Abstract
IPF is a progressive fibrotic lung disease whose pathogenesis remains incompletely understood. We have previously discovered pathologic mesenchymal progenitor cells (MPCs) in the lungs of IPF patients. IPF MPCs display a distinct transcriptome and create sustained interstitial fibrosis in immune deficient mice. However, the precise pathologic alterations responsible for this fibrotic phenotype remain to be uncovered. Quantitative mass spectrometry and interactomics is a powerful tool that can define protein alterations in specific subcellular compartments that can be implemented to understand disease pathogenesis. We employed quantitative mass spectrometry and interactomics to define protein alterations in the nuclear compartment of IPF MPCs compared to control MPCs. We identified increased nuclear levels of PARP1, CDK1, and BACH1. Interactomics implicated PARP1, CDK1, and BACH1 as key hub proteins in the DNA damage/repair, differentiation, and apoptosis signaling pathways respectively. Loss of function and inhibitor studies demonstrated important roles for PARP1 in DNA damage/repair, CDK1 in regulating IPF MPC stemness and self‐renewal, and BACH1 in regulating IPF MPC viability. Our quantitative mass spectrometry studies combined with interactomic analysis uncovered key roles for nuclear PARP1, CDK1, and BACH1 in regulating IPF MPC fibrogenicity.
Keywords: apoptosis, differentiation, DNA damage, idiopathic pulmonary fibrosis (IPF), ingenuity pathway analysis, mesenchymal progenitor cells (MPCs), nuclear fraction, quantitative mass spectrometry
Abbreviations
- IPF
idiopathic pulmonary fibrosis
- MPC
mesenchymal progenitor cell
- IPA
ingenuity pathway analysis
- TMT
tandem mass tag
1. INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic and ultimately fatal disease characterized by a progressive fibrotic destruction of the gas‐exchange apparatus (2002; [45]). We have previously identified intrinsically fibrogenic mesenchymal progenitor cells (MPCs) in the human IPF lung that serve as a source for IPF fibroblasts [57, 58, 60]. Bulk and single cell RNA‐sequencing data demonstrate that IPF MPCs have a distinct transcriptome and display a fibrogenic phenotype that is characterized by nonresolving interstitial lung fibrosis in a mouse xenograft model. While this suggests that IPF MPCs have undergone epigenetic changes that confer them with their fibrogenic phenotype, the precise molecular mechanisms underlying their pathological phenotype remain incompletely understood. The nucleus of the eukaryotic cell is an active site for a variety of cellular processes important for normal cell function. Abnormalities in nuclear proteins and chromatin organization can alter key cellular processes leading to cellular dysfunction and are hallmarks of many diseases [19, 50, 52]. In this regard, we previously identified differences in nuclear CD44 levels in IPF MPCs that play an important role in their pathologic function [62]. We hypothesize that IPF MPC fibrogenicity involves the abnormal location and/or quantity of nuclear proteins that function as pivotal hubs in the protein interactome and control fibrogenic behavior.
Defining variations in the quantity and location of proteins in cells with different phenotypes is an important step in biomedical studies whose objective is to understand the role of protein localization in disease pathogenesis [43]. Protein function can vary depending upon the location within a cell and may lead to alterations in cell phenotype. Spatial proteomics is an evolving powerful technology whose objective is to define the proteome in specific subcellular compartments [35, 43]. Approaches to define the spatial proteome include high‐throughput imaging to visualize proteins within a cell or subcellular compartment and quantitative mass spectrometry, to identify subcellular protein networks by organelle profiling or interactomics [4–7, 9, 22, 38].
Here we isolated proteins within the nuclear compartment of IPF and control MPCs and employed quantitative mass spectrometry to define differences in their nuclear proteome. The nuclear protein profile identified differences in the signature pattern between IPF and control MPCs. Ingenuity pathway analysis (IPA) of the nuclear proteome implied differences in DNA damage/repair, differentiation/stemness, and apoptosis signaling pathways between IPF and control MPCs. Interactomic analysis identified PARP, CDK1, and BACH1 as critical nodes within several key nuclear protein networks that may regulate these differences. Loss of function and inhibitor studies demonstrated important roles for PARP1 in IPF MPC DNA damage/repair; CDK1 in regulating IPF MPC stemness and self‐renewal; and BACH1 in regulating IPF MPC viability. Our quantitative mass spectrometry studies combined with interactomic analysis uncovered key roles for nuclear PARP1, CDK1, and BACH1 in regulating IPF MPC fibrogenicity.
STATEMENT OF SIGNIFICANCE
IPF MPCs display a fibrogenic phenotype but the molecular mechanism underlying their pathologic phenotype remain incompletely understood. Proteomic analysis of the IPF MPC nuclear proteome identified PARP, CDK1, and BACH1 as critical nodes within the DNA damage/repair, differentiation, and apoptosis signaling pathways respectively; providing evidence that these nuclear proteins function as pivotal hubs in the protein interactome that control their fibrogenic behavior.
2. RESULTS
2.1. The IPF MPC nuclear proteome differs from control MPCs
Nuclear fractions were isolated from IPF and control MPCs using the NE‐PER Nuclear and Cytoplasmic Extraction kit [51]. Nuclear fraction protein purity was augmented by including an additional wash from the recommended protocol. To analyze the purity of the resulting nuclear fractions, Western blot analysis was performed using antibodies to two predominantly cytoplasmic proteins, LDH and GAPDH. Following the additional wash step, only small amounts of LDH and GAPDH could be detected in the nuclear fractions (Figure S1A). We next subjected IPF and control MPC nuclear fractions to SDS‐PAGE. Silver staining of the SDS‐PAGE gel demonstrated clear differences in the nuclear protein profile between IPF and control MPCs (Figure S1B). Proteins from the nuclear fractions were then identified and quantified by TMT (Tandem Mass Tag) mass spectrometry. Global proteomic analyses of IPF and control MPCs identified and quantified 3989 nuclear proteins. Significant differences were observed for 1466 nuclear proteins, corresponding to 36% of all proteins identified (Figure S1C). Of these 3989 nuclear proteins, 64% were unchanged, 21% were increased in IPF MPCs, and 15% were decreased in IPF MPCs compared to control MPCs.
Following a published protocol [31], IPA pathway analysis was applied to our proteomics data and revealed key differences between IPF and control MPCs in 284 biological functions,186 key upstream hub proteins and 45 signaling networks. Biological function analysis identified DNA damage/repair, differentiation, and apoptosis as several of the most altered cell functions in IPF MPCs compared to control MPCs (Figure 1A, Table S1). Of note, this data is consistence with our prior RNA‐sequencing studies defining the IPF MPC transcriptome and the identification of biological functions associated with their distinct transcriptome [8, 57, 58, 60].
FIGURE 1.

Ingenuity pathway analysis of the IPF and control MPC nuclear proteome. Proteins identified from IPF and control MPC nuclear fractions with relative quantification were applied to Ingenuity Pathway Analysis (IPA). (A) Top cell functions associated with genes from IPF and control MPC dataset identified by IPA. (B) Top canonical pathways associated with proteins from IPF and control MPC dataset identified by IPA. (C) Top upstream regulators associated with proteins from IPF and control MPC dataset identified by IPA. Cell functions, upstream regulators or signal pathways identified are represented on the y‐axis. The x‐axis corresponds to the –log of the P‐value (Fisher's exact test) and the orange points on each cell function, upstream regulator, or signal pathway bar represent the ratio of the number of proteins in a given pathway that meet the cutoff criteria, divided by the total number of proteins that map to that cell function upstream regulator, or signal pathway. (D) PARP1, CDK1, and BACH1 protein levels from IPF MPC nuclear and cytoplasmic fractions were quantified by Western blot analysis. Lamin and GAPDH served as a loading controls for nuclear and cytoplasmic fractions, respectively. (E) Immunocytochemical analysis was performed on IPF MPCs to analyze PARP1, CDK1, and BACH1 subcellular location. DAPI staining was used to identify the nucleus
Forty‐five signaling pathways were identified as significantly altered between IPF and control MPCs. Included in this list were the PARP signaling pathway which regulates DNA damage/repair, cyclin, and cell cycle regulation pathway which regulates cell differentiation, and apoptosis signaling pathway that governs cell viability (Figure 1B, Table S2).
Among the 186 upstream nuclear hub molecules identified, SMARCA4 (Brg1) was identified. Recently, we discovered that a CD44/Brg1 nuclear complex regulates IPF MPC self‐renewal [62]. Importantly, our quantitative proteomics analysis identified nuclear PARP1, CDK1, and BACH1 as key upstream proteins positioned at pivotal hubs within DNA damage/repair, differentiation, and apoptosis signaling networks, respectively. PARP1 is within the PARP signaling pathway, CDK1 is within the cyclins and cell cycle regulation pathway, and BACH1 is within the apoptosis signaling pathway. While the role of PARP1, CDK1, and BACH1 in IPF pathogenesis remains to be elucidated, these data suggest that differences in these hub proteins may play key roles in differentially regulating IPF MPC function (Figure 1C, Table S3).
To verify the nuclear location of PARP1, CDK1, and BACH1 in IPF MPCs, we performed Western blot analysis on IPF MPC nuclear and cytoplasmic fractions and immunocytochemistry studies. Western blot analysis of nuclear and cytoplasmic fractions demonstrated that PARP1, CDK1, and BACH1 were predominantly found in our nuclear fractions (Figure 1D). Immunocytochemical analysis also demonstrated the presence of PARP1, CDK1, and BACH1 in the nucleus of IPF MPCs (Figure 1E). Of note, while PARP1, CDK1, and BACH1 predominantly localized to the nucleus of IPF MPCs, they were not exclusively localized to the nucleus, but were also present in the cytoplasm. This is consistent with prior work indicating that PARP1, CDK1, and BACH1 can be found both in the nucleus and cytoplasm [16, 26, 59, 61, 63].
Since our interactomic analysis identified PARP1, CDK1, and BACH1 as key hub proteins within the DNA damage/repair, cell differentiation, and apoptosis signaling networks, respectively, we next sought to validate our quantitative mass spectrometry combined with interactomic analysis. To do this we: (1) quantified nuclear PARP1, CDK1, and BACH1 proteins levels in IPF and control MPCs; and (2) performed loss of function and/or inhibitor studies to examine their role in regulating key IPF MPC fibrogenic functions.
2.2. PARP1, a key hub molecule in the DNA damage/repair network is increased in the nucleus of IPF MPCs and plays a role in regulating the IPF MPC's DNA damage and repair response
We next sought to validate our spatial proteomics data suggesting that alterations in key nuclear proteins in IPF MPCs result in dysfunctional IPF MPC behavior. Ingenuity Pathway Analysis identified DNA damage/repair as a key biologic function altered in IPF MPCs. Interactomic analysis suggested that PARP1 is a key upstream hub protein in the IPF MPC DNA damage/repair network (Figure 2A). To validate our quantitative mass spectrometry work, we quantified nuclear PARP1 levels by Western Blot analysis. PARP1 nuclear protein expression was increased in IPF MPCs compared to control (Figure 2B).
FIGURE 2.
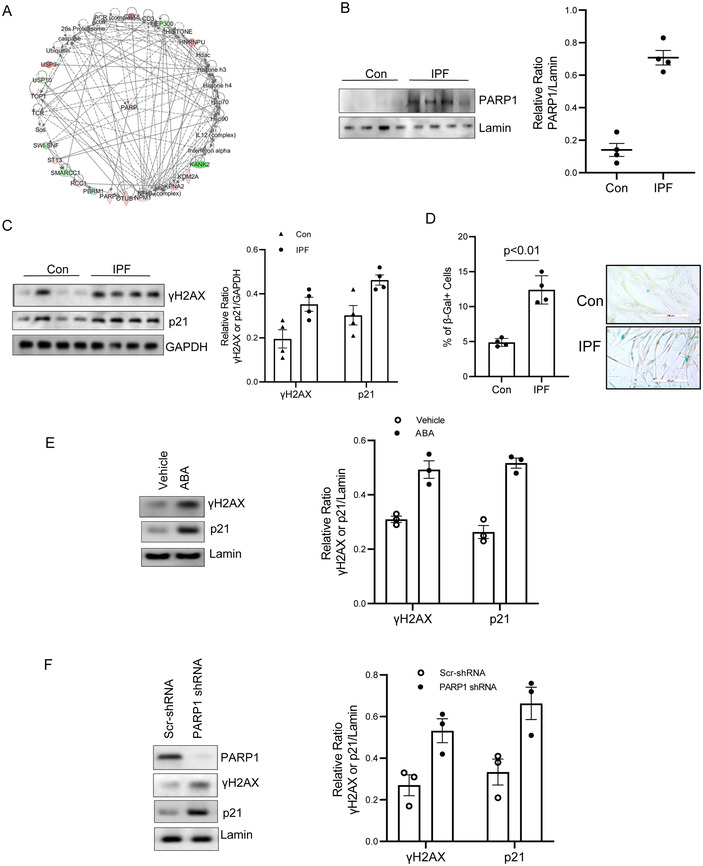
PARP1, a key hub molecule in the DNA damage/repair network is increased in the nucleus of IPF MPCs and plays a role in regulating the IPF MPC's DNA damage and repair response. (A) Network of proteins related to DNA damage identified by IPA of the IPF MPC nuclear protein data base. (B) PARP1 protein levels from IPF and control MPC (Con) nuclear fractions were quantified by Western blot analysis. Lamin served as a loading control. N = 4, each of control and IPF cell lines (C202, C130, C157, C279, IPF 424, IPF442, IPF327, IPF 259). Densitometry values are shown in the right hand graph. (C) p21 and γH2AX levels in IPF and control MPCs (Con) were quantified by Western blot analysis. GAPDH served as loading control. N = 4, each of control and IPF cell lines (C202, C130, C157, C279, IPF 424, IPF442, IPF327, IPF 259). Densitometry values are shown in the right hand graph. (D) IPF and control (Con) senescent MPCs were identified by β‐galactosidase staining. Quantification of IPF and control MPC senescent cells (left panel). Representative photomicrograph of β‐galactosidase staining of IPF and control MPCs (right panel). β‐galactosidase positive = blue stained cells. N = 4, each of control and IPF cell lines (C202, C130, C157, C279, IPF 424, IPF442, IPF327, IPF 259). (E) IPF MPCs were treated with the PARP1 inhibitor 3‐aminobenzamide (ABA, 50 μM) or vehicle control (Con). p21 and γH2AX levels were analyzed by Western blot analysis (left panel). Lamin served as a loading control. Densitometry values (right hand panel). F. IPF MPCs were transduced with PARP1 or scrambled shRNA. PARP1, p21, and γH2AX levels were analyzed by Western blot analysis (left hand panel). Lamin served as a loading control. Densitometry values (right hand panel). IPF cell lines 442 and 327 were used in Figure E and F. All data are shown as mean ± SE. 3 technical replicates were performed for all experiments
We next sought to determine if there were differences in DNA damage in IPF MPCs compared to control MPCs. As markers of DNA damage we quantified γH2AX and p21 levels in IPF and control MPCs. Phosphorylation of the histone H2AX, termed γH2AX, is an initial step involved in the recruitment of DNA repair proteins to sites of double stranded DNA breaks [3, 29]. In addition, activation of the p53‐p21 pathway frequently occurs during DNA damage [29, 56]. Both γH2AX and p21 levels were increased in IPF MPCs compared to control (Figure 2C). Induction of cellular senescence is one of the consequences of DNA damage. Therefore, we also quantified the number of senescent IPF and control MPCs by β‐galactosidase staining. We found increased numbers of senescent IPF MPCs compared to control (Figure 2D). Taken together, these data demonstrate that IPF MPCs display higher levels of DNA damage than control MPCs and express higher levels of the DNA damage repair protein PARP1. This suggests that elevated PARP1 expression serves as a compensatory cellular response to increased DNA damage and that PARP1 may play a critical role in DNA damage repair in IPF MPCs.
Since our interactomic analysis identified PARP1 as a key hub protein in the IPF MPC DNA damage/repair network, we next analyzed the role of PARP1 in regulating DNA damage/repair in IPF MPCs. 3‐aminobenzamide is a PARP1 inhibitor [42, 48]. We treated IPF MPCs with 3‐aminobenzamide and analyzed γH2AX and p21 levels. When IPF MPCs were treated with 3‐aminobenzamide, γH2AX and p21 levels increased indicating that antagonism of PARP1 DNA repair activity augments IPF MPC DNA damage (Figure 2E). To confirm the PARP1 inhibitor result, we knocked down PARP1 using PARP1 shRNA. Similar to the PARP1 inhibitor, PARP1 knock down increased γH2AX and p21 levels (Figure 2F). Together, these data validate the quantitative mass spectrometry data identifying increased levels of nuclear PARP1 in IPF MPCs and indicate a key role for PARP1 in IPF MPC DNA damage repair.
2.3. CDK1, a key hub molecule in the differentiation network is increased in the nucleus of IPF MPCs and is associated with IPF MPC stemness and self‐renewal
Ingenuity Pathway Analysis of our quantitative proteomic data also identified cellular differentiation as a key biologic function different between IPF and control MPCs. CDK1 plays key role in cellular differentiation. CDK1 controls the global epigenetic landscape in embryonic stem cells and decreased CDK1 activity is associated with differentiation. In cancer stem cells, CDK1 and its interaction with Sox2, Oct4 and Klf4 has been found to promote stemness [11, 30, 53]. Consistent with our quantitative proteomic analysis, we found increased nuclear CDK1 protein levels in IPF MPCs compared to control (Figure 3A).
FIGURE 3.

CDK1, a key hub molecule in the differentiation network is increased in the nucleus of IPF MPCs and is associated with IPF MPC stemness and self‐renewal. (A) IPF and control MPC (Con) CDK1 levels were quantified by Western blot analysis (left hand panel). Lamin served as loading control. Densitometry values (right hand panel). N = 4, each of control and IPF cell lines. (C202, C130, C157, C279, IPF 424, IPF442, IPF327, IPF 259). (B) Network of proteins related to cellular differentiation identified by IPA of the IPF MPC nuclear protein data base. (C) IPF MPCs were treated with IL8 (5 ng/mL) or vehicle control (Con) and CDK1 mRNA and protein levels were quantified by RT‐PCR (left panel) and Western blot analysis (middle panel). Densitometry values are shown in the right hand panel. IPF cell lines 327 and IPF442 were used. The experiment was replicated twice. (D and E) IPF MPCs were treated with CDK1 inhibitor Ro3306 (9 μM) or vehicle control (Con). Alternatively, IPF MPCs were treated with IL‐8 (5 ng/mL) plus CDK1 inhibitor Ro3306 or IL‐8 plus vehicle control (Con). Colony number (left hand panel) and size (right panel) were assessed using the colony forming assay and Image J analysis (D). Sox2 protein levels were quantified by Western blot analysis (E) (left hand panel). Densitometry values are shown in the right hand panel. (F and G). IPF MPCs were transduced with CDK1 shRNA or scrambled shRNA. Alternatively, IPF MPCs transduced with CDK1 or scrambled shRNA were treated with IL‐8. Self‐renewal was assessed using the colony forming assay (F). CDK1 and Sox2 expression (G) was quantified by RT‐PCR (left hand panel) and Western Blot analysis (middle panel). Densitometry values are shown in the right panel. Data in Figure D‐G are shown as mean ± SE. Three technical replicates were performed for all experiments
CDK1 is a key hub protein in the cell differentiation signaling network and interacts with a number of proteins that are in important in regulating cell differentiation (Figure 3B). In cancer stem cells, CDK1 has been found to promote stemness [11, 30, 53]. We have previously demonstrated that IL‐8 increases the expression of stemness markers in IPF MPCs and promotes IPF MPC self‐renewal. Since CDK1 regulates cell differentiation, we first examined the effect of the IL‐8 on CDK1 expression in IPF MPCs. We found that IL‐8 increased CDK1 expression (Figure 3C). We next examined the role of CDK1 in regulating IPF MPC self‐renewal. Treatment of IPF MPCs with the CDK1 inhibitor Ro3306 inhibited IPF MPC self‐renewal and also decreased IL‐8 mediated IPF MPC self‐renewal (Figure 3D). Sox 2 is a stemness marker [10, 11, 44]. We have previously found that Sox2 expression is increased in IPF MPCs compared to control MPCs and that IL‐8 treatment increases IPF MPC Sox2 expression [60]. Therefore, we next sought to determine whether CDK1 regulates Sox 2 expression. Treatment of IPF MPCs with the CDK1 inhibitor Ro3306 abrogated the increased expression of Sox2 in response to IL8 (Figure 3E). To verify the results of inhibition of CDK1 by Ro3306 on IPF MPC self‐renewal, we knocked down CDK1 using CDK1 shRNA and examined the ability of IPF MPCs to self‐renew in the colony forming assay. Knock‐down of CDK1 decreased IPF MPC self‐renewal compared to IPF MPCs transduced with scrambled shRNA (Figure 3F). Furthermore, knockdown of CDK1 also reduced IPF MPC self‐renewal in response to IL8 (Figure 3F). In contrast, IPF MPCs transduced with scrambled shRNA displayed a 30% increase in colony formation in response to IL8. Consistent with the CDK1 inhibitor Ro3306 result, knock‐down of CDK1 inhibited the IL‐8 mediated increase in Sox2 expression (Figure 3G). These data demonstrate that CDK1 regulates IPF MPC stemness and self‐renewal. Importantly, they validate the quantitative mass spectrometry results demonstrating that CDK1 expression is altered in IPF MPCs and support the concept that CDK1 is a key hub protein in the IPF MPC differentiation interactome.
2.4. Nuclear protein alterations affecting apoptosis pathways of IPF MPCs
BACH1 is a transcription factor that belongs to the leucine zipper factor family. BACH1 regulates cell proliferation and apoptosis [54, 55]. Our quantitative proteomic analysis indicated increased nuclear BACH1 expression in IPF MPCs compared to control MPCs. To validate our mass spectrometry result, we first examined BACH1 expression in IPF and control MPCs. Consistent with our quantitative proteomic results, we found that BACH1 nuclear protein expression was increased in IPF MPCs compared to control (Figure 4A). Analysis of the BACH1 interactome revealed that it is a key nodal upstream regulator in the apoptosis signaling network (Figure 4B). Therefore, we next examined whether there were differences in the level of apoptosis between IPF and control MPCs during routine culture by quantifying caspase 3 activity. We found that IPF MPCs displayed higher caspase 3 activity compared to control MPCs (Figure 4C).
FIGURE 4.
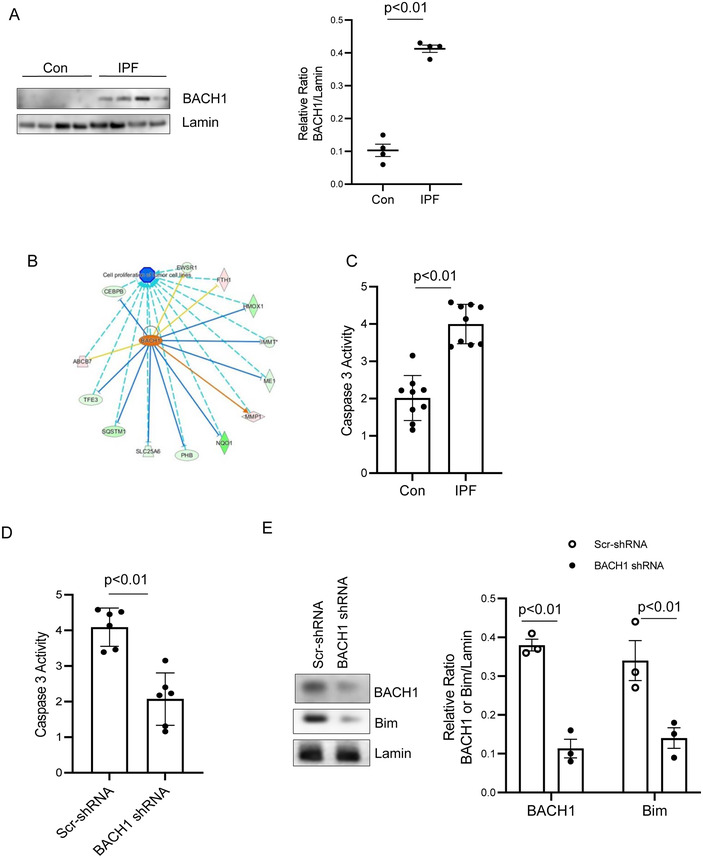
BACH1, a key hub molecule in the apoptosis signaling network is increased in the nucleus of IPF MPCs and is associated with IPF MPC viability. (A) IPF and control (Con) MPC BACH1 levels were analyzed by Western blot analysis (left hand panel). Lamin served as loading control. Densitometry values (right hand panel). N = 4, each of control and IPF cell lines. (C202, C130, C157, C279, IPF 424, IPF442, IPF327, IPF 259). (B) Network of proteins related to apoptosis identified by IPA of the IPF MPC nuclear protein data base. (C) IPF and control (Con) MPC apoptosis was quantified using a caspase 3 assay (Thermo Scientific, USA). N = 4; each of control and IPF cell lines (C202, C130, C157, C279, IPF 424, IPF442, IPF327, IPF 259). (D) IPF MPCs were transduced with BACH1 or scrambled shRNA. Cell apoptosis was quantified using a caspase3 assay. IPF MPC cell lines 442 and 458 were used. (E) BACH1 and Bim expression were quantified by Western Blot analysis. Lamin served as loading control. Densitometry values are shown in the right hand graph. All data are shown as mean ± SE. Three technical replicates were performed for all experiments.
To examine the role of BACH1 in regulating IPF MPC viability, we knocked down BACH1 using BACH1 shRNA and quantified caspase 3 activity. The level of caspase 3 activity in IPF MPCs transduced with BACH1 shRNA was reduced compared to IPF MPCs transduced with scrambled shRNA (Figure 4D), suggesting that high levels of BACH1 may diminish IPF MPC viability. BCL2L11 or Bim is a proapoptotic molecule in the BCL2 protein family and is a direct target of the BACH1 transcription factor [36]. Therefore, to begin to assess whether the ability of BACH1 to promote IPF MPC apoptosis might be mediated through Bim, we examined the effect of loss of BACH1 function on Bim expression. We found that knock‐down of BACH1 decreased Bim protein expression in IPF MPCs, suggesting a role for the BACH1/Bim axis in regulating IPF MPC viability (Figure 4E). Taken together, these data validate the quantitative mass spectrometry data identifying increased levels of nuclear BACH1 in IPF MPCs and suggest a key role for BACH1 in regulating IPF MPC viability.
3. DISCUSSION
IPF is a chronic and ultimately fatal disease characterized by a progressive decline in lung function (2002; [45]). We have previously identified intrinsically fibrogenic MPCs in the human IPF lung that: (i) cause nonresolving interstitial lung fibrosis in a humanized mouse xenograft model; (ii) are found concentrated in a highly cellular region on the perimeter of the fibroblastic focus in IPF lung tissue; and (iii) serve as one source for IPF fibroblasts [57, 58, 60, 62]. Bulk and single cell RNAseq studies indicate that IPF MPCs have a transcriptome that is distinct from control MPCs and that CD44 is a marker of fibrogenic MPCs [8]. We have found that nuclear CD44 interaction with the epigenetic modulator protein Brg1 is important in conferring IPF MPCs with their distinct transcriptome and cell‐autonomous fibrogenicity. Nevertheless, the precise molecular mechanisms underlying their pathological phenotype remain incompletely understood.
Abnormalities in nuclear proteins and chromatin organization can alter key cellular processes leading to cellular dysfunction and are hallmarks of many diseases [19, 50, 52]. Spatial proteomics is a powerful tool in biomedical research for defining the proteome in subcellular compartments, such as the nucleus [35, 43]. Quantitative mass spectrometry combined with interactomics permits both the identification and quantification of proteins and the ability to map them to specific signaling networks that regulate key cell functions [13, 17, 25, 64, 66]. To further delineate key alterations in IPF MPCs that underlie their pathologic phenotype, we purified nuclear proteins from IPF and control MPCs and subjected them to quantitative mass spectrometry combined with interactomic analysis. Three thousand nine hindered and eighty‐nine nuclear proteins were identified with greater than 1400 displaying different nuclear levels in IPF MPCs compared to control MPCs.
While the identification of 3989 proteins in the nucleus may be on the high side, according to the Human Protein Atlas, 6784 proteins have been experimentally detected in the nucleoplasm. However, 2796 nuclear proteins are supported by experimental evidence. It is important to note that many proteins found in the nucleoplasm may also be found in the cytoplasm due to nuclear‐cytoplasmic translocation. Because of the large number of proteins identified in our study, we performed additional analysis of the nuclear samples to examine the purity of the nuclear fractions. We used LDH and GAPDH as cytoplasmic markers. Despite including an additional wash step in our nuclear isolation protocol, we found the presence of small amounts of LDH and GAPDH in our nuclear fractions suggesting that our nuclear fractions may not be 100% pure. However, because some proteins may be found both in the nucleus and cytoplasm and because of nuclear‐cytoplasmic translocation, assessing purity of the nuclear sample by Western blot analysis may not be optimal. In this regard, LDH, which is found predominantly in the cytoplasm can also translocate to the nucleus [18]. In addition, we also analyzed vimentin and HIF1α. Both were present in our nuclear fractions. However, HIF1α can be found both in the nucleus and cytoplasm [12] and vimentin can be found in tight association with the nuclear membrane and within the nucleus of cells [21]. We interpret these results as suggesting that our nuclear sample preparation may not have resulted in a completely pure nuclear sample, but that our preparation resulted in enrichment for nuclear proteins.
Our proteomics experiment identified PARP1, CDK1, and BACH1 as nuclear proteins whose expression differed in IPF MPCs. Western blot analysis of nuclear and cytoplasmic fractions demonstrated that PARP1, CDK1, and BACH1 were predominantly found in our nuclear fractions. In addition, we performed immunocytochemistry experiments which demonstrate that PARP1, CDK1, and BACH1 are found predominantly in the nucleus of IPF MPCs. Consistent with our Western blot analysis results, some PARP1, CDK1, and BACH1 could also be detected in the cytoplasm of IPF MPCs. This is consistent with prior work indicating that PARP1, CDK1, and BACH1 can be found both in the nucleus and cytoplasm [16, 26, 63].
Since our interactomic analysis identified DNA damage/repair, differentiation, and apoptosis as several of the most altered cell functions in IPF MPCs, here we focused on PARP1, CDK1, and BACH1, regulators of DNA damage/repair, cell differentiation and apoptosis signaling, respectively. We demonstrate that IPF MPCs display higher levels of DNA damage and apoptosis, consistence with previous reports demonstrating that increased levels of DNA damage can promote apoptosis [27, 34]. It is likely that the increased levels of nuclear PARP1 identified by our quantitative mass spectrometry studies are in response to the increased levels of DNA damage in IPF MPCs. Our findings support the concept that PARP1 is a key protein regulating DNA damage/repair in IPF MPCs. This is consistent with prior studies linking PARP1 with DNA damage/repair [42, 65]. PARP1 plays a multifaceted role in DNA repair, facilitating single strand DNA break repair, double strand DNA repair, stabilization of replication forks and PARP1 also participates in chromatin modifications. The reason IPF MPCs display increased levels of DNA damage is unclear and will require further examination.
We have previously reported IPF MPCs display greater stemness marker expression and self‐renewal compared to control MPCs [60, 62]. Here we have discovered increased nuclear levels of CDK1 in IPF MPCs. Our interactomic analysis places CDK1 at center of regulation of differentiation in IPF MPCs. We demonstrate that CDK1 regulates IPF MPC self‐renewal and expression of the Sox2 stemness marker and loss of CDK1 function abrogates the IL‐8 mediated increase in IPF MPC self‐renewal and Sox2 expression. These data are consistent with prior work which has identified a central role for CDK1 in regulating cellular differentiation [14, 53]. CDK1 is required for the self‐renewal of pluripotent stem cells and decreased levels of CDK1 is associated with stem cell differentiation. Cancer stem cells also display high levels of CDK1 where it regulates stemness [23, 37, 39, 46].
In addition to elevated levels of DNA damage, we also found that IPF MPCs display a higher level of apoptosis compared to control MPCs. Our quantitative mass spectrometry identified elevated nuclear protein levels of the BACH1 transcription factor. Prior work has identified BACH1 as a key regulator of apoptosis in a variety of cell types including cancer cells [20, 24, 32, 33]. However, the role of BACH1 in apoptosis is complicated. While some studies implicate a role for BACH1 in mediating cell apoptosis, other work has linked BACH1 with DNA repair. In IPF MPCs, elevated BACH1 expression correlates with increased IPF MPC apoptosis. We demonstrate that knock‐down of BACH1 increases IPF MPC viability, indicating a role for BACH1 in regulating IPF MPC apoptosis. BLC2L11 or Bim is a direct target of BACH1. Bim is a proapoptotic protein of the BCL‐2 superfamily where it plays an essential role in initiating the intrinsic apoptosis pathway. Importantly, studies indicate that Bim plays a critical role in DNA damage‐induced apoptosis [15]. We show that knock‐down of BACH1 also decreases Bim expression. Since knock‐down of BACH1 decreases Bim expression and decreases the level of IPF MPC apoptosis, these data suggest a key role for the BACH1/Bim axis in regulating IPF MPC viability. Given the elevated levels of DNA damage in IPF MPCs, it is conceivable that the BACH1/Bim axis is playing a key role in IPF MPC damage‐induced apoptosis.
Taken together, our quantitative mass spectrometry studies combined with interactomic analysis suggest key roles for nuclear PARP1 in regulating DNA damage/repair; CDK1 in regulating IPF MPC stemness and self‐renewal; and BACH1 in regulating IPF MPC viability. In addition to these nuclear proteins, our quantitative mass spectrometry studies combined with interactomic analysis suggests that PML, SMARCA4, and SFPQ as well as other nuclear proteins positioned in key upstream signaling hubs may be worthy for further investigation for their potential role in regulating the pathologic behavior of IPF MPCs. We suggest that our IPF MPC nuclear proteomics data and IPA results are an excellent resource for future investigation of IPF MPC function. In conclusion, using quantitative mass spectrometry studies combined with interactomic analysis, we uncovered key roles for nuclear PARP1, CDK1, and BACH1 in regulating IPF MPC fibrogenicity.
4. METHODS
4.1. Primary mesenchymal cell lines
Six primary lung mesenchymal cell lines were established from six individual patients fulfilling diagnostic criteria for IPF including a pathological diagnosis of usual interstitial pneumonia (2000). Cell lines were derived from lungs, characterized as mesenchymal cells, and cultivated as previously described [57, 58, 62].
4.2. Isolation of mesenchymal progenitor cells
IPF and control mesenchymal progenitor cells were isolated from primary IPF and control mesenchymal cell cultures at passage 0 (initial isolate before subcultivation) through passage 4 [57]. For isolation of MPCs, primary IPF mesenchymal cells were labeled with mouse antihuman SSEA4 antibody conjugated to Alexa Fluor® 647 (Clone MC‐813‐70; Catalogue #560796; BD Biosciences, Franklin Lake, NJ, USA) and mouse antihuman CD44 conjugated to FITC (Clone IM7; Catalogue #103006; BioLegend, San Diego, CA, USA). Cells were sorted on a FACS Aria Cell Sorter (BD Biosciences). Cells that were SSEA4+ and CD44+ (relative to mouse IgG3 κ isotype control conjugated to Alexa Fluor® 647 and mouse IgM κ isotype control conjugated to FITC, respectively) (clone J606, catalogue #560803 BD Biosciences and catalogue #402207; BioLegend) were collected. For MPC isolation, the FACS Sorter gate was set to collect SSEA4 positive cells at the top 3% of CD44 expression.
4.3. Self–renewal assay
Single cell suspensions of IPF MPCs were incorporated into methylcellulose gels (Stemcell Technologies, Vancouver, Canada) and maintained in MSC SFM CTS (Thermo Scientific/Gibco, Rochford IL, USA) (37°C, 5% CO2; 1 week). Enumeration of colonies was performed microscopically and colony size was quantified by ImageJ. In some self‐renewal assays the cells were treated with the indicated concentrations of: i) recombinant IL‐8 (R & D Systems, Inc., Minneapolis, MN); ii) 3‐aminobenzamide (Sigma, USA); or Ro3306 (EMD Millipore, USA).
4.4. Apoptosis assay
Apoptosis was quantified using a caspase 3 activity apoptosis kit (Roche, USA) following the manufacturers’ instructions. Measurements were quantified using a SpectraMax M3 microplate reader (Molecular Devices).
4.5. Western blot and immunoprecipitation
Western blots were performed as previously described [58, 60]. Nuclear fractions were isolated from primary IPF and control MPCs using NE‐PER Nuclear and Cytoplasmic Extraction reagent (Thermo Scientific, USA) following manufacturer's instruction. The samples were centrifuged at 12,000 × g for 15 min at 4°C, and the lysates were precleared for 1 h at 4°C with protein A/G beads and immunoprecipitated for 2 h at 4°C with the appropriate primary antibody.
4.6. Real‐time reverse transcription PCR
Total RNA was extracted with the RNeasy minikit (Qiagen, USA). PCR reactions contained 10 μl miScript RT–PCR Reaction Mix (Qiagen, USA), 1 μl miScriptreverse transcriptase, 900 nM forward primer, 900 nM reverse primer, 250 nM probe and 50 ng RNA in 20 μl. Reactions were performed in a7900 HT Sequence Detector (Applied Biosystems) with a cycling protocol described before (Applied Biosystems) [59]. Primer sequences were as follows:
GAPDH Forward: 5′‐ TGTTGCCATCAATGACCCCTT‐3′
GAPDH Reverse: 5′‐CTCCACGACGTACTCAGCG‐3′
PARP1 Forward: 5′‐GGAAAGGGATCTACTTTGCCG‐3′
PARP1 Reverse: 5′‐TCGGGTCTCCCTGAGATGTG‐3′
CDK1 Forward: 5′‐ TGGATCTGAAGAAATACTTGGATTCTA ‐3′
CDK1 Reverse: 5′‐ CAATCCCCTGTAGGATTTGG ‐3′
BACH1 Forward: 5′‐TGTGCCAGAGGAAACAGTGAG‐3′
BACH1 Reverse: 5′‐TAGGCTTTCAAGACGCTGC‐3′
4.7. Detection of senescent cells
IPF and control MPCs at 50% confluency were fixed and stained using a β‐Galactosidase staining kit (Millipore, USA). Senescent cells were quantified using a phase contrast Leica microscope.
4.8. Plasmids/Constructs
For loss of function, PARP1, CDK1 and BACH1 were knocked down using shRNA (pGIPZ‐PARP1, pGIPZ‐CDK1 and pGIPZ‐BACH1 shRNA; ODT, Coralville, IA USA). Scrambled shRNA served as control.
4.9. Proteomics
Protein concentrations were determined in desalted samples with Bradford reagent (Bio‐Rad, Hercules, CA), and then samples containing equal amounts of protein (20 μg) were labeled with TMT reagent (Thermal Scientific, USA) as directed by the manufacturer's instructions. Peptide/protein isolation and identification were conducted as described previously. [47, 61]. TMT‐based MS was used to obtain proteomes from 6 samples (3 IPF and 3 control cell lines). LC‐MS data was acquired for each concatenated fraction using an Easy‐nLC 1000 HPLC (Thermo Scientific Inc., Waltham, MA) in tandem with a Thermo Fisher Orbitrap Fusion (Thermo Scientific Inc., Waltham, MA). Peptides were loaded directly onto a 75 cm × 100‐μm internal diameter fused silica PicoTip Emitter (New Objective, Woburn, MA) packed in‐house with ReproSil‐Pur C18‐AQ (1.9 μm particle, 120 Å pore; Dr. Maish GmbH Ammerbuch, Germany). The column was heated to 55°C and a flow rate of 300 μL/min was applied during the gradient. The gradient is as follows: 5% to 22% Buffer B (A: 0.1% formic acid in water, B: 0.1% formic acid in acetonitrile) for 45 min, 22% to 35% B for 25 min, and 35% to 95% B over 10 min. The column was mounted in a nanospray source directly in line with an Orbitrap Fusion mass spectrometer (Thermo Scientific). Spray voltage was 2.1 kV in positive mode, and the heated capillary was maintained at 275°C. The orbital trap was set to acquire survey mass spectra (380–1580 m/z) with a resolution of 60,000 at 100 m/z with automatic gain control (AGC) 1.0E6, 250 ms min injection. EASY‐IC was selected for internal mass calibration. The 12 most intense ions (2‐7 charged state) from the full scan were selected for fragmentation by higher‐energy collisional dissociation with normalized collision energy 35%, and detector settings of 60k resolution, AGC 5E4 ions, 250 ms maximum injection time and FT first mass mode fixed at 110 m/z. Dynamic exclusion was set to 40s with a 10 ppm high and low‐mass tolerance.
4.10. Database searching for protein detection
The tandem mass spectra were analyzed using Sequest (XCorr Only) in Proteome Discoverer 2.4.0.305 (Thermo Fisher Scientific, Waltham, MA). We used the Uniprot human Universal Proteome (UP000005640) sequence database from July 12, 2019 merged with the common lab contaminant protein database from, with a total of 174234 entries, for the database searching. The Sequest search parameters included: trypsin enzyme, fragment ion mass tolerance of 0.1 Da, precursor ion tolerance 10 ppm, carbamidomethyl cysteine as a fixed modification; pyroglutamic acid from glutamine, deamidation of asparagine, oxidation of methionine, N‐terminal protein acetylation and TMT 10plex for lysine and peptide N‐termini as variable modifications.
4.11. Relative protein quantification
Scaffold Q+ (v4.9, Proteome Software Inc., Portland, OR) was used for relative quantification of proteins. Peptide identifications were accepted if they could be established at greater than 89.0% probability to achieve an FDR less than 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 5.0% probability to achieve an FDR less than 1.0% and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [40]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Channels were corrected for incomplete isotope incorporation in all samples according to the algorithm described in i‐Tracker [49]. Normalization was performed iteratively (across samples and spectra) on intensities, as described in Statistical Analysis of Relative Labeled Mass Spectrometry Data from Complex Samples Using ANOVA [41]. Medians were used for averaging. Spectra data were log‐transformed, pruned of those matched to multiple proteins, and weighted by an adaptive intensity weighting algorithm. Of 46922 spectra in the experiment at the given thresholds, 36, 422 (78%) were included in quantitation. Differentially expressed proteins were determined by applying Permutation Test with unadjusted significance level p < 0.05 corrected by the Benjamini–Hochberg method.
4.12. Ingenuity Pathway Analysis (IPA)
The nuclear protein list from the proteomic analysis was imported to the IPA (http://www.ingenuity.com, 2021). Functional analyses were performed to: (i) identify biological functions and/or diseases that were most significant to the dataset; (ii) identify conical pathways that were most significant to the dataset; and iii) the proteins associated with the altered biological functions and/or diseases. Fisher's exact test was used to calculate a p‐value determining the probability that each biological function and/or disease assigned to that dataset is due to chance alone [28, 64].
4.13. Statistical analysis
All experiments were performed at least in triplicate, and results were analyzed using the Student's t‐test or Two‐Way ANOVA (Except the Proteomics data processed as described above). The criterion for significance was p < 0.05. Numerical data is reported as means ± standard deviations.
4.14. Proteomics data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD032352.
AUTHOR CONTRIBUTIONS
Libang Yang conceived, designed, and directed the studies with some input from Craig A. Henke, Libang Yang, and Craig A. Henke wrote the manuscript with assistance from all the authors. Libang Yang and Hong Xia established MPC cell lines, cultured MPCs, performed flow cytometry for isolation of MPCs, performed RT‐PCR, Western blot analysis, performed gain and loss of function experiments, and immunohistochemistry. LeeAnn Higgins and Candace Guerrero Proteomics analysis. Hong Xia and Libang Yang Tissue collection, tissue section preparation and IHC. Adam Gilbertsen and Karen Smith designed and constructed expression constructs and some function study.
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
Supporting information
Supplemental information
Supplemental information
Supplemental information
Supplemental information
ACKNOWLEDGMENTS
The proteomics sample preparation, mass spectrometry analysis and database searching was performed at the Center for Mass Spectrometry and Proteomics in the department of Biochemistry, Molecular Biology and Biophysics at the University of Minnesota; supporting agencies are listed here: https://cbs.umn.edu/cmsp/about
This work was supported by National Institutes of Health grants R01 HL125227 to C.A.H. and funds provided by the O'Brien and Witowski families.
Yang, L. , Gilbertsen, A. , Smith, K. , Xia, H. , Higgins, L. A. , Guerrero, C. , & Henke, C. A. (2022). Proteomic analysis of the IPF mesenchymal progenitor cell nuclear proteome identifies abnormalities in key nodal proteins that underlie their fibrogenic phenotype. Proteomics, 22, e2200018. 10.1002/pmic.202200018
DATA AVAILABILITY STATEMENT
The dataset has been deposited to the ProteomeXchange Consortium.via the PRIDE partner proteomics repository, dataset identifier PXD032352.
REFERENCES
- 1. American Thoracic Society . (2000). Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS). Am J Respir Crit Care Med., 161(2 Pt 1), 646–664. [DOI] [PubMed] [Google Scholar]
- 2. AmericanThoracic Society/European Respiratory Society International MultidisciplinaryConsensus Classification of the Idiopathic Interstitial Pneumonias . (2002). This jointstatement of the American Thoracic Society (ATS), and the European RespiratorySociety (ERS) was adopted by the ATS board of directors, June 2001 and by theERS Executive Committee, June 2001. Am J Respir Crit Care Med., 165(2), 277–304. [DOI] [PubMed] [Google Scholar]
- 3. Adamsen, B. L. , Kravik, K. L. , & De Angelis, P. M. (2011). DNA damage signaling in response to 5‐fluorouracil in three colorectal cancer cell lines with different mismatch repair and TP53 status. International Journal of Oncology, 39, 673–682. [DOI] [PubMed] [Google Scholar]
- 4. Alanazi, B. , Munje, C. R. , Rastogi, N. , Williamson, A. J. K. , Taylor, S. , Hole, P. S. , Hodges, M. , Doyle, M. , Baker, S. , Gilkes, A. F. , Knapper, S. , Pierce, A. , Whetton, A. D. , Darley, R. L. , & Tonks, A. (2020). Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia. Leukemia, 34, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Albrethsen, J. , Knol, J. C. , Piersma, S. R. , Pham, T. V. , de Wit, M. , Mongera, S. , Carvalho, B. , Verheul, H. M. , Fijneman, R. J. , Meijer, G. A. , & Jimenez, C. R. (2010). Subnuclear proteomics in colorectal cancer: Identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Molecular & Cellular Proteomics, 9, 988–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babeu, J.‐P. , Wilson, S. D. , Lambert, É. , Lévesque, D. , Boisvert, F.‐M. , & Boudreau, F. (2019). Quantitative proteomics identifies DNA repair as a novel biological function for hepatocyte nuclear factor 4alpha in colorectal cancer cells. Cancers (Basel), 11, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bandhakavi, S. , Kim, Y. M. , Ro, S. H. , Xie, H. , Onsongo, G. , Jun, C. B. , Kim, D. H. , & Griffin, T. J. (2010). Quantitative nuclear proteomics identifies mTOR regulation of DNA damage response. Molecular & Cellular Proteomics, 9, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beisang, D. J. , Smith, K. , Yang, L. B. , Benyumov, A. , Gilbertsen, A. , Herrera, J. , Lock, E. , Racila, E. , Forster, C. , Sandri, B. J. , Henke, C. A. , & Bitterman, P. B. (2020). Single‐cell RNA sequencing reveals that lung mesenchymal progenitor cells in IPF exhibit pathological features early in their differentiation trajectory. Scientific Reports‐Uk, 10, 11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bouchal, P. , Dvorakova, M. , Roumeliotis, T. , Bortlicek, Z. , Ihnatova, I. , Prochazkova, I. , Ho, J. T. , Maryas, J. , Imrichova, H. , Budinska, E. , Vyzula, R. , Garbis, S. D. , Vojtěšek, B. , & Nenutil, R. (2015). Combined proteomics and transcriptomics identifies carboxypeptidase B1 and nuclear factor kappaB (NF‐kappaB) associated proteins as putative biomarkers of metastasis in low grade breast cancer. Molecular & Cellular Proteomics, 14, 1814–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen, C.‐ L. , Tsai, Y.‐S. , Huang, Y.‐H. , Liang, Y.‐J. , Sun, Y.‐Y. , Su, C.‐W. , Chau, G.‐Y. , Yeh, Y.‐C. , Chang, Y.‐S. , Hu, J.‐T. , & Wu, J.‐C. (2018). Lymphoid enhancer factor 1 contributes to hepatocellular carcinoma progression through transcriptional regulation of epithelial‐mesenchymal transition regulators and stemness genes. Hepatology Communications, 2, 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng, C. C. , Shi, L. H. , Wang, X. J. , Wang, S. X. , Wan, X. Q. , Liu, S. R. , Wang, Y. F. , Lu, Z. , Wang, L. H. , & Ding, Y. (2018). Stat3/Oct‐4/c‐Myc signal circuit for regulating stemness‐mediated doxorubicin resistance of triple‐negative breast cancer cells and inhibitory effects of WP1066. International Journal of Oncology, 53, 339–348. [DOI] [PubMed] [Google Scholar]
- 12. Chilov, D. , Camenisch, G. , Kvietikova, I. , Ziegler, U. , Gassmann, M. , & Wenger, R. H. (1999). Induction and nuclear translocation of hypoxia‐inducible factor‐1 (HIF‐1): Heterodimerization with ARNT is not necessary for nuclear accumulation of HIF‐1alpha. Journal of Cell Science, 112, 1203–1212. [DOI] [PubMed] [Google Scholar]
- 13. Cirillo, E. , Parnell, L. D. , & Evelo, C. T. (2017). A review of pathway‐based analysis tools that visualize genetic variants. Frontiers in Genetics, 8, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das, B. , Pal, B. , Bhuyan, R. , Li, H. , Sarma, A. , Gayan, S. , Talukdar, J. , Sandhya, S. , Bhuyan, S. , Gogoi, G. , Gouw, A. M. , Baishya, D. , Gotlib, J. R. , Kataki, A. C. , & Felsher, D. W. (2019). MYC regulates the HIF2alpha stemness pathway via nanog and Sox2 to maintain self‐renewal in cancer stem cells versus non‐stem cancer cells. Cancer Research, 79, 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delbridge, A. R. , Grabow, S. , & Strasser, A. (2017). Loss of BIM augments resistance of ATM‐deficient thymocytes to DNA damage‐induced apoptosis but does not accelerate lymphoma development. Cell Death and Differentiation, 24, 1987–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donizy, P. , Pietrzyk, G. , Halon, A. , Kozyra, C. , Gansukh, T. , Lage, H. , Surowiak, P. , & Matkowski, R. (2014). Nuclear‐cytoplasmic PARP1 expression as an unfavorable prognostic marker in lymph node‐negative early breast cancer: 15 year follow up. Oncology Reports, 31, 1777–87. [DOI] [PubMed] [Google Scholar]
- 17. Duan, Z. , Yuan, C. , Han, Y. , Zhou, L. , Zhao, J. , Ruan, Y. , Chen, J. , Ni, M. , & Ji, X. (2020). TMT‐based quantitative proteomics analysis reveals the attenuated replication mechanism of Newcastle disease virus caused by nuclear localization signal mutation in viral matrix protein. Virulence, 11, 607–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferriero, R. , Nusco, E. , De Cegli, R. , Carissimo, A. , Manco, G. , & Brunetti‐Pierri, N. (2018). Pyruvate dehydrogenase complex and lactate dehydrogenase are targets for therapy of acute liver failure. Journal of Hepatology, 69, 3235–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallagher, P. S. , Oeser, M. L. , Abraham, A.‐C. , Kaganovich, D. , & Gardner, R. G. (2014). Cellular maintenance of nuclear protein homeostasis. Cellular and Molecular Life Sciences, 71, 1865–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo, H. , Wang, Y. , Jia, W. , & Liu, L. (2021). MiR‐133a‐3p relieves the oxidative stress induced trophoblast cell apoptosis through the BACH1/Nrf2/HO‐1 signaling pathway. Physiological Research, 70, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartig, R. , Shoeman, R. L. , Janetzko, A. , Tolstonog, G. , & Traub, P. (1998). DNA‐mediated transport of the intermediate filament protein vimentin into the nucleus of cultured cells. Journal of Cell Science, 111(Pt 24), 3573–3584. [DOI] [PubMed] [Google Scholar]
- 22. Hiragami‐Hamada, K. , Tani, N. , & Nakayama, J. I. (2020). Proteomics‐based systematic identification of nuclear proteins anchored to chromatin via RNA. Methods in Molecular Biology, 2161, 89–99. [DOI] [PubMed] [Google Scholar]
- 23. Huang, Z. , Shen, G. , & Gao, J. (2021). CDK1 promotes the stemness of lung cancer cells through interacting with Sox2. Clinical & Translational Oncology, 23, 1743–1751. [DOI] [PubMed] [Google Scholar]
- 24. Jian, L. , Mei, Y. , Xing, C. , & Rongdi, Y. (2021). Haem relieves hyperoxia‐mediated inhibition of HMEC‐1 cell proliferation, migration and angiogenesis by inhibiting BACH1 expression. Bmc Ophthalmology [Electronic Resource], 21, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin, L. , Zuo, X. Y. , Su, W. Y. , Zhao, X. L. , Yuan, M. Q. , Han, L. Z. , Zhao, X. , Chen, Y. D. , & Rao, S. Q. (2014). Pathway‐based analysis tools for complex diseases: A review. Genomics, Proteomics & Bioinformatics, 12, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanezaki, R. , Toki, T. , Yokoyama, M. , Yomogida, K. , Sugiyama, K. , Yamamoto, M. , Igarashi, K. , & Ito, E. (2001). Transcription factor BACH1 is recruited to the nucleus by its novel alternative spliced isoform. Journal of Biological Chemistry, 276, 7278–7284. [DOI] [PubMed] [Google Scholar]
- 27. Kim, J. , Lee, S. , Kim, H. , Lee, H. , Seong, K. M. , Youn, H. , & Youn, B. (2021). Autophagic organelles in DNA damage response. Frontiers in Cell and Developmental Biology, 9, 668735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krämer, A. , Green, J. , Pollard, J. , & Tugendreich, S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics, 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuo, L. J. , & Yang, L. X. (2008). Gamma‐H2AX ‐ a novel biomarker for DNA double‐strand breaks. In Vivo (Athens, Greece), 22, 305–309. [PubMed] [Google Scholar]
- 30. Lee, S.‐H. , Chen, T.‐Y. , Dhar, S. S. , Gu, B. , Chen, K. , Kim, Y. Z. , Li, W. , & Lee, M. G. (2016). A feedback loop comprising PRMT7 and miR‐24‐2 interplays with Oct4, Nanog, Klf4 and c‐Myc to regulate stemness. Nucleic Acids Research, 44, 10603–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lepage, S. I. , Nagy, K. , Sung, H.‐K. , Kandel, R. A. , Nagy, A. , & Koch, T. G. (2016). Generation, characterization, and multilineage potency of mesenchymal‐like progenitors derived from equine induced pluripotent stem cells. Stem Cells and Development, 25, 80–89. [DOI] [PubMed] [Google Scholar]
- 32. Li, J. , Shiraki, T. , & Igarashi, K. (2012a). Bach1 as a regulator of mitosis, beyond its transcriptional function. Communicative & Integrative Biology, 5, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li, S. , Chen, T. , Zhong, Z. , Wang, Y. , Li, Y. , & Zhao, X. (2012b). microRNA‐155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in renal cancer cells. Molecular Medicine Reports, 5, 949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindemann, A. , Takahashi, H. , Patel, A. A. , Osman, A. A. , & Myers, J. N. (2018). Targeting the DNA damage response in OSCC with TP53 mutations. Journal of Dental Research, 97, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lundberg, E. , & Borner, G. H. H. (2019). Spatial proteomics: a powerful discovery tool for cell biology. Nature Reviews Molecular Cell Biology, 20, 285–302. [DOI] [PubMed] [Google Scholar]
- 36. Malishev, R. , Ben‐Zichri, S. , Oren, O. , Shauloff, N. , Peretz, T. , Taube, R. , Papo, N. , & Jelinek, R. (2021). The pro‐apoptotic domain of BIM protein forms toxic amyloid fibrils. Cellular and Molecular Life Sciences, 78, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menon, D. R. , & Fujita, M. (2019). A state of stochastic cancer stemness through the CDK1‐SOX2 axis. Oncotarget, 10, 2583–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Narula, K. , Choudhary, P. , Ghosh, S. , Elagamey, E. , Chakraborty, N. , & Chakraborty, S. (2019). Comparative nuclear proteomics analysis provides insight into the mechanism of signaling and immune response to blast disease caused by magnaporthe oryzae in rice. Proteomics, 19, e1800188. [DOI] [PubMed] [Google Scholar]
- 39. Neganova, I. , Tilgner, K. , Buskin, A. , Paraskevopoulou, I. , Atkinson, S. P. , Peberdy, D. , Passos, J. F. , & Lako, M. (2014). CDK1 plays an important role in the maintenance of pluripotency and genomic stability in human pluripotent stem cells. Cell Death & Disease, 5, e1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nesvizhskii, A. I. , Keller, A. , Kolker, E. , & Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry, 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- 41. Oberg, A. L. , Mahoney, D. W. , Eckel‐Passow, J. E. , Malone, C. J. , Wolfinger, R. D. , Hill, E. G. , Cooper, L. T. , Onuma, O. K. , Spiro, C. , Therneau, T. M. , & Bergen, III H. R. (2008). Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. Journal of Proteome Research, 7, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ordonez, L. D. , Hay, T. , Mcewen, R. , Polanska, U. M. , Hughes, A. , Delpuech, O. , Cadogan, E. , Powell, S. , Dry, J. , Tornillo, G. , Silcock, L. , Leo, E. , O'connor, M. J. , Clarke, A. R. , & Smalley, M. J. (2019). Rapid activation of epithelial‐mesenchymal transition drives PARP inhibitor resistance in Brca2‐mutant mammary tumours. Oncotarget, 10, 2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pankow, S. , Martínez‐Bartolomé, S. , Bamberger, C. , & Yates, J. R. (2019). Understanding molecular mechanisms of disease through spatial proteomics. Current Opinion in Chemical Biology, 48, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park, S.‐J. , Kim, J.‐G. , Kim, N. D. , Yang, K. , Shim, J. W. , & Heo, K. (2016). Estradiol, TGF‐beta1 and hypoxia promote breast cancer stemness and EMT‐mediated breast cancer migration. Oncology Letters, 11, 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raghu, G. , Collard, H. R. , Egan, J. J. , Martinez, F. J. , Behr, J. , Brown, K. K. , Colby, T. V. , Cordier, J.‐F. , Flaherty, K. R. , Lasky, J. A. , Lynch, D. A. , Ryu, J. H. , Swigris, J. J. , Wells, A. U. , Ancochea, J. , Bouros, D. , Carvalho, C. , Costabel, U. , Ebina, M. , … Schünemann, H. J. (2011). An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence‐based guidelines for diagnosis and management. American Journal of Respiratory and Critical Care Medicine, 183, 788–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ravindran Menon, D. , Luo, Y. , Arcaroli, J. J. , Liu, S. , Krishnankutty, L. N. , Osborne, D. G. , Li, Y. , Samson, J. M. , Bagby, S. , Tan, A.‐C. , Robinson, W. A. , Messersmith, W. A. , & Fujita, M. (2018). CDK1 interacts with Sox2 and promotes tumor initiation in human melanoma. Cancer Research, 78, 6561–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sandri, B. J. , Kaplan, A. , Hodgson, S. W. , Peterson, M. , Avdulov, S. , Higgins, L. , Markowski, T. , Yang, P. , Limper, A. H. , Griffin, T. J. , Bitterman, P. , Lock, E. F. , & Wendt, C. H. (2018). Multi‐omic molecular profiling of lung cancer in COPD. European Respiratory Journal, 52, 1702665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schacke, M. , Kumar, J. , Colwell, N. , Hermanson, K. , Folle, G. , Nechaev, S. , Dhasarathy, A. , & Lafon‐Hughes, L. (2019). PARP‐1/2 inhibitor olaparib prevents or partially reverts EMT induced by TGF‐beta in NMuMG Cells. International Journal of Molecular Sciences, 20, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shadforth, I. P. , Dunkley, T. P. , Lilley, K. S. , & Bessant, C. (2005). i‐Tracker: For quantitative proteomics using iTRAQ. Bmc Genomics [Electronic Resource], 6, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shen, F. , Kirmani, K. Z. , Xiao, Z. , Thirlby, B. H. , Hickey, R. J. , & Malkas, L. H. (2011). Nuclear protein isoforms: Implications for cancer diagnosis and therapy. Journal of Cellular Biochemistry, 112, 756–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sochacki, J. , Devalle, S. , Reis, M. , Mattos, P. , & Rehen, S. (2016). Generation of urine iPS cell lines from patients with Attention Deficit Hyperactivity Disorder (ADHD) using a non‐integrative method. Stem Cell Research, 17, 102–106. [DOI] [PubMed] [Google Scholar]
- 52. Uhler, C. , & Shivashankar, G. V. (2018). Nuclear mechanopathology and cancer diagnosis. Trends in Cancer, 4, 320–331. [DOI] [PubMed] [Google Scholar]
- 53. Villodre, E. S. , Felipe, K. B. , Oyama, M. Z. , Oliveira, F H. D. , Lopez, P. L. D. C. , Solari, C. , Sevlever, G. , Guberman, A. , & Lenz, G. (2019). Silencing of the transcription factors Oct4, Sox2, Klf4, c‐Myc or Nanog has different effect on teratoma growth. Biochemical and Biophysical Research Communications, 517, 324–329. [DOI] [PubMed] [Google Scholar]
- 54. Wang, W. , Feng, J. , Zhou, H. , & Li, Q. (2020). Circ_0123996 promotes cell proliferation and fibrosisin mouse mesangial cells through sponging miR‐149‐5p and inducing Bach1 expression. Gene, 761, 144971. [DOI] [PubMed] [Google Scholar]
- 55. Wang, X. , Liu, J. , Jiang, L. , Wei, X. , Niu, C. , Wang, R. , Zhang, J. , Meng, D. , & Yao, K. (2016). Bach1 induces endothelial cell apoptosis and cell‐cycle arrest through ROS generation. Oxidative Medicine and Cellular Longevity, 2016, 6234043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weyemi, U. , Redon, C. E. , & Bonner, W. M. (2016). H2AX and EMT: Deciphering beyond DNA repair. Cell Cycle, 15, 1305–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xia, H. , Bodempudi, V. , Benyumov, A. , Hergert, P. , Tank, D. , Herrera, J. , Braziunas, J. , Larsson, O. , Parker, M. , Rossi, D. , Smith, K. , Peterson, M. , Limper, A. , Jessurun, J. , Connett, J. , Ingbar, D. , Phan, S. , Bitterman, P. B. , & Henke, C. A. (2014). Identification of a cell‐of‐origin for fibroblasts comprising the fibrotic reticulum in idiopathic pulmonary fibrosis. American Journal of Pathology, 184, 1369–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xia, H. , Gilbertsen, A. , Herrera, J. , Racila, E. , Smith, K. , Peterson, M. , Griffin, T. , Benyumov, A. , Yang, L. , Bitterman, P. B. , & Henke, C. A. (2017). Calcium‐binding protein S100A4 confers mesenchymal progenitor cell fibrogenicity in idiopathic pulmonary fibrosis. Journal of Clinical Investigation, 127, 2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang, L. , Geng, Z. , Nickel, T. , Johnson, C. , Gao, L. , Dutton, J. , Hou, C. , & Zhang, J. (2016). Differentiation of human induced‐pluripotent stem cells into smooth‐muscle cells: Two novel protocols. Plos One, 11, e0147155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang, L. , Herrera, J. , Gilbertsen, A. , Xia, H. , Smith, K. , Benyumov, A. , Bitterman, P. B. , & Henke, C. A. (2018). IL‐8 mediates idiopathic pulmonary fibrosis mesenchymal progenitor cell fibrogenicity. American Journal of Physiology. Lung Cellular and Molecular Physiology, 314, L127–L136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang, L. , Rudser, K. , Golnik, A. , Wey, A. , Higgins, L. A. , & Gourley, G. R. (2016). Urine protein biomarker candidates for autism. Journal of Proteomics & Bioinformatics, S14, 1–7. [Google Scholar]
- 62. Yang, L. , Xia, H. , Smith, K. , Gilbertsen, A. , Beisang, D. , Kuo, J. , Bitterman, P. B. , & Henke, C. A. (2021). A CD44/Brg1 nuclear complex confers mesenchymal progenitor cells with enhanced fibrogenicity in idiopathic pulmonary fibrosis. JCI Insight, 6, e144652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang, W. , Cho, H. , Shin, H.‐Y. , Chung, J.‐Y. , Kang, E. S. , Lee, E.‐J. , & Kim, J.‐H. (2016). Accumulation of cytoplasmic Cdk1 is associated with cancer growth and survival rate in epithelial ovarian cancer. Oncotarget, 7, 49481–49497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu, J. , Gu, X. , & Yi, S. (2016). Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: Insights into wallerian degeneration. Frontiers in Cellular Neuroscience, 10, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu, S. , Wang, X. , Geng, P. , Tang, X. , Xiang, L. , Lu, X. , Li, J. , Ruan, Z. , Chen, J. , Xie, G. , Wang, Z. , Ou, J. , Peng, Y. , Luo, X. , Zhang, X. , Dong, Y. , Pang, X. , Miao, H. , Chen, H. , & Liang, H. (2017). Melatonin regulates PARP1 to control the senescence‐associated secretory phenotype (SASP) in human fetal lung fibroblast cells. Journal of Pineal Research, 63, e12405. [DOI] [PubMed] [Google Scholar]
- 66. Zecha, J. , Satpathy, S. , Kanashova, T. , Avanessian, S. C. , Kane, M. H. , Clauser, K. R. , Mertins, P. , Carr, S. A. , & Kuster, B. (2019). TMT labeling for the masses: A robust and cost‐efficient, in‐solution labeling approach. Molecular & Cellular Proteomics, 18, 1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information
Supplemental information
Supplemental information
Supplemental information
Data Availability Statement
The dataset has been deposited to the ProteomeXchange Consortium.via the PRIDE partner proteomics repository, dataset identifier PXD032352.


