Abstract
Aims
The role of a Acinetobacter johnsonii strain, isolated from a soil sample, in the biotransformation of bile acids (BAs) was already described but the enzymes responsible for these transformations were only partially purified and molecularly characterized.
Methods and Results
This study describes the use of hybrid de novo assemblies, that combine long‐read Oxford Nanopore and short‐read Illumina sequencing strategies, to reconstruct the entire genome of A. johnsonii ICE_NC strain and to identify the coding region for a 12α‐hydroxysteroid dehydrogenase (12α‐HSDH), involved in BAs metabolism. The de novo assembly of the A. johnsonii ICE_NC genome was generated using Canu and Unicycler, both strategies yielded a circular chromosome of about 3.6 Mb and one 117 kb long plasmid. Gene annotation was performed on the final assemblies and the gene for 12α‐HSDH was detected on the plasmid.
Conclusions
Our findings illustrate the added value of long read sequencing in addressing the challenges of whole genome characterization and plasmid reconstruction in bacteria. These approaches also allowed the identification of the A. johnsonii ICE_NC gene for the 12α‐HSDH enzyme, whose activity was confirmed at the biochemical level.
Significance and impact or the study
At present, this is the first report on the characterization of a 12α‐HSDH gene in an A. johnsonii strain able to biotransform cholic acid into ursodeoxycholic acid, a promising therapeutic agent for several diseases.
Keywords: 12α‐hydroxysteroid dehydrogenase, Acinetobacter johnsonii, bile acid 12α‐dehydroxylation, bioinformatic analysis, cholic acid, genome assembly, hybrid sequencing, plasmid
INTRODUCTION
Bile acids (BAs), their conjugates and salts are natural products and fundamental constituents of bile (Kritchevsky, 1971). Among them, chenodeoxycholic (CDCA) and 7‐OH epimer ursodeoxycholic acid (UDCA) have important pharmaceutical applications related to their ability to solubilize cholesterol gallstones (Crosignani et al., 1996; Salen et al., 1980). In terms of pharmacology, UDCA is considered better than CDCA in the treatment against biliary calculi, since it possesses high efficacy in the transformation and excretion of cholesterol and total absence of side effects (Roda et al., 1982). UDCA, combined with taurine to form tauroursodeoxycholic acid (TUDCA), is effective not only in treating hepatobiliary diseases but also Alzheimer's and Parkinson's disease (Duan et al., 2002) and it appears to be a promising therapeutic agent for inflammatory bowel disease (Bossche et al., 2017).
UDCA can be prepared from cholic (CA), CDCA or lithocholic acids as starting material and several chemical, chemoenzymatic and enzymatic routes have been proposed for its production (Tonin & Arends, 2018). The available chemical routes to obtain UDCA yield about 30% of final product and they require several protection and deprotection steps based on toxic and dangerous reagents, leading to the production of a series of waste products (Carrea et al., 1992; Eggert et al., 2014). For these reasons, numerous studies have described the development of less toxic microbial transformations or chemoenzymatic procedures for the synthesis of UDCA (Bortolini et al., 1997; Prabha & Ohri, 2006; Sutherland et al., 1982). These promising approaches led by several research groups allowed the development of biotransformations based on nonpathogenic, easy‐to‐manage, micro‐organisms and their enzymes (Tonin & Arends, 2018).
Depending on the structure of the starting material, UDCA synthesis needs different combinations of regio‐ and stereo‐selective enzymes that catalyse oxidation and reduction reactions. These enzymes belong to the group of hydroxysteroid dehydrogenases (HSDHs), a subgroup of NAD+‐ or NADP+‐dependent oxidoreductases, among which 12α‐HSDHs and 7α‐HSDHs seem very efficient in the quantitative oxidation of the C12‐OH function of cholic acid and of the C7‐OH function of chenodeoxycholic acid respectively (Eggert et al., 2014).
There are several bacterial HSDHs able to carry out the desired reactions (Ferrandi et al., 2020), for example 7α‐HSDHs were isolated from both aerobic and anaerobic bacteria and their use for the selective oxidation of BA, their salts or derivatives was patented (Gupta et al., 2009; Monti et al., 2012). In addition to these reported biotransformations, many additional 7α‐HSDHs have been discovered and reported over the past years from strains of Clostridium (Baron et al., 1991; Coleman et al., 1994; Franklund et al., 1990), Bacteroides (Bennett et al., 2003), Escherichia coli (Yoshimoto et al., 1991), Xanthomonas (Medici et al., 2002; Pedrini et al., 2006) or Pseudomonas sp. (Ueda et al., 2004).
Regarding 12α‐HSDH, a few enzymes have been found among strains of the genus Clostridium or Eggerthella. In particular, a NADP+‐dependent 12α‐HSDH has been detected in Clostridium leptum (Harris & Hylemon, 1978) and in Clostridium group P strain C 48–50 (Braun et al., 1991; Braun et al., 2011; Macdonald et al., 1979), whereas NAD+‐dependent 12α‐HSDH activity has been reported in Eubacterium lentum (now Eggerthella lenta) (Macdonald et al., 1977) and Clostridium perfringens (Macdonald et al., 1976). More recently, Curvularia lunata VKM F‐644 was shown to exhibit 12α‐hydroxysteroid dehydrogenase activity and formed 12‐keto‐lithocholic acid (12‐keto‐LCA) from deoxycholic acid (DCA) (Kollerov et al., 2016). Furthermore, an efficient approach has been recently reported for the oxidation of the 12α‐OH of bile acid exploiting the new NAD(H)‐dependent 12α‐HSDH from Rhodococcus ruber (Rr12α‐HSDH) (Shi et al., 2019).
Unlike 7α‐HSDHs genes, which are better known (Tang et al., 2019), genes encoding 12α‐HSDHs have not been clearly identified yet. The earliest described gene encoding 12α‐HSDH was based on the amino acid sequence deduced from the enzyme isolated from Clostridium sp. strain ATCC 29733 (Aigner et al., 2011; Macdonald et al., 1979). Subsequently, a bioinformatic analysis based on the Clostridium gene identified a putative 12α‐HSDH coding gene in C. hylemonae (accession no. WP_006441568.1) (Kisiela et al., 2012). More recently, a new phylogenetic analysis updates and extends previous work on 12α‐HSDH enzymes in human gut bacteria by introducing newly 12α‐HSDHs reported on members of the gut microbiota, obtained through culturomics approaches, such as Eggerthella strains, Firmicutes, Coriobacteriaceae family members of the Actinobacteria and gut Archaea, thus suggesting the potential role of the bile acid metabolism in several gut microbes (Doden et al., 2018).
Recently, a comparative genomic analysis of A. johnsonii isolated from clinical and environmental sources showed that A. johnsonii has great adaptability to different environments and that clinical‐derived isolates accumulate more genes associated with translational modification, whereas environmental‐derived isolates possess more genes related to substance degradation (Jia et al., 2021).
Despite all these studies, based on coding sequences (CDS) predicted by protein sequences, to date, 12α‐HSDH coding genes have yet to be completely described and no information is available on 12α‐HSDH gene(s) in Acinetobacter. Giovannini et al. (2008) reported the partial purification of 7α and 12α‐hydroxysteroid dehydrogenases from Acinetobacter calcoaceticus lwoffii (now A. johnsonii), a strain isolated from a soil sample of a chemical plant working with bile (Industria Chimica Emiliana, ICE, n.d.). The authors described a simple and very efficient application of these partially purified enzymes in the synthesis of UDCA. However, the genes for these enzymes were never isolated and cloned from this strain.
In this study, the whole‐genome sequence of the A. johnsonii ICE_NC strain was determined by combining Illumina and Oxford Nanopore technologies. Our approach resulted in the assembly of one chromosome plus one plasmid and allowed the identification of the gene for the 12α‐HSDH enzyme on the plasmid. This finding enabled us to clone the gene and characterize the 12α‐HSDH recombinant enzyme from this A. johnsonii ICE_NC strain.
MATERIALS AND METHODS
DNA isolation and preparation
The A. johnsonii strain (as described in Giovannini et al., 2008) was grown at 28°C in LB medium for 18 h. Whole genomic DNA was extracted from harvested cells using the GenElute™ Bacterial Genomic DNA Kit (Sigma), in accordance with the manufacturer's instructions. Purified DNA was quantified by spectrophotometer (Shimadzu BioSpec‐nano) and by Qubit (Life Technologies) fluorometric analyses.
Library preparation and sequencing
Nanopore MinION
Whole genome sequencing was performed on a MinION device loaded with a MinION Flow Cell (R9.4.1). One microgram of genomic DNA was first subjected to the NEBNext® Ultra™ II End Repair/dA‐Tailing module (New England Biolabs) for end repair and dA‐Tailing, in accordance with the manufacturer's instructions. Upon bead purification (AMPure XP beads, Beckman Coulter), DNA was ligated to adapters using Blunt/TA ligase (New England Biolabs), size selected with the AMPure XP beads and finally loaded on the MinION cell.
Illumina NextSeq
The library was generated from 20 ng of genomic DNA, in accordance with the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina protocol (New England Biolabs). Prior to library preparation, DNA was fragmented using NEBNext® dsDNA Fragmentase for 30 min. NEBNext® Multiplex Oligos for Illumina® (Dual Index Primers set 1) were used to produce the library and label it with specific molecular barcodes. Then, AMPure XP beads (Beckman Coulter) were used for library purification. Finally, the library was quantified using the High Sensitivity DNA Kit (Agilent Technologies) on the Bioanalyzer instrument (Agilent). Sequencing was performed with an Illumina NextSeq 500 sequencer with 2 × 150‐bp read, using NextSeq® 500/550 Mid Output Kit v2.
Read preparation
Nanopack's tools (De Coster et al., 2018) were used to manage the MinION raw reads: (i) NanoPlot (v.1.21.0) to assess read quality, (ii) NanoLyze (v.1.1.0) to remove reads mapping to the Lambda phage genome and (iii) NanoFilt (v.2.2.0) to filter fastq file based on a minimum quality cut‐off (>10) and minimum length (>2000 bp and >20,000 bp). From the reads obtained with the MinION sequencer we generated two different datasets named subset_2k and subset_20k retaining reads >2000 bp and >20,000 bp respectively.
Estimation of the Nanopore MinION error rate was performed aligning the 2D reads of Lambda phage spike‐in DNA (control DNA, CS, ONT) against the Lambda reference genome by using minimap2 (v.2.16‐r922; Li, 2018); results were obtained by parsing BAM files with SAMtools (Li et al., 2009) and using R scripts described in Fuselli et al. (2018).
Illumina read quality was assessed with FastQC (v.0.10.1 available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc). AdapterRemoval (v.2.3.0; Schubert et al., 2016) was used to remove Illumina adapters.
Genome assembly
Two length filters were applied to MinION data in order to exclude reads shorter than 2 and 20 Kbp respectively. The two resulting datasets were used to produce the de novo assemblies with two approaches that combine ONT data with Illumina data: Canu (v1.8; Koren et al., 2017), that generated de novo assemblies made up by long reads polished with short reads using Pilon (v. 1.23; Walker et al., 2014), followed by an Unicycler hybrid pipeline (v.0.4.8‐beta; Wick et al., 2017), that generates the assemblies from short reads. The long reads were then used to deconvolute the assembly graphs, decreasing the number of contigs.
Due to the circularity of the chromosome, we aligned each assembly against itself using BLASTN (NCBI/BLASTN at https://blast.ncbi.nlm.nih.gov/Blast.cgi; Zhang et al., 2000) to identify and remove potential duplicated regions located close to the ends of the linear scaffolds. Nanopore reads were aligned to each assembly and the coverage profile along the sequence was exploited to identify further residual misassemblies using Tigmint (Jackman et al., 2018).
The assemblies quality assessment was performed using assembly‐stats (https://github.com/sanger‐pathogens/assembly‐stats), that evaluates genome assemblies by computing various metrics, BUSCO (v. 5.0.0, Manni et al., 2021) to estimate the completeness and Merqury (v. 1.3, Rhie et al., 2020) to assess global completeness and assembly consensus quality value (QV).
Characterization of the complete genome and plasmid sequences
The final complete circular chromosome and the plasmid genome sequences of A. johnsonii ICE_NC obtained through Canu or Unicycler assembler were analysed using Prokka (Seemann, 2014). We compared the obtained gene content with five published A. johnsonii genomes (IC001 GCA_003952785.1, LXL_C1 GCA_003335165.1, M19 GCA_004337595.1, AJ14 GCA_021496325.1, DJ‐RED GCA_021432745.1) processed with the same gene annotation pipeline.
Additionally, a refined BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis was performed to identify the different contigs and the presence of sequences ascribable to the 12α‐hydroxysteroid dehydrogenase.
Presence of prophages inserted in the chromosome was detected with PHASTER (https://phaster.ca/) (Arndt et al., 2016), while CRISPR arrays and CRISPR‐associated genes (cas) were searched with CRISPRcasFinder (https://crisprcas.i2bc.paris‐saclay.fr/) (Couvin et al., 2018).
VFanalyzer (http://www.mgc.ac.cn/cgi‐bin/VFs/v5/main.cgi) (Liu et al., 2019), with the Virulence Factors Database, was used to find virulence factors across the chromosome and a BLASTP (version 2.10.0+; Altschul et al., 1997) analysis against the full Virulence Factors Database was performed to find putative virulence factors in the plasmid; eventually a ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) (Bortolaia et al., 2020; Camacho et al., 2009; Zankari et al., 2020) analysis was performed to search for potential antibiotic resistance genes.
Cloning and expression of the recombinant 12α‐HSDH
The coding region showing homology to the Clostridium 12α‐HSDH gene (Clostridium sp. ATCC 29733, accession nr ERJ00208) was cloned into the expression vector pLATE51 by means of the aLICator Ligation Independent Cloning and Expression Kit (Thermo Scientific). The following primers, designed to introduce the coding fragment into pLATE51 plasmid, forward 5′‐ GGTGATGATGATGACAAGAAAAATTTAAAAGACCAT ‐3′ and reverse 5′‐ GGAGATGGGAAGTCATTATCTTAATTTAATATCACC ‐3′, were used to amplify genomic Acinetobacter DNA using Herculase II Fusion DNA Polymerase (Agilent) in a touchdown thermal profile: predenaturation at 95°C for 3 mins, followed by 5 cycles at 95°C for 30 s, 65–55°C for 30 s (minus 2°C/cycle), 68°C for 2 mins, 5 cycles at 95°C for 30 s, 58–68°C for 20 s (plus 2°C/cycle), 68°C for 2 mins, 25 cycles at 95°C for 30 s, 68°C for 2 mins and a final extension at 68°C for 4 mins. The PCR fragment was gel purified by the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) and inserted into pLATE51 vector in accordance with the manufacturer's instructions. Recombinant plasmid clones, selected on LB broth agar plates supplemented with 100 μg/ml ampicillin, were then extracted by GenElute™ Plasmid Miniprep Kit (Sigma‐Aldrich) and verified by DNA sequencing (BMR Genomics, Italy). The pLATE51 vector, containing the recombinant 12α‐HSDH, once transferred to E. coli SHuffle cells (New England BioLabs), codes for a putative 12α‐HSDH polypeptide carrying an N‐terminal His‐tag for the specific purification by affinity chromatography.
E. coli SHuffle cells transformed with the 12α‐HSDH‐pLATE51 plasmid were grown in LB‐broth containing 100 μg/ml of ampicillin. An overnight culture was used to start a fresh culture (dilution 1:100) incubated at 30°C (180 rpm) for 4–6 hours until OD 0.6. The expression of the 12α‐HSDH was induced with IPTG at 0.4 mM final concentration for 16–21 h at 30°C.
For the lysis, pelleted cells were resuspended in 50 mM phosphate buffer pH 7.0 and treated at high pressure (1280 bar) by means of a French press (ThermoFisher) The extract was clarified by centrifugation at 13,000×g for 20 min at 4°C.
Purification of the His‐tagged recombinant enzyme was performed by affinity chromatography using His GraviTrap (GE Healthcare) columns in accordance with the manufacturer's instructions. After elution, the imidazole was removed by ultracentrifugation (Amicon), the buffer exchanged to 50 mM phosphate buffer pH 7 and the resulting enzyme used for assays. The purified protein mass was measured spectrophotometrically at 280 nm.
Enzyme assay
Enzymatic activity assay was performed using 1 ml of 50 mM selected buffer solution, 0.01 ml of 0.025 mM sodium cholate solution, 0.01 ml of 0.025 mM NAD+ solution and 0.1 ml (containing between 120 and 150 μg) of purified 12α‐HSDH. The absorbance was measured at 340 nm for 1 min in order to evaluate the production of NADH. The interval between readings was 5 seconds, for a total of 12 readings per sample. One international unit (U) of enzyme is defined as the amount required to produce 1 μmol of NADH per minute.
Evaluation of pH and temperature optimum
The study of the influence of pH on enzymatic activity was carried out at 25°C varying the pH in the range 4.0–10.0 (with 0.5 increases). Three types of 50 mM buffer solutions were used: acetate buffer (buffer range pH 4.0–5.5), phosphate buffer (pH range 6.0–8.0) and diethanolamine buffer (pH range 8.5–10.0).
The reaction was started by adding 0.5 U of 12α‐HSDH to 1 ml of the appropriate buffer at different pH from 4.0 to 10.0 containing 0.025 mM NAD+ and 0.025 mM sodium cholate and incubated for 1 min at 25°C. At the end of the incubation time, the activity was analysed as described above. The effect of the temperature was determined by incubating 0.5 U of 12α‐HSDH in 50 mM phosphate buffer pH 8.0 at temperatures in the range 5–50°C (5°C increments) for 1 min. After the incubation time, the enzyme activity was determined as described above.
Oxidation of sodium cholate with recombinant 12α‐HSDH
In 50 mM phosphate buffer solution (100 ml at pH 8) containing 0.2 mM NAD+, 60 mM sodium pyruvate, 1 mM dithiothreitol (DTT), 0.1 mM EDTA and 12 mM sodium cholate, 12α‐HSDH (2.5 U) was added. After the absorbance of the solution (at 340 nm) reached 2.0, lactate dehydrogenase (30 U) was added and the reaction was kept at 25°C overnight. The reaction mixture was acidified to pH 2 with 20% HCl, extracted with ethyl acetate (3 × 150 ml) and the combined organic layers, dried over anhydrous Na2SO4, were evaporated under reduced pressure. The crude reaction was purified on a chromatographic column (silica gel, ethyl acetate/cyclohexane/acetic acid 70/30/2 as eluent).
The product obtained was analysed by 1H‐NMR and the spectrum was compared with that reported in literature. The biotransformation yield was expressed as a percentage of the ratio between the moles of the product obtained compared to the moles of the substrate added in the reaction.
Compound data
12‐keto cholic acid (3α,7α‐dihydroxy‐12‐keto‐5‐cholan‐24‐oic acid): 1H NMR (CD3OD): (selected data) 𝛿 0.90 (d, J = 6.3 Hz, 3H, H‐21); 1.10 (s, 3H, H‐18); 1.13 (s, 3H, H‐19); 2.62 (dd, J = 13.3, 1H, H‐11); 3.42 (br, 1H, H‐3); 3.94 (brs, 1H, H‐7).
RESULTS
Sequencing
Results of whole‐genome sequencing of A. johnsonii ICE_NC from Oxford Nanopore MinION and Illumina Nextesq 500 platforms are summarized in Table S1. The MinION device generated 12.5 Gbp organized in 2,737,448 reads, with median read length of 2191 bp (longest read was 198,250 bp long) and median read quality of 11.3.
Illumina sequencing resulted in 1.3 Gbp of throughput, consisting of 9,211,819 paired‐end reads with median length of 141 bp and median read quality of 33. The median error rate of the Nanopore sequencing (Figure S1) computed using Lambda DNA, was 2.3% for single nucleotide variations (SNV), 1.2% for insertion (INS) and 2.1% for deletions (DEL).
These sequenced data have been submitted to the NCBI databases under Bioproject accession number: PRJNA791777.
Genome assembly
Since plasmids have a wide range of lengths, from roughly 1000 DNA base pairs to hundreds of thousands of base pairs, we decided to filter long reads applying two read‐length cutoffs: (i) the subset ‘2K’ (including reads longer than 2000 bp) was generated to avoid loss of plasmid material; (ii) the subset ‘20K’ (removing reads shorter than 20,000 bp) was produced to evaluate the performance of very long reads in the whole chromosome assembly. Canu and Unicycler‐hybrid were both applied on these subsets to generate different assemblies.
The assembly of the Oxford Nanopore's MinION reads with Canu 1.8 (Table 1) gave almost equal outputs for the two subsets, in term of length of first and second longest contig, but the total number of contigs was quite different: the assembly of subset ‘2K’ originated a total of 13 contigs, while the subset ‘20K’ produced only three contigs (Table 1). For both assemblies, the longest contig, a sequence of about 3,583,300 bp (slightly different for the two subsets, Table 1), corresponds to the genome of A. johnsoni ICE_NC; the second contig, of about 207,000 bp, represents the plasmid sequence. For both assemblies, a BLASTN analysis of the remaining smaller contigs against the reference chromosome (GCA_003952785.1) revealed that they were part of the main chromosomal contig.
TABLE 1.
Assemblies features of a. johnsonii ICE_NC
| Sequencing platforms | OxfordNanopore‐MinION | Illumina NextSeq 500 + OxfordNanopore‐MinION | ||
|---|---|---|---|---|
| Assembly method | Canu v. 1.8 + Pilon v.1.23 | Unicycler v.0.4.8‐beta | ||
| Subset | 2K | 20K | 2K | 20K |
| Finishing quality | Scaffold | Scaffold | Closed complete genome + plasmid | Closed complete genome + plasmid |
| Number of contigs | 13 | 3 | 2 | 2 |
| Number of N's | 0 | 0 | 0 | 0 |
| Total length bp | 4,121,247 | 3,974,922 | 3,701,113 | 3,701,113 |
| GC% | 41.17 | 41.21 | 41.24 | 41.24 |
| N50 | 3,674,325 | 3,674,351 | 3,583,655 | 3,583,655 |
| Complete BUSCOs | 98.3% | 98.3% | 98.6% | 98.6% |
| Merqury | 96.7% | 96.7% | 68% | 68% |
The de novo Unicycler‐hybrid assembly of the Illumina sequence reads plus the base‐called Oxford Nanopore reads resulted in a complete circular chromosome of 3,583,655 bp plus a plasmid sequence of 117,458 bp identical for both subsets analysed (Table 1).
Analysis performed with QUAST confirmed that all assemblies did not contain any gaps (i.e. no N's, Table 1). Furthermore, BUSCO and Merqury estimate over the Canu assemblies provided a value of gene completeness of 98.3% and a global completeness equal to 96.7%, respectively. The Unicycler assemblies were characterized by a slightly higher completeness, having a comparable BUSCO value (98.6%) but notably decreased global completeness estimated by Merqury (68%). Canu's assembly had a QV of 41.6, while the value for the Unicycler assembly was 68.
These assembly data have been submitted to the NCBI databases under accession number: CP090416–CP090417 (chromosome and plasmid respectively).
Characterization of the complete genome and plasmid sequences
The final complete chromosome sequences of A. johnsonii ICE_NC, obtained through Canu or Unicycler assemblies, showed a genome length in the range of variation of other A. johnsonii assemblies (Table 2). For plasmid assembly, a BLASTN alignment between the two 20K subsets obtained with Canu and Unicycler showed that the Canu contig of 207 Kb was actually a chimaera produced by the merging of two sequences highly similar to the contig of 117 Kb obtained from Unicycler assembly. Thus, we annotated and characterized only the 117 Kbp plasmid sequence obtained from the 20K Unicycler subset.
TABLE 2.
Annotation features of a. johnsonii ICE_NC genome using Prokka v.1.13.7
| Assembly | Canu20k chromosome | Unicycler20k chromosome | Unicycler20k plasmid | IC001 a | LXL_C1 a | M19 a | AJ14 a | DJ_RED a |
|---|---|---|---|---|---|---|---|---|
| Bases | 3,583,342 | 3,583,655 | 117,458 | 3,610,322 | 3,398,706 | 3,749,210 | 3,586,280 | 3,360,823 |
| ORFs predicted | 3395 | 3418 | 125 | 3502 | 3243 | 3649 | 3357 | 3173 |
| ORFs code for known protein | 2585 | 2582 | 90 | 2671 | 2475 | 2641 | 2481 | 2407 |
| ORFs code for hypothetical proteins | 810 | 836 | 35 | 831 | 768 | 1008 | 876 | 766 |
| RNA genes | 111 | 110 | – | 110 | 109 | 110 | 111 | 109 |
| tRNA | 89 | 88 | – | 88 | 87 | 88 | 89 | 87 |
| tmRNA | 1 | 1 | – | 1 | 1 | 1 | 1 | 1 |
| rRNA | 21 | 21 | – | 21 | 21 | 21 | 21 | 21 |
IC001 GCA_003952785.1, LXL_C1 GCA_003335165.1, M19 GCA_004337595.1, AJ14 GCA_021496325.1, DJ‐RED GCA_021432745.1.
Complete genome annotation
The annotation of the chromosomal sequences obtained with Prokka for the two 20 k Canu or Unicycler assemblies revealed a total number of genes, CDSs and RNA genes that was consistent with the literature (Jia et al., 2021) and with what observed on other A. johnsonii genomes (Table 2).
Prophage
Using the software PHASTER putative prophage regions were detected in both chromosome assemblies (Table 3). In the Canu assembly two regions were recognized as intact prophages (score > 90), while the Unicycler assembly contained only one intact sequence, Entero Arya NC 031048, the same identified within the Canu's assembly.
TABLE 3.
Regions identified with PHASTER as intact prophages. The position in the genome, length, number of CDSs, most common tag and percentage of GC are indicated for each putative prophage
| Assembly | Canu20k intact region 1 | Canu20k intact region 2 | Unicycler20k intact region 1 |
|---|---|---|---|
| Region length (bp) | 5852 | 36,998 | 37,010 |
| Score | 110 | 120 | 110 |
| Total proteins | 11 | 58 | 57 |
| Region position | 993,621–999,473 | 3,558,700–3,595,698 | 877,031–914,041 |
| Most common prophage | Stx2 c Stxa F451 NC 049924 | Entero Arya NC 031048 | Entero Arya NC 031048 |
| GC (%) | 46.27 | 40.30 | 40.29% |
CRISPR‐Cas system
The analysis of both chromosome assemblies with CRISPRFinder did not reveal any CRISPR array.
Antibiotic resistances
The analysis of the chromosomes with Resfinder uncovered one gene related to antibiotic resistance in both assembled sequences. The gene codes for a putative class D (blaOXA‐309) β‐lactamase protein. The enzymes of class D, known as oxacillinases (OXAs), are largely distributed among Gram negatives. The OXAs have a wide genetic diversity and broad heterogeneity in β‐lactame hydrolysis spectrum (Poirel et al., 2010).
Virulence factors
The genomic sequences of A. johnsonii ICE_NC were analysed with VFfinder against the genus Acinetobacter. Several putative virulence factors were detected and the most represented categories were as follows: iron uptake (28 items), biofilm formation (14 items) and immune evasion (8 items). These results were shared for both types of assemblies.
Plasmid annotation
As the plasmid assembly by Canu turned out to be a chimaera, only the plasmid sequence reconstructed by Unicycler was annotated and characterized. A genome atlas of this annotation is depicted in Figure 1.
FIGURE 1.
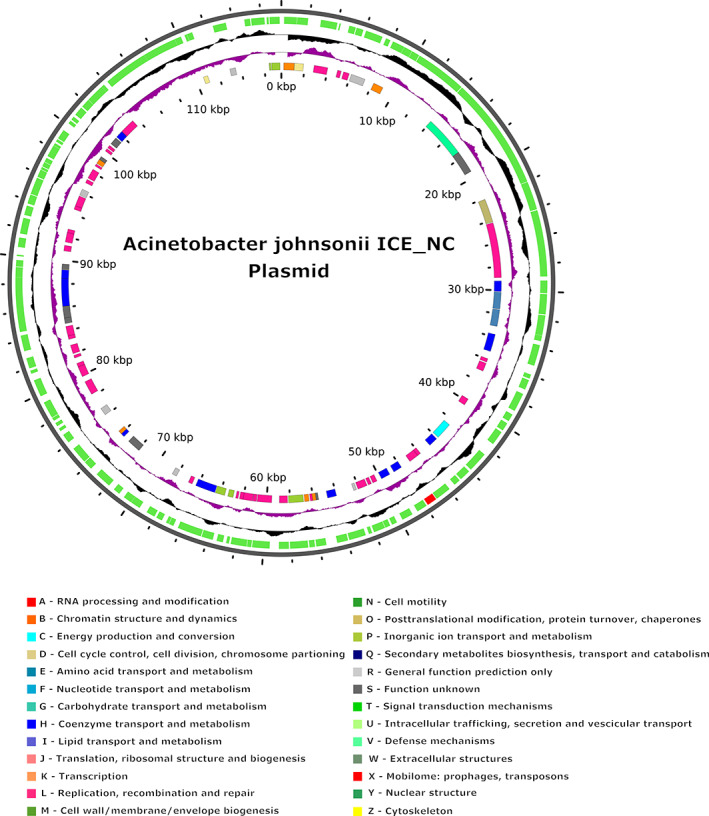
Features of the A. johnsonii ICE_NC plasmid built with the hybrid assembly annotated with Prokka, using the online server GView. Labelling from the outside to inside: Backbone; CDSs (coloured in green with 12α‐HSDH highlighted in red); GC content (GC‐rich towards outside, GC‐poor towards inside); GC skew (excess of guanine over cytosine towards outside, and vice versa) and CDSs coloured by COG categories (if assigned).
The plasmid annotation with Prokka (Table 3) revealed a total of 125 genes, all classified as CDSs, three putative genes and 61 ‘hypothetical proteins’. The characterization of the plasmid sequence revealed the presence of a putative prophagic region of 33,041 bp, with a GC content of 37.89% identified as Paenib Tripp NC 028930 and of 33 protein sequences corresponding to virulence factors. No CRISPR elements or antibiotic resistance genes were detected in the plasmid sequence.
12α‐HSDH identification and cloning
The characterization of the complete genome and plasmid sequences prompted us to search for the 12α‐HSDH coding gene, since the strain possesses such activity, as already confirmed (Giovannini et al., 2008). The presence of a 12α‐HSDH gene in the Acinetobacter genome was investigated by protein BLAST using the predicted protein sequence of the Clostridium 12α‐HSDH gene as a query. While no significant similarity was found along the chromosome, a good candidate was identified in the plasmid. The DNA fragment (NCBI accession number OM333240) coded for a polypeptide of 261 residues with a calculated molecular weight of 27.7 kDa. The amino acid sequence showed 41% identity with the Clostridium 12α‐HSDH protein (Figure 2). When compared to other known bacterial proteins the putative Acinetobacter 12α‐HSDH amino acid sequence was found highly similar (up to 94%) to several ‘Short‐Chain Dehydrogenases/Reductases’ identified (but not yet characterized) in different bacterial genomes such as Psychrobacter immobilis, a Gammaproteobacterium belonging to the same family of Acinetobacter, the Moraxellaceae (Figure 3). No information is available whether these other 12α‐HSDH‐coding genes are localized on the chromosome or in plasmids. Other known Acinetobacter whole genomes do not possess 12α‐HSDH‐coding genes (data not shown).
FIGURE 2.

Amino acid sequence alignment of a. johnsonii 12α‐HSDH with the 12α‐HSDH from Clostridium sp. ATCC 29733. Identical residues are highlighted in black, while conservative substitutions are shaded.
FIGURE 3.
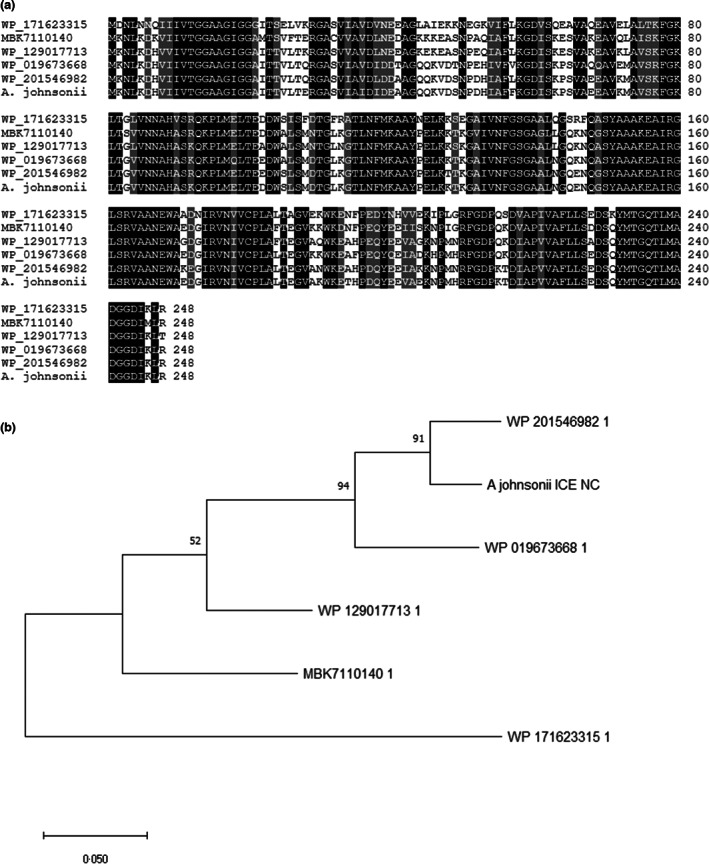
(a) Multiple alignment of the amino acid sequence of A. johnsonii 12α‐HSDH with other SDR, putative 12α‐HSDH, from Empedobacter stercoris (WP_171623315), Bacteroidetes bacterium (MBK7110140), Gelidibacter gilvus (WP_129017713), Psychrobacter lutiphocae (WP_019673668) and Psychrobacter immobilis (WP_201546982). Identical residues are highlighted in black, while conservative substitutions are shaded. (b) Maximum likelihood phylogenetic tree (bootstrap: 1000) of the same proteins obtained by MEGA‐X (Kumar et al., 2018).
To test the function of the predicted polypeptide, the coding region of the putative 12α‐HSDH gene was amplified from the Acinetobacter genome with specific primers, cloned into a His‐tag expression vector and, upon sequencing, expressed as a recombinant protein in E. coli cells. His‐Tag allowed the purification of the recombinant protein, which showed a molecular size compatible with the calculated one (27 kDa) as judged by SDS‐PAGE (Figure 4a).
FIGURE 4.
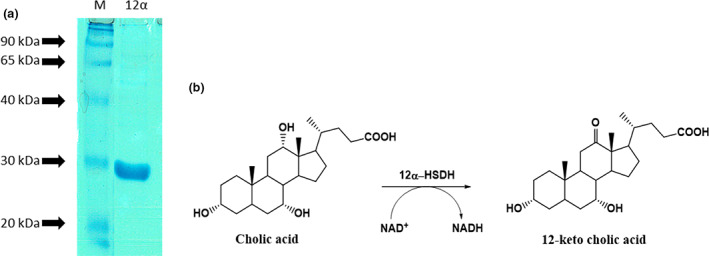
(a) SDS‐PAGE analysis of the A. johnsonii 12α‐HSDH recombinant protein expressed in E. coli cells and purified with His‐affinity chromatography. M: Molecular weight marker. (b) Schematic reaction of the oxidation reaction at position C‐12𝛂.
Enzymatic activity in oxidation of sodium cholate with recombinant 12α‐HSDH
The purified recombinant 12α‐HSDH was then functionally tested on sodium cholate to confirm its NAD+‐dependent oxidative specific activity (Figure 4b).
Based on the change in NADH absorbance measured at 340 nm, using 0.1 ml of enzyme we verified that 0.277 mmoles of cholic acid had been oxidized in 1 min. For this reason, we can affirm that the recombinant polypeptide exhibits the expected enzymatic activity, thus confirming the effective specificity and regio‐stereoselectivity of the catalysed reaction, analogously to the partially purified enzyme (Giovannini et al., 2008, data not shown).
Evaluation of the pH‐optimum
The dependence of the recombinant 12α‐HSDH enzymatic activity on pH changes was evaluated in a pH range 4.0–10.0 at 25°C. As shown in Figure S2, the pH‐optimum for the recombinant purified 12α‐HSDH was 8.0.
The relative percentage activity was calculated on the basis of the maximum absorbance variation obtained. The data collected show that at acidic pH values the activity is very low (below 20%). The activity progressively increases from pH 6 reaching a maximum at pH 8.
By further increasing the pH of the reaction, the activity of the enzyme rapidly decreases reaching a relative activity value of about 20% at pH 10.
According to Giovannini et al. (2008) the pH‐optimum of the enzyme purified from the wild‐type micro‐organism was 8.5, a value very similar to the one attributed to the recombinant purified 12α‐HSDH.
Effect of temperature
The effect of temperature on the activity of purified 12α‐HSDH on sodium cholate was studied in the range from 5 to 50°C. As shown in Figure S3, the tests revealed that at low temperatures (5–10°C) the relative enzymatic activity was lower than 21%. The activity then rapidly rises at higher temperatures, reaching a maximum at 25°C. Between 25°C and 35°C, the activity stayed at around 50%. It then drastically decreased until, at 50°C, there was no detectable enzymatic activity, probably due to enzymatic denaturation. 25°C is also the temperature used in the oxidation reaction of sodium cholate with the partially purified 12α‐HSDH studied by Giovannini et al. (2008).
Evaluation of the reaction of C12α‐oxidation catalysed by recombinant 12α‐HSDH
The recombinant purified 12α‐HSDH enzyme was also tested in a quantitative biotransformation of sodium cholate to 12‐keto cholic acid. The enzyme lactate dehydrogenase was used as a coupled recycling system for the cofactor, by reducing pyruvate to lactic acid it can regenerate the NAD+ allowing its use in catalytic and nonstoichiometric quantities. The reaction was carried out at pH 8, 25°C overnight. After extraction in organic solvent, the reaction mixture was subsequently dried and analysed by 1H NMR. By comparing the spectrum obtained (Figure S4a) with those of the reference products (cholic acid and 12‐keto cholic acid, Figure S4b,c), it was possible to calculate the ratio of the height of the integrals of characteristic peaks and consequently to calculate their percentage ratio. In particular, we considered the double doublet at 2.62 ppm related to the CHs at position C11 and characteristic of 12‐keto cholic acid and the singlet at 3.94 ppm related to the CH‐OH group at position C12 of cholic acid.
From the comparison of the integrals of the 1H‐NMR spectrum of the starting substrate with that of the product obtained, the biotransformation yielded a 98% of 12‐keto cholic acid derivative, a result comparable to that obtained with the enzyme partially purified from the wild‐type strain (Giovannini et al., 2008). This yield was confirmed by product purification, performed on a chromatographic column, 0.513 g of 12‐keto cholic acid (1.176 mMol) and 0.0098 g of cholic acid (0.024 mMol).
DISCUSSION
Taken together, these results illustrate the added value of long read sequencing in addressing the whole genome characterization and the plasmid reconstruction challenge, and also demonstrate the ability to produce a highly accurate description of the 12α‐HSDH gene in A. johnsonii ICE_NC.
Up to now, 45 genomes of A. johnsonii appear available on the National Center for Biotechnology Information (NCBI) database, but only 16 of them, isolated from multiple sources (clinical or environmental isolates) were assembled at a complete level. A comparative genomic analysis performed on 16 isolates (Jia et al., 2021) demonstrated that to adapt to human host and to the high selective pressure of antibacterial agents, clinical‐derived strains accumulate more functional genes associated with translational modification, β‐lactamase and defence mechanisms. Members of the gastrointestinal microbiota have evolved 12α‐HSDHs capable of oxidizing and epimerizing the 12α‐hydroxyl group from host CA. However, no gene(s) encoding 12α‐HSDH has been identified in A. johnsonii.
The strain of A. johnsonii ICE_NC (previously named A. calcoaceticus lwoffii) was isolated from a soil sample collected at the ICE production plant (ICE, Industria Chimica Italiana) and it appeared to contain the enzymes able to perform the chemo‐enzymatic synthesis of UDCA starting from CA (i.e. 7α‐ and 12α‐HSDH) as judged by biochemical analyses with partially purified enzymes (Giovannini et al., 2008). Using hybrid de novo genome assemblies, we were able to reconstruct and annotate one chromosome of about 3.6 Mb and one 117 kb‐long plasmid.
The utility of the approach used in this study was tested to identify and clone the gene for the A. johnsonii 12α‐HSDH. The availability of the annotation for both the chromosome and the plasmid allowed the identification of the putative gene by sequence homology. Upon cloning and expression as recombinant protein, we were able to accurately describe and characterize the 12α‐HSDH gene/enzyme in A. johnsonii ICE_NC. Combining long and short reads allowed the reconstruction of the chromosomal and plasmid sequence of A. johnsonii ICE_NC overcoming the challenges posed by the repetitive content, typical of bacterial genomes, and by the fact that plasmid sequences are sometimes even shared with the chromosomal DNA (Arredondo‐Alonso et al., 2017; de Toro et al., 2014).
At present, this is the first report of the identification of a 12α‐HSDH coding gene in A. johnsonii strain, that allowed the cloning and expression in E. coli of the A. johnsonii ICE_NC gene. Such information might provide important insight into the chemo‐enzymatic procedures for the synthesis of the therapeutic bile acid UDCA as the expression in E. coli allows the production of these enzymes in sufficient quality and quantity for the industrial preparation of UDCA.
AUTHOR CONTRIBUTIONS
A.B., G.B., E.T. and C.S. developed the main idea and designed the project. N.F. and S.C. performed the experiments and analysed the data. N.F. and A.C. performed bioinformatics analysis. S.C. and E.T. planned and interpreted the 12α‐HSDH characterization analyses and enzymatic assays. A.B. and G.B. performed the Nanopore MinION sequencing. S.S. performed Illumina NextSeq sequencing. G.B. conceived and performed 12α‐HSDH gene identification and cloning. E.T., C.S. and S.S. contributed experimental resources. C.S., G.B. and N.F. drafted the manuscript. All authors contributed, revised the article and approved the final version for publication.
FUNDING INFORMATION
The genome sequencing and experimental research costs were covered by FAR UniFe funds assigned to C.S., S.S., E.T. and G.B.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1
Figure S1
Figure S2
Figure S3
Figure S4
ACKNOWLEDGEMENT
We acknowledge Dr. Morena De Bastiani, University of Ferrara, for the valuable technical assistance. Open Access Funding provided by Universita degli Studi di Ferrara within the CRUI‐CARE Agreement.
Favale, N. , Costa, S. , Scapoli, C. , Carrieri, A. , Sabbioni, S. & Tamburini, E. et al. (2022) Reconstruction of Acinetobacter johnsonii ICE_NC genome using hybrid de novo genome assemblies and identification of the 12α‐hydroxysteroid dehydrogenase gene. Journal of Applied Microbiology, 133, 1506–1519. Available from: 10.1111/jam.15657
Nicoletta Favale and Stefania Costa have equally contributed to this research.
Contributor Information
Andrea Benazzo, Email: bnzndr@unife.it.
Giovanni Bernacchia, Email: bhg@unife.it.
DATA AVAILABILITY STATEMENT
Data available on request from the authors.
REFERENCES
- Aigner, A. , Gross, R. , Schmid, R. , Braun, M. & Mauer, S. (2011) Novel 12α‐hydroxysteroid dehydrogenases, production and use thereof. US patent 20110091921A1.
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. et al. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt, D. , Grant, J.R. , Marcu, A. , Sajed, T. , Pon, A. , Liang, Y. et al. (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Research, 44, W16–W21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo‐Alonso, S. , Willems, R.J. , van Schaik, W. & Schürch, A.C. (2017) On the (im)possibility of reconstructing plasmids from whole‐genome short‐read sequencing data. Microbial Genomics, 3, e000128. 10.1099/mgen.0.000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, S.F. , Franklund, C.V. & Hylemon, P.B. (1991) Cloning, sequencing, and expression of the gene coding for bile acid 7α‐hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Journal of Bacteriology, 173, 4558–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J. , McKnight, S.L. & Coleman, J.P. (2003) Cloning and characterization of the NAD‐dependent 7α‐hydroxysteroid dehydrogenase from Bacteroides fragilis. Current Microbiology, 47, 475–484. [DOI] [PubMed] [Google Scholar]
- Bortolaia, V. , Kaas, R.F. , Ruppe, E. , Roberts, M.C. , Schwarz, S. , Cattoir, V. et al. (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. The Journal of Antimicrobial Chemotherapy, 75, 3491–3500. 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolini, O. , Medici, A. & Poli, S. (1997) Biotransformations on steroid nucleus of bile acids. Steroids, 62, 564–577. 10.1016/S0039-128X(97)00043-3 [DOI] [PubMed] [Google Scholar]
- Bossche, L.V.D. , Borsboom, D. , Devriese, S. , Welden, S.V. , Holvoet, T. , Devisscher, L. et al. (2017) Tauroursodeoxycholic acid protects bile acid homeostasis under inflammatory conditions and dampens Crohn's disease‐like ileitis. Laboratory Investigation, 97, 519–529. [DOI] [PubMed] [Google Scholar]
- Braun, M. , Link, H. , Liu, L. , Schmid, R.D. & Weuster‐Botz, D. (2011) Biocatalytic process optimization based on mechanistic modeling of cholic acid oxidation with cofactor regeneration. Biotechnology and Bioengineering, 108, 1307–1317. [DOI] [PubMed] [Google Scholar]
- Braun, M. , Lünsdorf, H. & Bückmann, A.F. (1991) 12α‐hydroxysteroid dehydrogenase from clostridium group P, strain C 48–50. European Journal of Biochemistry, 196, 439–450. [DOI] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrea, G. , Pilotti, A. , Riva, S. , Canzi, E. & Ferrari, A. (1992) Enzymatic synthesis of12‐ketoursodeoxycholic acid from dehydrocholic acid in a membrane reactor. Biotechnology Letters, 14, 1131–1135. [Google Scholar]
- Coleman, J.P. , Hudson, L.L. & Adams, M.J. (1994) Characterization and regulation of the NADP‐linked 7α‐hydroxysteroid dehydrogenase gene from Clostridium sordellii. Journal of Bacteriology, 176, 4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvin, D. , Bernheim, A. , Toffano‐Nioche, C. , Touchon, M. , Michalik, J. , Néron, B. et al. (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Research, 46, W246–W251. 10.1093/nar/gky425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosignani, A. , Battezzati, P.M. , Setchell, K.D.R. , Invernizzi, P. , Covini, G. , Zuin, M. et al. (1996) Tauroursodeoxycholic acid for treatment of primary biliary cirrhosis. A dose‐response study. Digestive Diseases and Sciences, 41, 809–815. 10.1007/BF02213140 [DOI] [PubMed] [Google Scholar]
- De Coster, W. , D'Hert, S. , Schultz, D.T. , Cruts, M. & Van Broeckhoven, C. (2018) NanoPack: visualizing and processing long‐read sequencing data. Bioinformatics, 34, 2666–2669. 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Toro, M. , Garcilláon‐Barcia, M.P. & De La Cruz, F. (2014) Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli plasmids. Microbiology Spectrum, 2(6), PLAS‐0031‐2014. 10.1128/microbiolspec.PLAS-0031-2014 [DOI] [PubMed] [Google Scholar]
- Doden, H. , Sallam, L.A. , Devendran, S. , Ly, L. , Doden, G. , Daniel, S.L. et al. (2018) Metabolism of oxo‐bile acids and characterization of recombinant 12α‐hydroxysteroid dehydrogenases from bile acid 7α‐dehydroxylating human gut bacteria. Applied and Environmental Microbiology, 84, e00235‐18. 10.1128/AEM.00235-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, W.M. , Rodrigues, C.M. , Zhao, L.R. , Steer, C.J. & Low, W.C. (2002) Tauroursodeoxycholic acid improves the survival and function of nigral transplants in a rat model of Parkinson's disease. Cell Transplantation, 11, 195–205. [PubMed] [Google Scholar]
- Eggert, T. , Bakonyi, D. & Werner Hummel, W. (2014) Enzymatic routes for the synthesis of ursodeoxycholic acid. Journal of Biotechnology, 191, 11–21. 10.1016/j.jbiotec.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Ferrandi, E.E. , Bertuletti, S. , Monti, D. & Riva, S. (2020) Hydroxysteroid dehydrogenases: an ongoing story. European Journal of Organic Chemistry, 2020, 4463–4473. [Google Scholar]
- Franklund, C.V. , de Prada, P. & Hylemon, P.B. (1990) Purification and characterization of a microbial, NADP‐dependent bile acid 7‐hydroxysteroid dehydrogenase. The Journal of Biological Chemistry, 265, 9842–9849. [PubMed] [Google Scholar]
- Fuselli, S. , Baptista, R.P. , Panziera, A. , Magi, A. , Guglielmi, S. , Tonin, R. et al. (2018) A new hybrid approach for MHC genotyping: high‐throughput NGS and long read MinION nanopore sequencing, with application to the non‐model vertebrate alpine chamois (Rupicapra rupicapra). Heredity, 121, 293–303. 10.1038/s41437-018-0070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini, P.P. , Grandini, A. , Perrone, D. , Pedrini, P. , Fantin, G. & Fogagnolo, M. (2008) 7alpha‐ and 12alpha‐hydroxysteroid dehydrogenases from Acinetobacter calcoaceticus lwoffii: a new integrated chemo‐enzymatic route to ursodeoxycholic acid. Steroids, 73, 1385–1390. 10.1016/j.steroids.2008.06.013 [DOI] [PubMed] [Google Scholar]
- Gupta, A. , Tschentscher, A. & Bobkova, M. (2009) Process for the enantioselective reduction and oxidation, respectively, of steroids. US 20090280525 A1. U.S. Patent.
- Harris, J.N. & Hylemon, P.B. (1978) Partial purification and characterization of NADP‐dependent 12alpha‐hydroxysteroid dehydrogenase from Clostridium leptum. Biochimica et Biophysica Acta, 528, 148–157. [DOI] [PubMed] [Google Scholar]
- ICE . ICE (Industria Chimica Emiliana) extracts and purifies bile acids from raw materials (ox and pig bile).
- Jackman, S.D. , Coombe, L. , Chu, J. , Warren, R.L. , Vandervalk, B.P. , Yeo, S. et al. (2018) Tigmint: correcting assembly errors using linked reads from large molecules. BMC Bioinformatics, 19, 1–10. 10.1186/s12859-018-2425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, J. , Liu, M. , Feng, L. & Wang, Z. (2021) Comparative genomic analysis reveals the evolution and environmental adaptation of Acinetobacter johnsonii . Gene, 808, 145985. 10.1016/j.gene.2021.145985 [DOI] [PubMed] [Google Scholar]
- Kisiela, M. , Skarka, A. , Ebert, B. & Maser, E. (2012) Hydroxysteroid dehydrogenases (HSDs) in bacteria: a bioinformatic perspective. The Journal of Steroid Biochemistry and Molecular Biology, 129, 31–46. [DOI] [PubMed] [Google Scholar]
- Kollerov, V.V. , Lobastova, T.G. , Monti, D. , Deshcherevskaya, N.O. , Ferrandi, E.E. , Fronza, G. et al. (2016) Deoxycholic acid transformations catalyzed by selected filamentous fungi. Steroids, 107, 20–29. [DOI] [PubMed] [Google Scholar]
- Koren, S. , Walenz, B.P. , Berlin, K. , Miller, J.R. , Bergman, N.H. & Phillippy, A.M. (2017) Canu: scalable and accurate long‐read assembly via adaptive k‐mer weighting and repeat separation. Genome Research, 27, 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritchevsky, D. (1971) Chemistry of bile acids. In: Nair, P.P. & Kritchevsky, D. (Eds.) The bile acids: chemistry, physiology and metabolism, Vol. 1. New York: Plenum Press, pp. 1–10. [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. & Tamura, K. (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 34, 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Zheng, D. , Jin, Q. , Chen, L. & Yang, J. (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Research, 47, D687–D692. 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, I.A. , Jellet, J.F. & Mahony, D.E. (1979) 12α‐hydroxysteroid dehydrogenase from clostridium group P strain C48‐50 ATCC #29733: partial purification and characterization. Journal of Lipid Research, 2, 234–239. [PubMed] [Google Scholar]
- Macdonald, I.A. , Mahony, D.E. , Jellet, J.F. & Meier, C.E. (1977) NAD‐dependent 3α‐ and 12α‐hydroxysteroid dehydrogenase activities from Eubacterium lentum ATCC no. 25559. Biochimica et Biophysica Acta, 489, 466–476. [DOI] [PubMed] [Google Scholar]
- Macdonald, I.A. , Meier, E.C. , Mahony, D.E. & Costain, G.A. (1976) 3α, 7α‐ and 12α‐hydroxysteroid dehydrogenase activities from Clostridium perfringens . Biochimica et Biophysica Acta, 450, 142–153. [DOI] [PubMed] [Google Scholar]
- Manni, M. , Berkeley, M.R. , Seppey, M. , Simão, F.A. & Zdobnov, E.M. (2021) BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molecular Biology and Evolution, 38, 4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici, A. , Pedrini, P. , Bianchini, E. , Fantin, G. , Guerrini, A. , Natalini, B. et al. (2002) 7α‐OH epimerisation of bile acids via oxido‐reduction with Xanthomonas maltophilia. Steroids, 67, 51–56. [DOI] [PubMed] [Google Scholar]
- Monti, D. , Ferrandi, E.E. , Riva, S. & Polentini, F. , (2012) New process for the selective oxidation of bile acids, their salts or derivatives. WO 2012131591 A1. WO Patent.
- Pedrini, P. , Andreotti, E. , Guerrini, A. , Dean, M. , Fantin, G. & Giovannini, P.P. (2006) Xanthomonas maltophilia CBS 897.97 as a source of new 7β‐ and 7α‐hydroxysteroid dehydrogenases and cholylglycine hydrolase: improved biotransformations of bile acids. Steroids, 71, 189–198. [DOI] [PubMed] [Google Scholar]
- Poirel, L. , Naas, T. & Nordmann, P. (2010) Diversity, epidemiology, and genetics of class D beta‐lactamases. Antimicrobial Agents and Chemotherapy, 54, 24–38. 10.1128/AAC.01512-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabha, V. & Ohri, M. (2006) Review: bacterial transformations of bile acids. World Journal of Microbiology and Biotechnology, 22, 191–196. 10.1007/s11274-005-9019-y [DOI] [Google Scholar]
- Rhie, A. , Walenz, B.P. , Koren, S. & Phillippy, A.M. (2020) Merqury: reference‐free quality, completeness, and phasing assessment for genome assemblies. Genome Biology, 21, 245. 10.1186/s13059-020-02134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda, E. , Bazzoli, F. , Labate, A.M. , Mazzella, G. , Roda, A. , Sama, C. et al. (1982) Ursodeoxycholic acid vs. chenodeoxycholic acid as cholesterol gallstone‐dissolving agents: a comparative randomized study. Hepatology (Hoboken, NJ, US), 2, 804–810. 10.1002/hep.1840020611 [DOI] [PubMed] [Google Scholar]
- Salen, G. , Colalillo, A. , Verga, D. , Bagan, E. , Tint, G. & Shefer, S. (1980) Effect of high and low doses of ursodeoxycholic acid on gallstone dissolution in humans. Gastroenterology, 78, 1412–1418. [PubMed] [Google Scholar]
- Schubert, M. , Lindgreen, S. & Orlando, L. (2016) AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Research Notes, 9, 88. 10.1186/s13104-016-1900-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics, 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Shi, S.C. , You, Z.‐N. , Zhou, K. , Chen, Q. , Pan, J. , Qian, X.‐L. et al. (2019) Efficient synthesis of 12‐Oxochenodeoxycholic acid using a 12α‐hydroxysteroid dehydrogenase from Rhodococcus ruber . Advanced Synthesis and Catalysis, 361, 4661–4668. [Google Scholar]
- Sutherland, J.D. , Macdonald, I.A. & Forrest, T.P. (1982) The enzymic and chemical synthesis of ursodeoxycholic and chenodeoxycholic acid from cholic acid. Preparative Biochemistry, 12, 307–321. 10.1080/00327488208065679 [DOI] [PubMed] [Google Scholar]
- Tang, S. , Pan, Y. , Lou, D. , Ji, S. , Zhu, L. , Tan, J. et al. (2019) Structural and functional characterization of a novel acidophilic 7α‐hydroxysteroid dehydrogenase. Protein Science, 28, 910–919. 10.1002/pro.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonin, F. & Arends, I.W.C.A. (2018) Latest development in the synthesis of ursodeoxycholic acid (UDCA): a critical review. Beilstein Journal of Organic Chemistry, 14, 470–483. 10.3762/bjoc.14.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, S. , Oda, M. , Imamura, S. & Ohnishi, M. (2004) Molecular and enzymatic properties of 7α‐hydroxysteroid dehydrogenase from pseudomonas sp. B‐0831. Journal of Biological Macromolecules, 4, 33–38. [Google Scholar]
- Walker, B.J. , Abeel, T. , Shea, T. , Priest, M. , Abouelliel, A. , Sakthikumar, S. et al. (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One, 9, e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, R.R. , Judd, L.M. , Gorrie, C.L. & Holt, K.E. (2017) Completing bacterial genome assemblies with multiplex MinION sequencing. Microbial Genomics, 3, e000132. 10.1099/mgen.0.000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, T. , Higashi, H. , Kanatani, A. , Lin, X.S. , Nagai, H. , Oyama, H. et al. (1991) Cloning and sequencing of the 7α‐hydroxysteroid dehydrogenase gene from Escherichia coli HB101 and characterization of the expressed enzyme. Journal of Bacteriology, 173, 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari, E. , Allesøe, R. , Joensen, K.G. , Cavaco, L.M. , Lund, O. & Aarestrup, F.M. (2020) PointFinder: a novel web tool for WGS‐based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. The Journal of Antimicrobial Chemotherapy, 72, 2764–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Schwartz, S. , Wagner, L. & Miller, W. (2000) A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7, 203–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figure S1
Figure S2
Figure S3
Figure S4
Data Availability Statement
Data available on request from the authors.


