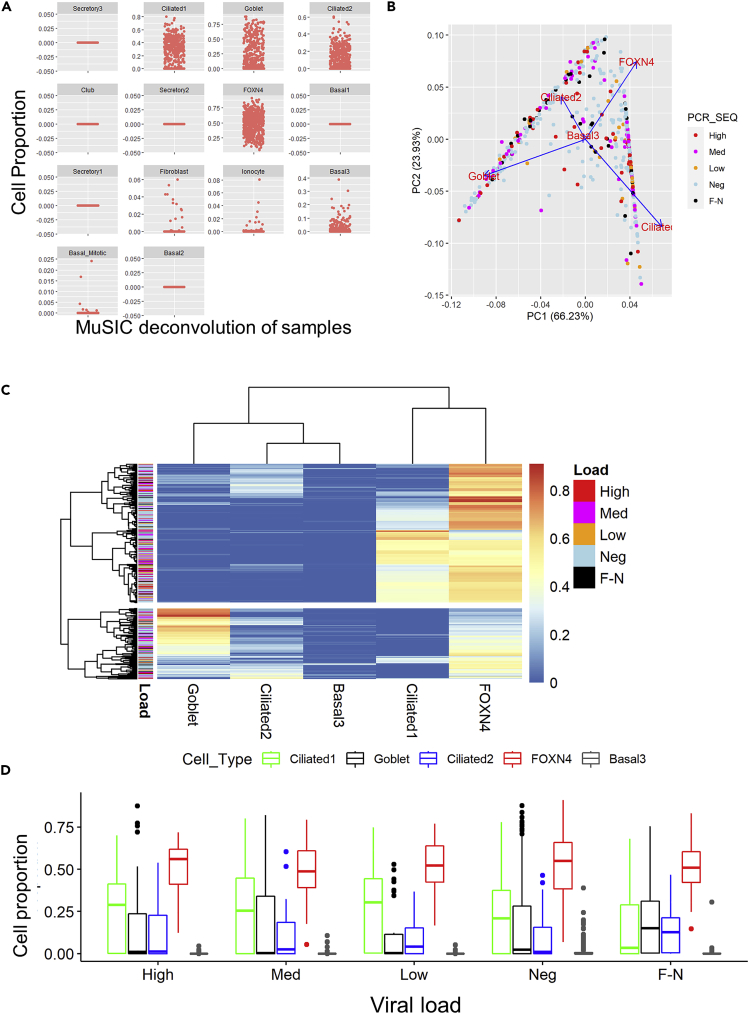
Figure 6.
MuSiC deconvolution of RNA-Seq data does not show differential cell populations in different viral load samples
Sequence data were deconvolved comparing to a nasal brush and upper airway and lung cell single cell reference data set (Lukassen et al., 2020).
(A) Identification of cell proportion in samples based on deconvolution of data.
(B) Principal component analysis of cell proportion data as denoted by PCR viral load and sequence identification of SARS-CoV-2 infection (SARS-CoV-2 Viral Load) status of samples.
(C) Heatmap of cell proportions (x-axis) by sample ID (y axis). The color denotes the relative proportion of the different cell types in the samples. The color bar by the samples denotes their viral load status.
(D) Box plot of airway cell proportion by sample type. No sample showed significant enrichment (2-way ANOVA). Data shown are median +/− quartiles of cell proportions of high (52), med (81) low (33) Neg (275) and FN (42) samples.