Abstract
The aerobic yeast Kluyveromyces lactis and the predominantly fermentative Saccharomyces cerevisiae share many of the genes encoding the enzymes of carbon and energy metabolism. The physiological features that distinguish the two yeasts appear to result essentially from different organization of regulatory circuits, in particular glucose repression and gluconeogenesis. We have isolated the KlCAT8 gene (a homologue of S. cerevisiae CAT8, encoding a DNA binding protein) as a multicopy suppressor of a fog1 mutation. The Fog1 protein is a homologue of the Snf1 complex components Gal83p, Sip1p, and Sip2p of S. cerevisiae. While CAT8 controls the key enzymes of gluconeogenesis in S. cerevisiae, KlCAT8 of K. lactis does not (I. Georis, J. J. Krijger, K. D. Breunig, and J. Vandenhaute, Mol. Gen. Genet. 264:193–203, 2000). We therefore examined possible targets of KlCat8p. We found that the acetyl coenzyme A synthetase genes, KlACS1 and KlACS2, were specifically regulated by KlCAT8, but very differently from the S. cerevisiae counterparts. KlACS1 was induced by acetate and lactate, while KlACS2 was induced by ethanol, both under the control of KlCAT8. Also, KlJEN1, encoding the lactate-inducible and glucose-repressible lactate permease, was found under a tight control of KlCAT8.
When the yeast Saccharomyces cerevisiae grows on glucose-containing media, energy is produced by glycolysis and fermentation even if oxygen is available, because mitochondrial function is repressed by glucose. A large number of genes encoding enzymes essential for the catabolism of alternate carbon sources are turned off. This regulatory mechanism, known as glucose repression (6, 13), affects several other metabolic pathways, such as gluconeogenesis and the tricarboxylic acid cycle, or peroxisomal functions. Glucose repression is a major regulatory device of carbon metabolism in S. cerevisiae, a species in which fermentation is the preferred form of energy acquisition.
In Kluyveromyces lactis, like in many other aerobic yeasts, the metabolic response to glucose is different from that of S. cerevisiae. Respiration is not glucose repressed, and fermentative and oxidative metabolism can take place simultaneously. Glucose repression does exist, however. A number of enzymes required for alternate carbohydrate metabolism have been shown to be subject to glucose repression to various degrees (5, 10, 12, 15, 19, 35). The mechanism of this repression in K. lactis is the object of this study.
We have recently isolated two genes of K. lactis, FOG1 and FOG2 (16). They are homologous to the components of the glucose-responding Snf1 complex of S. cerevisiae. Fog2p is structurally and functionally homologous to the Snf1 kinase. Fog1p belongs to the family of regulatory proteins Gal83p, Sip1p, and Sip2p, which interact with Snf1p. FOG1 is a unique gene whose deletion leads to the inability to grow on certain carbon sources, while in S. cerevisiae the gal83Δ sip1Δ sip2Δ triple mutation does not display any phenotype (11, 33).
In the attempt at isolating genes involved in this regulatory circuit, we looked for suppressors that, when overexpressed, would be able to reverse one or more phenotypes of the fog1 mutant. In this paper, we report that one of the multicopy suppressors isolated was the KlCAT8 gene, which encodes a transcription activator, reinforcing the view that KlCat8p is functioning downstream of the Fog1-Fog2 complex in a way analogous to the Cat8-Snf1 interaction in S. cerevisiae. In S. cerevisiae, CAT8 encodes a DNA-binding protein which is essential for growth on nonfermentable carbon sources (17) and is also required for derepression of genes involved in gluconeogenesis. Several targets of the Cat8p in S. cerevisiae are known: the FBP1 and PCK1 genes, both required for the gluconeogenic pathway (17, 23); ICL1 and MLS1, key genes for the glyoxylate cycle (31, 8); ACR1 and JEN1, encoding carriers of succinate and lactate, respectively (3, 4); ACS1, required for utilization of acetate (18); IDP2, encoding the NADP-dependent cytosolic isocitrate dehydrogenase which regenerates NADPH under nonfermentative growth conditions (4); MDH2, encoding the malate dehydrogenase cytoplasmic isoenzyme (26). Cat8p-dependent glucose derepression of these genes is mediated by cis-acting elements (carbon source responsive elements [CSRE]) present in all of their promoters. CAT8 is itself glucose regulated, activated by Snf1p, and repressed by Mig1p (24, 25, 32).
As is the case for S. cerevisiae, the Klcat8 null mutant was unable to utilize ethanol, acetate, and lactate, but in contrast it grew on glycerol, indicating that KlCAT8 is not involved in gluconeogenesis control. Indeed, it has recently been reported that a Klcat8 mutation has no influence on the regulation of the gluconeogenic genes, KlFBP1 and KlPCK1 (14). Also, a partial reduction of the malate synthase and isocitrate lyase activities has been observed for the ΔKlcat8 mutant, although it is not clear whether the reduced level of these enzymes is sufficient to explain the impaired growth of the ΔKlcat8 mutant on ethanol, lactate, and acetate (14).
The conspicuous difference between K. lactis and S. cerevisiae in the responses to glucose suggests that the KlCat8p has its specific range of target genes. We will show that KlCat8p is necessary for the full induction of both genes, KlACS1 and KlACS2, encoding the two isoforms of acetyl coenzyme A (acetyl-CoA) synthetase. Moreover, KlCat8p is required for the expression of KlJEN1 gene, which encodes lactate permease.
MATERIALS AND METHODS
Strains and media.
The K. lactis strain JA6 (α ade1 ade2 trp1-1 uraA) has previously been described (5). The mutant ΔKlcat8 was obtained by gene disruption (27) of the KlCAT8 gene and insertion of the URA3 marker in cells of the JA6 strain. The JA6Δfog1 and the JA6/M207 (fog2) strains, derived from the JA6 strain, have been previously described (16). Escherichia coli strain JM83 [ara Δ(lac-proAB) rpsL (=strA) φ80 lacZΔM15] was used for plasmid propagation and maintenance.
The complete medium contained 1% Bacto-yeast extract and 2% Bacto-peptone (Difco). The minimal medium (YNB) contained 7 g of yeast nitrogen base without amino acids (Difco)/liter supplemented with appropriate amino acids and bases. Various carbon sources were added at 2% except when indicated otherwise.
Transcript analysis.
Total RNA was prepared by extraction with hot acidic phenol (1). Northern analysis was carried out according to the protocol of Sherman et al. (30).
Probes.
KlACS1, KlACS2, and KlJEN1 probes were obtained by PCR amplification, with K. lactis genomic DNA as template.
The KlSDH1 probe corresponded to the 2.2-kb BamHI-SphI fragment derived from the plasmid p3AS (M. Saliola, unpublished data).
The amount of RNA loaded on the gel was estimated by hybridization with a K. lactis actin gene probe (KlACT1) (9). All the probes were labeled with [α-32P]dCTP by the rediprime DNA labeling system (Amersham).
Miscellaneous.
Published procedures were used for the transformation of K. lactis (2) and E. coli (20), for the preparation of yeast DNA (21), for the isolation and the purification of plasmids from E. coli, for agarose gel electrophoresis, for Southern blot analysis (28), and for DNA sequence analysis (29).
RESULTS
Increased KlCAT8 dosage suppresses fog1 mutant defects.
To identify proteins that interact with the Fog1-Fog2 complex, we have sought the genes that, at increased dosage, could overcome the defects in Fog1p function. We transformed a fog1 null mutant with a K. lactis genomic library constructed in the KEp6 multicopy vector (34). Clones were selected for their ability to grow on respiratory carbon sources, since the fog1 mutant is unable to utilize nonfermentable carbon sources. One of these clones, able to grow on ethanol, glycerol, and lactate, contained the plasmid pSF1/5A with an insert of about 6 kbp. This plasmid also suppressed the fog2 mutation, as expected if the suppressor gene had a role downstream of the Fog1p-Fog2p complex. The DNA insert in the plasmid contained a single open reading frame which was identical to the recently reported KlCAT8 gene (14). We concluded that overexpression of KlCAT8 can compensate for the loss of at least two of the proteins of the Fog complex. The KlCAT8 gene product was 40% homologous to, and able to substitute in vivo for, S. cerevisiae Cat8p (14).
Impaired utilization of carbon sources in Klcat8 null mutant.
A Klcat8 null mutant was constructed by replacing about 80% of its coding region, including the zinc finger motif, by the URA3 marker sequence. The plasmid containing the disrupted KlCAT8 gene was linearized and introduced by transformation into the JA6 strain. One clone out of the 350 Ura+ transformants was unable to grow on minimal medium supplemented with 1% ethanol. Southern analysis of this clone confirmed the correct integration of the deleting cassette into the KlCAT8 locus (data not shown).
The phenotype of this mutant was tested with other nonfermentable carbon sources. Besides on ethanol, the mutant was unable to grow on lactate, succinate, and acetate like the S. cerevisiae cat8 mutant (Table 1), but it did grow on glycerol. These data agree with those of Georis et al. (14), except that for our Klcat8 null mutant, the growth on the above-mentioned respiratory substrates was not only reduced but completely absent. The ability of the ΔKlcat8 mutant to grow on glycerol indicated that the blocked function in the ΔKlcat8 mutant is not gluconeogenesis per se, consistent with the fact that the two key enzymes of gluconeogenesis, PCK1 and FBP1, were not dependent on the Cat8p function in K. lactis (14). However, the addition of a small amount of glucose (0.02%) allowed the ΔKlcat8 strain to grow on ethanol, acetate, succinate, and lactate (Table 1), suggesting that, in this condition, utilization of the above-mentioned carbon sources can escape from KlCAT8 regulation.
TABLE 1.
Colony sizes of cells of mutant strains JA6Klcat8 and JA6fog2 on different carbon sourcesa
| Carbon source | Colony size (μm)
|
||
|---|---|---|---|
| JA6 wt | JA6Klcat8 | JA6fog2 | |
| 1% glycerol | 652 | 522 | − |
| 1% succinate | 634 | − | − |
| 1% ethanol | 738 | − | − |
| 1% acetate | 728 | − | − |
| 1% lactate | 682 | − | − |
| 0.02% glucose | − | − | − |
| 0.02% glucose + 1% succinate | 828 | 699 | − |
| 0.02% glucose + 1% ethanol | 850 | 674 | − |
| 0.02% glucose + 1% acetate | 886 | 829 | − |
| 0.02% glucose + 1% lactate | 768 | 772 | − |
| 0.02% glucose + 1% glycerol | 970 | 926 | − |
| 1% acetate + 1% glycerol | ND | ND | 224* |
| 1% ethanol + 1% glycerol | ND | ND | − |
Briefly, 102 cells were spread on solid minimal media supplemented with different carbon sources. Colony sizes were measured after 4 days. All the values are the average of five independent measurements. −, colony not detectable; *, measurement after 7 days; ND, not determined; wt, wild type.
The role of KlCAT8 is exclusively regulatory and is not directly involved in the respiratory mechanism. In cells of the ΔKlcat8 strain, neither respiration nor cytochrome composition were affected (data not shown).
KlCAT8 is necessary for full induction of KlACS1 and KlACS2 genes.
In order to find possible targets of KlCat8p, we focused our attention on genes involved in the utilization of acetate and ethanol. The first step of acetate utilization requires its activation to acetyl-CoA by the acetyl-CoA synthetases. This enzyme is also involved in ethanol utilization, since ethanol is oxidized to acetate by alcohol dehydrogenase and acetaldehyde dehydrogenase. Acetyl-CoA can then enter the tricarboxylic acid cycle. Two K. lactis genes, KlACS1 and KlACS2, which are homologous to S. cerevisiae ACS1 and ACS2 genes encoding the acetyl-CoA synthetases, have been isolated and characterized (35) (KlACS1, EMBL accession no. AF061265; KlACS2 EMBL accession no. AF134491).
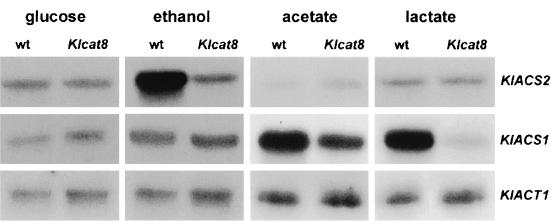
The effects of Klcat8 disruption on the transcription of these two genes were examined under various growth conditions. Total RNA was extracted for Northern analysis from cells grown at low glucose concentration (0.3%) and after a medium shift from 2% glucose to 2% ethanol, 2% lactate, or 2% acetate. This medium shift was necessary because the ΔKlcat8 mutant was unable to grow on these substrates. Results are shown in Fig. 1. In cells of the wild-type strain JA6, transcription of KlACS1 was enhanced in ethanol, acetate, and lactate media, compared to that in glucose medium. The highest level of expression was observed with acetate and lactate. The responses of KlACS2 were quite contrasting to those of KlACS1. KlACS2 was specifically induced by ethanol and poorly expressed in both lactate and acetate.
FIG. 1.
Effect of Klcat8 mutation on KlACS1 and KlACS2 mRNA level. Northern blot analysis of the KlACS1 and KlACS2 transcripts from JA6 (wt) and from JA6ΔKlcat8 (Klcat8) strains. Total RNA was prepared from cells grown in YNB medium supplemented with 0.3% glucose or after a medium shift from 2% glucose to 2% ethanol, from 2% glucose to 2% acetate, or from 2% glucose to 2% lactate. RNA was extracted 4 h after the medium shift, electrophoresed, and hybridized with labeled probes corresponding to KlACS1, KlACS2, and KlACT1 genes. Each lane contained 20 μg of total RNA.
In glucose medium, both KlACS1 and KlACS2 mRNA levels were low and unaffected by the ΔKlcat8 mutation. However, the ethanol induction of KlACS2 as well as the lactate induction of KlACS1 were prevented in the mutant (reduction of about 75%). Acetate induction of KlACS1 was only partially affected by Klcat8 mutation (reduction of about 35%).
Role of Fog2p on expression of ACS genes.
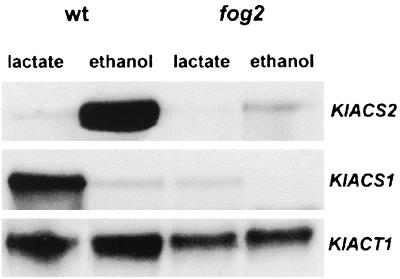
In S. cerevisiae, CAT8 is upregulated by Snf1 kinase (6, 13). If a similar signaling cascade of glucose repression is conserved in K. lactis, we would expect an upstream control of Fog2p-KlSnf1p on the expression of ACS genes. The fog2 mutation would affect the transcription of KlACS1 and KlACS2. Indeed, as shown in Fig. 2, after a medium shift from 2% glucose to 2% lactate, the expression of KlACS1 was abolished in a fog2 mutant. Also, in a shift from 2% glucose to 2% ethanol, the ethanol induction of KlACS2 was almost totally abolished in fog2 mutant.
FIG. 2.
Effect of fog2 mutation on KlACS1 and KlACS2 mRNA levels. Northern blot analysis of the KlACS1 and KlACS2 transcripts from JA6 (wt) and from JA6/M207 (fog2) strains. Total RNA was prepared from cells grown in YNB medium after a medium shift experiment from 2% glucose to 2% ethanol or 2% lactate. RNA was extracted 4 h after the medium shift. Hybridization was carried out with labeled probes corresponding to KlACS1, KlACS2, and KlACT1 genes. Each lane contained 20 μg of total RNA.
The fog2 mutant does not grow on any respiratory substrates, glycerol included. As shown in Table 1, growth on glycerol was possible only when complemented with acetate. These results indicate that the inability of the fog2 mutant to grow on glycerol is due to a defect in the production and/or activation of acetate. We therefore analyzed the role of Fog2p in the transcription of ACS genes. After a medium shift from glucose to glycerol, both ACS genes were equally expressed in the isogenic series of strains, the wild type, fog2CAT8, and FOG2cat8 (data not shown). Therefore, the inability of fog2 mutant to grow on glycerol is not due to an impaired expression of ACS genes. Yet, unidentified factor(s) should be involved in glycerol activation.
Role of KlCat8p on expression of succinate dehydrogenase gene.
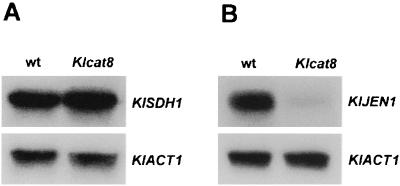
K. lactis cells grow on succinate. ΔKlcat8 mutant cells do not. The step at which this block occurs is not known. As shown in Fig. 3A, the transcription of the succinate dehydrogenase gene KlSDH1 is not affected in mutant cells. The target(s) of KlCAT8 may be another component(s) of the respiratory complex II or other steps, such as succinate transport. In S. cerevisiae, a cat8 mutation is known to affect the mitochondrial succinate-fumarate transporter gene ACR1 (3, 22). The presence in K. lactis of an ACR1 counterpart has not been reported so far. The control of succinate metabolism in K. lactis is still an open question.
FIG. 3.
Effect of Klcat8 mutation on succinate dehydrogenase and on KlJEN1 transcripts. Total RNA was prepared from JA6 (wt) and from JA6ΔKlcat8 (Klcat8) strains 4 h after a medium shift from 2% glucose to 2% ethanol. RNA was hybridized with labeled probes for KlSDH1 and KlACT1 genes (A) and for KlJEN1 and KlACT1 genes (B). Each lane contained 20 μg of total RNA.
KlCAT8 is required for expression of lactate permease gene JEN1
Lactate utilization in S. cerevisiae requires the induced expression of JEN1, a CAT8-regulated gene that encodes a specific lactate-proton symporter permease (4, 7). Therefore, we asked whether the lack of lactate utilization in ΔKlcat8 mutant cells was due to an impaired induction of lactate permease. A gene highly homologous to the JEN1 gene of S. cerevisiae has recently been identified (M. Bolotin, personal communication). A probe corresponding to the putative KlJEN1 gene was prepared by PCR amplification, and the level of the corresponding mRNA was analyzed during a metabolic shift from glucose to lactate. As shown in Fig. 3B, the amount of the KlJEN1 transcript was strongly reduced in ΔKlcat8 mutant cells, indicating that KlCat8p is required for the lactate uptake process.
DISCUSSION
Several pieces of evidence show that there is a striking difference between K. lactis and S. cerevisiae in the regulation of major carbon metabolism, despite a high degree of conservation of individual gene sequences involved. Concerning the genes participating in glucose repression and/or derepression, it has been demonstrated that FOG2 (KlSNF1), encoding a highly conserved serine/threonine protein kinase, performs a function similar to that of the SNF1 gene in S. cerevisiae. The product of the unique FOG1 gene belongs to the protein family comprising Gal83, Sip1, and Sip2 and is absolutely required for the correct functioning of the Fog1-Fog2 complex in K. lactis (16). In order to find out possible targets of this complex, we looked for genes that, when overexpressed, could compensate for fog1 defects.
Two multicopy suppressors of fog1 which restored growth on respiratory substrates were isolated. One was the KlCAT8 gene studied here. The other suppressor gene was found to be coding for a leucine zipper protein (presently under investigation). Despite the structural and functional similarity between KlCat8p and its S. cerevisiae counterpart, the phenotypic consequences of their mutations are clearly different. The Klcat8 disruption mutant, like the cat8 mutant of S. cerevisiae, was unable to utilize ethanol, acetate, lactate, and succinate but was capable of growing on glycerol. This result shows that gluconeogenesis is not impaired in the K. lactis mutant. Consistent with this, it has recently been demonstrated that the activation of KlPCK1 and KlFBP1, two key genes required for gluconeogenesis, was independent of KlCat8p function (14), and KlCat8p was required for the glyoxylate cycle enzymes (malate synthase and isocitrate lyase). However, the reduced activities of these enzymes in the mutant could not fully explain the complete absence of growth of the Klcat8 mutant on ethanol, acetate, succinate, and lactate. Therefore, we were led to search for other possible targets of KlCat8p.
First, the role of KlCat8p in the expression of the KlACS1 and KlACS2 genes was investigated, because the utilization of acetate and ethanol requires acetyl-CoA synthetase activity. The two KlACS genes were found to be transcribed at a low level in glucose-grown wild-type cells, as described by Zeeman et al. (36). KlACS1 was induced by a medium shift to ethanol, acetate, and lactate whereas KlACS2 was induced only by ethanol. Both of these inductions were prevented by the Klcat8 mutation. These results demonstrate that the two KlACS genes were differently controlled by KlCat8p. This situation is different from that of S. cerevisiae, where only ACS1 gene expression is regulated by a carbon source under CAT8 control. This control is supposed to be mediated by the CSRE of the target gene promoters. This CSRE motif is not known in K. lactis genes studied so far.
The two KlACS genes appear to have different metabolic roles in the response to carbon sources: KlACS2 is necessary for growth on ethanol, and KlACS1 is necessary for acetate utilization. Since the utilization of lactate does not require acetyl-CoA synthetase activity, according to the current theory, we are not able to explain why lactate is an inducer of KlACS1 and why KlCat8p is required for this induction. The growth defect of Klcat8 null mutant cells on ethanol and acetate could be ascribed to the impairment of the specific induction of KlACS genes by these substrates in combination with the deficiency of the glyoxylate cycle. The growth defect of Klcat8 mutant cells on lactate is due to at least two deficient steps: the absence of induction of the lactate permease gene KlJEN1 (which could be sufficient) and the impaired glyoxylate cycle.
The control by Fog2p on the expression of KlACS genes indicates that the KlCAT8 gene is under the control of Fog2 kinase and supports the idea that the signaling cascade of glucose repression is essentially conserved between S. cerevisiae and K. lactis. However, more importantly, the spectrum of individual target genes of CAT8 is not the same in these two yeasts, which differ in lifestyle with respect to carbon sources. The response of ACS genes is illustrative of this difference.
ACKNOWLEDGMENTS
We would like to thank Iliana Ferrero for encouragement and helpful discussions. We are grateful to Monique Bolotin for communicating the unpublished KlJEN1 sequence and to Micheline Wésolowski-Louvel for providing the genomic library.
This work was supported by a grant from Ministero Università e Ricerca Scientifica e Tecnologica–Università di Parma Cofin 1999.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1994. [Google Scholar]
- 2.Bianchi M M, Falcone C, Chen X J, Wésolowski-Louvel M, Frontali L, Fukuhara H. Transformation of the yeast Kluyveromyces lactis by new vectors derived from the 1,6 μm circular plasmid pkD1. Curr Genet. 1987;12:185–192. [Google Scholar]
- 3.Bojunga N, Kotter P, Entian K D. The succinate/fumarate transporter Acr1p of Saccharomyces cerevisiae is part of the gluconeogenic pathway and its expression is regulated by Cat8p. Mol Gen Genet. 1998;260:453–461. doi: 10.1007/s004380050916. [DOI] [PubMed] [Google Scholar]
- 4.Bojunga N, Entian K D. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p) Mol Gen Genet. 1999;262:869–875. doi: 10.1007/s004380051152. [DOI] [PubMed] [Google Scholar]
- 5.Breunig K D. Glucose repression of LAC genes expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet. 1989;216:422–427. doi: 10.1007/BF00334386. [DOI] [PubMed] [Google Scholar]
- 6.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 7.Casal M, Paiva S, Andrade R P, Gancedo C, Leao C. The lactate proton symport of Saccharomyces cerevisiae is encoded by JEN1. J Bacteriol. 1999;181:2620–2623. doi: 10.1128/jb.181.8.2620-2623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspary F, Hartig A, Schuller H J. Constitutive and carbon source-responsive promoter elements are involved in the regulated expression of the Saccharomyces cerevisiae malate synthase gene MLS1. Mol Gen Genet. 1997;255:619–627. doi: 10.1007/s004380050536. [DOI] [PubMed] [Google Scholar]
- 9.Deshler J O, Larson G P, Rossi J J. Kluyveromyces lactis maintains Saccharomyces cerevisiae intron-encoded splicing signals. Mol Cell Biol. 1989;9:2208–2213. doi: 10.1128/mcb.9.5.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson R C, Markin J S. Physiological studies of β-galactosidase induction in Kluyveromyces lactis. J Bacteriol. 1980;142:777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erikson J R, Johnston M. Genetic and molecular characterization of GAL83: its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics. 1993;135:655–664. doi: 10.1093/genetics/135.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero I, Rossi C, Landini M P, Puglisi P P. Role of the mitochondrial protein synthesis in the catabolite repression of the petite negative yeast Kluyveromyces lactis. Biochem Biophys Res Commun. 1978;80:340–348. doi: 10.1016/0006-291x(78)90682-4. [DOI] [PubMed] [Google Scholar]
- 13.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georis I, Krijger J J, Breunig K D, Vandenhaute J. Differences in regulation of yeast gluconeogenesis revealed by Cat8p-independent activation of PCK1 and FBP1 genes in Kluyveromyces lactis. Mol Gen Genet. 2000;264:193–203. doi: 10.1007/s004380000314. [DOI] [PubMed] [Google Scholar]
- 15.Goffrini P, Ficarelli A, Ferrero I. Hexokinase activity is affected in mutants of Kluyveromyces lactis resistant to glucose repression. Microbiology. 1995;141:441–447. [Google Scholar]
- 16.Goffrini P, Ficarelli A, Donnini C, Lodi T, Puglisi P P, Ferrero I. FOG1 and FOG2 genes, required for the transcriptional activation of glucose-repressible genes of Kluyveromyces lactis, are homologous to GAL83 and SNF1 of Saccharomyces cerevisiae. Curr Genet. 1996;29:316–326. [PubMed] [Google Scholar]
- 17.Hedges D, Proft M, Entian K D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kratzer S, Schuller H J. Transcriptional control of the yeast acetyl-CoA synthetase gene, ACS1, by the positive regulators CAT8 and ADR1 and the pleiotropic repressor UME6. Mol Microbiol. 1997;26:631–641. doi: 10.1046/j.1365-2958.1997.5611937.x. [DOI] [PubMed] [Google Scholar]
- 19.Lodi T, O'Connor D, Goffrini P, Ferrero I. Isolation and characterization of the KlDLD gene encoding the carbon catabolite repressible mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase in Kluyveromyces lactis. Mol Gen Genet. 1994;244:622–629. doi: 10.1007/BF00282752. [DOI] [PubMed] [Google Scholar]
- 20.Mandel J K, Higa S. Calcium dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 21.Nasmyth K A, Reed S I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2121. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmieri L, Lasorsa F M, De Palma A, Palmieri F, Runswick M J, Walker J E. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 1997;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- 23.Proft M, Grzesitza D, Entian K D. Identification and characterization of regulatory elements in the phosphoenolpyruvate carboxykinase gene PCK1 of Saccharomyces cerevisiae. Mol Gen Genet. 1995;246:367–373. doi: 10.1007/BF00288610. [DOI] [PubMed] [Google Scholar]
- 24.Rahner A, Scholer A, Martens E, Gollwitzer B, Schuller H L. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 1996;24:2331–2337. doi: 10.1093/nar/24.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randez-Gil F, Bojunga N, Proft M, Entian K-D. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol. 1997;17:2502–2510. doi: 10.1128/mcb.17.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth S, Schuller H-J. Cat8 and Sip4 mediate regulated transcriptional activation of the yeast malate dehydrogenase gene MDH2 by three carbon source-responsive promoter elements. Yeast. 2001;18:151–162. doi: 10.1002/1097-0061(20010130)18:2<151::AID-YEA662>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein R J. One step disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 31.Scholer A, Schuller H J. A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3613–3622. doi: 10.1128/mcb.14.6.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent O, Carlson M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998;17:7002–7008. doi: 10.1093/emboj/17.23.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Jiang R, Carlson M. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase. EMBO J. 1994;13:5878–5886. doi: 10.1002/j.1460-2075.1994.tb06933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wésolowski-Louvel M, Tanguy-Rougeau C, Fukuhara H. A nuclear gene required for the expression of the linear DNA-associated killer system in the yeast Kluyveromyces lactis. Yeast. 1988;4:71–81. doi: 10.1002/yea.320040108. [DOI] [PubMed] [Google Scholar]
- 35.Zachariae W, Kuger P, Breunig K D. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9. Nucleic Acids Res. 1993;21:69–77. doi: 10.1093/nar/21.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeeman A M, Kuyper M, Pronk J T, van Dijken J P, Steensma H Y. Regulation of pyruvate metabolism in chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast. 2000;16:611–620. doi: 10.1002/(SICI)1097-0061(200005)16:7<611::AID-YEA558>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]