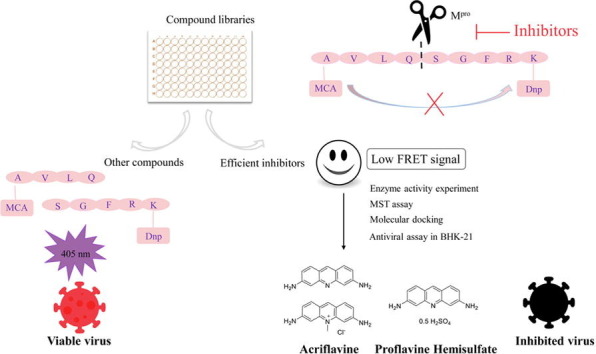
Graphical abstract
Keywords: SARS-CoV-2, Mpro inhibitor, Acriflavine, Proflavine hemisulfate
Abstract
The evolving SARS-CoV-2 epidemic buffets the world, and the concerted efforts are needed to explore effective drugs. Mpro is an intriguing antiviral target for interfering with viral RNA replication and transcription. In order to get potential anti-SARS-CoV-2 agents, we established an enzymatic assay using a fluorogenic substrate to screen the inhibitors of Mpro. Fortunately, Acriflavine (ACF) and Proflavine Hemisulfate (PRF) with the same acridine scaffold were picked out for their good inhibitory activity against Mpro with IC50 of 5.60 ± 0.29 μM and 2.07 ± 0.01 μM, respectively. Further evaluation of MST assay and enzymatic kinetics experiment in vitro showed that they had a certain affinity to SARS-CoV-2 Mpro and were both non-competitive inhibitors. In addition, they inhibited about 90 % HCoV-OC43 replication in BHK-21 cells at 1 μM. Both compounds showed nano-molar activities against SARS-CoV-2 virus, which were superior to GC376 for anti-HCoV-43, and equivalent to the standard molecule remdesivir. Our study demonstrated that ACF and PRF were inhibitors of Mpro, and ACF has been previously reported as a PLpro inhibitor. Taken together, ACF and PRF might be dual-targeted inhibitors to provide protection against infections of coronaviruses.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a pandemic sweeping the world and had become the focus of global health [1], [2]. Although the marketing of inactivated vaccines, attenuated vaccines, and mRNA vaccines effectively blocked the SARS-CoV-2 pandemic and significantly reduced the incidence [3], [4], the increased infectivity caused by the high-frequency gene mutation of SARS-CoV-2, especially the global spread of the Delta variant and Omicron variant, has bought another round of the storm [5], [6]. It is urgent to develop anti-coronavirus drugs with multi-scale mechanisms.
As a cysteine protease, Mpro (main protease, also named 3C-like protease, 3CLpro) regulates viral RNA replication and transcription, has relatively conservative evolution in pathogenic β-coronavirus, and shares significant similarity in the catalytic dyad, His41 and Cys145. The active form of Mpro is a homodimer structure, and its N-finger residues authentic integrity is crucial for proteases activity. Besides, there is a lack of homologous protease in humans, which promotes it as one of the ideal targets for developing anti-coronavirus drugs [7], [8], [9], [10].
Large numbers of Mpro inhibitors have been reported with a variety of screening methods, including virtual screening, FRET technology, cell model screening, and so on [11], [12]. Among these, the in-silico techniques are currently the most widely used and convenient strategy. Typically, scientists used candidate targets to extensively screen the FDA-approved drug databases, natural product databases, clinical trial library, and previously reported coronavirus inhibitors, combined with the molecular dynamics and pharmacological validation to explore their pharmacodynamics and potential binding mechanisms, which can be determined to identify the potential inhibitors or drug repurposing of Mpro [13], [14], [15]. At this stage, a variety of SARS-CoV-2 inhibitors have been found by this strategy, such as Ebselen [16], Baicalein [17], [18], Plumbagin [19], Ginkgolic acid [20], [21], Theaflavin 3-gallate [22], Theasinensin-D, Oolonghomobisflavan-A [23], DSPD-2/5/6 [24]. However, these inhibitors need to be further validated and researched to obtain candidacy for clinical trials.
In the present study, we performed an enzymatic assay using a fluorogenic substrate to screen the inhibitors of Mpro. Acriflavine (ACF) and Proflavine hemisulfate (PRF) were identified as micromolar-range inhibitors. On this basis, MST assay, enzyme activity experiment in vitro, molecular docking, and antiviral activity assay were conducted in-depth research. In summary, this study comprehensively elaborated that the ACF and PRF can be developed as good candidates for anti-coronaviral drugs in vitro and provided useful guidance for its drug repurposing in SARS-CoV-2 therapy.
2. Material and methods
The cDNA of SARS-CoV-2 Mpro (GenBank:MN908947.3) was cloned into pGEX-6P-1 vector (GE Healthcare, Cat.No:27-4597-01, USA). SARS-CoV-2 Mpro gene was constructed by Tsingke Biotechnology Company, China. Human rhinovirus (HRV) 3C protease was obtained from Prof. Li Yan, Huazhong University of Science and Technology. The fluorogenic substrate and compounds were purchased from meilunbio®, China. The ligand library contained 2817 compounds from ZINC database (https://zinc.docking.org/), as well as 1066 compounds separated from traditional Chinese herbals.
2.1. Expression and purification of Mpro
To obtain authentic SARS-CoV-2 Mpro, four amino acids (AVLQ) were added between the GST-tag and N-foreign DNA fragment and eight amino acids (GPHHHHHH) were added to the C-terminus when we constructed the plasmid. The plasmid was then transformed into TSINGKE TSC-E03 TSR2566 Chemically Competent Cells for protein expression.
The signal clone was pre-cultured at 37 °C in 100 ml Luria Broth (LB) medium with ampicillin (100 mg/mL) overnight, and then transferred into 4 L LB medium with ampicillin (100 mg/L). 4 mM isopropyl-d-thiogalactoside (IPTG) was adding until the A 600 value to 0.8. After 16 h incubated at 18 ℃, the cells were harvested, resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 % Triton-X100, 2 mM DTT), and lysed by Continuous High Pressure Cell Disrupter with 1000 bar at a time for 5 rounds on ice. Then cell debris was removed by centrifugation at 20,000 rpm, 4 °C for 30 min. The supernatants were loaded onto a Ni-NTA (GE Healthcare) affinity column and washed with buffer A1 (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM imidazole) and buffer A2 (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20 mM imidazole) for five column volumes, respectively. 0.4 mg/mL HRV-3C protease was then added to samples of buffer A2 for cleaving the C-terminal eight amino acids at 4 ˚C overnight. The resulting protein was loaded onto a Ni-NTA column to separate the His-tag-label protein or non-His-tag-label protein by buffer A1. The resulting protein sample was further purified to Superdex 75 10/300GL loaded by AKTA-pure (GE Healthcare). The purity of the recombinant protein was collected and confirmed by 15 % SDS-PAGE (Supplementary Information, S1). The pro-prepared protein with 2 mM DTT was stored at −80 ℃ for enzymatic inhibition assay and MST assay.
2.2. Enzymatic inhibition assay
An enzymatic assay using a fluorogenic substrate was applied to measure the inhibitory activity of compounds on Mpro. Firstly, the correctness of the enzyme activity system in this experiment was determined by assaying the effects of two reported positive drugs GC376 [25] and carmofur [9]. Secondly, a large-scale preliminary screening was launched, and the inhibition activities of candidate compounds were finally confirmed.
The compounds were dissolved in DMSO (20 μM final concentration), mixed with the reaction system (150 nM SARS-CoV-Mpro, 50 mM Tris-HCl, pH 7.3, 1 mM EDTA) and then incubated at 30 °C for 15 min. The reaction was initiated by adding fluorogenic substrate (MCA-AVLQSGFR(Dnp)-Lys-NH2) at a final concentration of 22 μM in 96-well black non-detachable plate. After that, the fluorescence signal at 320 nm (excitation)/405 nm (emission) were monitored every 2 s for 15 min by BioTEK Synergy H1 fluorescence spectrophotometer. The equivalent Mpro, fluorogenic substrate DMSO and positive control GC376 and carmofur were assayed as control simultaneous.
When the inhibition activity of compounds is up to 50 %, the complexes were picked out to determine the half-maximal inhibitory (IC50) again. In detail, the complexes were assayed at ten different concentration between 0 and 40 µM, and the concentration of the enzyme was 150 nM. To further determine the inhibition mode by enzymatic kinetics study, the complexes were assayed at six different concentrations between 0 and 20 µM, and concentrations of enzyme was 150 nM, the substrates were assayed at eight different concentration between 0.78125 and 200 µM, respectively. The hydrolysis of the substrate was monitored again, and the rate of hydrolysis was determined in the linear range.
For each compound experiment was performed in triplicate. The IC50 values were calculated by plotting the average percentage inhibition against inhibitor concentration and fitting the data in GraphPad Prism 7.
2.3. Microscale thermophoresis (MST) assay
The pro-protein was diluted with buffer B (20 mM HEPES, pH 7.3, 1 mM EDTA) and kept constant at 10 µM. The tested compounds were diluted in proper concentration for the test. After incubating 100 µL pro-protein and isometric fluorochrome at room temperature lucifugally for 30 min, then loaded into Monolith™ (Germany) standard-treated capillaries to collect samples with a Norm. The fluorescence value was 2000–2500 AU. After that, the same amount of compounds and labeled samples were measured at 25 °C after 15 min incubation, while laser power was set to 20 % or 40 % using 30 sec on-time, and the LED power was set to 100 %. All experiments were repeated three times for each measurement. The dissociation constant Kd values were fitted by using the NT analysis.
2.4. Structure-based molecular docking
PDB ID: 6YB7/6LU7 in RCSB Protein Data Bank (https://www.pdb.org) is the best-resolution form of ligand-free and ligand-induced monomers in SARS-CoV-2 Mpro. Before docking, protein structures were optimized using Protein Preparation Wizard (Schrodinger), polar hydrogens and Gasteriger charges were added using AutodockTools (Vision-1.5.7).
The skeleton was acquired from PubChem (Compound CID: 7099, https://pubchem.ncbi.nlm.nih.gov). All torsions were chosen and Gasteriger charges were added using AutodockTools (Vision-1.5.7).
The active sites were defined according to the literature [26]. The first is target the “cryptic site” (CS): Lys5, Met6, Pro108, Gly109, Arg131, Trp218, Phe219, Tyr239, Glu240, Leu271, Leu272, Leu287, Glu288, Asp289, Glu290, Arg298, Gln299 and Val303. The second is target the “dimerization site” (DS): Arg4, Met6, Ser10, Gly11, Glu14, Asn28, Ser139, Phe140, Ser147, Glu166, Glu290 and Arg298. The third is the “ligand-induced substrate binding site” (ISBS): His41, Met49, Tyr54, Gly143, Ser144, Cys145, Phe140, Leu141, Asn142, His163, Met165, Glu166, Leu167, Pro168, His172, Phe185, Asp187, Gln189, Thr190, Ala191, Gln192.
In the case of CS and DS, 6YB7 was the chosen receptor, while in the case of ISBS, the 6LU7 was chosen. The grid-line of points in X-Y-Z dimension is 126-126-84 with 0.336 spacing at the center of X: 11.522, Y: 7.344, Z: −4.732 for CS. While the grid-line of points in the X-Y-Z dimension is 66-88-102 with 0.386 spacing at the center of X: 3.639, Y: −0.005, and Z: 4.985 for DS. Moreover, the grid-line of points in the X-Y-Z dimension is 56-56-70 with 0.464 spacing at the center of X: −15.106, Y: 12.610, and Z: 68.479 for ISBS. The docking calculation was performed in AutoDock Vina (Version-1.1.2), 2D ligand–protein interaction diagrams were generated by PoseView in Proteins Plus (https://proteins.plus/), and the 3D interaction diagrams were predicted in Pymol.
2.5. HCoV-OC43 and SARS-CoV-2 antiviral assay
The HCoV-OC43 antiviral assay was performed routinely by plaque assay as previously described with slight modification [27], [28]. Briefly, BHK-21 cells were seeded in 6-well plates and cultured overnight. Then cells were infected with 100 plaque-forming units (PFU) HCoV-OC43 in the presence of compounds with different concentrations. After incubation at 33 °C for 2 h, the supernatant was removed and cells were supplemented with DMEM containing different concentrations of the compounds, 1.2 % Avicel (FMC Biopolymer, USA) and 2 % FBS. After 3 to 4 days post infection, BHK-21 cells were fixed and stained with 4 % formaldehyde containing 1 % crystal violet, and the number of plaques was counted after rinsing with water.
For testing the anti-SARS-CoV-2 efficacy of acriflavine and proflavine hemisulfate, plaque assay was conducted in a Biosafety Level 3 (BSL-3) laboratory of Fudan University as previously described [29]. Vero-E6 cells were first seeded into a 96-well plate. After cultured overnight, these two compounds were serially diluted in DMEM medium and incubated with authentic SARS-CoV-2 viruses for 30 min. The mixture was subsequently applied to the Vero-E6 cells and further incubated for 2 h. Subsequently, 1 % methyl cellulose (Sigma, USA) was added followed by culture for further 72 h. Finally, PBS containing 4 % paraformaldehyde and 1 % crystal violet was added for fixation and staining, and then plaques were counted after rinsing with water.
3. Results
3.1. Screening of SARS-CoV-2 Mpro inhibitors
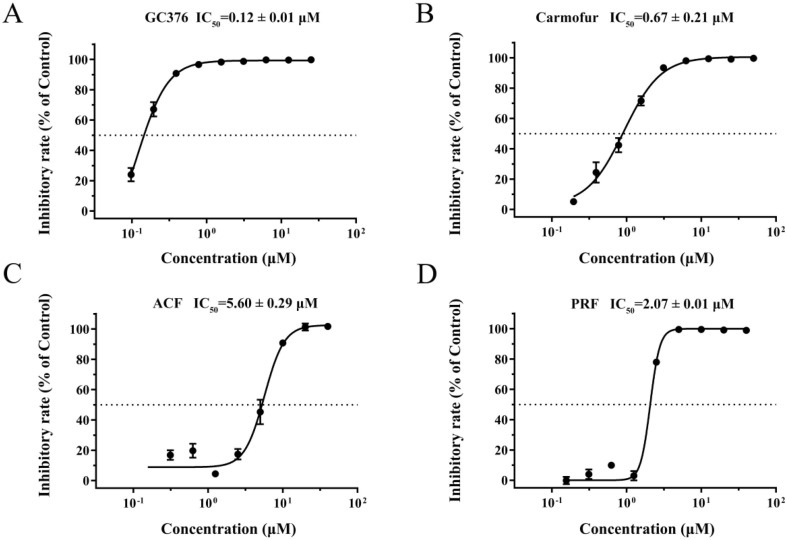
The enzymatic activity was measured by time-dependent kinetics using a fluorogenic substrate MCA-AVLQSGFR(Dnp)-Lys-NH2 to identify the potential inhibitors of SARS-CoV-2 Mpro. The relative fluorescence unit (RFU) was used to measure the amount of substrate depletion. Based on that, the activity of Mpro could be tested through the measurement of Km and Vmax values. The specific experimental protocol has been reported in detail [9]. We first determined the previously reported potent inhibitors of Mpro, GC376, and carmofur. The dose–response curve was shown in Fig. 1 A and B, the IC50 values of GC376 and carmofur were 0.12 μM and 0.67 μM, respectively. 2817 FDA-approved drugs and several natural products were first screened as the protocol described above. To our surprise, ACF and PRF showed an encouraging inhibitory effect with IC50 values of 5.60 μM and 2.07 μM (Fig. 1 C, D), respectively. In brief, the results of the enzymatic assay implied that ACF and PRF would be developed as anti-SARS-CoV-2 reagents (Fig. 1).
Fig. 1.
Inhibition of the enzymatic activity of SARS-CoV-2 Mpro by enzymatic assay. A, GC376 against SARS-CoV-2 Mpro. B, Carmofur against SARS-CoV-2 Mpro. C, Representative curve for ACF against SARS-CoV-2 Mpro. D, Representative curve for PRF against SARS-CoV-2 Mpro. All data are shown as mean ± SD, n = 3.
3.2. Inhibition mode assays supported by enzymatic kinetic and MST
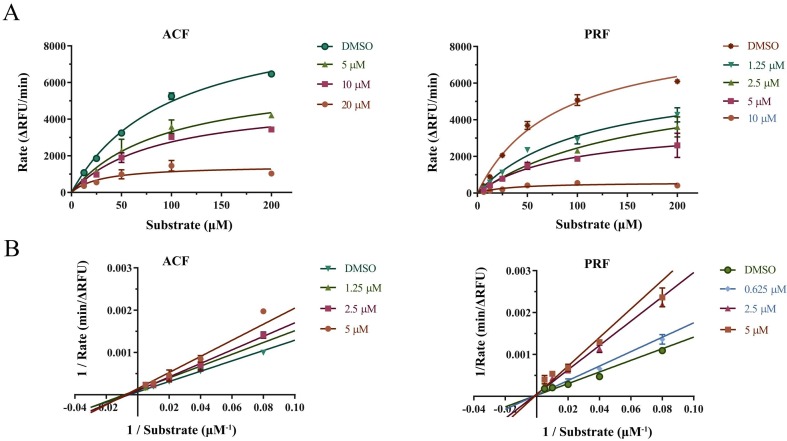
To further validate the inhibition mode of the ACF and PRF with Mpro, the enzyme kinetic parameters were determined. After the enzymatic reaction proceeded for about 15 min, it can be observed that a large amount of substrate was consumed. As shown in Fig. 2 , it was observed that their inhibition activities had a good correlation with the concentration. The point we would like to raise was ACF with the constant Km and concentration-dependent-decreased Vmax, implying that ACF non-competitively inhibited Mpro. Similarly, in the study of inhibition mode measurement of PRF, Km basically remained unchanged with the increase of compound concentration, while Vmax decreased, indicating that PRF also non-competitively inhibited Mpro (Table 1 ).
Fig. 2.
Inhibition mode of ACF and PRF. A, Reaction rate of the fluorogenic substrate MCA-AVLQSGFR(Dnp)-Lys-NH2 catalyzed by SARS-CoV-2 Mpro in the presence of different concentration compounds. B, Enzyme kinetics curve by Lineweaver-Burk. All data are shown as mean ± SD, n = 3.
Table 1.
The maximum reaction velocity (Vmax) and constant (Km) during the process of the Enzymatic-inhibitions reaction.
| Compound (μM) | Kinetic parameter | 0 | 1.25 | 2.5 | 5 |
|---|---|---|---|---|---|
| ACF | Vmax (ΔRFU/min) | 9877 | 9273 | 6245 | 6397 |
| Km (μM) | 99.01 | 115.80 | 70.60 | 93.44 |
| Compound (μM) | Kinetic parameter | 0 | 0.625 | 2.5 | 5 |
|---|---|---|---|---|---|
| PRF | Vmax (ΔfRFU/min) | 8897 | 8888 | 6491 | 3740 |
| Km (μM) | 79.74 | 113.00 | 164.10 | 89.94 |
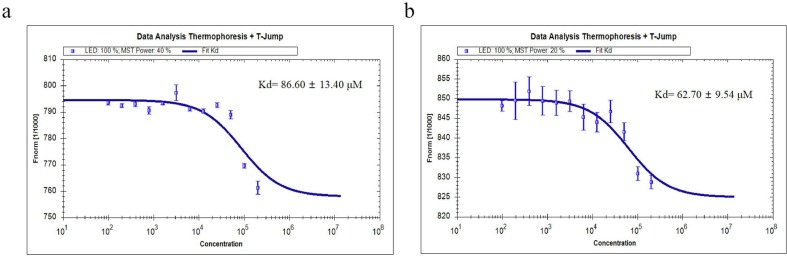
Furthermore, MST assay had been used to identify the binding affinity between compounds and Mpro. ACF and PRF demonstrated that the Kd values were 86.60 ± 13.14 μM and 62.70 ± 9.54 μM, respectively (Fig. 3 ). The Kd was used to describe the binding affinity: the smaller Kd, the higher the affinity of the inhibitor to a target, the firmer the complexes. The encouraging Kd provided the groundwork for the potent inhibition. More than this, it was highly likely that the inhibition mode could be associated with the Kd. ACF and PRF were both non-competitive inhibitors, they had an affinity with the free Mpro and enzyme-substrate (ES) complexes. There was less interference of substrate with its binding to the target, therefore ACF and PRF had strong affinity with Mpro in vitro.
Fig. 3.
The binding affinity between compounds and of SARS-CoV-2 Mpro measured with MST assay. A, ACF. B, PRF. The error bars represent the SD of each data point calculated from three independent thermophoresis measurements, n = 3.
3.3. Molecular docking
Non-competitive inhibitors would bind to enzyme in sites rather than substrate binding sites (SBS). To better understand the binding mechanism between the compound and protein, we presumed its binding sites would deviate from the active pocket.
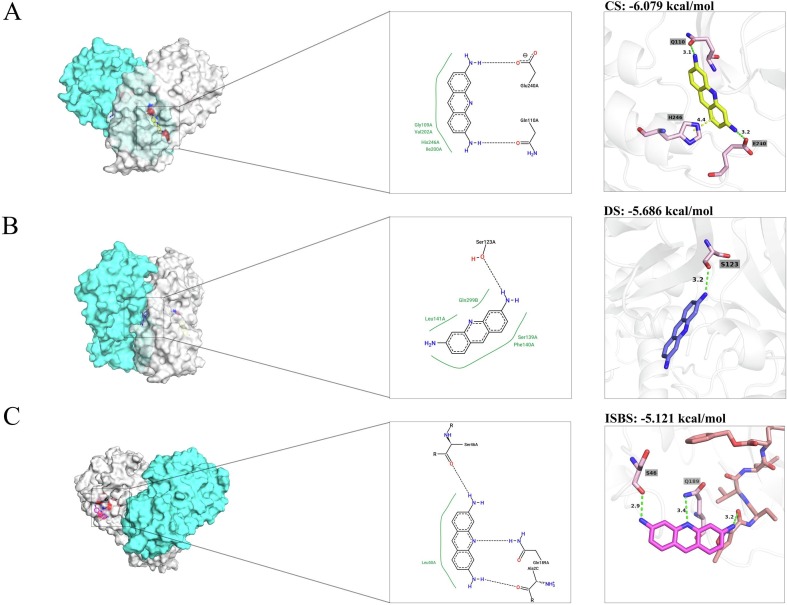
The present study revealed that there are three candidate-binding sites for inhibiting the Mpro. The first strategy is target the “cryptic site” (CS), which is a candidate allosteric site. The second strategy is target the “dimerization site” (DS), which could interrupt the dimeric conformation and inactive Mpro. The third is bound to the complex with the present of the substrate, named “ligand-induced substrate binding site” (ISBS). Thus, three plausible docking strategies were adopted and results were given as Fig. 4 .
Fig. 4.
Interaction analysis of Proflavine (ACF) with SARS-CoV-2 Mpro proteasea. A, The enlarged view of the “cryptic site” (CS). B, The enlarged view of “dimerization site” (DS). C, The enlarged view of the “ligand-induced substrate binding site” (ISBS). Note: Left panel: The overview of ACF in different sites. PDB ID: 6YB7 is shown in surface with two protomer, while protomer A is in cyans, protomer B is in gray. Middle panel: Two dimensions (2D) ACF-Mpro interaction diagram. Binding interactions of ACF to Mpro as analyzed by Proteins Plus. Right panel: The 3D visualization of interaction diagram. Both ligands and interacting residues are shown as sticks, while protein is depicted as cartoon. Hydrogen bonds are represented by the green-dashed line and pi-pi interaction are shown in yellow-dashed line. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To our surprise, ACF was fitted well in three models, which presented the energy to CS (−6.079 kcal/mol), DS (−5.686 kcal/mol), and ISBS (−5.121 kcal/mol). For CS, the amino-terminal formed two hydrogen bonds with Gln110, and Glu240. Benzene contacted imidazole on His246 via pi-pi interaction. For DS, ACF was fitted well between the junctions of protomer, while it bound to Ser123 with one hydrogen, showing disrupted the interplay of dimerization stability. For ISBS, the amino-terminal formed hydrogen bonds with Ser46 and the inhibitor N3. Remarkably, the 10-N of ACF were involved the hydrogen with Gln189. Gln189 is utilized as one of the key residue of Mpro for influencing the connecting loop between Domain II and III [30], [31]. Darunavir, ritonavir, and saquinavir etc. had the comparable interactions with Gln189 [32], which implied that our compound had a broad possibility to bind to ISBS. Different docking strategies provided conformational variation in different sites. Taken together, these results indicated that there were diverse interaction patterns of acridine skeleton recognition by SARS-CoV-2 Mpro.
3.4. Antiviral assay
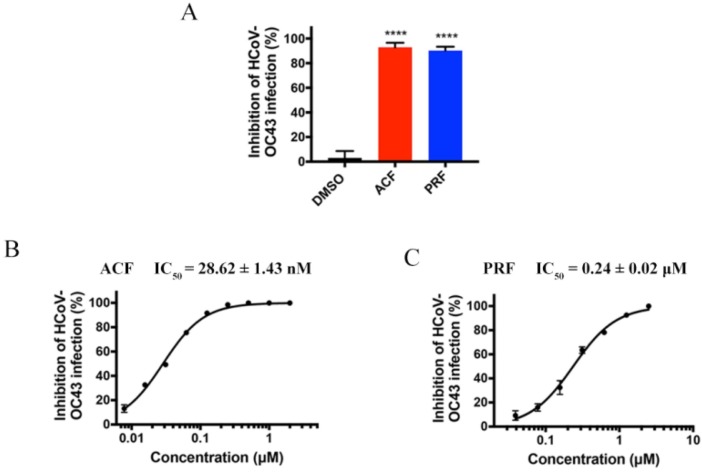
It’s imperative to evaluate the antiviral activity of ACF and PRF against human β-coronavirus. One of the classical human β-coronaviruses, HCoV-OC43, was first used to evaluate the antiviral activity by plaque assay. HCoV-OC43 can replicate efficiently in BHK-21 cell, which was used as a host for virus [27], [28], [33]. Particularly, at a concentration of 1 μM, the inhibitory rate of ACF and PRF on OC43 reached about 90 %. Especially, ACF and PRF showed inhibition of HCoV-OC43 replication, with an IC50 value of 28.62 ± 1.43 nM and 0.24 ± 0.02 μM, respectively (Fig. 5 ), which were stated to be more effective than the positive control, GC376 (36.95 ± 2.92 nM).
Fig. 5.
Antiviral activities of the drug leads against HCoV-OC43. A, Antiviral activities of ACF and PRF against HCoV-OC43 at 1 μM. B, Dose–response curves for ACF against HCoV-OC43. C, Dose–response curves for PRF against HCoV-OC43. All data are shown as mean ± SD, n = 3. Probability (p) values were calculated by the unpaired two-tailed Student’s-t-test between the compound and DMSO (**** p < 0.0001).
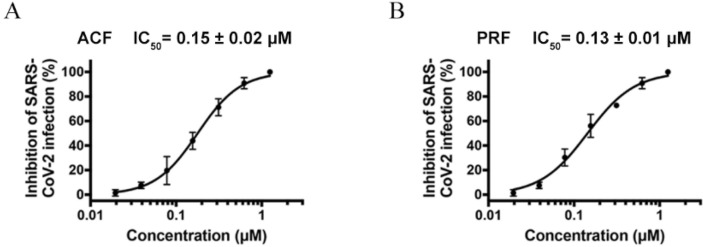
Authentic SARS-CoV-2 was also used to evaluate the antiviral activity of ACF and PRF. In the experiment, the IC50 values of ACF and PRF against SARS-CoV-2 were 0.15 ± 0.02 μM and 0.13 ± 0.01 μM, respectively (Fig. 6 ), which showed the equivalent with the standard molecule remdesivir (0.19 ± 0.05 μM). The result was encouraging since nanomole-level antiviral activities opened a solid avenue for anti-SARS-CoV-2.
Fig. 6.
Antiviral activities of the drug leads against SARS-CoV-2. Dose–response curves for ACF (A) and PRF (B) against SARS-CoV-2. All data are shown as mean ± SD, n = 3.
4. Discussion
The COVID-19 epidemic caused by SARS-CoV-2 infection has always been highly infectious since its outbreak [34]. The emergence of SARS-CoV-2 variants further enhances its infectivity and pathogenicity and reduces the protection of vaccines [10]. Hence, it is of great significance to develop anti-coronavirus drugs. The evolutionarily conserved Mpro plays an important role in regulating the RNA replication and transcription of the virus, and there is no protease similar to Mpro in the human body. Therefore, Mpro is a promising therapeutic target for the development of anti-coronavirus drugs.
A variety of screening methods have been reported for the screening and discovery of small molecule inhibitors of Mpro, including virtual screening [35], FRET technology [36], [37], [38], [39], cell model screening[31], and phenotypic screening [40], etc. We constructed a nine-peptide-fluorogenic molecular as the substrate of Mpro, which recognize the sequence of -AVLQ/SGFRK- (the cleavage site is indicated by /) [41]. The value of RFU is applied to characterizing the activity of candidate compounds. Of note, since the fluorescent molecules are easily quenched, they need to be freshly prepared. In addition, some natural products may have fluorescent properties and can cause interference in the RFU value detection of the FRET screening. It is necessary to use the physical and chemical information of the database in time to eliminate the interference of these compounds effectively to improve the screening efficiency. Furthermore, considering that the enzyme kinetic reaction of Mpro is easily affected by environment and temperature, to maintain the stability and repeatability of the FRET screening model, it is necessary to add DTT for containing Cys145 more stable and ensure the consistency of the FRET reaction conditions as much as possible [42].
Based on FRET technology, ACF and PRF with the same acridine scaffold were screened out, and showed promising inhibition of HCoV-OC43 replication in BHK-21. Their inhibition abilities are approximately same as that of Shuanghuanglian and Ebselen [9], [17]. Remarkably, the IC50 values of ACF and PRF have reached the micron mole level, which were superior to GC376 for anti-HCoV-43, and equivalent to the standard molecule remdesivir, suggesting that they have the potential for drug repurposing. In addition, ACF has been reported to have a strong inhibitory effect on SARS-CoV-2 papain-like protease (PLpro) with an IC50 of 1.66 μM. The antiviral activity results of ACF in cell and mouse models showed that the combination of ACF and remdesivir had a strong synergistic effect on inhibiting viral replication. Moreover, the researchers successfully resolved the co-crystal structure of ACF-PLpro by X-ray crystallography. Combined with our study, we suspect that ACF and PRF might be a dual-target inhibitor of SARS-CoV-2, including but not limited to Mpro and PLpro, which explains why ACF shows excellent antiviral activity [43]. Generally, Mpro and PLpro are two proteases that function in the replication and packaging of new generation viruses and can handle the translation of peptides from genomic RNA to structural or non-structural proteins during viral replication. Similarly, Disulfiram and Ebselen [44], [45], [46], undergoing clinical trials, also inhibited the hydrolysis of these two proteolytic enzymes, which suggests that ACF is worthy of further study.
By confirming the enzyme kinetics, it was determined that both ACF and PRF were non-competitive inhibitors. And MST assay demonstrated that they both have a certain affinity with SARS-CoV-2 Mpro. Fortunately, with the simple scaffold, visual docking mechanism, and encouraging antiviral activity in vitro, ACF and PRF were elaborated as good candidates for the development of anti-coronaviral drugs.
5. Conclusion
In summary, we combined FRET technology, enzyme kinetics, MST, molecular docking, and cell-based antiviral assay, and finally ACF and PRF were found to be a new type of Mpro small molecule inhibitor with encouraging inhibitory activity. ACF and PRF were determined to be non-competitive inhibitors. Two compounds exhibited a concentration-dependent inhibition pattern against Mpro. Of note, our study opened a new avenue for exploring new uses of acridine scaffolds, and demonstrated that ACF was a dual-target candidate, which was a complementary to its target. As the epidemic is raging around the world, we proposed that ACF and PRF still have potential for the treatment of coronaviruses.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We particularly thank Prof. Li Yan from Huazhong University of Science and Technology for grants of plasmids. Heartfelt thanks to the support and technical guidance of members in Biosafety Level 3 Laboratory of Fudan University, especially Dr. Di Qu, Xia Cai, Gaowei Hu, Qian Wang, Zhiping Sun, Yutang Li and Jing Pu.
Statements and Declarations
Funding
This work was supported by National Natural Science Foundation of China (NSFC) (No. 82141216), Chunhui Program-Cooperative Research Project of the Ministry of Education, Liaoning Province Natural Science Foundation (No. 2020-MZLH-31), Shenyang Young and Middle-aged Innovative Talents Support Program (RC210446), National Key Research and Development Program of China (2021YFC2300703) and the Shanghai Science and Technology Committee (STCSM) Science and Technology Innovation Program (No. 21S11902600).
Ethics approval
No need approval.
Competing interests
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2022.106185.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M., Urquiza J., Ramírez D., Alonso C., Campillo N.E., Martinez A. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63(21):12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 2.V’Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.COVID-19 vaccines, Drugs and Lactation Database (LactMed), National Library of Medicine (US), Bethesda (MD), 2006.
- 4.Pandey S.C., Pande V., Sati D., Upreti S., Samant M. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sci. 2020;256:117956. doi: 10.1016/j.lfs.2020.117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxena S.K., Kumar S., Ansari S., Paweska J.T., Maurya V.K., Tripathi A.K., Abdel‐Moneim A.S. Characterization of the novel SARS‐CoV‐2 Omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 2022;94(4):1738–1744. doi: 10.1002/jmv.27524. [DOI] [PubMed] [Google Scholar]
- 6.B.1.1.529 (Omicron) Variant – United States, December 1–8, 2021. MMWR Morb. Mortal Wkly. Rep. 2021;70(50):1731–1734. doi: 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong M., Su H., Zhao W., Xie H., Shao Q., Xu Y. What coronavirus 3C-like protease tells us: from structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 2021;41(4):1965–1998. doi: 10.1002/med.21783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anirudhan V., Lee H., Cheng H., Cooper L., Rong L. Targeting SARS-CoV-2 viral proteases as a therapeutic strategy to treat COVID-19. J. Med. Virol. 2021;93(5):2722–2734. doi: 10.1002/jmv.26814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 10.Xie X.D., Hu L.C., Xue H., Xiong Y., Panayi A.C., Lin Z., Chen L., Yan C.C., Zhou W., Mi B.B., Liu G.H. Prognosis and treatment of complications associated with COVID-19: a systematic review and meta-analysis. Acta Mater. Med. 2022;1(1):124–137. doi: 10.15212/AMM-2022-0002. [DOI] [Google Scholar]
- 11.Song Z.J., Nik Nabil W.N., Xi Z.C., Xu H.X. Current global status and future development of traditional Chinese medicine in the prevention and treatment of coronavirus disease 2019. World J. Tradit. Chin. Med. 2021;7(2):155–166. doi: 10.4103/wjtcm.wjtcm_43_20. [DOI] [Google Scholar]
- 12.Li M.X., Yang Y.Y., Yang L., Zheng M.Z., Li J., Chen L.X., Li H. Progress of traditional Chinese medicine treating COVID-19. World J. Tradit. Chin. Med. 2021;7(2):167–183. doi: 10.4103/wjtcm.wjtcm_68_20. [DOI] [Google Scholar]
- 13.Amin S.A., Jha T. Fight against novel coronavirus: a perspective of medicinal chemists. Eur. J. Med. Chem. 2020;201:112559. doi: 10.1016/j.ejmech.2020.112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R., Hu Q., Wang H., Zhu G., Wang M., Zhang Q., Zhao Y., Li C., Zhang Y., Ge G., Chen H., Chen L. Identification of Vitamin K3 and its analogues as covalent inhibitors of SARS-CoV-2 3CLpro. Int. J. Boil. Macromol. 2021;183:182–192. doi: 10.1016/j.ijbiomac.2021.04.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Y., Zhu G.H., Zhang Y.N., Hu Q., Wang H.N., Yu H.N., Qin X.Y., Guan X.Q., Xiang Y.W., Tang H., Ge G.B. Flavonoids in Ampelopsis grossedentata as covalent inhibitors of SARS-CoV-2 3CLpro: inhibition potentials, covalent binding sites and inhibitory mechanisms. Int. J. Boil. Macromol. 2021;187:976–987. doi: 10.1016/j.ijbiomac.2021.07.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haritha C.V., Sharun K., Jose B. Ebselen, a new candidate therapeutic against SARS-CoV-2. Int. J. Surg. 2020;84:53–56. doi: 10.1016/j.ijsu.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zandi K., Musall K., Oo A., Cao D., Liang B., Hassandarvish P., Lan S., Slack R.L., Kirby K.A., Bassit L., Amblard F., Kim B., AbuBakar S., Sarafianos S.G., Schinazi R.F. Baicalein and baicalin inhibit SARS-CoV-2 RNA-dependent-RNA polymerase. Microorganisms. 2021;9(5):893. doi: 10.3390/microorganisms9050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Yu S., Li Y., Liang X., Su M., Li R. Medical significance of uterine corpus endometrial carcinoma patients infected with SARS-CoV-2 and pharmacological characteristics of plumbagin. Front Endocrinol. (Lausanne) 2021;12:714909. doi: 10.3389/fendo.2021.714909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Cui Q., Cooper L., Zhang P., Lee H., Chen Z., Wang Y., Liu X., Rong L., Du R. Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 2021;11(1):45. doi: 10.1186/s13578-021-00564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong Y., Zhu G.H., Wang H.N., Hu Q., Chen L.L., Guan X.Q., Li H.L., Chen H.Z., Tang H., Ge G.B. Discovery of naturally occurring inhibitors against SARS-CoV-2 3CL(pro) from Ginkgo biloba leaves via large-scale screening. Fitoterapia. 2021;152:104909. doi: 10.1016/j.fitote.2021.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauhan M., Bhardwaj V.K., Kumar A., Kumar V., Kumar P., Enayathullah M.G., Thomas J., George J., Kumar B.K., Purohit R., Kumar A., Kumar S. Theaflavin 3-gallate inhibits the main protease (Mpro) of SARS-CoV-2 and reduces its count in vitro. Sci. Rep. 2022;12(1):13146. doi: 10.1038/s41598-022-17558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39(10):3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11(1):4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiménez-Avalos G., Vargas-Ruiz A.P., Delgado-Pease N.E., Olivos-Ramirez G.E., Sheen P., Fernández-Díaz M., Quiliano M., Zimic M. COVID-19 Working Group in Perú, Comprehensive virtual screening of 4.8 k flavonoids reveals novel insights into allosteric inhibition of SARS-CoV-2 MPRO. Sci. Rep. 2021;11(1):15452. doi: 10.1038/s41598-021-94951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Wang X., Shi H., Zou P. Montelukast inhibits HCoV-OC43 infection as a viral inactivato. Viruses. 2022;14(5):861. doi: 10.3390/v14050861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Chen Y., Shi H., Zou P. Erythromycin estolate is a potent inhibitor against HCoV-OC43 by directly inactivating the virus particle, frontiers in cellular and infection. Microbiology. 2022:930. doi: 10.3389/fcimb.2022.905248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z., Xu W., Chen Z., Fu W., Zhan W., Gao Y., Zhou J., Zhou Y., Wu J., Wang Q., Zhang X. An ultrapotent pan-β-coronavirus lineage B (β-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope. Protein & cell. 2022;13(9):655–675. doi: 10.1007/s13238-021-00871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komatsu T.S., Okimoto N., Koyama Y.M., Hirano Y., Morimoto G., Ohno Y., Taiji M. Drug binding dynamics of the dimeric SARS-CoV-2 main protease, determined by molecular dynamics simulation. Sci. Rep. 2020;10(1):16986. doi: 10.1038/s41598-020-74099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63(9):4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J.W., Xiong Y., Wang F., Zhang F.M., Yang X., Lin G.Q., Tian P., Ge G., Gao D. Discovery of 9,10-dihydrophenanthrene derivatives as SARS-CoV-2 3CLpro inhibitors for treating COVID-19. Eur. J. Med. Chem. 2022;228:114030. doi: 10.1016/j.ejmech.2021.114030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S., Tan W. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93(12):e00023–e00119. doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee R., Perera L., Tillekeratne L.M.V. Potential SARS-CoV-2 main protease inhibitors. Drug Disc. Today. 2021;26(3):804–816. doi: 10.1016/j.drudis.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiou W.C., Hsu M.S., Chen Y.T., Yang J.M., Tsay Y.G., Huang H.C., Huang C. Repurposing existing drugs: identification of SARS-CoV-2 3C-like protease inhibitors. J. Enzyme Inhib. Med. Chem. 2021;36(1):147–153. doi: 10.1080/14756366.2020.1850710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu M., Uchil P.D., Li W., Zheng D., Terry D.S., Gorman J., Shi W., Zhang B., Zhou T., Ding S., Gasser R., Prévost J., Beaudoin-Bussières G., Anand S.P., Laumaea A., Grover J.R., Liu L., Ho D.D., Mascola J.R., Finzi A., Kwong P.D., Blanchard S.C., Mothes W. Real-time conformational dynamics of SARS-CoV-2 spikes on virus particles. Cell Host Microbe. 2020;28(6):880–891.e8. doi: 10.1016/j.chom.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorshkov K., Vasquez D.M., Chiem K., Ye C., Tran B.N., de la Torre J.C., Moran T., Chen C.Z., Martinez-Sobrido L., Zheng W. A SARS-CoV-2 nucleocapsid protein TR-FRET assay amenable to high-throughput screening. ACS Pharmacol. Transl. Sci. 2022;5(1):8–19. doi: 10.1101/2021.07.03.450938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jans D.A., Wagstaff K.M. The broad spectrum host-directed agent ivermectin as an antiviral for SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:163–172. doi: 10.1016/j.bbrc.2020.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., Benner S.E., Patel E.U., Littlefield K., Laeyendecker O., Shoham S., Sullivan D., Casadevall A., Pekosz A., Nardin A., Fehlings M., Tobian A.A., Quinn T.C. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131(5):e145476. doi: 10.1172/jci145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuck C.P., Chen C., Ke Z., Wan D.C., Chow H.F., Wong K.B. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur. J. Med. Chem. 2013;59:1–6. doi: 10.1016/j.ejmech.2012.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are non-specific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 2020;3(6):1265–1277. doi: 10.1101/2020.09.15.299164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Napolitano V., Dabrowska A., Schorpp K., Mourão A., Barreto-Duran E., Benedyk M., Botwina P., Brandner S., Bostock M., Chykunova Y., Czarna A., Dubin G., Fröhlich T., Hölscher M., Jedrysik M., Matsuda A., Owczarek K., Pachota M., Plettenburg O., Potempa J., Rothenaigner I., Schlauderer F., Slysz K., Szczepanski A., Greve-Isdahl Mohn K., Blomberg B., Sattler M., Hadian K., Popowicz G.M., Pyrc K. Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses. Cell Chem. Biol. 2022;29(5):774–784.e8. doi: 10.1016/j.chembiol.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma C., Tan H., Choza J., Wang Y., Wang J. Validation and invalidation of SARS-CoV-2 main protease inhibitors using the Flip-GFP and Protease-Glo luciferase assays. Acta Pharm. Sin. B. 2022;12(4):1636–1651. doi: 10.1016/j.apsb.2021.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin M.H., Moses D.C., Hsieh C.H., Cheng S.C., Chen Y.H., Sun C.Y., Chou C.Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nogara P.A., Omage F.B., Bolzan G.R., Delgado C.P., Aschner M., Orian L., Teixeira Rocha J.B. In silico Studies on the Interaction between Mpro and PLpro From SARS-CoV-2 and Ebselen, its Metabolites and Derivatives. Mol. Inform. 2021;40(8) doi: 10.1002/minf.202100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.