Abstract
Shiga toxin‐producing E. coli (STEC) are zoonotic foodborne pathogens of outmost importance and interest has been raised in recent years to define the potential zoonotic role of wildlife in STEC infection. This study aimed to estimate prevalence of STEC in free‐ranging red deer (Cervus elaphus) living in areas with different anthropisation levels and describe the characteristics of strains in order to evaluate the potential risk posed to humans. Two‐hundred one deer faecal samples collected in 2016–2018 from animals of Central Italian Alps were examined by bacteriological analysis and PCR screening of E. coli colonies for stx1, stx2 and eae genes. STEC strains were detected in 40 (19.9%) deer, with significantly higher prevalence in offspring than in yearlings. Whole genome analysis was performed to characterise a subset of 31 STEC strains. The most frequently detected serotype was O146:H28 (n = 10, 32.3%). Virulotyping showed different stx subtypes combinations, with stx2b‐only (n = 15, 48.4%) being the most prevalent. All STEC lacked the eae gene but harbored additional virulence genes, particularly adhesins, toxins and/or other colonisation factors also described in STEC isolated from disease in humans. The most frequently detected genes were astA (n = 22, 71%), subAB (n = 21, 68%), iha (n = 26, 83.9%) and lpfA (n = 24, 77%). Four hybrid STEC/Enterotoxigenic E. coli strains were also identified. According to the most recent paradigm for pathogenicity assessment of STEC issued by the European Food Safety Authority, our results suggest that red deer are carriers of STEC strains that may have zoonotic potential, regardless of the anthropisation levels. Particular attention should be drawn to these findings while handling and preparing game meat. Furthermore, deer may release STEC in the environment, possibly leading to the contamination of soil and water sources.
Keywords: STEC, virulence features, wild ruminants, wild ungulates, zoonoses
1. INTRODUCTION
The populations of wild ungulates, especially red deer (Cervus elaphus), are increasing their density and distribution across Europe (Dias et al., 2019). This, alongside habitat fragmentation caused by human activities, increases spatial aggregations and wildlife conflicts with humans, livestock and other animal species, possibly generating interfaces that might be important routes for the transmission of infectious agents (Galiero et al., 2018).
In recent years, the popularity of wild game meat consumption has also increased (Costa et al., 2016). The role of wild ungulates as reservoir of foodborne pathogens, including Shiga toxin‐producing Escherichia coli (STEC), has been previously reported (Kruse et al., 2004, EFSA and ECDC, 2019). STEC are zoonotic foodborne pathogens of outmost importance: in 2018, countries from the European Union and European Economic Area reported a total of 9633 cases of STEC infection in humans (EFSA & ECDC, 2019). Clinical condition associated with STEC infection in humans varies from mild intestinal discomfort to severe life‐threatening conditions (such as haemolytic uremic syndrome –HUS – and kidney failure) (FAO/WHO, 2018). STEC belonging to serogroups O26, O103, O111, O145 and O157 are the most frequently detected among patients with HUS and are termed as top‐five STEC serogroups (EFSA and ECDC, 2019). According to the most recent pathogenicity assessment of STEC issued by European Food Safety Authority (EFSA), the most severe clinical pictures in humans are associated with certain Shiga‐toxin subtypes and STEC strains carrying genes whose products enable attachment to intestinal epithelial cells, regardless of serotype (EFSA BIOHAZ Panel et al., 2020 ).
The transmission of STEC strains to humans occurs primarily through consumption of contaminated food products of animal origin (Espinosa et al., 2018). Ruminants, especially cattle, are the natural reservoir of STEC and shed the microorganism in faeces (Caprioli et al., 2005).
STEC strains have been isolated from faeces of wild ungulates, especially deer species, in different European countries, the United States, and Japan (Dias et al., 2019; Szczerba‐Turek et al., 2020). Long‐term shedding of STEC from deer faeces has been observed (Topalcengiz et al., 2020). STEC have been detected also in carcasses of hunted wild ruminants (Sauvala et al., 2019) and they may cause severe diseases in humans, although rarely (Díaz‐Sánchez et al., 2013; FAO/WHO, 2018).
There is scant information about the prevalence and zoonotic potential of the STEC population in territories where wildlife develops in areas with different levels of anthropisation, as those included in National parks. The aims of this study were (i) to evaluate the prevalence and spatial distribution of STEC in free‐ranging red deer in the Stelvio National park (SNP, Italy) where different levels of anthropisation occur, over two consecutive Winter seasons (2016–2017 and 2017–2018) and (ii) to describe the major virulence features of STEC isolates in order to define the zoonotic potential of wildlife‐associated STEC.
2. MATERIALS AND METHODS
2.1. Study area and deer sampling
The study area was located in the Lombardy sector of the SNP, Central Italian Alps (46°27′ N, 10°25′ E), where free‐ranging red deer (Cervus elaphus), roe deer (Capreolus capreolus), chamois (Rupicapra rupicapra) and ibex (Capra ibex) are present. Furthermore, cattle and small ruminant herds share alpine pastures in Summer season. The SNP has been functionally divided in 11 different areas (identified as A‐L), included within three macroareas with different density levels of presence of humans and domestic animals (low, medium, high), based on the proportion of human settlements and agricultural landscapes (Figure 1). Briefly, as previously reported (Formenti et al., 2015), in the low anthropised macroarea (772 ha) human settlements and agricultural landscapes around small villages constitute 7% of the whole surface, in the high anthropised macroarea (707 ha) they are much more widely distributed and represent 32% of the total surface and in medium macroarea (1200 ha) the density level of anthropisation is intermediate compared to previous ones. Radio‐tracking of animals suggest limited deer movements among different areas of the SNP territory.
FIGURE 1.
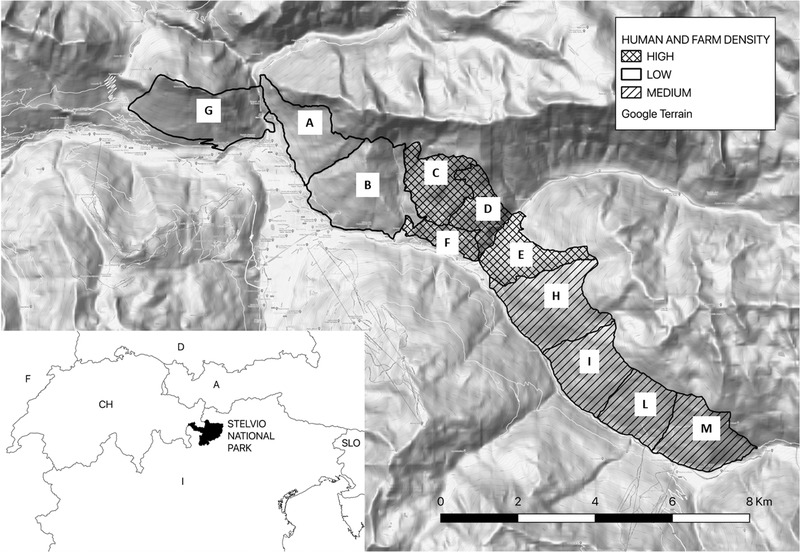
Macroareas and areas of the Stelvio Natural Park, Italy according to human and domestic animals density level
Red deer increased over the recent years in the SNP, causing intense browsing impact leading to damage of forest vegetation and cross transmission of pathogens with domestic ruminants (Galiero et al., 2018). To reduce deer density, an official culling plan has been authorised by Istituto Superiore per la Protezione e la Ricerca Ambientale (ISPRA), the Italian Ministry of Environment (Prot. 48585/T‐A25‐Ispra), in the Lombardy sector of the Park starting from 2011 and 170 individuals/year is the maximum level of the culling plan in autumn–winter 2017–2021 (Pedrotti et al., 2017). Between the 2014 and 2017, the census estimated a red deer population of 1,792 individuals in SNP with a mean density of 9.5 individuals/km2 in autumn (Pedrotti et al., 2017). The animals investigated for the presence of STEC described here were culled for management purposes and were not sacrificed for research purposes specific to this study. The red deer included in this study were all wild and free‐living and were selectively culled during the 2016–2017 and the 2017–2018 Winter seasons.
After selective culling, all animals were immediately brought to the check point of the SNP for inspection by veterinarians. Culling site (area of SNP), sex and age were recorded for each animal. Animals were classified as calves (< 1 year old), yearlings (1 year old) and adults (≥ 2 years old). Faecal samples were collected directly from the rectum. Samples were stored at −20°C immediately after collection. Frozen samples were brought to the laboratory monthly for further processing.
2.2. Bacteriological analysis and isolation of STEC strains
One g of each faecal sample was enriched into 9 mL of buffered peptone water (BPW; Microbiol Diagnostici, Cagliari, Italy) in aseptic conditions and incubated overnight at 37°C.
The BPW enrichment cultures of the samples were plated on Levine‐eosin methylene blue agar and incubated at 37°C for 18–24 hours. Ten single typical E. coli colonies were selected from each sample. DNA was extracted using the boiling method from each colony. Extracted DNAs from the 10 colonies of single animals were pooled and tested using previously published PCR protocols for stx1 and stx2 genes to confirm the presence of STEC (Hu et al., 1999). As the PCR protocol used did not amplify the stx2f gene subtype, a separate reaction was set up to identify the strains producing this toxin subtype as described elsewhere (Scheutz et al., 2012).
A faecal sample with a negative stx PCR result in pooled colonies was considered negative. In the presence of stx positive pooled samples, DNA from single colonies composing the pools was further analysed using the same PCR protocols. In the presence of the same stx profile observed in the single colonies composing a pool, one STEC colony/animal was isolated and further subjected to characterisation. The strains possessing stx1 and/or stx2 genes were confirmed as E. coli using biochemical analysis (API® systems, bioMérieux, Florence, Italy) and were eventually designated as STEC in this study. A faecal sample was considered as STEC‐positive based on the isolation of a single E. coli colony positive for the presence of stx genes. The isolated STEC strains were assayed for the presence of the eae gene by conventional PCR (Paton & Paton, 1998 ).
2.3. Whole genome sequencing, strains characterisation and phylogenetic analysis
Thirty‐one STEC strains out of those initially isolated from STEC‐positive faeces of free‐ranging red deer in the SNP were successfully regained after long‐term storage and were subjected to whole genome sequencing (WGS) and analysis. In detail, total DNA was extracted from 2 mL of overnight cultures in Trypticase soy broth (TSB) using the E.Z.N.A. Bacterial DNA kit (Omega Bio‐tek, Norcross, GA, USA) and sequenced on an Ion Torrent S5 platform (Life Technologies, Carlsbad, USA). The template preparation and sequencing run were performed with the ION 520/530 KIT‐OT2 following the manufacturer's instructions for 400 bp DNA libraries (NEBNext® Fast DNA Fragmentation & Library Prep Set for Ion Torrent, New England Biolabs, Ipswich, Massachusetts, USA) on ION 530 chips.
All the bioinformatics analyses were performed on the public ARIES Galaxy server (Knijn et al., 2020) applying the PHANtAsTiC 1.0 pipeline (https://github.com/aknijn/phantastic‐galaxy). Briefly, prior to the pipeline FastQC v0.11.9 was used for quality check and Trimmomatic v0.36 (Bolger et al., 2014) for trimming the raw reads. The contigs were assembled from trimmed data using SPAdes version 3.12.0 (Bankevich et al., 2012), followed by the tool ‘Filter SPAdes repeats’ Galaxy Version 1.0.1 (https://github.com/phac‐nml/galaxy_tools/tree/master/tools/filter_spades_repeats), using default parameters in the two steps.
The assembled contigs were used for the determination of the serotype by alignment against the database of the reference O and H antigens‐associated genes sequences (Joensen et al., 2015), through the NCBI BLAST + blastn algorithm v2.9.0+ (Camacho et al., 2009). The virulence genes content was determined through the Patho_typing tool v1.0 (https://github.com/B‐UMMI/patho_typing), developed by the INNUENDO project (Llarena et al., 2018), using the E. coli virulence genes database (Joensen. et al, 2014). The stx‐subtyping was performed through the Shiga toxin typer tool v2.0 (https://github.com/aknijn/shigatoxin‐galaxy), an optimised blastn search against the sequences database of stx subtypes developed by the Statens Serum Institut (https://bitbucket.org/genomicepidemiology/virulencefinder_db/src/master/stx.fsa).
Additionally, the presence of tia gene was determined using NCBI BLAST+blastn tool querying the assembled contigs versus tia gene (Acc. No U20318.1).
Multilocus sequence typing (MLST) of the STEC isolates was carried out in silico according to the scheme proposed by Wirth et al. (2006).
Subtyping of the enterohaemolysin coding‐gene was performed by carrying out in silico PCR‐RFLP as previously described (Michelacci et al., 2018).
The whole‐genome sequences (WGS) of the 31 STEC strains have been uploaded on the National Center for Biotechnology Information public repository (https://www.ncbi.nlm.nih.gov; Study Accession no. PRJNA643386).
2.4. Data analyses
A Pearson's chi‐square test was used to evaluate the association of STEC presence with sex, age and SNP macroarea of culled red deer. Statistical comparisons were conducted using SPSS v.15.0 software, using p < 0.05 as threshold for statistical significance.
3. RESULTS
3.1. Occurrence of STEC in red deer
Faecal samples were collected from a total of 201 red deer. The characteristics of the sampled deer are reported in Table 1. Forty faecal samples out of 201 red deer faeces showed stx PCR positive results in pooled colonies. All the STEC colonies composing a pool showed the same stx profile and one STEC colony per animal was obtained for all the 40 faecal samples that were PCR‐positive in pooled colonies. Results showed a STEC prevalence in red deer of 19.9% (95% CI:14.4–25.4). Four STEC isolates harbored stx1 and stx2 genes, whereas 23 and 13 possessed stx2 and stx1 only, respectively. The stx‐2f gene and the intimin coding gene (eae) were never detected by PCR or WGS.
TABLE 1.
Characterisation of the red deer analyzed in this study
| Age | Sex | Season | STEC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP area (macroarea) | No. | Calf | Yearling | Adult | Female | Male | 2016–2017 | 2017–2018 | No. stx‐positive faecal sample |
| A (low) | 12 | 1 | 4 | 7 | 6 | 6 | 2 | 10 | 3 |
| B (low) | 10 | 4 | 2 | 4 | 9 | 1 | 2 | 8 | 4 |
| C (high) | 15 | 4 | 0 | 11 | 11 | 4 | 2 | 13 | 2 |
| D (high) | 54 | 24 | 6 | 24 | 30 | 24 | 15 | 39 | 15 |
| E (high) | 26 | 5 | 3 | 18 | 13 | 13 | 10 | 16 | 3 |
| F (high) | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| G (low) | 25 | 5 | 7 | 13 | 15 | 10 | 6 | 19 | 5 |
| H (medium) | 26 | 7 | 6 | 13 | 17 | 9 | 12 | 14 | 3 |
| I (medium) | 7 | 1 | 1 | 5 | 4 | 3 | 3 | 4 | 1 |
| L (medium) | 17 | 10 | 1 | 6 | 13 | 4 | 9 | 8 | 2 |
| M (medium) | 8 | 2 | 1 | 5 | 5 | 3 | 4 | 4 | 2 |
| Total | 201 | 63 | 31 | 107 | 123 | 78 | 65 | 136 | 40 |
The results of the WGS‐based characterisation of a set of 31 STEC strains isolated in this study and successfully regained after long‐term storage are reported in Table 2 in grouped form and in Table S1 in extended form. Results showed the detection of seven different stx subtypes combinations among the isolates, with stx2b‐only being the most frequent pattern.
TABLE 2.
Serotype, sequence type and number of isolates harbouring virulence genes among the 31 STEC strains from red deer analyzed by whole genome sequencing
| O146:H28 ST738 (n = 10) | O113:H4 ST10 (n = 4) | O187:H28 ST200 (n = 4) | O91:H14 ST33 (n = 3) | O91:H21 ST442 (n = 1) | O27:H30 ST53 (n = 3) | O104:H7 ST1817 (n = 1) | O174:H8 ST13 (n = 1) | O178:H19 ST443 (n = 1) | ONT:H49 ST5418 (n = 3) | Total (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shiga‐toxin pattern † | stx1a | 2 | 3 | 5 (16.1) | ||||||||
| stx1c | 2 | 1 | 3 (9.7) | |||||||||
| stx2b | 10 | 1 | 3 | 1 | 15 (48.4) | |||||||
| stx2g | 4 | 4 (12.9) | ||||||||||
| stx1a+stx2a | 1 | 1 (3.2) | ||||||||||
| stx1a+stx2b | 1 | 1 (3.2) | ||||||||||
| stx1c+stx2b | 2 | 2 (6.5) | ||||||||||
| Genes involved in colonisation ‡ | eae | 0 | ||||||||||
| iha | 10 | 4 | 3 | 1 | 3 | 1 | 1 | 3 | 26 (83.9) | |||
| saa | 3 | 1 | 4 (12.9) | |||||||||
| lpfA | 10 | 4 | 3 | 1 | 1 | 1 | 1 | 3 | 24 (77) | |||
| tia | 4 | 3 | 1 | 3 | 1 | 1 | 13 (41.9) | |||||
| aggR | 0 | |||||||||||
| aaiC | 1 | 3 | 4 (12.9) | |||||||||
| aat | 0 | |||||||||||
| Genes encoding toxins § | ehxA A | 4 | 3 | 1 | 1 | 1 | 10 (32.3) | |||||
| ehxA D | 4 | 4 (12.9) | ||||||||||
| astA | 10 | 4 | 4 | 1 | 3 | 22 (71) | ||||||
| subAB | 10 | 4 | 3 | 1 | 3 | 21 (67.7) | ||||||
| sta1 | 4 | 4 (12.9) | ||||||||||
| ltcA | 0 |
stx, Shiga toxin subtype.
eae, intimin; iha, encoding the adherence‐conferring protein Iha, a homologue of Vibrio cholera IrgAiron‐regulated gene A homolog adhesion similar to V. cholerae; saa, STEC agglutinating adhesin; lpfA, long polar fimbriae closely related to LPF of Salmonella enterica serovar Typhimuriuman; tia, invasion determinant of the subtilase‐encoding pathogenicity island of LEE‐negative STEC; aggR, transcriptional activator of aggregative adherence fimbria I expression of EAEC; aaiC, EAEC aggR‐activated island C; aat, EAEC‐associated anti‐aggregation transporter.
ehxA A, enterohemolysin‐encoding gene subtype A; ehxA D, enterohemolysin‐encoding gene subtype D; astA, EAEC heat‐stable toxin; subAB, subtilase cytotoxin locus; sta1, ETEC heat‐stable enterotoxin 1; ltcA, ETEC heat‐labile enterotoxin.
3.2. Molecular serotyping and multi‐locus sequence typing
The 31 STEC isolates showed 10 different serotypes and 10 sequence types (Figure 2; Table 2), comprising 9 O‐groups and 9 H‐types. For three isolates the O‐group could not be determined (ONT).
FIGURE 2.
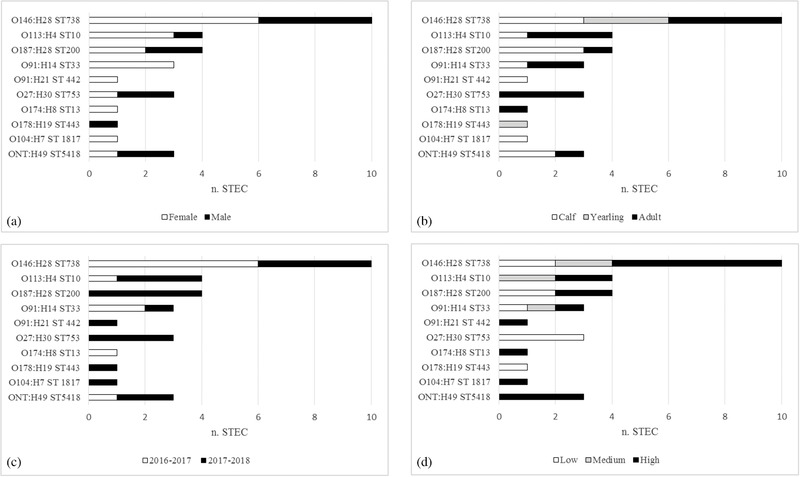
Distribution of 31 STEC serotypes and sequence types from red deer of Stelvio National Park according to sex (a), age (b), winter season of collection (c) and human and domestic animals density level macroareas (d)
Ten (32.3%) strains belonged to serotype O146:H28, the most common among the STEC strains isolated in this study. The other two most frequently detected serotypes were O113:H4 and O187:H28, observed in four (12.9%) STEC strains each. Isolates with the same serotype belonged to the same sequence type (Tables 2 and S1).
3.3. Virulence genes
The presence of genes encoding virulence factors assessed by WGS of the 31 STEC strains is reported in Table 2.
A total of 10 virulence genes reported in STEC and other diarrhoeagenic E. coli were detected among the isolates and there was a median of 5 virulence genes per isolate (range: 3–7). In particular, the number of virulence genes additional to stx in each STEC varied between two and six. Our study showed different virulence genes profiles that were specific for each STEC serotype.
As described above, all the analyzed STEC isolates lacked the eae gene but possessed one or more putative STEC‐associated colonisation factors including those encoded by the genes iha, lpfA, saa (Toma et al., 2004) and the tia gene (Michelacci et al., 2013 ). In particular, the iha adhesion gene was the most prevalent and was present in the majority of the isolates, followed by the lpfA gene.
The aaiC gene described in some Enteroaggregative E. coli (EAEC) strains (Dudley et al., 2006) was detected in four strains harboring stx1 gene. However, the aggR and the aat genes, typically associated with EAEC, were not detected.
Beside the stx genes, all but one isolate possessed at least one of the following toxin genes described in STEC: ehxA, subAB (Paton et al., 2004) and astA. The majority of the isolates (n = 22, 71%) contained two or more of these genes whereas the remaining eight strains (25.8%) possessed only one of them. The astA gene was the most frequently detected followed by the subAB. Two of the six described subtypes of the enterohemolysin‐encoding gene (ehxA) (Cookson et al., 2007) were detected. These included the subtype A and D.
The sta1 gene encoding for the heat‐stable enterotoxin 1 typically associated with Enterotoxigenic E. coli (ETEC) was detected in four strains all belonging to O187:H28 whereas the ltcA gene coding for ETEC heat‐labile enterotoxin was never detected.
3.4. Distribution of STEC strains and risk factors
STEC prevalence was significantly higher in offspring compared to yearlings (p = 0.02). STEC‐positive calves were detected in all anthropised macroareas of the SNP. No significant differences were observed for STEC presence considering the sex of animals and the winter season of collection (Table 3).
TABLE 3.
Number and percentage of red deer with STEC according to age, sex, anthropisation level of the SNP (macroarea) and winter season of collection
| Variable | Category | No. tested | No. with STEC (% STEC prevalence; 95% C.I.) |
|---|---|---|---|
| Age | Calves | 63 | 19 (30.2; 18.8–41.5)* |
| Yearling | 31 | 2 (6.5; 0.1–15.1)* | |
| Adult | 107 | 19 (17.8; 10.5–25.0) | |
| Sex | Female | 123 | 25 (20.3; 13.2–27.4) |
| Male | 78 | 15 (19.2; 10.5–28.0) | |
| Anthropsation level SNP | Low | 47 | 12 (25.5; 13.1–38.0) |
| Medium | 58 | 8 (13.8; 4.9–22.7) | |
| High | 96 | 20 (20.8; 12.7–29.0) | |
| Winter season of collection | 2016–2017 | 65 | 12 (18.5; 9.0–27.9) |
| 2017–2018 | 136 | 28 (20.6; 13.8–27.3) |
Significant difference between categories of the same variable (p < 0.05).
The distribution of the 31 STEC serotypes and sequence types according to sex, age, winter season of collection and macroarea is reported in Figure 2.
4. DISCUSSION
The gastrointestinal tract of ruminants is the main natural reservoir of STEC (Caprioli et al., 2005). While the role of farmed ruminants is well recognised in the epidemiology of STEC disease in humans, evidences are accumulating that wildlife, including wild ruminants, may be also an important reservoir of foodborne pathogens including STEC (Espinosa et al., 2018; Szcerba‐Turek et al., 2020). Of particular interest is the study of the dynamics of the STEC population in territories where wildlife develops in areas with different levels of anthropisation, as those included in the Stelvio National Park.
The present study showed that wild populations of red deer in Stelvio National Park carried STEC strains and may shed these pathogens in the environment during winter, with prevalence similar to cattle (FAO/WHO, 2018 ). The high STEC prevalence observed is in accordance with previous studies on red deer (Espinosa et al., 2018). Our results did not confirm previous reports showing that STEC prevalence in red deer was significantly higher in areas with presence of livestock (Díaz‐Sánchez et al., 2013). On the contrary, STEC shedding especially occurred in young animals and in all macroareas, regardless the anthropisation level. In turn, this suggests a possible role of this species as STEC carrier confirming the importance of wildlife in STEC shedding dynamics (Singh et al., 2015; Topalcengiz et al., 2020).
The high frequency of stx2 (especially stx2b) genes and the absence of the eae gene in all strains isolated indicates that red deer from this study mainly carry LEE‐negative STEC strains, confirming previous reports showing that eae‐negative STEC are apparently a feature of STEC typically shed by red deer (FAO/WHO, 2018; Dias et al., 2019). The majority of STEC strains isolated from red deer in this study belonged to serogroup O146, similarly with what reported in previous studies (Sánchez et al., 2009; Díaz‐Sánchez et al., 2013). This is an emerging STEC serogroup in the EU (ECDC surveillance atlas of infectious diseases, https://atlas.ecdc.europa.eu/public/index.aspx). Two hundred‐twenty STEC O146 infections have been reported to ECDC in 2019 to have occurred in the EU/EEA including two HUS cases in children from Sweden and Austria. Unfortunately, the characterisation of the H antigen was not available in the ECDC dataset. STEC strains of O146:H28 serotype had also been isolated from human illness (Michelacci et al., 2013).
STEC belonging to the top‐five STEC serogroups were not detected in our study, confirming their rare prevalence in deer (Dias et al., 2019). However, we have not used serogroup‐specific tests as the immunomagnetic enrichment for the detection and isolation of STEC and this may have accounted for the absence of the top‐five STEC serogroups among strains isolated in the red deer population.
The most recent risk assessment exercises (FAO/WHO, 2018) and pathogenicity assessment of STEC (EFSA BIOHAZ Panel, 2020) agree that STEC serogroups are not indicative of virulence. Serogroups are rather to be considered as markers for epidemiological surveillance, whereas the determination of the Stx subtypes harboured by a strain is now considered the main feature for STEC implication to severe disease. Strains producing stx2a are most consistently associated with HUS, even in the absence of adherence factors. Among the other Stx subtypes, stx1a, stx2c and stx2d have also been implicated in cases of bloody diarrhoea and HUS, but their association with HUS requires the presence of adherence‐conferring genes and is not as definitive nor conclusive, and other factors may affect disease outcome (EFSA BIOHAZ Panel, 2020).
In our study, one STEC strain obtained from a yearling from an area with low level of anthropisation harboured the gene stx2a. This finding is interesting, as the presence of stx2a gene has been rarely detected in STEC from red deer from other countries (Szczerba‐Turek et al., 2020). Remarkably, this STEC strain also harboured several additional virulence genes (stx1a, ehxA type A, saa, iha, lpfA, tia) reinforcing the virulence potential of this isolate. This Stx2a‐producing strain belonged to O178:H19 serotype and STEC belonging to this serotype have been rarely described in STEC from human disease in the EU (https://atlas.ecdc.europa.eu/public/index.aspx).
All the STEC strains isolated in this study, regardless the stx2 subtype, also encoded adhesins, toxins and possessed other genes involved in the colonisation of the host gut, already described in STEC isolated from disease in humans (Toma et al., 2004; Michelacci et al., 2013 ). The red deer isolates belonging to the serogroups O113 and O91 hosted the genes encoding the enterohemolysin‐coding gene (ehxA) subtype A, a subtype already described in eae‐negative STEC associated with severe disease in humans (Cookson et al., 2007).
We also identified STEC O187:H28 in four animals carrying the rare stx2g subtype, the ehxA type D, the lpfA and the gene astA common in STEC, coupled with the presence of the sta1 gene, encoding the heat stable enterotoxin typically produced by ETEC. Hybrid E. coli pathotypes with enhanced virulence from different pathotypes represent emerging health threats. A hybrid STEC/ETEC similar to those identified in red deer, of serotype O187:H28, harbouring the genes stx2g and the genes encoding two different heat stable enterotoxin‐coding genes, sta4 and sta5, has been described recently from a small child with diarrhoea in Sweden (Bai et al., 2019). The identification of these strains encoding cross‐pathotype features in red deer calves from four of the areas sampled in this study suggests that this STEC population is likely circulating in the study area.
It should be considered that the approach used in this study, including freezing and storing faecal matter at −20°C, the testing of single colonies per faecal sample and characterising by WGS only one isolated colony per sample, may have led to underestimate the variability of the STEC strains circulating in the deer population in the study area. Additionally, only 31 isolates from the 40 originally isolated from the study population were recovered after the long term storage and this may have also limited the observation of the whole picture, although the prevalence estimated is in line with that described elsewhere (Espinosa et al, 2018 ).
Further work based on WGS of all STEC colonies detected in each faecal sample is necessary to define if red deers shed more than one STEC strain. The influence of warm seasons on the increase of faecal shedding of STEC in the red deer population and the dynamics affecting the circulation of STEC in wild ruminants in the area should also be evaluated.
Overall, our data confirmed that wild ruminants are a natural reservoir of STEC and suggest that the STEC strains investigated and circulating in red deer population in the study area may pose a threat for human health. Unfortunately, STEC surveillance in humans in Italy is based on the identification of the HUS cases only and no information is available on the circulation of STEC causing less severe illness. Ad hoc studies would be therefore necessary to assess whether STEC strains with similar characteristics to those observed in the deers’ isolates have shown up in the human population living close to the SNP.
Altogether, considering that following the culling plan in the SNP 37.3 tons of deer meat have been introduced into the food chain close to the SNP area in Italy between 2011 and 2016 (Pedrotti et al., 2017), our findings strengthen the need of observance of good hygiene practice while preparing game meat (Sauvala et al., 2019; Topalcengiz et al., 2020).
Moreover, it should be mentioned that the STEC contamination of soils from faeces of wild ruminants may be a source of contamination of the environment, including water bodies that in turn may cause exposure of humans to these pathogens (Caprioli et al., 2005). This points the attention to the possible role of wild ruminants in the environmental pathways of spreading of STEC infections.
Emphasis should be focused on the interconnection of STEC presence with the level of anthropisation and the possible overlap with areas where livestock farming activities may account for the inter‐species transfer of isolates and the shuffling of virulence features among strain belonging to different pathotypes.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest associated with this study.
AUTHOR CONTRIBUTIONS
All authors reviewed, revised and approved the final manuscript and have contributed significantly to the work.
ETHICAL APPROVAL
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required, and ethical statement is not applicable as sample collection from animals has been gathered after animals were culled for management purposes according to the official culling plan to reduce red deer density that has been authorised by Istituto Superiore per la Protezione e la Ricerca Ambientale (ISPRA), the Italian Ministry of Environment (Prot. 48585/T‐A25‐Ispra), in the Lombardy sector of the Park starting from 2011 (Pedrotti et al., 2017). Therefore animals were not sacrificed for research purposes specific to this study.
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGEMENTS
This work was partially funded by University of Milan (CUP G46D15001850005 Piano di Sostegno alla Ricerca UNIMI 2017, Identification of hot spots for zoonosis transmission in Stelvio National Park, principal investigator Luzzago C.).
Open Access Funding provided by Universita degli Studi di Milano within the CRUI‐CARE Agreement.
Lauzi, S. , Luzzago, C. , Chiani, P. , Michelacci, V. , Knijn, A. , Pedrotti, L. , Corlatti, L. , Buccheri Pederzoli, C. , Scavia, G. , Morabito, S. , & Tozzoli, R. (2022). Free‐ranging red deer (Cervus elaphus) as carriers of potentially zoonotic Shiga toxin‐producing Escherichia coli . Transboundary and Emerging Diseases, 69, 1902–1911. 10.1111/tbed.14178
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bai, X. , Zhang, J. , Ambikan, A. , Jernberg, C. , Ehricht, R. , Scheutz, F. , Xiong, Y. , & Matussek, A. (2019). Molecular characterization and comparative genomics of clinical hybrid Shiga toxin‐producing and enterotoxigenic Escherichia coli (STEC/ETEC) strains in Sweden. Scientific Reports, 9, 5619. 10.1038/s41598-019-42122-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A. A. , Dvorkin, M. , Kulikov, A. S. , Lesin, V. M. , Nikolenko, S. I. , Pham, S. , Prjibelski, A. D. , Pyshkin, A. V. , Sirotkin, A. V. , Vyahhi, N. , Tesler, G. , Alekseyev, M. A. , & Pevzner, P. A. (2012). SPAdes: A new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computational Biology: A Journal of Computational Molecular Cell Biology, 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England), 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. , & Madden, T. L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics, 10, 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli, A. , Morabito, S. , Brugère, H. , & Oswald, E. (2005). Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Veterinary Research, 36, 289–311. 10.1051/vetres:2005002 [DOI] [PubMed] [Google Scholar]
- Cookson, A. L. , Bennett, J. , Thomson‐Carter, F. , & Attwood, G. T. (2007). Molecular subtyping and genetic analysis of the enterohemolysin gene (ehxA) from Shiga toxin‐producing Escherichia coli and atypical enteropathogenic E. coli . Applied and Environmental Microbiology, 73, 6360–6369. 10.1128/AEM.00316-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, H. , Mafra, I. , Oliveira, M. B. P. P. , & Amaral, J. S. (2016). Game: Types and composition. Caballero F.T.B., Finglas P.M., & Toldrá F. (Eds.), Encyclopedia of food and health (pp. 177–183). Oxford: Academic Press. [Google Scholar]
- Dias, D. , Caetano, T. , Torres, R. T. , Fonseca, C. , & Mendo, S. (2019). Shiga toxin‐producing Escherichia coli in wild ungulates. The Science of the Total Environment, 651, 203–209. 10.1016/j.scitotenv.2018.09.162 [DOI] [PubMed] [Google Scholar]
- Díaz‐Sánchez, S. , Sánchez, S. , Herrera‐León, S. , Porrero, C. , Blanco, J. , Dahbi, G. , Blanco, J. E. , Mora, A. , Mateo, R. , Hanning, I. , & Vidal, D. (2013). Prevalence of Shiga toxin‐producing Escherichia coli, Salmonella spp. and Campylobacter spp. in large game animals intended for consumption: Relationship with management practices and livestock influence. Veterinary Microbiology, 163, 274–281. 10.1016/j.vetmic.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Dudley, E. G. , Thomson, N. R. , Parkhill, J. , Morin, N. P. , & Nataro, J. P. (2006). Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli . Molecular Microbiology, 61, 1267–1282. 10.1111/j.1365-2958.2006.05281.x [DOI] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (European Food Safety Authority Panel on Biological Hazards) , Koutsoumanis, K. , Allende, A. , Alvarez Ordóñez, A. , Bover Cid, S. , Chemaly, M. , Davies, R. , De Cesare, A. , Herman, L. , Hilbert, F. , Lindqvist, R. , Nauta, M. , Peixe, L. , Ru, G. , Simmons, M. , Skandamis, P. , Suffredini, E. , Jenkins, C. , Pires, S. M. , … Bolton, D. (2020). Pathogenicity assessment of Shiga toxin‐producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA Journal, 18, 5967. 10.2903/j.efsa.2020.5967 [DOI] [Google Scholar]
- EFSA, ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) (2019). The European Union One Health 2018 Zoonoses report. EFSA Journal, 17, 5926. 10.2903/j.efsa.2019.5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa, L. , Gray, A. , Duffy, G. , Fanning, S. , & McMahon, B. J. (2018). A scoping review on the prevalence of Shiga‐toxigenic Escherichia coli in wild animal species. Zoonoses and Public Health, 65, 911–920. 10.1111/zph.12508 [DOI] [PubMed] [Google Scholar]
- FAO/WHO (World Health Organization & Food and Agriculture Organization of the United Nations) (2018). Shiga toxin‐producing Escherichia coli (STEC) and food: Attribution, characterization, and monitoring: Report. World Health Organization. Retrieved from https://apps.who.int/iris/handle/10665/272871 [Google Scholar]
- Formenti, N. , Trogu, T. , Pedrotti, L. , Gaffuri, A. , Lanfranchi, P. , & Ferrari, N. (2015). Toxoplasma gondii infection in Alpine Red Deer (Cervus elaphus): Its spread and effects on fertility. Plos One, 10, e0138472. 10.1371/journal.pone.0138472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiero, A. , Leo, S. , Garbarino, C. , Arrigoni, N. , Russo, S. , Giacomelli, S. , Bianchi, A. , Trevisiol, K. , Idrizi, I. , Daka, G. , Fratini, F. , Turchi, B. , Cerri, D. , & Ricchi, M. (2018). Mycobacterium aviumsubsp. paratuberculosis isolated from wild red deer (Cervus elaphus) in Northern Italy. Veterinary Microbiology, 217, 167–172. 10.1016/j.vetmic.2018.03.015 [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, Q. , & Meitzler, J. C. (1999). Rapid and sensitive detection of Escherichia coli O157:H7 in bovine faeces by a multiplex PCR. Journal of Applied Microbiology, 87, 867–876. 10.1046/j.1365-2672.1999.00938.x [DOI] [PubMed] [Google Scholar]
- Joensen, K. G. , Scheutz, F. , Lund, O. , Hasman, H. , Kaas, R. S. , Nielsen, E. M. , & Aarestrup, F. M. (2014). Real‐time whole‐genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli . Journal of Clinical Microbiology, 52, 1501–1510. 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen, K. G. , Tetzschner, A. M. , Iguchi, A. , Aarestrup, F. M. , & Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole‐genome sequencing data. Journal of Clinical Microbiology, 53, 2410–2426. 10.1128/JCM.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijn, A. , Michelacci, V. , Orsini, M. , & Morabito, S. (2020). Advanced research infrastructure for experimentation in genomicS (ARIES): A lustrum of Galaxy experience. Bioinformatics, Retrieved from http://biorxiv.org/lookup/doi/10.1101/2020.05.14.095901 [Google Scholar]
- Kruse, H. , Kirkemo, A. M. , & Handeland, K. (2004). Wildlife as source of zoonotic infections. Emerging Infectious Diseases, 10, 2067–2072. 10.3201/eid1012.040707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llarena, A.‐K. , Ribeiro‐Gonçalves, B. F. , Silva, D. N. , Halkilahti, J. , Machado, M. P. , Da Silva, M. S. , Jaakkonen, A. , Isidro, J. , Hämäläinen, C. , Joenperä, J. , Borges, V. , Viera, L. , Gomes, J. P. , Correia, C. , Lunden, J. , Laukkanen‐Ninios, R. , Fredriksson‐Ahomaa, M. , Bikandi, J. , Millan, R. S. , … Rossi, M. (2018). INNUENDO: A cross sectoral platform for the integration of genomics in the surveillance of food‐borne pathogens. EFSA Supporting Publications, 15, 1498E. 10.2903/sp.efsa.2018.EN-1498 [DOI] [Google Scholar]
- Michelacci, V. , Maugliani, A. , Tozzoli, R. , Corteselli, G. , Chiani, P. , Minelli, F. , Gigliucci, F. , Arancia, S. , Conedera, G. , Targhetta, C. , Pierasco, A. , Collini, L. , Parisi, A. , Scavia, G. , & Morabito, S. (2018). Characterization of a novel plasmid encoding F4‐like fimbriae present in a Shiga‐toxin producing enterotoxigenic Escherichia coli isolated during the investigation on a case of hemolytic‐uremic syndrome. International Journal of Medical Microbiology, 308, 947–955. 10.1016/j.ijmm.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Michelacci, V. , Tozzoli, R. , Caprioli, A. , Martínez, R. , Scheutz, F. , Grande, L. , Sánchez, S. , & Morabito, S. (2013). A new pathogenicity island carrying an allelic variant of the Subtilase cytotoxin is common among Shiga toxin producing Escherichia coli of human and ovine origin. Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases, 19, E149–E156. 10.1111/1469-0691.12122 [DOI] [PubMed] [Google Scholar]
- Paton, A. W. , Srimanote, P. , Talbot, U. M. , Wang, H. , & Paton, J. C. (2004). A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli . The Journal of Experimental Medicine, 200, 35–46. 10.1084/jem.20040392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton, A. W. , & Paton, J. C. (1998). Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. Journal of Clinical Microbiology, 36, 598–602. 10.1128/JCM.36.2.598-602.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti, L. , Gugiatti, A. , & Corlatti, L. (2017). Piano di conservazione e gestione del cervo nel settore Lombardo del Parco Nazionale dello Stelvio, Settembre 2017. Rapporto di sintesi delle attività di controllo numerico 2011–2016 e proposta di piano di controllo numerico delle popolazioni di cervo del UG Alta Valtellina‐Quinquennio 2017–2021. Article in Italian. [Google Scholar]
- Sánchez, S. , García‐Sánchez, A. , Martínez, R. , Blanco, J. , Blanco, J. E. , Blanco, M. , Dahbi, G. , Mora, A. , de Mendoza, J. H. , Alonso, J. M. , & Rey, J. (2009). Detection and characterisation of Shiga toxin‐producing Escherichia coli other than Escherichia coli O157:H7 in wild ruminants. Veterinary Journal (London, England : 1997), 180, 384–388. 10.1016/j.tvjl.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Sauvala, M. , Laaksonen, S. , Laukkanen‐Ninios, R. , Jalava, K. , Stephan, R. , & Fredriksson‐Ahomaa, M. (2019). Microbial contamination of moose (Alces alces) and white‐tailed deer (Odocoileus virginianus) carcasses harvested by hunters. Food Microbiology, 78, 82–88. 10.1016/j.fm.2018.09.011 [DOI] [PubMed] [Google Scholar]
- Scheutz, F. , Teel, L. D. , Beutin, L. , Piérard, D. , Buvens, G. , Karch, H. , Mellmann, A. , Caprioli, A. , Tozzoli, R. , Morabito, S. , Strockbine, N. A. , Melton‐Celsa, A. R. , Sanchez, M. , Persson, S. , & O'Brien, A. D. (2012). Multicenter evaluation of a sequence‐based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. Journal of Clinical Microbiology, 50, 2951–2963. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. , Sha, Q. , Lacher, D. W. , Del Valle, J. , Mosci, R. E. , Moore, J. A. , Scribner, K. T. , & Manning, S. D. (2015). Characterization of enteropathogenic and Shiga toxin‐producing Escherichia coli in cattle and deer in a shared agroecosystem. Frontiers in Cellular and Infection Microbiology, 5, 29. 10.3389/fcimb.2015.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerba‐Turek, A. , Siemionek, J. , Socha, P. , Bancerz‐Kisiel, A. , Platt‐Samoraj, A. , Lipczynska‐Ilczuk, K. , & Szweda, W. (2020). Shiga toxin‐producing Escherichia coli isolates from red deer (Cervus elaphus), roe deer (Capreolus capreolus) and fallow deer (Dama dama) in Poland. Food Microbiology, 86, 103352. 10.1016/j.fm.2019.103352 [DOI] [PubMed] [Google Scholar]
- Toma, C. , Martínez Espinosa, E. , Song, T. , Miliwebsky, E. , Chinen, I. , Iyoda, S. , Iwanaga, M. , & Rivas, M. (2004). Distribution of putative adhesins in different seropathotypes of Shiga toxin‐producing Escherichia coli . Journal of Clinical Microbiology, 42, 4937–4946. 10.1128/JCM.42.11.4937-4946.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalcengiz, Z. , Jeamsripong, S. , Spanninger, P. M. , Persad, A. K. , Wang, F. , Buchanan, R. L. , Lejeune, J. , Kniel, K. E. , Jay‐Russell, M. T. , & Danyluk, M. D. (2020). Survival of shiga toxin‐producing Escherichia coli in various wild animal feces that may contaminate produce. Journal of Food Protection, 83, 1420–1429. 10.4315/JFP-20-046 [DOI] [PubMed] [Google Scholar]
- Wirth, T. , Falush, D. , Lan, R. , Colles, F. , Mensa, P. , Wieler, L. H. , Karch, H. , Reeves, P. R. , Maiden, M. C. J. , Ochman, H. , & Achtman, M. (2006). Sex and virulence in Escherichia coli: An evolutionary perspective. Molecular Microbiology, 60(5), 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


