN1, visual neuropiles 1 (lamina) of the main eye pathway; MEVN2, visual neuropiles 2 (medullae) of the main eye pathway; MEOT, main optic track; OES, esophagus; OpNe, optic nerves; OpNeML, optic nerves of the left main eye; OpNeSR, optic nerves of the right secondary eyes; Pg, pedipalp ganglion; SEVN1, visual neuropiles 1 (lamina) of the secondary eye pathway; SEOT, secondary optic track; and SOG, subesophageal ganglion. Scale bars: 0.5 mm.
4. DISCUSSION
The usefulness of micro‐CT for the visualization of internal organs and tissues has been extensively tested and confirmed in arthropods (Lipke et al., 2015; Sombke et al., 2015; Castejón et al., 2018; Dirks‐Mulder et al., 2019; Pramanik et al., 2020; Faulwetter et al., 2012; Sakurai & Ikeda 2019; Swart et al., 2016; Lesciotto et al., 2020; Sombke et al., 2019; Dederichs et al., 2019; Rivera‐Quiroz et al., 2021; Steinhoff et al., 2017; Stafstrom et al., 2017; Steinhoff et al., 2018, 2020; Alba‐Alejandre et al., 2019; Alba‐Alejandre et al., 2018). Furthermore, its application for identifying and reconstructing peripheral and central nervous tissue and other systems in spiders has also been explored (Lipke et al., 2015; Dederichs et al., 2019; Rivera‐Quiroz et al., 2021; Steinhoff et al., 2017; Stafstrom et al., 2017; Steinhoff et al., 2018, 2020; Lin et al., 2021). Most of these arthropod studies have relied on the use of iodine as a contrast agent. The use of PTA has been examined in a wide array of organisms from plants to marine invertebrates and mammal tissue, producing results that match or surpass in quality and detail those achieved using iodine (Dirks‐Mulder et al., 2019; Pramanik et al., 2020; Faulwetter et al., 2012; Sakurai & Ikeda, 2019; Swart et al., 2016; Lesciotto et al., 2020). Nonetheless, this staining technique has not been commonly applied in studies of terrestrial arthropods such as spiders.
One of the reasons why PTA has barely been used in this group might be the slow tissue penetration time compared to iodine. Our tests of PTA under different times and conditions demonstrated that our two vacuum assisted approaches were extremely efficient in obtaining penetration similar to passively absorbed PTA in ca. 20–50% of the time (Figure 2). Still, this method takes more time than the commonly used iodine staining and some parts of the specimen remain understained even after several days (Figure 1). Although a direct comparison of the image quality might be prone to subjectivity from the observer, we consider that PTA offers considerably higher contrast and resolution of the different tissues and organs, facilitating easy observation and identification (Figures 4 and 5). Furthermore, this is achieved without drying, helping to preserve the specimen and reducing the introduction of artifacts created by tissue shrinkage during the drying process (Figure 2). The minimal impact on the tissue facilitates the generation of more accurate 3D reconstructions of the internal organs without spatial artifacts and volumetric distortion (Figure 6).
Our results show that the quality of the iodine samples visualization is considerably improved after CPD by comparison to the scans in EtOH (Figure 3). Still, the morphology and volumes of the soft tissue are severely affected. Our analyses of the supraesophageal ganglion found a mean volume reduction of 45% with the least impacted samples losing more than 30% of their volume (Table 2). Although we kept all settings constant, we did find a difference between the two rounds of CPD with sample AD_08 loosing 70% of its brain volume. This could be due to an error on the CPD calibration or during the process itself (e.g., a faster release of CO2 or an interruption on the cycles). Still both the t‐test and Mann–Whitney test showed a significant difference in the SOG volumes before and after CPD. The visualization and interpretation of the SOG morphology is also affected creating artifacts like the separation of the visual neuropiles from the rest of the brain (Figures 3(d) and 3(f)). Similar affectations of the brain morphology can be observed in literature (see Figures 2 and 4; Stafstrom et al., 2017). Furthermore, CPS is detrimental to the long‐term preservation of spider specimens archived in natural history collections. PTA‐stained samples scanned in EtOH offered similar results in image quality and tissue identifiability to iodine‐stained samples after CPD (compare Figures 3(a) and 3(h)). Notably, the PTA protocol described here allows specimens to remain in alcohol continuously avoiding the creation of shrinkage artifacts and aiding the proper preservation of the specimens. Furthermore, useful observations can be done without the need for a specific fixation or preservation of the specimens. This opens up the possibility of scanning old or rare material that may be difficult to find outside of museum collections. To properly study the morphological diversity of the nervous systems and the many interesting phenomena that might be related to brain architecture (e.g., eye reduction, sexual dimorphism, miniaturization, sociality, etc.), it is especially important to have appropriate and well considered collections of samples that may include taxa that are rarely collected or difficult to obtain. So far, only a few studies have dealt with the diversity of neuroarchitecture in spiders (Saint‐Remy, 1890; Long, 2016, 2021). We anticipate that this could be a character rich system with potential taxonomic and phylogenetic applications.
Our PTA‐based approach enhances the contrast between the samples tissue and the surrounding medium providing clearer visualizations than the commonly used iodine staining in ethanol. It also reduces negative impacts on specimen preservation by eliminating the use of a drying step by comparison with similar protocols in arthropods (e.g., Stafstrom et al., 2017; Alba‐Tercedor et al., 2021). This is an especially compelling argument for studying material sourced from natural history collections (Rivera‐Quiroz & Miller, 2021). Micro‐CT is not meant to replace other visualization methods like histological preparations (e.g., immunohistochemistry, bodian silver) but offers a rapid way to observe and compare internal anatomical characters of rare and non‐model taxa with minimal impact on the specimens. Micro‐CT will continue to provide valuable insights for the study of the evolution of nervous organs such as the optic nerves, brain, and the CNS (Harzsch 2006; Strausfeld & Andrew 2011; Strausfeld et al., 2020) along with countless other internal anatomical features (e.g. muscles, venom glands, silk glands, digestive and respiratory systems, among many others).
AUTHOR CONTRIBUTIONS
A. R. conceived the ideas and designed methodology; A. R. and J. M. collected the data and participated on the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
CONFLICT OF INTEREST
The authors declare that there are no potential sources of conflict of interest.
TRANSPARENT PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/cne.25343
Supporting information
Supporting information
ACKNOWLEDGMENTS
Thanks to Rob Langelaan and Dirk van der Marel for their help obtaining micro‐CT scans and their suggestions on the protocol. Thanks to Martin Rücklin for facilitating the use of Avizo software. Thanks to Davinia Arguedas for her help finding and collecting the A. diadematus specimens used in this work. Thanks to Vincent Merckx and Naturalis Understanding Evolution research group for the support to obtain the scans. Thanks to the editor and two anonymous reviewers for their useful comments and suggestions. Thanks to Naturalis Biodiversity Center for providing funding for the first author through a Martin & Temminck Fellowship.
Rivera‐Quiroz, F. A. , & Miller, J. A. (2022). Micro‐CT visualization of the CNS: Performance of different contrast‐enhancing techniques for documenting the spider brain. Journal of Comparative Neurology, 530, 2474–2485. 10.1002/cne.25343
DATA AVAILABILITY STATEMENT
All relevant data were made accessible in the present publication. The original raw scan files can be made available upon reasonable request.
REFERENCES
- Alba‐Alejandre, I. , Alba‐Tercedor, J. , & Hunter, W. B. (2020). Anatomical study of the female reproductive system and bacteriome of Diaphorina citri Kuwayama, (Insecta: Hemiptera, Liviidae) using micro‐computed tomography. Scientific Reports, 10(1), 1–14. Nature Publishing Group. 10.1038/s41598-020-64132-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba‐Alejandre, I. , Alba‐Tercedor, J. , & Vega, F. E. (2019). Anatomical study of the coffee berry borer (Hypothenemus hampei) using micro‐computed tomography. Scientific Reports, 9(1), 1–16. Nature Publishing Group. 10.1038/s41598-019-53537-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba‐Alejandre, I. , Hunter, W. B. , & Alba‐Tercedor, J. (2018). Micro‐CT study of male genitalia and reproductive system of the Asian citrus psyllid, Diaphorina citri Kuwayama, 1908 (Insecta: Hemiptera, Liviidae). PLoS One, 13(8), e0202234. Public Library of Science:e0202234. 10.1371/JOURNAL.PONE.0202234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba‐Tercedor, J. , Hunter, W. B. , & Alba‐Alejandre, I. (2021). Using micro‐computed tomography to reveal the anatomy of adult Diaphorina citri Kuwayama (Insecta: Hemiptera, Liviidae) and how it pierces and feeds within a citrus leaf. Scientific Reports, 11(1), 1–30. Nature Publishing Group. 10.1038/s41598-020-80404-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, K. S. , & Barth, F. G. (1984). Neuroanatomy of the central nervous system of the wandering spider, Cupiennius salei (Arachnida, Araneida). Zoomorphology, 104, 344–539. 10.1007/BF00312185 [DOI] [Google Scholar]
- Castejón, D. , Alba‐Tercedor, J. , Rotllant, G. , Ribes, E. , Durfort, M. , & Guerao, G. (2018). Micro‐computed tomography and histology to explore internal morphology in decapod larvae. Scientific Reports, 8(1), 14399. 10.1038/s41598-018-32709-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dederichs, T. M. , Müller, C. H. G. , Sentenská, L. , Lipke, E. , Uhl, G. , & Michalik, P. (2019). The innervation of the male copulatory organ of spiders (Araneae) ‐ A comparative analysis. Frontiers in Zoology, 16(39), 1–14. 10.1186/s12983-019-0337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks‐Mulder, A. , Ahmed, I. , uit het Broek, M. , Krol, L. , Menger, N. , Snier, J. , van Winzum, A. , de Wolf, A. , van't Wout, M. , Zeegers, J. J. , Butôt, R. , Heijungs, R. , van Heuven, B. J. , Kruizinga, J. , Langelaan, R. , Smets, E. F. , Star, W. , Bemer, M. , & Gravendeel, B. (2019). Morphological and molecular characterization of orchid fruit development. Frontiers in Plant Science, 10, 137. Frontiers Media S.A.: 10.3389/fpls.2019.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulwetter, S. , Dailianis, T. , Vasileiadou, A. , & Arvanitidis, C. (2012). Investigation of contrast enhancing techniques for the application of Micro‐CT in marine biodiversity studies. In SkyScan MicroCT User Meeting 2012 .
- Hanstrom, B. (1921). Uber die Histologie und vergleichende Anatomie der Sehganglien und Globuli der Araneen. Kunglica Svenska Vetenskapsakademiens Handligar, 61, 1–39. [Google Scholar]
- Hanstrom, B. (1923). Further notes on the central‐nervous system of arachnids: scorpions, phalangids, and trap‐door spiders. Journal of Comparative Neurology, 35(4), 249–274. [Google Scholar]
- Harzsch, S. (2006). Neurophylogeny: Architecture of the nervous system and a fresh view on arthropod phylogeny. Integrative and Comparative Biology, 46(2), 162–194. Integr Comp Biol. 10.1093/icb/icj011 [DOI] [PubMed] [Google Scholar]
- Hill, D. E. (2006). The structure of the central nervous system of jumping spiders of the genus Phidippus (Araneae: Salticidae). Oregon State University, 1–94. [Google Scholar]
- Keklikoglou, K. , Faulwetter, S. , Chatzinikolaou, E. , Wils, P. , Brecko, J. , Kvaček, J. , Metscher, B. , & Arvanitidis, C. (2019). Micro‐computed tomography for natural history specimens: a handbook of best practice protocols. European Journal of Taxonomy, 522, 1–55. 10.5852/ejt.2019.522 [DOI] [Google Scholar]
- Lesciotto, K. M. , Motch Perrine, S. M. , Kawasaki, M. , Stecko, T. , Ryan, T. M. , Kawasaki, K. , & Richtsmeier, J. T. (2020). Phosphotungstic acid‐enhanced microCT: Optimized protocols for embryonic and early postnatal mice. Developmental Dynamics, 249(4), 573–585. John Wiley and Sons Inc. 10.1002/dvdy.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. W. , Lopardo, L. , & Uhl, G. (2021). Diversification through gustatory courtship: an X‐ray micro‐computed tomography study on dwarf spiders. Frontiers in Zoology, 18(1), 1–33. BioMed Central: 10.1186/s12983-021-00435-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke, E. , Hammel, J. U. , & Michalik, P. (2015). First evidence of neurons in the male copulatory organ of a spider (Arachnida, Araneae). Biology Letters, 11, 20150465. 10.1098/rsbl.2015.0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S. M. (2016). Spider brain morphology & behavior. University of Massachusetts Amherst, 239. [Google Scholar]
- Long, S. M. (2021). Variations on a theme: Morphological variation in the secondary eye visual pathway across the order of Araneae. Journal of Comparative Neurology, 529(2), 259–280. 10.1002/cne.24945 [DOI] [PubMed] [Google Scholar]
- Metscher, B. D. (2009). Micro CT for comparative morphology: Simple staining methods allow high‐contrast 3D imaging of diverse non‐mineralized animal tissues. BMC Physiology, 9(1), 11. 10.1186/1472-6793-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik, D. , Dorst, N. , Meesters, N. , Spaans, M. , Smets, E. , Welten, M. , & Gravendeel, B. (2020). Evolution and development of three highly specialized floral structures of bee‐pollinated Phalaenopsis species. EvoDevo, 11(1), 16. BioMed Central: 10.1186/s13227-020-00160-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera‐Quiroz, F. A. , & Miller, J. A. (2021). Old brains in alcohol: The usability of legacy collection material to study the spider neuroarchitecture. Diversity, 13(11), 601. 10.3390/d13110601 [DOI] [Google Scholar]
- Rivera‐Quiroz, F. A. , Petcharad, B. , & Miller, J. A. (2021). First records and three new species of the family Symphytognathidae (Arachnida, Araneae) from Thailand, and the circumscription of the genus Crassignatha Wunderlich, 1995. ZooKeys, 2021(1012), 21–53. 10.3897/zookeys.1012.57047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint‐Remy, G. (1890). Contribution a l’étude du cerveau chez les arthropodes trachéates. Faculté des Sciences de Paris, 274. [Google Scholar]
- Sakurai, Y. , & Ikeda, Y. (2019). Development of a contrast‐enhanced micro computed tomography protocol for the oval squid (Sepioteuthis lessoniana) brain. Microscopy Research and Technique, 82(11), 1941–1952. Wiley‐Liss Inc.:jemt.23363. 10.1002/jemt.23363 [DOI] [PubMed] [Google Scholar]
- Schmid, A. , & Becherer, C. (1999). Distribution of histamine in the CNS of different spiders. Microscopy Research and Technique, 44, 81–93. 10.1002/(SICI)1097-0029(19990115/01)44:2/3<81::AID‐JEMT3>3.0.CO;2‐O [DOI] [PubMed] [Google Scholar]
- Sombke, A. , Klann, A. E. , Lipke, E. , & Wolf, H. (2019). Primary processing neuropils associated with the malleoli of camel spiders (Arachnida, Solifugae): A re‐evaluation of axonal pathways. Zoological Letters, 5(1), 26. 10.1186/s40851-019-0137-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sombke, A. , Lipke, E. , Michalik, P. , Uhl, G. , & Harzsch, S. (2015). Potential and limitations of X‐Ray micro‐computed tomography in arthropod neuroanatomy: A methodological and comparative survey. Journal of Comparative Neurology, 523(8), 1281–1295. 10.1002/cne.23741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom, J. A. , Michalik, P. , & Hebets, E. A. (2017). Sensory system plasticity in a visually specialized, nocturnal spider. Scientific Reports, 7, 46627. 10.1038/srep46627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff, P. O. M. , Liedtke, J. , Sombke, A. , Schneider, J. M. , & Uhl, G. (2018). Early environmental conditions affect the volume of higher‐order brain centers in a jumping spider. Journal of Zoology, 304(3), 182–192. 10.1111/jzo.12512 [DOI] [Google Scholar]
- Steinhoff, P. O. M. , Sombke, A. , Liedtke, J. , Schneider, J. M. , Harzsch, S. , & Uhl, G. (2017). The synganglion of the jumping spider Marpissa muscosa (Arachnida: Salticidae): Insights from histology, immunohistochemistry and microCT analysis. Arthropod Structure and Development, 46(2), 156–170. 10.1016/j.asd.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Steinhoff, P. O. M. , Uhl, G. , Harzsch, S. , & Sombke, A. (2020). Visual pathways in the brain of the jumping spider Marpissa muscosa . Journal of Comparative Neurology, 528(11), 1883–1902. 10.1002/cne.2486 [DOI] [PubMed] [Google Scholar]
- Strausfeld, N. J. , & Andrew, D. R. (2011). A new view of insect‐crustacean relationships I. Inferences from neural cladistics and comparative neuroanatomy. Arthropod Structure and Development, 40(3), 276–288. Arthropod Struct Dev. 10.1016/j.asd.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Strausfeld, N. J. , Barth, F. G. , Weltzien, P. , & Barth, F. G. (1993). Two visual systems in one brain: Neuropils serving the principal eyes of the spider Cupiennius salei . Journal of Comparative Neurology, 328(1), 63–75. 10.1002/cne.903280105 [DOI] [PubMed] [Google Scholar]
- Strausfeld, N. J. , Wolff, G. H. , & Sayre, M. E. (2020). Mushroom body evolution demonstrates homology and divergence across Pancrustacea. ELife, 9, e52411. eLife Sciences Publications Ltd. 10.7554/eLife.52411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart, P. , Wicklein, M. , Sykes, D. , Ahmed, F. , & Krapp, H. G. (2016). A quantitative comparison of micro‐CT preparations in Dipteran flies. Scientific Reports, 6(1), 1–12. Nature Publishing Group. 10.1038/srep39380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubick, D. , Paquin, P. , Cushing, P. E. , & Roth, V. (2017). Spiders of North America an identification manual. In Ubick D., Paquin P., Cushing P. E., & Roth V. (Eds.), American Arachnological Society (2nd ed.) Keene, New Hampshire, American Arachnological Society, 425. [Google Scholar]
- Wickham, H. (2016). Ggplot2 elegant graphics for data analysis. Springer. Use R! Cham: Springer International Publishing. 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Wickham, H. , François, R. , Henry, L. , & Müller, K. (2021). dplyr: A Grammar of Data Manipulation. 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
All relevant data were made accessible in the present publication. The original raw scan files can be made available upon reasonable request.