Abstract
Several characteristics of Mycobacterium tuberculosis (e.g., conserved genome and low growth rate) have severely restricted the study of the microorganism. The discovery of IS6110 raised hopes of overcoming these obstacles. However, our knowledge of this IS element is relatively limited; even its two basic characteristics (transposition mechanism and target site selection) are far from well understood. In this study, IS6110 insertions in ipl loci (iplA and iplB) in two collections of clinical isolates of M. tuberculosis from different geographic locations, one from Scotland and the other from Thailand, were investigated. Five different IS6110 insertions in the loci were identified: ipl-4::IS6110, ipl-5::IS6110, ipl-11::IS6110, ipl-12::IS6110, and ipl-13::IS6110. An attempt to establish the phylogenetic relationship of the isolates containing these insertions was unsuccessful, suggesting that some of these insertions may have arisen from more than one event. This possibility is further supported by the observation that IS6110 copies existed in the same site but with different orientations in different isolates, and the insertion site of ipl-1::IS6110 harbored IS6110 copies in both iplA and iplB in different strains. All these suggest the independent occurrence of IS6110 insertions at the same sites of the genome of M. tuberculosis in different clinical isolates. The implications of this finding are discussed.
Mycobacterium tuberculosis, the causative agent of tuberculosis, infects one-third of humans and causes 3 million deaths annually (27). Bacterial insertion sequences (IS) are transposable genetic elements present in multiple copies in a genome and capable of movement to new locations in the genome. They have a wide range of applications in the studies of evolution of bacterial genomes, bacterial genetics, and dissemination of antibiotic resistance. IS elements exhibit variable degrees of specificity in the selection of insertion sites on the genome, with some being highly specific and others quite random. Many, however, are between these two extremes (6, 11, 17, 23).
The relatively higher rate of IS transposition on genomes than that of mutations in structural genes and other loci has elicited strong interest in the applications of IS as genetic markers to study bacterial population genetics and phylogeny, especially for species with conserved genomes (e.g., M. tuberculosis) and strains under the level of subspecies (2, 15). IS6110, a member of the IS3 family, was identified in the M. tuberculosis complex (5, 13, 32, 36). It is usually present in multiple copies in the genome, and this along with other characteristics has led to its use as a powerful genetic marker for strain differentiation (12, 19, 27, 32). Application of IS6110-based restriction fragment length polymorphism (RFLP) analysis has dramatically advanced our knowledge of the molecular epidemiology of M. tuberculosis. Identification of the Beijing family of M. tuberculosis strains is one example. The family is a group of genetically closely related strains which show the following characteristics: (i) they harbor 15 to 20 copies of IS6110, and more than two-thirds of them are at the same genomic sites in these strains; (ii) they are identical in spoligotyping (based on a polymorphic repetitive sequence), polymorphic GC-repetitive sequence typing (based on a dispersed polymorphic sequence), and IS1081 RFLP pattern; and (iii) they have been found in parts of the world other than China and its adjacent countries, but all have more than 80% similarity in their IS6110 RFLP patterns (33).
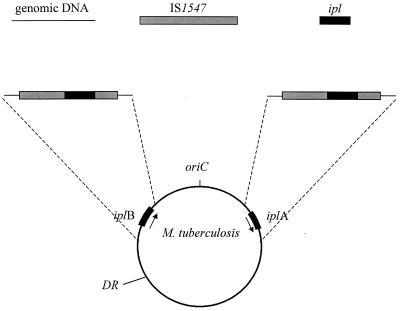
Although the general diversity of IS6110 RFLP patterns observed in M. tuberculosis isolates suggests its insertion at random on the genome, some genomic regions are preferential loci for its insertion (9, 12, 14, 20, 26); the locus ipl is one of them (9). Further investigation of ipl has revealed that it is a part of an insertion sequence (IS1547) and is usually present at two genomic locations (iplA and iplB) (Fig. 1) (8). In addition, a high frequency of IS6110 insertions in these loci is observed (9, 14). It is, however, not clear whether IS6110 insertions in the same DNA sequence sites in these loci resulted from single or multiple events. Clarification of this point would have a wide range of implications, as the typing of M. tuberculosis isolates using IS6110 RFLP analysis depends on related strains having the same or nearly the same insertion patterns. However, if an IS6110 insertion resulted from multiple events rather than one event, it would greatly reduce the power of the IS6110 RFLP technique. In this study, we present evidence that IS6110 insertions at the same DNA sequence sites (rather than loci) could occur in independent events.
FIG. 1.
Schematic illustration of locations of iplA, iplB, and the direct repeat (DR) in the genome of M. tuberculosis. ipl is a part of IS1547, as detailed in the text; arrows indicate the expression direction of open reading frame 1 of the putative transposase of IS1547.
MATERIALS AND METHODS
Strains and DNA preparation.
A total 112 of clinical M. tuberculosis isolates were used in this study, including 102 isolates collected in Scotland (10) and 10 Beijing family isolates (B104, B303, B401, B508, B530, B543, B556, B568, B595, and B605) collected in Thailand (24). DNA preparation was carried out according to a recommended method (30).
IS6110 RFLP analysis.
According to the recommended method (30), the M. tuberculosis genomic DNA was digested with PvuII, subjected to agarose gel electrophoresis, and then blotted onto a nylon membrane. After hybridization with the probe containing the partial IS6110 DNA sequence labeled with dUTP-digoxigenin (Boehringer Mannheim GmbH, Mannheim, Germany), the membrane was subjected to the digoxigenin detection procedure. The result was analyzed with the computer program GelCompar (version 4.0; Applied Maths, Kortrijk, Belgium) (10).
Conventional PCR, long PCR, and DNA sequencing.
Conventional PCR, long PCR, and DNA sequencing were used to identify different IS6110 insertion sites. Both conventional PCR and long PCR (Boehringer Mannheim GmbH) were carried out on a thermocycler (WellTemp, Cambridge, United Kingdom); DNA was sequenced using an Applied Biosystems 377A automated DNA sequencer with a Prism Ready Mix kit based on AmpliTaq CS polymerase (ABI, Warrington, United Kingdom) (7). The primers used in this study were designed from the DNA sequence (EMBL/GenBank/DDBJ accession no. Y13470): P3 (5′-GCCGATTCCACTCACCCAGTC-3′), P4 (5′-CGCAAAGTGAGCCAGACACCA-3′), P5 (5′-TCGCGGGAGTTGAAGTTGTTG-3′), P6 (5′-GGGTCAGTGCGATGCGGTGTA-3′), P7 (5′-GCTCCCATCCCGGTGTGGTCG-3′), and P8 (5′-TGACGTTCCTTTGCTACACCG-3′) (8).
DNA sequence analysis.
Programs in the GCG package (version 8.1) used for DNA sequence analyses were GAP (21), BESTFIT, and PUBLISH.
RESULTS
bj allele.
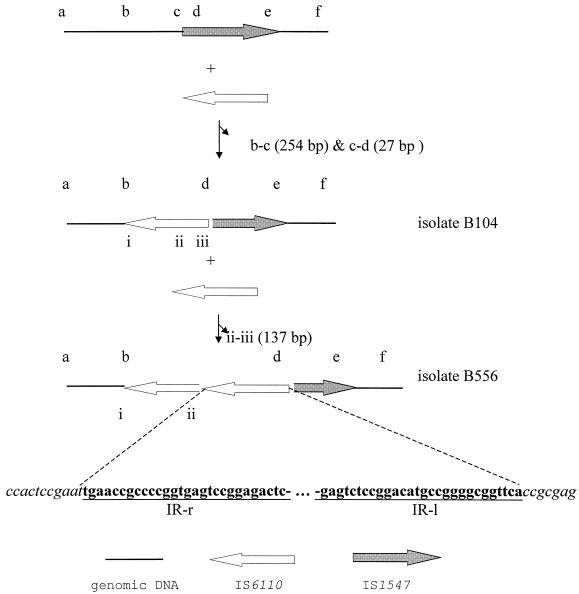
Conventional PCR, long PCR, and DNA sequencing were applied to investigate the presence of IS6110 insertion in the iplB locus of the 10 Beijing family isolates. A copy of IS6110 with an orientation opposite to the direction of the IS1547 copy was observed in isolate B104 (Fig. 2) (8). Compared with the IS6110-free iplB locus (accession no. Y16254) (8), there was a deletion of 281 bp of DNA sequence at the insertion site of this IS6110, comprising 27 bp from IS1547 and 254 bp from its upstream flanking sequence (Fig. 2). This IS6110 insertion was designated ipl-11::IS6110 (accession no. Y17219), and since all 10 Beijing family isolates carried it, it was also named the bj allele.
FIG. 2.
Polymorphisms and evolution of iplB loci in the Beijing family isolates. An IS6110 element inserted into the IS6110-free iplB locus with a deletion of 281 bp in the insertion site, generating the construct known as isolate B104. Based on this structure, a new IS6110 element was observed in isolate B556. The DNA sequences of the conjunctions at iplB of isolate B556 are presented. IR-r, inverted repeat-right; IR-l, inverted repeat-left.
Intriguingly, not only did 1 of the 10 isolates (isolate B556) harbor a second copy of IS6110 (ipl-12::IS6110; accession no. Y17220) in its iplB which was immediately adjacent to and in the same orientation as that in ipl-11::IS6110, but there was also a deletion of 137 bp from the IR-l (inverted repeat-left) end of the upstream IS6110 element (Fig. 2). Among the 10 isolates, isolates B530 and B568 had the same iplB structure as B104 and the others had the same iplB structure as isolate B556.
Isolates containing ipl-5::IS6110 and the bj allele in both the Scottish collection and the Beijing family strains.
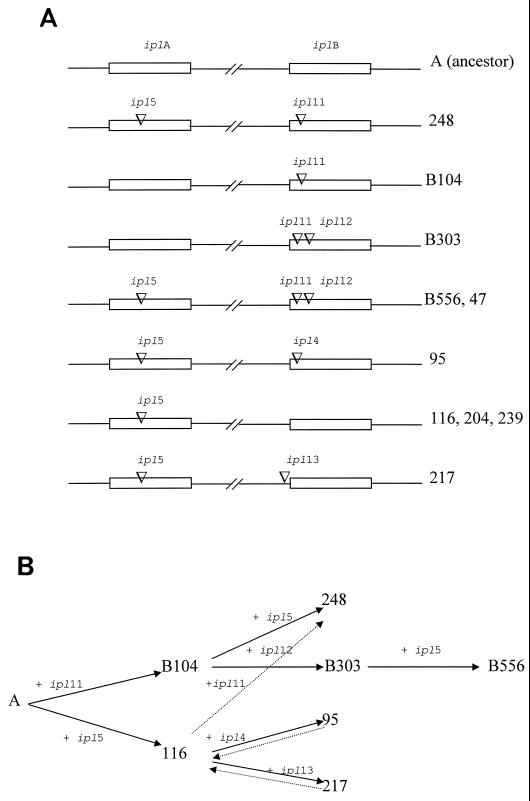
The use of ipl-5::IS6110 (accession no. X98155) in this study was initiated by the observation that at the iplA locus of the 10 isolates, some contained ipl-5::IS6110, such as isolate B556, and others were IS6110 free, such as isolates B104 and B303 (Fig. 3). The ipl-5::IS6110 insertion had previously been identified in iplA in seven isolates collected in Scotland (isolates 47, 95, 116, 204, 217, 239, and 248) (9). The question then arose whether the IS6110 insertion in ipl-5::IS6110 identified in the Scottish isolates and in the Beijing family isolates was derived from a single insertion event or multiple events. To clarify this, the status of IS6110 insertions in iplB of the seven Scottish isolates was examined, and five different alleles were identified: IS6110-free iplB (isolates 116, 204, and 239), ipl-11::IS6110 (isolate 248), both ipl-11::IS6110 and ipl-12::IS6110 (isolate 47), ipl-4::IS6110 (isolate 95), and ipl-13::IS6110 (isolate 217) (Fig. 3A).
FIG. 3.
(A) IS6110 insertions in iplA and iplB loci of representative strains in this study. Numbers on the right indicate M. tuberculosis isolates; IS6110 insertions are shown by their ipl loci. (B) The most probable phylogeny of these isolates was deduced from the IS6110 insertions, although some unlikely acquisitions of IS6110 were noted (dotted lines), and the details are discussed in the text.
Two isolates (47 and 248) from Scotland harbored the bj allele, and their IS6110 banding patterns showed that isolate 47 had 15 IS6110 copies whereas isolate 248 had 11. Compared with the IS6110 RFLP patterns of the Beijing family, isolate 47 had more than 80% of Dice coefficient, and isolate 248 had only about 60%. According to the criteria for identifying the members of the Beijing family (33), it is most likely that isolate 47 is a member of the Beijing family. Why strain 248 has the bj allele is unclear at the moment.
Dilocus linkage analysis.
As there is no evidence to date of transferring genomic materials between M. tuberculosis strains (28, 35), a lineage analysis of strains based on particular mutations should enable us to infer whether an identical mutation arose by being inherited from their common ancestor or by occurring repeatedly. Based on the polymorphisms of iplA and iplB loci in terms of different IS6110 insertions revealed above, the representative isolates with ipl-5::IS6110 and their structures of iplA and iplB loci are presented in Fig. 3A. A lineage analysis of these isolates (Fig. 3B) could indicate that an ancestor strain (strain A), which was presumably IS6110 free at both iplA and iplB, could diverge into two lineages by the insertions of IS6110 at ipl-11::IS6110 in iplB and at ipl-5::IS6110 in iplA, which could explain the structures observed in isolates B104 and 116, respectively. Isolate B104 could acquire a further IS6110 at ipl-5::IS6110 in iplA, generating the structure as in isolate 248. In another divergence, the isolate B104 could obtain an IS6110 at ipl-12::IS6110 in iplB, producing the structure in isolate B303; the latter could further gain an IS6110 copy in iplA (ipl-5::IS6110), giving the structure seen in isolate B556. Isolate 116 could further diverge into two lineages as represented by isolates 95 and 217 by acquisition of the alleles ipl-4::IS6110 and ipl-13::IS6110, respectively. This proposed lineage of these isolates can make sense only under the assumption that the unique IS6110 insertion in ipl-5::IS6110 allele would not originate from the same insertion event, and it seems likely that they were generated by three independent insertion events: one in isolate 248, one in isolate 116, and one in isolate B556. It could, however, be assumed that the structure of ipl-5::IS6110 was due to one insertion event rather than different events; then more difficulties in establishing the association between these isolates could be encountered.
Other direct evidence for the existence of preferential insertion sites for IS6110.
In addition to the suggestions from the above dilocus analysis, there is other direct evidence for the existence of preferential insertion sites of IS6110 in the genome. Firstly, ipl-4::IS6110 (accession no. X98154), an IS6110 insertion at iplB, was initially identified in isolate 91 (9). This insertion site was also found in another clinical isolate (isolate F6) to be occupied by an IS6110 copy but in the orientation opposite that of the IS6110 element in ipl-4::IS6110 of isolate 91, designated ipl-8::IS6110 (accession no. X95799). This finding is consistent with the observation that the insertions of IS30 in the genome of Escherichia coli was found in both orientations in the same genomic sites in different strains (1).
Secondly, in spite of the different nomenclature for iplA and iplB, they are virtually identical in terms of DNA sequence but differ in specific location in the M. tuberculosis genome (Fig. 1). ipl-1::IS6110 is the most commonly identified insertion in ipl and is usually an allele of iplA (9). It is in the same site in iplB in which an IS6110 was found in strains H37Rv and H37Ra (7).
DISCUSSION
All bacterial strains are genetically related to one another, and these relationships should be able to be reconstructed via phylogenetic analyses based on their genotypes. A genetic marker is crucial and should not only reveal a large amount of polymorphism and be selectively neutral but also rarely undergo recombination, because frequent recombination would randomize the associations between the strains. In recent years, IS elements have been widely used as genetic markers for the study of epidemiology, ecology, population genetics, and phylogeny of bacteria due to their high rates of transposition on genomes relative to mutations of structure genes and other loci (2, 15, 22, 29, 34). So far, however, studies of bacterial populations based on IS elements as genetic markers have yielded contradictory results with respect to association with other phenotypic and genetic markers (2, 15).
In this study, IS6110 insertions in ipl loci were investigated in clinical isolates of M. tuberculosis isolated from different geographic locations. Five different IS6110 insertions in the loci were identified: ipl-4::IS6110, ipl-5::IS6110, ipl-11::IS6110, ipl-12::IS6110, and ipl-13::IS6110. An attempt to establish the phylogenetic relationship of the isolates containing these insertions failed, suggesting that some of these insertions may have arisen from more than one event. In addition, IS6110 copies were observed in one site in both orientations in different isolates (ipl-4::IS6110 in isolate 91 and ipl-8::IS6110 in isolate F6). Furthermore, ipl-1::IS6110, the most common insertion site in ipl, was observed to harbor IS6110 copies in both iplA and iplB; it is most likely that these insertions happened in independent events. Finally, it is also difficult to explain why about 86% of isolates were found to harbor different IS6110 insertions in their ipl loci (9). All these suggest the existence of preferential insertion sites on the genome for IS6110. The influence of this will be more or less similar to that of horizontal transfer or recombination of genetic materials. Consequently, the association of strains based on IS6110 elements could be erroneous. These characteristics of IS elements may be responsible for the lack of success in establishing phylogenetic relationships of E. coli populations by using the IS elements (15) and for the limitation of application of the IS6110 RFLP technique only in the field of “short-time” epidemiological issues (31).
No IS element selects its target sites absolutely randomly; some IS elements show strong target site selectivity, while others show less strong selectivity (3, 6). For instance, IS4 always inserts at the same site in the galactosidase operon of E. coli (17, 18), whereas IS1 inserts fairly randomly on the genomes of E. coli (23); however, the majority of IS elements show variable degrees of preference in insertion sites for their transposition. Therefore, it would be wise to take precautions when using an IS element as a genetic marker to study bacterial population genetics and phylogeny unless its characteristics are well-known.
IS6110 RFLP analysis has been recommended as a standard method to distinguish M. tuberculosis isolates (30), but it is a labor-intensive and time-consuming approach, and various methods based on IS6110 insertion sites on the genomes have been developed to characterize M. tuberculosis strains (4, 16, 25, 28). Based on the finding from this study plus the observation that IS6110 can also mediate deletions of its flanking DNA sequences (7), a conclusion based on particular insertion sites should be approached with caution.
There are several limitations to this study. Firstly, our conclusion is based on preferential IS6110 insertion sites (ipl). Strictly speaking, a conclusion can be applied only to the sample from which the conclusion is drawn. Therefore, whether this conclusion can be applied to IS6110 insertions in nonpreferential loci has yet to be clarified. Secondly, the isolates used in this study were collected in two different geographic locations, which might not be sufficient to represent the M. tuberculosis population throughout the world, leading to biased conclusions. Thirdly, the evolutionary scheme proposed in Fig. 3 is the one we believe to be most likely, but that does not mean that other routes are impossible. It is also worth mentioning here that the proposed route does not reflect the effects of IS6110 deletions.
ACKNOWLEDGMENTS
We thank T. H. Pennington for his academic advice and support. DNA sequence analysis benefited from SEQNET, the SERC facility (Daresbury, United Kingdom). We also acknowledge the two anonymous reviewers for their comments and suggestions.
This study was financially supported by Chest, Heart and Stroke Scotland, The Scottish Office Department of Health, and the Milner Scholarship of the University of Aberdeen.
REFERENCES
- 1.Arber W. Elements in microbial evolution. J Mol Evol. 1991;33:4–12. doi: 10.1007/BF02100190. [DOI] [PubMed] [Google Scholar]
- 2.Baquar N, Burnens A, Stanley J. Comparative evaluation of molecular typing of strains from a national epidemic due to Salmonella brandenburg by rRNA gene and IS200 probes and pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1876–1880. doi: 10.1128/jcm.32.8.1876-1880.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd E F, Hartl D L. Nonrandom location of IS1 elements in the genomes of natural isolates of Escherichia coli. Mol Biol Evol. 1997;14:725–732. doi: 10.1093/oxfordjournals.molbev.a025812. [DOI] [PubMed] [Google Scholar]
- 4.Butler W R, Haas W H, Crawford J T. Automated DNA fingerprinting analysis of Mycobacterium tuberculosis using fluorescent detection of PCR products. J Clin Microbiol. 1996;34:1801–1803. doi: 10.1128/jcm.34.7.1801-1803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 6.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 7.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Z, Doig C, Morrison N, Watt B, Forbes K J. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex, and its association with IS6110. J Bacteriol. 1999;181:1021–1024. doi: 10.1128/jb.181.3.1021-1024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Z, Morrison N, Doig C, Watt B, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiandt M, Szybalski W, Malamy M H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119:223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- 12.Hermans P W, van Soolingen D, Bik E M, de Haas P E, Dale J W, van Embden J D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermans P W, van Soolingen D, Dale J W, Schuitema A R, McAdam R A, Catty D, van Embden J D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurepina N E, Sreevatsan S, Plikaytis B B, Bifani P J, Connell N D, Donnelly R J, van Sooligen D, Musser J M, Kreiswirth B N. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber Lung Dis. 1998;79:31–42. doi: 10.1054/tuld.1998.0003. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence J G, Dykhuizen D E, DuBose R F, Hartl D L. Phylogenetic analysis using insertion sequence fingerprinting in Escherichia coli. Mol Biol Evol. 1989;6:1–14. doi: 10.1093/oxfordjournals.molbev.a040531. [DOI] [PubMed] [Google Scholar]
- 16.Loeffelholz M J, Thompson C J, Gaunt D D, Koontz F P, Gilchrist M J. Polymerase chain reaction typing of nonviable Mycobacterium tuberculosis isolates. Diagn Microbiol Infect Dis. 1996;26:149–151. doi: 10.1016/s0732-8893(96)00214-3. [DOI] [PubMed] [Google Scholar]
- 17.Malamy M H. Some properties of insertion mutations in the lac operon. In: Beckwith J R, Ziper D, editors. The lactose operon. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1970. pp. 359–373. [Google Scholar]
- 18.Malamy M H, Fiandt M, Szybalski W. Electron microscopy of polar insertions in the lac operon of Escherichia coli. Mol Gen Genet. 1972;119:207–222. doi: 10.1007/BF00333859. [DOI] [PubMed] [Google Scholar]
- 19.McAdam R A, Hermans P W, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D, Dale J W. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 20.McHugh T D, Gillespie S H. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J Clin Microbiol. 1998;36:1410–1413. doi: 10.1128/jcm.36.5.1410-1413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 22.Odaert M, Berche P, Simonet M. Molecular typing of Yersinia pseudotuberculosis by using an IS200-like element. J Clin Microbiol. 1996;34:2231–2235. doi: 10.1128/jcm.34.9.2231-2235.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohtsubo H, Nyman K, Doroszkiewicz W, Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981;292:640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- 24.Palittapongarnpim P, Luangsook P, Tansuphaswadikul S, Chuchottaworn C, Prachaktam R, Sathapatayavongs B. Restriction fragment length polymorphism study of Mycobacterium tuberculosis in Thailand using IS6110 as probe. Int J Tuber Lung Dis. 1997;1:370–376. [PubMed] [Google Scholar]
- 25.Patel S, Wall S, Saunders N A. Heminested inverse PCR for IS6110 fingerprinting of Mycobacterium tuberculosis strains. J Clin Microbiol. 1996;34:1686–1690. doi: 10.1128/jcm.34.7.1686-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampson S L, Warren R M, Richardson M G, van der Spuy D, van Helden P D. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79:349–359. doi: 10.1054/tuld.1999.0218. [DOI] [PubMed] [Google Scholar]
- 27.Small P M, van Embden J D. Molecular epidemiology of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 569–582. [Google Scholar]
- 28.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Threlfall E J, Torre E, Ward L R, Davalos-Perez A, Rowe B, Gibert I. Insertion sequence IS200 fingerprinting of Salmonella typhi: an assessment of epidemiological applicability. Epidemiol Infect. 1994;112:253–261. doi: 10.1017/s0950268800057666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen D, de Haas P E, Blumenthal R M, Kremer K, Sluijter M, Pijnenburg J E, Schouls L M, Thole J E, Dessens-Kroon M W, van Embden J D, Hermans P W. Host-mediated modification of PvuII restriction in Mycobacterium tuberculosis. J Bacteriol. 1996;178:78–84. doi: 10.1128/jb.178.1.78-84.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Soolingen D, Qian L, de Haas P E, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werner S, Jording D, Simon R, Puhler A. Insertion sequence (IS) elements as natural constituents of the genomes of the Gram-negative Rhizobiaceae and their use as a tool in ecological studies. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Reading, United Kingdom: Society for General Microbiology; 1995. pp. 89–110. [Google Scholar]
- 35.Zainuddin Z F, Dale J W. Does Mycobacterium tuberculosis have plasmids? Tubercle. 1990;71:43–49. doi: 10.1016/0041-3879(90)90060-l. [DOI] [PubMed] [Google Scholar]
- 36.Zainuddin Z F, Dale J W. Polymorphic repetitive DNA sequences in Mycobacterium tuberculosis detected with a gene probe from a Mycobacterium fortuitum plasmid. J Gen Microbiol. 1989;135:2347–2355. doi: 10.1099/00221287-135-9-2347. [DOI] [PubMed] [Google Scholar]