Abstract
Tumor associated macrophages (TAMs) suppress the cancer immune response and are a key target for immunotherapy. The effects of ruthenium and rhodium complexes on TAMs have not been well characterized. To address this gap in the field, a panel of 22 dirhodium and ruthenium complexes were screened against three subtypes of macrophages, triple-negative breast cancer and normal breast tissue cells. Experiments were carried out in 2D and biomimetic 3D co-culture experiments with and without irradiation with blue light. Leads were identified with cell-type-specific toxicity toward macrophage subtypes, cancer cells, or both. Experiments with 3D spheroids revealed complexes that sensitized the tumor models to the chemotherapeutic doxorubicin. Cell surface exposure of calreticulin, a known facilitator of immunogenic cell death (ICD), was increased upon treatment, along with a concomitant reduction in the M2-subtype classifier arginase. Our findings lay a strong foundation for the future development of ruthenium- and rhodium-based chemotherapies targeting TAMs.
Keywords: antitumor agents, immunotherapy, macrophage, rhodium, ruthenium, transition metals
Graphical Abstract
Introduction
The human immune system involves a complex interplay between chemical signals and biological responses. Macrophages play an essential role in humans and the immune response, including maintenance of tissue homeostasis through breakdown of senescent cells, mediation of the inflammatory immune response, wound healing, and foreign pathogen detection.[1] In addition to their essential roles in normal health, macrophages also fuel the progression of human diseases, including those caused by chronic inflammation, such as atherosclerosis[2] and cancer.[3]
Macrophages display a high degree of cellular plasticity and are able to make drastic changes in their behavior depending on their environment.[4] The classical definition for macrophage subtyping holds that M0 or undifferentiated macrophages can differentiate into two forms; either pro-inflammatory (M1) or anti-inflammatory (M2) macrophages. M1 macrophages are responsible for pathogen detection, triggering inflammation via release of pro-inflammatory cytokines, and cytotoxicity through the production of reactive oxygen and nitrogen species.[4] M2 macrophages, on the other hand, are immuno-suppressive and act to reduce inflammation caused from M1 macrophages.[4–5] The balance between M1 and M2 polarization affects not only the local inflammatory milieu, but also influences adaptive immune priming.
Cancer is the second leading cause of death in the US, and progresses in large part due to the inability of the host immune system to eliminate transformed cells and differentiate between cancer and its own healthy cells.[6] The most aggressive cancer cells secrete chemical signals, also known as chemokines, to recruit M0 macrophages into tumor tissue.[7] Upon accumulation at the tumor site, M0 macrophages further differentiate to either the M1 or M2 subtype, which are known collectively as tumor associated macrophages (TAMs). Tumors with high numbers of M2 TAMs are associated with poor prognoses and more severe outcomes.[8] Furthermore, M2 TAMs display an immunosuppressive capacity by expressing programmed death ligands 1 and 2 (PD-L1/2) which in turn bind to the PD-1 receptor on killer T-cells, suppressing their capacity to kill tumor cells.[9] M2 TAMs also downregulate levels of pro-inflammatory M1 macrophages through mediation of polyamine signaling and metabolism, reducing pro-immune inflammation at the tumor site.[5]
The first immunotherapy for the treatment of cancer began in the 1980’s with the FDA approval of the cytokine interferon alpha 2 (IFN-α2).[10] More recently, immune checkpoint therapies targeting the CTLA-4 and PD-1 pathways have spawned a new era and interest in harnessing the immune system to fight cancer.[11] Likewise, due to their outsized role in tumor progression, macrophages have also been identified as prime targets for anti-tumor immunotherapies.[12] There are two main strategies for targeting macrophages. The first is macrophage repolarization or switching the TAM phenotype from the immunosuppressive M2 state to the anti-tumor M1 state through chemical signaling.[13] The second approach involves macrophage depletion, or simply macrophage-specific toxicity. Common treatment modalities for macrophage depletion include nanoparticles[14] and photosensitizing agents.[15] Macrophage-specific photodynamic therapy (PDT), represents an exciting opportunity for anti-tumor activity.[15] PDT is a photochemical process, which unlike macrophage depletion, allows for spatiotemporal control over cell killing.
Historically, metal complexes have played a major role in cancer therapy. Cisplatin, and its derivatives, are currently used to treat roughly 50 % of human cancers. Unfortunately, platinum agents show low specificity towards cancer versus normal cells, leading to harrowing side effects in patients. Inspired by the success of platinum drugs and the need to discover more selective and less toxic metal-based compounds, recent efforts to develop new anti-cancer therapeutics have focused on other late transition metals, including ruthenium[16] and rhodium.[17] Ruthenium complexes have many attractive properties for PDT and photochemotherapy (PCT)[18] due to their high dark-to-light ratios for cancer cell death.[19] In addition, dirhodium paddlewheel complexes have been known for over 40 years to show promising activity in cancer cells, including cytostatic behavior[20] and more recently photo-induced damage to DNA.[21,17a] Altogether hundreds of research articles have been published concerning the evaluation synthesis of Ru and Rh complexes as anti-cancer agents.[22] These compounds have undergone rigorous testing in a myriad of different in vitro and in vivo models of human cancers, including breast, prostate, lung, colon, skin, bladder and brain.[22a,b] Several ruthenium-based drugs have entered human clinical trials.[23] Given the large role immune cells and signaling play in cancer progression and survival, the effects of metal compounds on immune cells, including macrophages, are still fragmentary.[22c] This is especially true for Ru and Rh complexes whose effects on macrophages has been reported minimally.[24]
Herein we describe the evaluation of a library of ruthenium- and rhodium-based complexes against M0, M1, and M2 murine macrophages. To provide points of comparison, this library was also screened against the human triple-negative breast cancer (TNBC) cell line MDA-MB-231 and the normal human breast epithelial line MCF-10 A. After screening the full library, in the dark and under visible light irradiation to uncover compounds with PDT/PCT effects, rich structure-activity relationships were revealed, and leads were identified. From EC50 determinations in 2D cell culture experiments, we discovered complexes with specific toxicity toward macrophages versus cancer cells and normal cells, as well as compounds that are specific for killing immunosuppressive M2 macrophages versus non-polarized M0 macrophages or pro-inflammatory M1 cells. Furthermore, several compounds in our series showed high selectivity for killing cells under light versus dark conditions. A subset of the most promising compounds from 2D studies was carried into 3D cell co-culture experiments, where spheroids containing macrophages and TNBC cells were used to mimic the in vivo tumor microenvironment, modeling the signaling between TAMs and cancer cells that modulates the phenotype of these populations. Key functional markers were probed to identify the transition between M1 and M2 macrophages, as well as tumor cell surface calreticulin exposure, a classifier of immunogenic cell death (ICD) that can lead to long-lasting anti-tumor immunity. These studies revealed that Ru and Rh metal-based agents can kill M2 macrophages in 3D culture and sensitize tumor spheroids to the cancer drug doxorubicin, an inducer of ICD. Furthermore, imaging experiments confirmed that the lead metal complexes increase calreticulin translocation to the cancer cell surface. This study opens new avenues for metalloimmunotherapy with rhodium and ruthenium compounds.
Results and Discussion
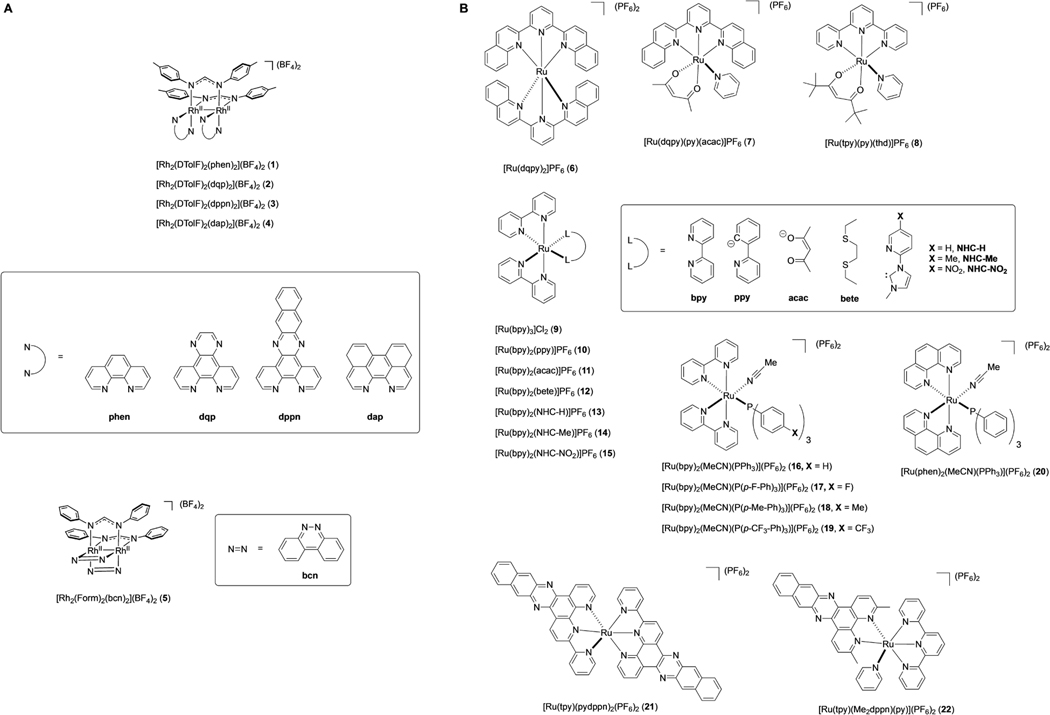
Compounds 1–22 (Figure 1) were selected for this study because they represent a diverse group of dinuclear rhodium and mononuclear ruthenium complexes. A wide net was cast because structure activity relationships (SAR) are notoriously difficult to predict for metal complexes. Rh2(II,II) paddlewheel complexes 1,[25] 2–3,[26] 4,[27] and 5[28] are derived from bidentate or tridentate polypyridyl ligands. Compounds 6[29] and 7[30] are mononuclear Ru(II) complexes containing the ligand dqpy (2,6-di(quinolin-2-yl)pyridine), which supports photosensitization and/or photorelease chemistry,[31] depending on the nature of the other mono- or bidentate ligands. Complex 8[31] carries the tridentate ligand tpy (2,2′:6′,2′′-terpyridine) and the bidentate ligand 2,2,6,6-tetramethylheptane-3,5-dione. However, unlike complexes containing the Ru(dqpy) fragment, which are largely unexplored in biological applications, there are many examples of those with the Ru(tpy) unit that show photosensitization and/or photorelease properties in vitro and in vivo.[32] Analogs 9,[33] 10,[34] 11,[35] 12,[36] and 13–15[37] all contain the fragment Ru(bpy)2, a molecular scaffold found in many bioactive Ru(II) complexes that show photorelease, photosensitization and, in many cases, luminescence.[38] Compounds 16[39] and 17–19[40] are photoactive Ru(II) complexes that release monodentate nitriles, complex 19 induces apoptosis in MDA-MB-231 cells upon irradiation with blue light[41] and 20[42] is a phosphine-containing complex containing the related fragment Ru(phen)2 (phen = 1,10-phenanthroline). Compound 21[43] is a homoleptic Ru(II) complex coordinated by the tridentate ligand pydppn (3-(pyridin-2-yl)benzo[i]dipyrido[3,2-a:2′,3′-c]phenazine), which carries an extended π-system that facilitates photochemical protein-DNA crosslinking in cells.[44] Finally compound 22[45] contains the Ru(tpy) fragment and the sterically encumbered ligand Me2dppn (3,6-dimethylbenzo[i]dipyrido[3,2-a:2′,3′-c]phenazine), which facilitates photochemical release of monodentate pyridine ligands bound to ruthenium, as well as generation of 1O2 for PCT/PDT dual-action behavior.[46]
Figure 1.
Structures and formulas of dirhodium paddlewheel complexes 1–5 (A) and ruthenium complexes 6–22 (B).
General compound screen
Compounds 1–22 were screened at a single concentration in cell viability assays against a panel of five different cell types; M0, M1, and M2 murine macrophages, MDA-MB-231 triple-negative breast cancer cells, and the normal breast epithelial line, MCF-10 A. Cell types were chosen to identify hit compounds with specific toxicity towards either macrophages of a certain polarization state, cancer cells, or normal cells. Identifying leads with cell-line specific toxicity has major clinical implications. For example, selective macrophage depletion is a viable strategy for cancer treatment.[47] Cancer-cell specific compounds are also important for reducing side-effects and preventing toxicity in normal tissue.[48] Compounds were evaluated with or without light irradiation. Specifically, compounds 1–22 (10 μM) were incubated with all cell types for 1 h, then left in the dark or irradiated with blue light (λirr=460–470 nm, tirr=20 min, 56 J/cm2) and viability was assessed using the MTT assay 72 h after irradiation was ceased to identify photoactive PCT or PDT compounds.
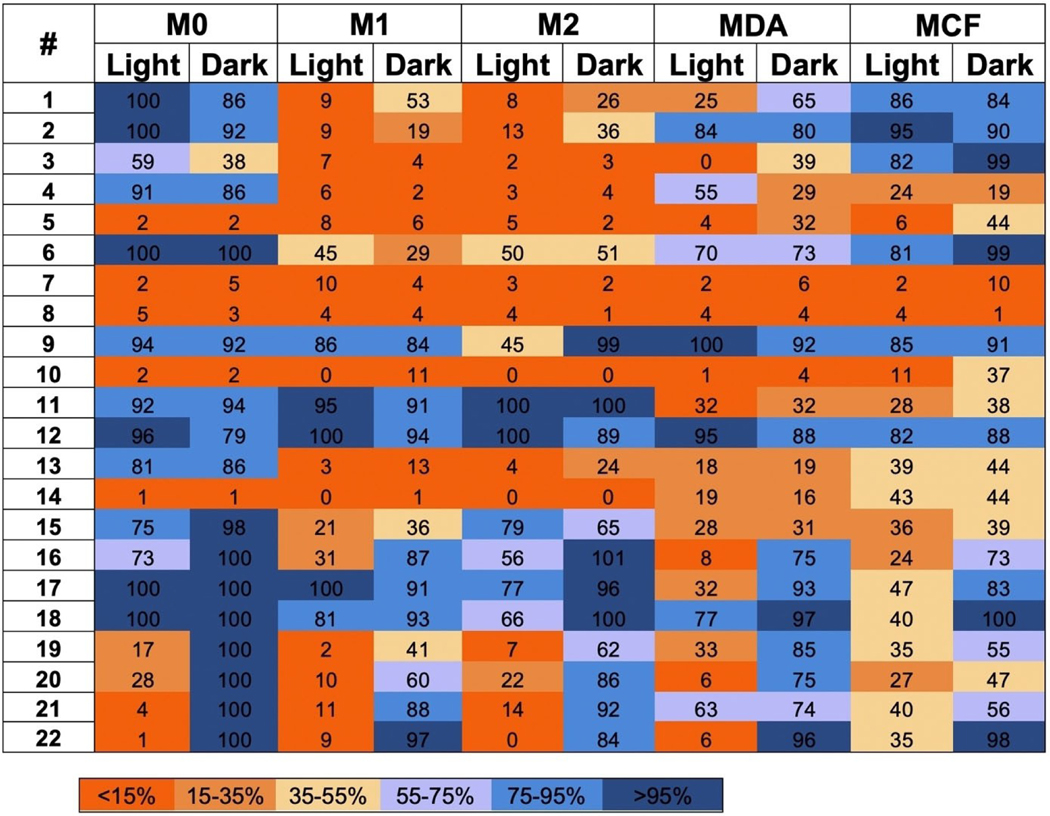
Results of the screening experiment are shown in heat map form in Figure 2. Numerical data in Figure 2 are percent viabilities (vs. vehicle control as 100 %) from a single experiment but are representative of three independent experiments. Entries are color-coded in gradient form from orange to blue transitioning from low to high viabilities. Compounds 1–4 are a closely related series of dirhodium complexes with two formamidinate (form−) bridging ligands and two neutral chelating ligands, which display potent toxicity toward both lines of polarized macrophages, with 1 and 2 displaying some amount of light activated toxicity. Compounds 1–3 display a lack of selectivity toward non-cancerous cells while 4 shows toxicity towards normal MCF-10 A cells. Therefore 1–3 were selected as candidates for EC50 determination. Compound 5 is also a Rh2(II,II) complex bridged by two form− moieties that also possesses two additional neutral bridging ligands, which shows potent toxicity across all cell lines and was selected for further investigation.
Figure 2.
Heat map for light and dark cell viabilities of 5 cell lines treated with compounds 1–22. Cells were treated with compound 1–22 (10 μM) and either irradiated (λirr=460–470 nm, tirr=20 min, 56 J/cm2) or left in the dark. Cells were incubated for 72 h before viabilities were evaluated by MTT assay. Values in Figure 2 are represented as % viability vs. vehicle control with the same cell type. Values range from low viabilities (<15 %: bright orange) to high viability (>95 %: dark blue). Light alone did not decrease viability vs. non-irradiated controls. Viability data are averages from quadruplicate wells. Data are representative from three independent experiments. MDA: MDA-MB-231; MCF: MCF-10 A.
Ruthenium compounds 6 and 7 bear the tridentate ligand dqpy and show rich structure-activity relationships. Ruthenium compound 6 is essentially non-toxic against all five cell lines, independent of light irradiation. Compound 7 bears an acetylacetonate (acac−) ligand and was selected for further investigation; compounds of this type show favorable cellular toxicity properties and cancer cell selectivity.[49] Compound 8, which is closely related to 7, bearing a 2,2,6,6-tetramethyl-3,5-heptanedione (thd) ligand displayed potent toxicity across all cell lines and was also selected for further investigation. Compounds 9–19 all contain the Ru(bpy)2 fragment, with two monodentate ligands or one bidentate ligand occupying the other coordination sites. Compound 11 is related to 7 and contains the acac− ligand, yet 11 displays no notable toxicity toward any of the cell lines. Compounds 12 and 9 likely suffer from poor cellular uptake as a result of bearing two bpy ligands, as evidenced by their lack of activity. Furthermore, 9 is known to be an excellent photosensitizer,[50] yet is inactive in cells, likely due to its poor internalization. Compound 10 is closely related to 9, bearing a deprotonated phenylpyridine ligand rather than bpy found in 19, which reduces the overall charge of 10 to + 1. This reduction in charge results in greater lipophilicity in 10 vs. 9, lending to better cellular penetration and therefore more potent toxicity. Compounds 13–15 bear anionic N-heterocyclic carbene (NHC−) ligands, resulting in a reduction of charge by one unit compared to 9. These compounds are closely related, varying only at the meta position on the pyridine ring of the NHC− ligand, yet their properties are remarkably different. Both 13 and 14 show similarities in their toxicities toward polarized macrophage and towards the non-immune cell lines, yet 14 shows great potency toward non-polarized macrophages that 13 does not. Based on this finding, compound 14 was selected for further investigation. Comparatively, complex 15 bearing a nitro group was mostly inactive in the viability screen. Compounds 16–20 represent a panel of Ru(bpy)2 containing complexes with a variety of triphenylphosphine (TPP) ligands. Compound 16 displays some capacity for light-induced toxicity, however 20, a bisphen compound, improves upon the light-activated toxicity of 16. Therefore, 20 was selected for future experimentation. Compounds 18 and 17 are mostly ineffective across all lines. Compound 19, however displays potent light activated toxicity, specifically in macrophages, and was therefore selected for further investigation. Ruthenium compound 21 shows excellent light activated toxicity specifically toward M1 and M2 macrophages and was also selected for further study. Lastly, 22 is a well-known dual-action compound, capable of both photo-dissociation and photosensitization. Compound 22 displays potent light-activated killing among each macrophage line and in the cancer cell line, while displaying a lack of selectivity toward the non-cancerous MCF-10 A line. Therefore 22, was selected for further investigation.
EC50 determination
After screening was complete, eleven compounds were selected for rigorous EC50 determinations against murine macrophages (M0, M1 and M2), MDA-MB-231 and MCF-10 A cells (Table 1). Cells were incubated with each compound (25–0.1 μM) for 1 h, media was aspirated and replenished, then cells were irradiated with blue light (λirr = 460–470 nm, tirr = 20 min, 56 J/cm2) or left in the dark for 20 min, and viabilities were determined by MTT assay 72 h later. A media aspiration step was incorporated before irradiation in these EC50 experiments to exclude false positives, compounds incapable of traversing the cell membrane that produce cell-permeable and cytotoxic photoproducts. Compounds 1 and 2 were largely inactive under these conditions. However, compound 3 was not toxic toward MCF-10 A at concentrations <25 μM, but it did display specificity towards polarized macrophages (M1 and M2) and MDA-MB-231 cells. Thus, 3 was considered a lead. Compound 5 killed MCF-10 A cells at lower concentrations than the other cell lines. Although 7 did not show evidence of light activation, it did show sub-micromolar EC50 values against all cell lines except the normal MCF-10 A line. Compound 8 displayed sub-micromolar EC50 values across all cell lines, independent of light irradiation, but was not specific. Each of the previously mentioned compounds displayed little to no light-enhanced toxicity, showing phototherapeutic indices (PI) values of 3 or less (Table S1 in Supporting Information). Compounds 19 and 20 were not active under these conditions and showed EC50 values >25 μM. Consistent with a prior report which suggested 21 is not readily taken up by cells,[44b] compound 21 was not active in MDA-MB-231 and MCF-10 A cells. However, 21 showed excellent ability to kill macrophages in a light-activated fashion, especially M2 macrophages (light EC50 = 0.7 ± 0.2 μM, PI>36) (Table S1). Lastly, compound 22 showed light-induced toxicity across all cell lines, showing the least potency in MCF-10 A cells and the most potency in polarized macrophages. Compound 22 showed excellent PI values of >25, >100 and >45 in M0, M1, and M2 macrophages respectively, and a respectable PI value of 7.4 in MDA-MB-231 cells (Table S1), making 22 a prime candidate to carry over into 3D cell culture experiments.
Table 1.
EC50 Values (μM) of Metal Complexes against Various Cell Lines.[a]
| Entry | Compound | M0 Light | Dark | M1 Light | Dark | M2 Light | Dark | MDA-MB-231 Light | Dark | MCF-10A Light | Dark |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 1 | 1 | 22 ± 5 | > 25 | 24 ± 1 | > 25 | 20 ± 5 | > 25 | 16 ± 5 | 15 ± 4 | > 25 | > 25 |
| 2 | 2 | 16 ± 3 | > 25 | 12 ± 3 | > 25 | 13 ± 1 | > 25 | > 25 | > 25 | > 25 | > 25 |
| 3 | 3 | 20 ± 5 | > 25 | 24 ± 1 | 15 ± 4 | 20 ± 4 | 24 ± 1.0 | 8.6 ± 2 | 9.3 ± 0.5 | > 25 | > 25 |
| 4 | 5 | 3.0 ± 2.1 | 3.4 ± 0.6 | 1.5 ± 0.9 | 1.8 ± 0.8 | 1.4 ± 0.7 | 3.0 ± 0.5 | 8.4 ± 1.1 | 8.4 ± 1.8 | 2.1 ± 1.2 | 2.9 ± 2.1 |
| 5 | 7 | 0.2 ± 0.04 | 0.3 ± 0.1 | 0.45 ± 0.2 | 0.99 ± 0.1 | 0.40 ± 0.11 | 1.7 ± 1.0 | 0.58 ± 0.07 | 0.67 ± 0.1 | 2.1 ± 1.1 | 1.7 ± 0.3 |
| 6 | 8 | 0.25 ± 0.14 | 0.75 ± 0.3 | 1.1 ± 0.1 | 0.89 ± 0.3 | 0.46 ± 0.2 | 0.88 ± 0.3 | 1.7 ± 0.7 | 2.8 ± 0.5 | 0.90 ± 0.2 | 1.2 ± 0.2 |
| 7 | 14 | 2.3 ± 1.9 | 1.7 ± 1.0 | 4.6 ± 1.2 | 13 ± 5 | 8.2 ± 2.6 | 17 ± 3 | 1.8 ± 0.8 | 1.4 ± 0.5 | 3.1 ± 0.4 | 7.8 ± 0.9 |
| 8 | 19 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 |
| 9 | 20 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 | > 25 |
| 10 | 21 | 1.4 ± 0.4 | > 25 | 2.2 ± 0.4 | > 25 | 0.7 ± 0.2 | > 25 | > 25 | > 25 | > 25 | > 25 |
| 11 | 22 | 1.0 ± 0.1 | > 25 | 0.23 ± 0.14 | > 25 | 0.55 ± 0.1 | > 25 | 4.6 ± 0.5[b] | 34 ± 3[b] | 7.0 ± 2.1 | > 25 |
Cells of each type were exposed to experimental treatments in media ranging from 25 μM-100 nM for 1 h. Following treatment, media was aspirated and replaced with fresh media. Cells were then either irradiated with blue (460/70 nm) light for 20 min or wrapped in foil and incubated for 72 h before relative viabilities were determined by MTT assay. EC50 values were obtained using the Igor Pro graphing software. Data are averages of three independent experiments using quadruplicate wells, errors are standard deviations.
Previously reported by Toupin et al.[51]
3D cell viability assays
Compounds 1, 3, 21 and 22 showed favorable properties in 2D culture and were moved forward to 3D experiments. Unlike 2D monolayer culture, where in vivo features are not reproduced, 3D spheroids mimic solid tumors by including interaction with the extracellular matrix (ECM), cell polarity, and cell-to-cell contacts, thus providing a more accurate context in which to evaluate potential drug candidates.[52] Furthermore, incorporating tumor cells and macrophages into heterogeneous 3D spheroids mimics cytokine signaling between the two cell types that drives macrophage polarization to the M2 state, mimicking TAMs in solid tumors. Finally, TAMs have been shown to reduce the efficacy of some chemotherapeutics in the tumor microenvironment, leading to chemoresistance.[53] Doxorubicin is a commonly employed breast cancer chemotherapeutic agent associated with chemoresistance and cardiotoxicity. Drugs that synergize with doxorubicin have the potential to mitigate these effects by making doxorubicin active at lower doses. In the following experiments, doxorubicin was evaluated in combination with lead complexes to determine whether the effect of doxorubicin can be exacerbated upon cell-type specific depletion using a metal complex.
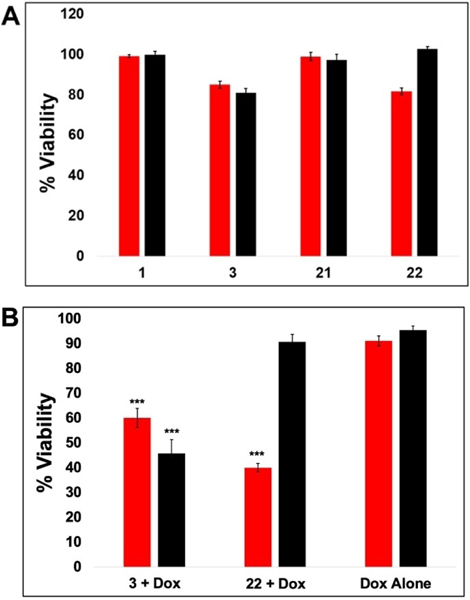
Tumor spheroids were established on cultrex by combining MDA-MB-231 cells and murine bone marrow derived macrophages (BMM). Cells were incubated for 72 h after seeding to allow for spheroid formation. After 72 h, cells were treated with each compound (25 μM) for 24 h before being rinsed, followed by light irradiation (λirr=460–470 nm, tirr=20 min, 56 J/cm2). Viability was determined after 72 h using the alamar blue fluorescence assay. Interestingly, compounds 1 and 21 displayed little to no capacity to reduce cell viability in the co-culture experiment at 25 μM concentration and were not selected for further experimentation, which disagreed with the data from 2D experiments. However, compounds 3 and 22 did show a cytotoxic effect, with data for 3 displaying similar potency under light and dark conditions and data for 22 indicating light-activated toxicity (Figure 3A). Importantly, compounds 3 and 22 (25 μM) reduced cell viability in combination with low, non-lethal concentrations of doxorubicin (Figure 3B). Spheroids treated with doxorubicin (1.25 μM) alone displayed nearly 100 % viability versus vehicle control spheroids (Figure 3B). However, the combination of 25 μM 3 and 1.25 μM doxorubicin decreased viability to ∼ 50 % of the control, independent of light irradiation, indicating synergistic toxicity. Furthermore, compound 22 (25 μM) slightly reduced cell viability only when irradiated with light and showed a reduction in viability to about 50 % when cells were co-treated with 1.25 uM doxorubicin in combination with light. Interestingly, the results of the 2D experiments determined that 3 was able to selectively kill cancer cells while 22 was more selective towards polarized macrophages, yet when used in combination with doxorubicin, a similar effect was observed for both compounds. With these promising results, 3 and 22 were selected for further examination by flow cytometry and fluorescence microscopy to accurately characterize effects on MDA-MB-231 cells vs. macrophages in the heterogeneous mixture and to probe macrophage polarization and immunogenic cell death.
Figure 3.
Co-culture Macrophage/TNBC 3D Spheroids Viability Assays. A: 3D tumor spheroids treated with 25 μM 1,3,21, or 22 for 24 h in the presence (red) or absence (black) of 20 min blue light (460/70 nm) irradiation. Viability was assessed by alamar blue assay 72 h after light treatment B: 3D tumor spheroids treated with 1.25 μM doxorubicin and either 25 μM 3, 22, or vehicle for 24 h in the presence (red) or absence (black) of 20 min blue light (460/70 nm) irradiation. Viability was assessed by alamar blue assay 72 h after light treatment. Statistical significance was determined against doxorubicin alone *** P <0.01.
Flow cytometry
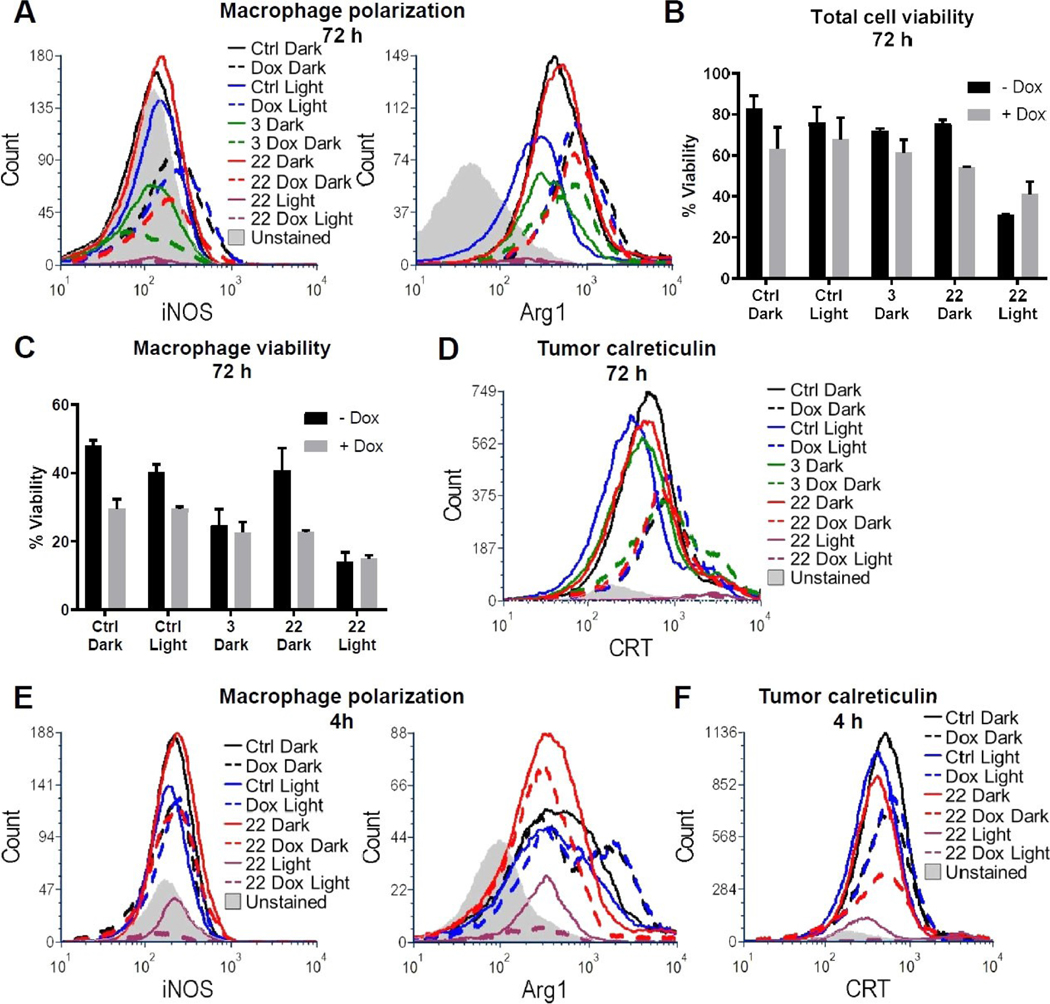
For flow cytometry experiments, spheroids established over 72 h period were treated with 3 or 22 (25 μM), either alone or in combination with doxorubicin for 24 h. After 24 h, the treatment media was aspirated and replaced with vehicle. Where indicated (denoted as “Light”), spheroids were irradiated (λirr=460–470 nm, tirr=20 min, 56 J/cm2) or left in the dark and after 72 h cells were harvested, dissociated, and stained for flow cytometric analysis. Markers for polarized macrophages (M1: iNOS and M2: arginase 1), for immunogenic cell death (calreticulin), and a general viability dye were used. Initial results indicated that the macrophage population was almost entirely polarized to the M2 state in all conditions as indicated by the relatively high arginase signal and the relatively low iNOS levels (Figure 4A). This finding is consistent with the literature, which suggests that tumor cells polarize recruited macrophages toward the M2 phenotype. Interestingly, arginase signal was increased in samples dosed with doxorubicin, a finding also consistent with the literature suggesting tumor cells induce macrophage polarization to the M2 subtype as a mechanism of chemotherapy resistance.[54] Compound 3 alone did not significantly affect overall viability (Figure 4B, Figure S1A), consistent with the results of the 3D spheroid assay; however, contrary to the alamar blue assay, overall viability was not significantly decreased in samples treated with 3 in combination with doxorubicin. There was a clear effect on macrophage viability in the combination of 3 and doxorubicin (Figure 4C, Figure S1B) as well as a small but notable increase in tumor cell calreticulin expression (Figure 4D). Based on our results in Figure 3, where irradiation did not enhance compound 3’s toxicity, we did not combine compound 3 with light irradiation for the flow cytometry experiments. In the dark, 22+ doxorubicin showed an increase in the non-viable tumor cell population by nearly 20 % vs. the control (Figure 4B), whereas the results of previous experiments showed no evidence of toxicity for the combo in the dark. At the 72 h time point following irradiation, 22 either alone or in combination with doxorubicin showed highly effective killing, yielding low viable cell counts (1,969 viable tumor cells in 22 Light and 1,506 viable tumor cells in 22 Dox Light groups), although calreticulin was elevated in remaining detectable cells (Figure 4D, Figure S1C). Therefore, we repeated the experiment utilizing a 4 h incubation time post-irradiation rather than 72 h in an effort to capture the effect on cells prior to necrotic death and lysis over the 72 h time period.[32b] Compound 22 showed clear light-activated toxicity with or without doxorubicin (Figure S2A). While both conditions did display relatively equivalent levels of increase in the overall non-viable macrophage population (Figure S2B), arginase levels were muted in samples treated with the combination of 22 and doxorubicin vs. doxorubicin alone either with or without light (Figure 4E). This finding suggests that the combination with 22 reduces doxorubicin-induced M2 macrophages. Furthermore, treatment with 22, doxorubicin and light also produced the largest observed tumor cell calreticulin signal among all conditions (Figure 4F, Figure S2C), which is consistent with our 72 h treatment results (Figure 4D, Figure S1C), altogether suggesting this combination may have optimal M2 macrophage killing with immunogenic cell death induction.
Figure 4.
Flow cytometry assessment of macrophage polarization and immunogenic cell death. 3D tumor spheroids were treated with vehicle, 25 μM 3 or 22 for 24 h, with or without 1.25 μM doxorubicin, with and without exposure to 20 min blue light (460/70 nm) irradiation as indicated. Spheroids were dissociated 72 h (A–D) or 4 h (E–F) after light treatment and stained for flow cytometric analysis. The unstained sample for all histograms is 3,000 unstained cells from the dissociated Ctrl Dark sample at the appropriate time point. A: Macrophage polarization after 72 h was determined by intracellular iNOS (M1) or Arg1 (M2) detection in F4/80 + macrophages. Viability was assessed after 72 h on the (B) total cell and (C) F4/80 + macrophage populations. Results are shown as the average +/− SD from two independent biological replicates. D: Surface calreticulin exposure levels were detected on CD45-/F4/80-tumor cells 72 h after treatment. E: Macrophage polarization was assessed as in (A) 4 h after treatment. F: Tumor cell surface calreticulin exposure was assessed as in (D) 4 h after treatment.
Confocal imaging
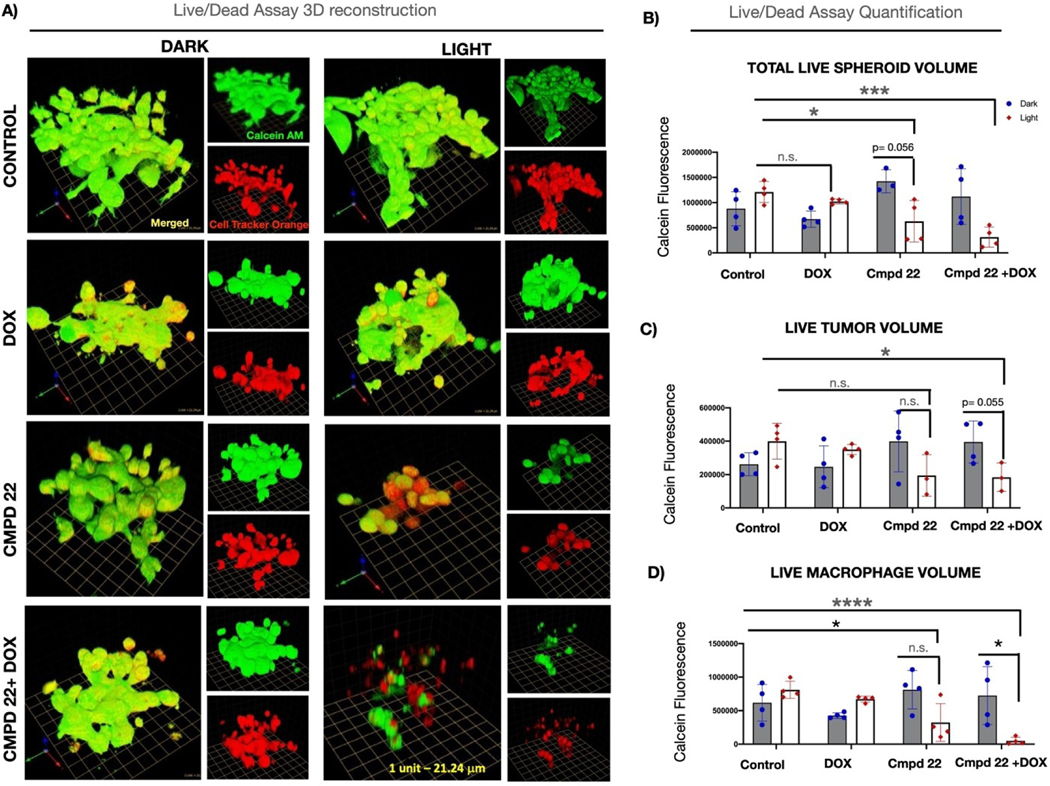
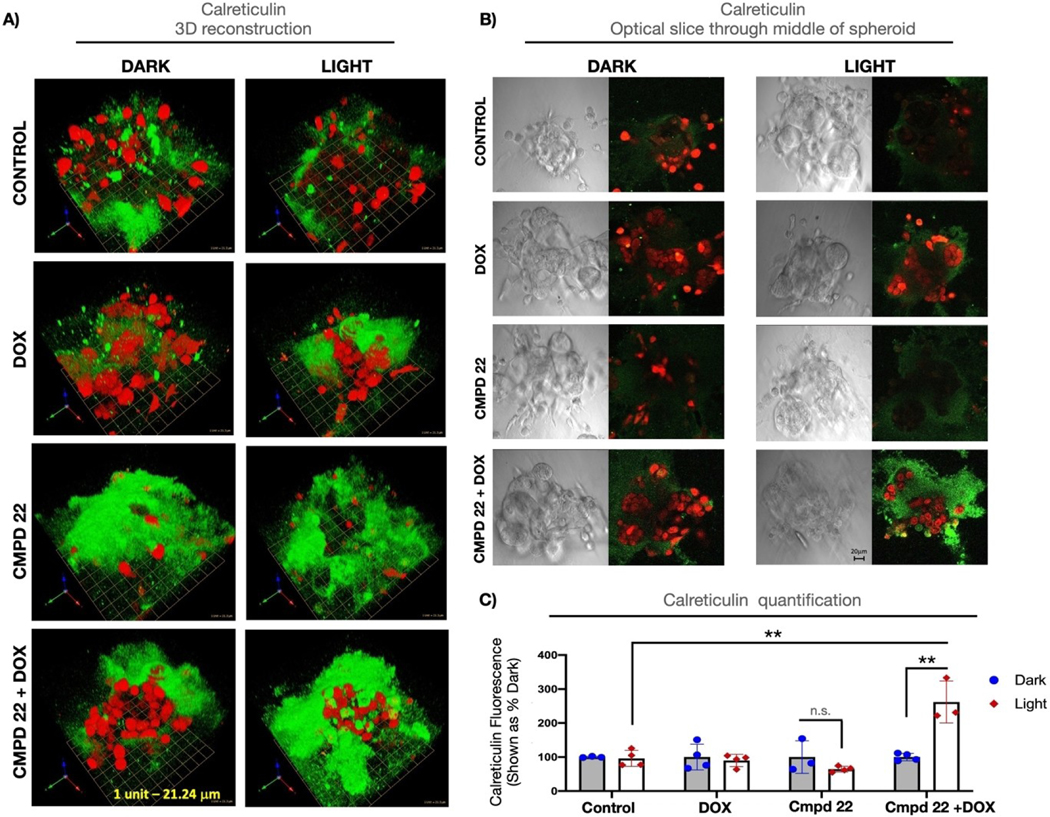
Compound 22 was selected for further experimentation after the flow cytometry results showed that it was able to deplete macrophages selectively, decrease arginase levels (M2 macrophage marker), and increase calreticulin signals (ICD marker). Confocal microscopy was utilized to quantify the overall viability as well as the calreticulin levels. Spheroids were treated with 22 (25 μM) either alone or in combination with 1.25 μM doxorubicin for 24 h. After 24 h the treatment media was aspirated and replaced with vehicle. Spheroids were irradiated (λirr=460–470 nm, tirr=20 min, 56 J/cm2) or left in the dark and incubated for 4 h before imaging. Calcein AM staining of live cultures revealed that the combination of 22 and doxorubicin was lethal to cells that were irradiated when compared to all other conditions including 22 alone in the light (Figure 5A,B). Cell Tracker Orange labeling was used to distinguish calcein AM-positive macrophages from calcein AM-positive tumor cells (Figure 5C,D). Quantification of calcein fluorescence in these two cell populations revealed that the combination of 22 and doxorubicin was particularly effective in killing the macrophages. Calreticulin signal was measured next across all conditions to assess for indicators of ICD (Figure 6). The combination of 22 and doxorubicin under light conditions showed clear evidence of calreticulin translocation to the cell surface. In particular, the pericellular region at the tumor spheroid/ECM interface showed heavy staining for calreticulin (Figure 6A,B). Signals from calreticulin in this region were quantified in Figure 6C, which shows that the combination of 22, doxorubicin and light produced the largest amount of extracellular calreticulin. Taking the alamar blue 3D viability data, flow cytometry, and imaging data together, it is clear that the combination of 22, doxorubicin and light depletes immunosuppressive macrophages and tumor cells, and leads to the highest expression of calreticulin, a key signaling event in ICD. Although the detection of extracellular calreticulin shows great potential for achieving immunogenicity, further studies in immune competent animal models will be needed to prove that our compounds are immunogenic.
Figure 5.
Assessment of viability of 3D cultures exposed to 22 and Doxorubicin. A: 3D reconstruction of Calcein AM-stained 3D MDA MB 231/Macrophage spheroids treated with 22 (Cmpd 22), doxorubicin (DOX) or combination of the two (Cmpd 2 + DOX); green: Calcein AM-positive live tumor cells; red: Cell Tracker Orange-labeled macrophages; yellow: live macrophages; merged 3D image and separate Calcein AM and Orange Tracker Red images are shown for each condition; 1 unit in the scale grid corresponds to 21.24 μm. B: Quantification of all Calcein AM-positive (live) cells in the spheroid [tumor cells (green) +macrophages(yellow)] shown as integrated fluorescence intensity; C: Quantification of live tumor cells (green) shown as integrated fluorescence intensity; D: Quantification of live macrophages (yellow) shown as integrated fluorescence intensity. Data are shown as individual values from at least 3 different spheroids/condition and are representative of 3 biological replicates. Statistical values were determined by student t-test. * p<0.05; ***p<0.001; ****p<0.0001; n.s. – not significant.
Figure 6.
Quantification of immunofluorescence staining for calreticulin in 3D tumor/macrophage spheroids. A: 3D reconstruction of Calreticulin staining (green) in 3D MDA MB 231/Macrophage spheroids treated with 22 (Compound 22), doxorubicin (DOX) or combination of the two (Compound 2 + DOX); red: Cell Tracker Orange-labeled macrophages; 1 unit in the scale grid corresponds to 21.24 μm. B: DIC (differential interference contrast) images of the middle slice (left panels) depicting morphology of the spheroids and middle slice depiction of calreticulin staining (right panels; green: calreticulin staining, red: Cell Tracker Orange-labeled macrophages); scale bar indicates 20 μm. C: Quantification of calreticulin staining shown as sum of surface fluorescence intensities throughout the Z-stack. Data are shown as calreticulin fluorescence in light-irradiated sample relative to its non-irradiated (dark) control. Individual values from at least 3 different spheroids/condition are depicted and are representative of 3 biological replicates. Statistical values were determined by student t-test. ** p<0.01; n.s. – not significant.
Prior knowledge concerning Ru and Rh metal complexes and their effects on macrophages is limited compared to other metal complexes. In particular, gold compounds have been studied for their immunogenic effects and effects on TAMs. Recent work with Au(III) complexes,[55] among others,[22c,56] exemplifies the utility of gold complexes towards TAMs. Unlike gold, rhodium and ruthenium complexes have yet to be tested on macrophages in settings that mimic the tumor microenvironment. Prior work with Ru compounds and macrophages is limited mainly to carbon monoxide releasing molecules (CORMs) where released CO, rather than Ru, is likely the active agent.[24b–d,g,i] Rh CORMs have also been assessed for their ability to affect macrophage polarization through modulation of NO signaling.[24f] Furthermore, in the pursuit of assessing their anti-leishmania activity, both Ru and Rh complexes have been shown to display toxicity against macrophages.[24a,h] These limited examples, which relate Ru and Rh complexes to macrophages, have not assessed compound activity in TAMs. In the present study, Ru and Rh complexes showed macrophage toxicity in biomimetic 3D co-culture, the ability to sensitize tumor spheroids to chemotherapy, and to initiate signaling events consistent with ICD. This study clearly establishes a new role for Ru and Rh in targeting TAMs.
Conclusions
As our understanding of the complexity of the tumor microenvironment has improved with time it is becoming increasingly imperative that efficacy of any potential anti-cancer agents should be evaluated against multiple components of the tumor; chief among these being the macrophages, a critical cell type shown to play a major role in tumor survival and metastasis. This report aimed to assess the ability of mono- and di-nuclear rhodium and ruthenium complexes for their capacity to elicit a specific toxic response in macrophages, cancer cells, or healthy tissue. A panel of 22 previously synthesized and fully characterized metal complexes was used at a single concentration to assess for toxic effects across five cell types both in the presence and absence of light irradiation. A group of 11 compounds, which showed promising attributes in the general screen, were then used to perform EC50 determination across all cell lines. EC50 determination narrowed the field further for complexes which would be brought into 3D cell culture experiments. Tumor spheroids consisting of MDA-MB-231 breast cancer cells co-cultured with bone marrow derived murine macrophages were utilized for 3D experimentation in an effort to more closely resemble the true nature of the tumor microenvironment. It is well documented that macrophages render chemotherapy less effective at killing tumor tissue.[53] Therefore, tumor spheroids were treated with two complexes, 3 and 22, in tandem with doxorubicin, a known chemotherapeutic. Both complexes were able to significantly reduce cancer viability as compared to doxorubicin alone, suggesting sensitization to treatment. Cell surface exposure of calreticulin, a known facilitator of ICD, was also increased upon treatment, suggesting the mechanism of killing may promote adaptive immune priming. In addition to tumor cell death, macrophages were found to be targeted, with a concomitant reduction in the M2 classifier arginase. Collectively, this approach may release immune suppression and support anti-tumor T cell activation. Future studies will evaluate this potential in immune competent animal models. In summation, this study makes a strong case for the utility of Ru and Rh anti-cancer complexes against TAMs and support their development into real-word therapies.
Supplementary Material
Acknowledgements
These studies were partially supported by the NSF (CHE-2102508 to C.T. and J.K.) and NIH (NCI R01 CA251394 to I.P.). J.K. and H.G. acknowledge Wayne State University (Rumble Fellow-ship to N.T.) and Detroit Medical Center Foundation Research Grant (H.G.). We thank the Microscopy, Imaging and Cytometry Resources Core (MICR) for assistance with confocal imaging and flow cytometry analyses. MICR is supported, in part, by NIH Center grant P30 CA22453 to the Karmanos Cancer Institute and R50 CA251068–01 to Dr. Moin, Wayne State University.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting information for this article is available on the WWW under https://doi.org/10.1002/chem.202104430
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Lichtnekert J, Kawakami T, Parks WC, Duffield JS, Curr. Opin. Pharmacol. 2013, 13, 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hansson GK, Engl N. J. Med. 2005, 352, 1685–1695. [DOI] [PubMed] [Google Scholar]
- [3] a).Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P, Nat. Rev. Clin. Oncol. 2017, 14, 399–416; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, Soong DYH, Cotechini T, Anur P, Lin EY, Fidanza A, Lopez-Yrigoyen M, Millar MR, Urman A, Ai Z, Spellman PT, Hwang ES, Dixon J, Wiechmann L, Coussens LM, Smith HO, Pollard JW, Cancer Cell 2019, 35, 538–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sica A, Mantovani A, J. Clin. Invest. 2012, 122, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Latour YL, Gobert AP, Wilson KT, Amino Acids 2020, 52, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Houghton AN, Guevara-Patiño JA, J. Clin. Invest. 2004, 114, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Argyle D, Kitamura T, Front. Immunol. 2018, 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao X, Qu Y Sun J, Wang J, Liu X, Wang F, Zhang H, Wang W, Ma X, Gao X, Zhang S, Oncotarget 2017, 8, 30576–30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9] a).Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z, J. Biomed. Sci. 2019, 26, 78; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zheng Y, Fang Y-C, Li J, Onco. Lett. 2019, 18, 5399–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Herndon TM, Demko SG, Jiang X, He K, Gootenberg JE, Cohen MH, Keegan P, Pazdur R, Oncologist 2012, 17, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Buchbinder EI, Desai A, Am. J. Clin. Oncol. 2016, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, Krishnan V, Hatakeyama J, Dorigo O, Barkal LJ, Weissman IL, Nature 2019, 572, 392–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lyle DB, Breger JC, Baeva LF, Shallcross JC, Durfor CN, Wang NS, Langone JJ, J. Biomed. Mater. Res. 2008, 93A, 893–904. [DOI] [PubMed] [Google Scholar]
- [14] a).Qian Y, Qiao S, Dai Y, Xu G, Dai B, Lu L, Yu X, Luo Q, Zhang Z, ACS Nano 2017, 11, 9536–9549; [DOI] [PubMed] [Google Scholar]; b) Kim H, Kim Y, Kim I-H, Kim K, Choi Y, Theranostics 2014, 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Demidova TN, Hamblin MR, Int. J. Immunopathol. Pharmacol. 2004, 17, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16] a).Gandioso A, Purkait K, Gasser G, Chimia 2021, 75, 845–855; [DOI] [PubMed] [Google Scholar]; b) Chellan P, Sadler PJ, Chem. Eur. J. 2020, 26, 8676–8688; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cole HD, Roque JA, Lifshits LM, Hodges R, Barrett PC, Havrylyuk D, Heidary D, Ramasamy E, Cameron CG, Glazer EC, McFarland SA, Photochem. Photobiol. Sci. 2022, 98, 73–84; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Murray BS, Dyson PJ, Curr. Opin. Chem. Biol. 2020, 56, 28–34; [DOI] [PubMed] [Google Scholar]; e) Thota S, Rodrigues DA, Crans DC, Barreiro EJ, J. Med. Chem. 2018, 61, 5805–5821; [DOI] [PubMed] [Google Scholar]; f) Knoll JD, Turro C, Coord. Chem. Rev. 2015, 282–283, 110–126; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Monro S, Colon KL, Yin H, Roque J III, Konda P, Gujar S, Thummel RP, Lilge L, Cameron CG, McFarland SA, Chem. Rev. 2019, 119, 797–828; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Hess J, Huang H, Kaiser A, Pierroz V, Blacque O, Chao H, Gasser G, Chem. Eur. J. 2017, 23, 9888–9869; [DOI] [PubMed] [Google Scholar]; i) Chow MJ, Licona C, Pastorin G, Mellitzer G, Ang WH, Gaiddon C, Chem. Sci. 2016, 7, 4117–4124; [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Lameijer LN, Ernst D, Hopkins SL, Meijer MS, Askes SHC, Le Dévédec SE, Bonnet S, Angew. Chem. Int. Ed. 2017, 56, 11549–11553; Angew. Chem. 2017, 129, 11707–11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a).Aguirre JD, Angeles-Boza AM, Chouai A, Pellois J-P, Turro C, Dunbar KR, J. Am. Chem. Soc. 2009, 131, 11353–11360; [DOI] [PubMed] [Google Scholar]; b) Truong D, Sullivan MP, Tong KKH, Steel TR, Prause A, Lovett JH, Andersen JW, Jamieson SMF, Harris HH, Ott I, Weekley CM, Hummitzsch K, Söhnel T, Hanif M, Metzler-Nolte N, Goldstone DC, G Hartinger C, Inorg. Chem. 2020, 59, 3281–3289; [DOI] [PubMed] [Google Scholar]; c) Graf M, Siegmund D, Metzler-Nolte N, Sünkel K, Böttcher H-C, Inorg. Chim. Acta 2019, 487, 9–14. [Google Scholar]
- [18] a).Poynton FE, Bright SA, Blasco S, Williams DC, Kelly JM, Gunnlaugsson T, Chem. Soc. Rev. 2017, 46, 7706–7756; [DOI] [PubMed] [Google Scholar]; b) White JK, Schmehl RH, Turro C, Inorg. Chim. Acta 2017, 454, 7–20; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Heinemann F, Karges J, Gasser G, Acc. Chem. Res. 2017, 50, 2727–2736; [DOI] [PubMed] [Google Scholar]; d) Bonnet S, Dalton Trans. 2018, 47, 10330–10343. [DOI] [PubMed] [Google Scholar]
- [19].Ghosh G, Colon KL, Fuller A, Sainuddin T, Bradner E, McCain J, Monro SMA, Yin H, Hetu MW, Cameron CG, McFarland SA, Inorg. Chem. 2018, 57, 7694–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20] a).Erck A, Sherwood E, Bear JL, Kimbal AP, Cancer Res. 1976, 36, 2205–2209; [PubMed] [Google Scholar]; b) Rao PN, Smith ML, Pathak S, Howard RA, Bear JL, Nat J. Cancer Institute. 1980, 64, 905–912. [PubMed] [Google Scholar]
- [21].Angeles-Boza AM, Bradley PM, Fu PK-L, Wicke SE, Bacsa J, Dunbar KR, Turro C, Inorg. Chem. 2004, 43, 8510–8519. [DOI] [PubMed] [Google Scholar]
- [22] a).Lee SY, Kim CY, Nam T-G, Drug Des. Dev. Ther. 2020, 14, 5375–5392; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Máliková K, Masaryk L, Štarha P, Inorganics 2021, 9, 26; [Google Scholar]; c) Englinger B, Pirker C, Heffeter P, Terenzi A, Kowol CR, Keppler BK, Berger W, Chem. Rev. 2019, 119, 1519–1624; [DOI] [PubMed] [Google Scholar]; d) Thota S, Rodrigues DA, Crans DC, Barreiro EJ, J. Med. Chem. 2018, 61, 5805–5821; [DOI] [PubMed] [Google Scholar]; e) Popolin C, Cominetti MR, Med. Chem. 2017, 17, 1435–1441. [DOI] [PubMed] [Google Scholar]
- [23] a).Rausch M, Dyson PJ, Nowak-Sliwinska P, Adv. Ther. 2019, 2, 1900042. [Google Scholar]
- [24] a).Costa MS, Concalves YG, Nunes DCO, Napolitano DR, Maia PIS, Rodrigues RS, Rodrigues VM, von Poelhsitz G, Yoneyama KAG, J. Inorg. Biochem. 2017, 175, 225–231; [DOI] [PubMed] [Google Scholar]; b) Lee DW, Shin HY, Jeong JH, Han J, Ryu S, Nakahira K, Moon J-S, Biochem. Biophys. Res. Commun. 2017, 493, 957–963; [DOI] [PubMed] [Google Scholar]; c) Yamamoto-Oka H, Mizuguchi S, Toda M, Minamiyama Y, Takemura S, Shibata T, Cepinskas G, Nishiyama N, Inflammopharmacology 2017, 26, 435–445; [DOI] [PubMed] [Google Scholar]; d) Eun-Young Choi S-HC, Hyeon J-Y, Choi J-I, Choi S, Kim S-J, Eur. J. Pharmacol. 2015, 764, 22–29; [DOI] [PubMed] [Google Scholar]; e) Liu SL-J, Lin S, Chan DS-J, Vong CT, Hoi PM, Wong C-Y, Ma D-L, Leung CH, J. Inorg. Biochem. 2014, 140, 23–28; [DOI] [PubMed] [Google Scholar]; f) Moragues ME, Brines R, Terencio M, Sancenón F, Martínez-Máñez R, Alcaraz M, Inorg. Chem. 2013, 52, 13806–13808; [DOI] [PubMed] [Google Scholar]; g) Sawle P, Foresti R, Mann BE, Johnson TR, Green CJ, Motterlini R, Br. J. Pharmacol. 2009, 145, 800–810; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Rodriguez-Cabezas MN, Mesa-Valle CM, Azzouz S, Moraleda-Lindez V, Craciunescu D, Gutierrez-Rios MT, De Frutos MI, Osuna A, Pharmacol. 2001, 63, 112–119; [DOI] [PubMed] [Google Scholar]; i) Otterbein LE, Bach FH, Alam J, Soares M, Lu HT, Wysk M, Davis RJ, Flavell RA, Choi AMK, Nat. Med. 2000, 6, 422–428; [DOI] [PubMed] [Google Scholar]; j) Aresta M, De Fazio M, Fumarulo R, Giordano D, Pantaleo R, Riccardi S, Biochem. Biophys. Res. Commun. 1982, 104, 121–125. [DOI] [PubMed] [Google Scholar]
- [25].White TA, Witt SE, Li Z, Dunbar KR, Turro C, Inorg. Chem. 2015, 54, 10042–10048. [DOI] [PubMed] [Google Scholar]
- [26].Li Z, Leed NA, Dickson-Karn NM, Dunbar KR, Turro C, Chem. Sci. 2014, 5, 727–737. [Google Scholar]
- [27].Turro C, Dunbar KR, “ Multimetallic Systems for the Photocatalytic Production of Fuels from Abundant Sources”, can be found under https://www.osti.gov/servlets/purl/1430648/, 2018.
- [28].Whittemore TJ, Xue C, Huang J, Gallucci JC, Turro C, Nat. Chem. 2020, 12, 180–185. [DOI] [PubMed] [Google Scholar]
- [29].Thummel R, Jahng Y, Inorg. Chem. 1986, 25, 2527–2534. [Google Scholar]
- [30].Al-Afyouni MH, Rohrabaugh TN, Al-Afyouni KF, Turro C, Chem. Sci. 2018, 9. 6711–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Loftus LM, Al-Afyouni KF, Turro C, Chem. Eur. J. 2018, 24, 11550–11553. [DOI] [PubMed] [Google Scholar]
- [32] a).Toupin N, Steinke SJ, Nadella S, Li A, Rohrabaugh TN, Samuels ER, Turro C, Sevrioukova IF, Kodanko JJ, J. Am. Chem. Soc. 2021, 24, 9191–9205; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Toupin NP, Nadella S, Steinke SJ, Turro C, Kodanko JJ, Inorg. Chem. 2020, 59, 3919–3933; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) van Rixel VHS, Ramu V, Auyeung AB, Beztsinna N, Leger DY, Lameijer LN, Hilt ST, Le Dévédec SE, Yildiz T, Betancourt T, Gildner MB, Hudnall TW, Sol V, Liagre B, Kornienko A, Bonnet S, J. Am. Chem. Soc. 2019, 141, 18444–18454; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Arora K, Herroon M, Al-Afyouni MH, Toupin NP, Rohrabaugh TN, Loftus LM, Podgorski I, Turro C, Kodanko JJ, J. Am. Chem. Soc. 2018, 140, 14367–14380; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Lameijer LN, Hopkins SL, Brevé TG, Askes SHC, Bonnet S, Chem. Eur. J. 2016, 22, 18484–18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bossmann SH, Turro C, Schnabel C, Pokhrel MR, Payawan LM, Baumeister B, Woerner M, J. Phys. Chem. B 2001, 105, 5374–5382. [Google Scholar]
- [34].Muro-Small ML, Yarnell JE, McCusker CE, Castellano FN, Eur. J. Inorg. Chem. 2012, 2012, 4004–4011. [Google Scholar]
- [35].Xiao X, Sakamoto J, Tanabe M, Yamazaki S, Yamabe S, Matsumura- Inoue T, J. Electroanal. Chem. 2002, 527, 33–40. [Google Scholar]
- [36].Garner RN, Joyce LE, Turro C, Inorg. Chem. 2011, 50, 4384–4391. [DOI] [PubMed] [Google Scholar]
- [37].Kender WT, Turro C, J. Phys. Chem. A 2019, 123, 2650–2660. [DOI] [PubMed] [Google Scholar]
- [38] a).Respondek T, Sharma R, Herroon MK, Garner RN, Knoll JD, Cueny E, Turro C, Podgorski I, Kodanko JJ, ChemMedChem 2014, 9, 1306–1315; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Howerton BS, Heidary DK, Glazer EC, J. Am. Chem. Soc. 2012, 134, 8324–8327; [DOI] [PubMed] [Google Scholar]; c) Respondek T, Garner RN, Herroon MK, Podgorski I, Turro C, Kodanko JJ, J. Am. Chem. Soc. 2011, 133, 17164–17167; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Byrne A, Burke CS, Keyes TE, Chem. Sci. 2016, 7, 6551–6562; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Zayat L, Salierno M, Etchenique R, Inorg. Chem. 2006, 45, 1728–1731. [DOI] [PubMed] [Google Scholar]
- [39].Litke SV, Ershov AY, Meyer TJ, J. Phys. Chem. A 2011, 115, 14235–14242. [DOI] [PubMed] [Google Scholar]
- [40].Marmion ME, Takeuchi KJ, J. Am. Chem. Soc. 1988, 110, 1472–1480. [Google Scholar]
- [41].Lanquist AP, Gupta S, Al-Afyouni KF, Al-Afyouni M, Kodanko JJ, Turro C, Chem. Sci. 2021, 12, 12056–12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huang EK, Cheung W, Chan SL, Sung HHY, Williams ID, Leung W, Organometallics 2013, 32, 4483–4489. [Google Scholar]
- [43].Liu Y, Hammett R, Lutterman DA, Joyce LE, Thummel R, Turro C, Inorg. Chem. 2009, 48, 375–385. [DOI] [PubMed] [Google Scholar]
- [44] a).Liu Y, Hammitt R, Lutterman DA, Joyce LE, Thummel RP, Turro C, Inorg. Chem. 2009, 48, 375–385; [DOI] [PubMed] [Google Scholar]; b) Zhao R, Hammitt R, Thummel RP, Liu Y, Turro C, Snapka RM, Dalton Trans. 2009, 251, 10926–10931; [DOI] [PubMed] [Google Scholar]; c) Sun Y, El Ojaimi M, Hammitt R, Thummel RP, Turro C, J. Phys. Chem. B 2010, 114, 14664–14670. [DOI] [PubMed] [Google Scholar]
- [45].Knoll JD, Albani BA, Turro C, Chem. Commun. 2015, 51, 8777–8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Knoll JD, Albani BA, Turro C, Acc. Chem. Res. 2015, 48, 2280–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, Jones T, Jucknischke U, Scheiblich S, Kaluza K, Gorr IH, Walz A, Abiraj K, Cassier PA, Sica A, Gomez-Roca C, de Visser KE, Italiano A, Le Tourneau C, Delord J, Levitsky H, Blay J, Ruttinger D, Cancer Cell 2014, 25, 846–859. [DOI] [PubMed] [Google Scholar]
- [48].Schirrmacher V, Int. J. Oncol. 2018, 54, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gupta S, Vandevord JM, Loftus LM, Toupin N, Al-Afyouni MH, Rohrabaugh TN, Turro C, Kodanko JJ, Inorg. Chem. 2021, 60, 18964–18974. [DOI] [PubMed] [Google Scholar]
- [50].Demas JN, Harris EW, McBride RP, J. Am. Chem. Soc. 1977, 99, 3547–3551. [Google Scholar]
- [51].Toupin NP, Nadella S, Steinke SJ, Turro C, Kodanko JJ, Inorg. Chem. 2020, 59, 3919–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52] a).Lovitt CJ, Shelper TB, Avery VM, Assay Drug Dev. Technol. 2013, 11, 435–448; [DOI] [PubMed] [Google Scholar]; b) Elliott NT, Yuan F, J. Pharm. Sci. 2011, 100, 59–74. [DOI] [PubMed] [Google Scholar]
- [53].Shree T, Olson OC, Elie BT, Kester JC, Garfall AL, Simpson K, Bell-McGuinn KM, Zabor EC, Brogi E, Joyce JA, Genes Dev. 2011, 25, 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang Z, Sun C, Li C, Jiao X, Griffin BB, Dongol S, Wu H, Zhang C, Cao W, Dong R, Yang X, Zhang Q, Kong B, Front. Oncol. 2020, 10, 1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang J, Jiang M, Li S, Zhang Z, Sun H, Yang F, Liang H, J. Med. Chem. 2021, 64, 6777–6791. [DOI] [PubMed] [Google Scholar]
- [56] a).Jurgens S, Casini A, Int. J. Chem. Sci. 2017, 71, 92–101; [Google Scholar]; b) Zetterström CK, Jiang W, Wähämaa H, Östberg T, Aveberger A, Schierbeck H, Lotze MT, Andersson U, Pisetsky DS, Harris HE, Leukocyte Biol J. 2008, 83, 31–38; [DOI] [PubMed] [Google Scholar]; c) Han S, Kim K, Kim H, Kwon J, Lee Y, Lee C, Song Y, Lee S, Ha N, Kim K, Arch. Pharmacal Res. 2008, 31, 67–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.