Abstract
Among the products that are expressed when Mycobacterium tuberculosis undergoes hypoxic shiftdown to nonreplicating persistence (NRP) is the alpha-crystallin chaperone protein homologue (Acr). This expression coincides with the previously reported appearance of a respiratory type of nitrate reductase activity, the increase in glycine dehydrogenase activity, and the production of a unique antigen, URB-1. In a timed sampling study, using a slowly stirred oxygen depletion culture model, we have demonstrated that the hspX mRNA that codes for Acr protein as well as the protein itself is induced just as the bacilli enter the microaerophilic NRP stage 1 (NRP-1). In contrast to the induction observed for hspX mRNA, levels of 16S rRNA, fbpB mRNA (encoding the 85B alpha antigen), and aroB mRNA (encoding dehydroquinate synthase) demonstrate relatively small to no change upon entering NRP-1. Acr protein was shown to be identical to URB-1 by Western analysis with anti-URB-1 antibody. The fact that antibody to Acr is found in a high percentage of tuberculosis patients suggests that the hypoxic shiftdown of tubercle bacilli to the NRP state that occurs in vitro, resulting in production of the alpha-crystallin protein, occurs in vivo as well. Simultaneous abrupt increases in hspX mRNA and Acr protein suggest that Acr protein expression is controlled at the level of transcription.
A major impediment to the control and eradication of tuberculosis is the ability of the tubercle bacillus to persist for long periods in a nonreplicating state in which it does not appear to be susceptible to the action of many antimycobacterial agents (18). More than one-third of the world's population is infected with Mycobacterium tuberculosis, and reactivation of latent disease is of increasing importance. This is especially true among individuals with declining immune systems, including the elderly and persons with AIDS (18). An increased understanding of such latency is needed in the development of new treatment strategies. It has been proposed that adaptation to depletion of available oxygen plays a significant role in the ability of M. tuberculosis to enter this state of nonreplicating persistence (NRP) (17). Abrupt depletion of oxygen is known to cause death and autolysis of tubercle bacilli (19), but gradual depletion induces a shiftdown to a state in which the bacilli can survive microaerophilic and anaerobic conditions (22), such as those that occur in granulomatous or necrotic lesions of the lung. A model of this process utilizing slowly stirred sealed cultures has been developed which demonstrates that the shiftdown of M. tuberculosis occurs in two stages, a microaerophilic nonreplicating stage, referred to as NRP-1, followed by an anaerobic nonreplicating stage, NRP-2, and events that occur during the microaerophilic stage appear to be critical to their ultimate hypoxic survival (20).
Several physiologic changes occur when aerobically growing bacilli are exposed to gradual limitations of available oxygen and enter into NRP-1. These include termination of replication and DNA synthesis (20) but with a continuing enlargement of the cells associated with cell wall thickening (3, 20), at least a 10-fold increase in activity of an enzyme that reductively aminates glyoxylate to glycine (glycine dehydrogenase), with concomitant regeneration of NAD from NADH (20, 22), appearance of a respiratory type of nitrate reductase activity (21), and production of a novel antigen, originally designated URB-1 (23). The URB-1 antigen has been considered to correspond to the 16-kDa alpha-crystallin chaperone protein homologue (Acr), although this has never been established (3, 25, 26).
Acr protein is of special interest not only because of its physiologic functions in tubercle bacilli (22, 26) but also because there is immunologic evidence that this protein is produced by tubercle bacilli in the human host (16, 24). This protein is produced only after exposure of tubercle bacilli to hypoxia and nitric oxide precursors in vitro but not to a number of other stress conditions (8, 25). It is also produced after ingestion by ThP-1 macrophage-like cells (26). All of these observations suggest that the hypoxic in vitro model of shiftdown reflects events that occur as M. tuberculosis shifts down to the nonreplicating state in vivo as well. This has prompted us to explore the actual induction of Acr protein and its mRNA in the slowly stirred hypoxic shiftdown model and compare the hspX mRNA expression profile to genomic DNA content and levels of 16S rRNA (a stable and abundant structural RNA). In addition, levels of fbpB and aroB mRNA were examined in this model. The fbpB gene encodes the 85B protein (alpha antigen), which functions as a mycolyl transferase in cell wall biosynthesis and is a dominant protein in mycobacterial culture filtrates (1) and which is presently a target in vaccine development strategies (11). The aroB gene encodes dehydroquinate synthase, an enzyme (AroB) involved in aromatic amino acid biosynthesis, and is believed to be an essential gene (9).
MATERIALS AND METHODS
Cultivation of M. tuberculosis in the hypoxic shiftdown model.
The H37Rv strain of M. tuberculosis was grown in tubes of Dubos Tween-albumin broth (DTA) (Difco, Detroit, Mich.) in the slowly stirred 0.5 headspace ratio (HSR) model of Wayne and Hayes (20). Briefly, aerobically growing broth cultures were inoculated to a set of 20- by 125-mm screw-cap culture tubes containing 17 ml of DTA and an 8-mm Teflon-coated magnetic stirring bar, resulting in a headspace of 8.5 ml of air; i.e., the volumetric ratio of air to medium was 0.5. The calculated initial optical density at 580 nm (OD580) was 0.004. The tightly sealed cultures were placed in a rack on a low-speed magnetic tissue culture stirrer at 120 rpm in a 37°C incubator. At designated times individual cultures were removed, the OD580 was measured, and 15 ml of the culture was concentrated to a 1.0-ml volume by centrifugation and quickly frozen and stored at −70°C until it was used for isolation of nucleic acids.
Isolation of mycobacterial DNA and RNA.
Nucleic acids were isolated from freshly thawed culture samples that were processed simultaneously. RNA was isolated by organic extraction coupled with mechanical disruption as described by DesJardin et al. (4, 7). DNA was isolated from the same aliquots as used for RNA isolation, using a differential extraction protocol of the interface and organic portions remaining after RNA isolation (5). Controls for efficiency of DNA and RNA recovery, consisting of standardized broth cultures of M. tuberculosis with known colony counts, were included with each series of isolations (7). Two independent nucleic acid isolations were performed per culture sample, and the nucleic acids were stored at −70°C until tested.
Quantification of nucleic acids using the ABI 7700 (TaqMan). (i) TaqMan technology.
Amplification and quantification of specific mycobacterial DNA and cDNA products were performed using the real-time PCR format of the ABI Prism 7700 Sequence Detection System (ABI/Perkin-Elmer, Foster City, Calif.). Sequences of the oligonucleotide primers and detector probe (TaqMan probes) specific for each target are listed in Table 1. The internal oligonucleotide TaqMan probes were fluorescently labeled on the 5′ end with 5-carboxyfluoroscein (FAM) and N,N,N′,N′-tetramethyl-6-carboxyrhodamine (TAMRA) on the 3′ end (Synthegen, Houston, Tex.). When the two dyes are in close proximity, as with an intact oligonucleotide probe, TAMRA acts as a quencher for FAM by absorbing at the FAM emission spectra. The 5′ exonuclease activity of Taq polymerase degrades the internally hybridizing probe during the course of PCR (13), and degradation of the probe leads to separation of these two dyes in solution, with a subsequent increase in the level of fluorescence in the reaction mixture. The amount of fluorescence measured in a sample is proportional to the amount of specific PCR product generated (5, 10, 12). The TAMRA emission spectrum was used to standardize for background fluorescence (18). The threshold value of fluorescence for a positive PCR (cycle threshold, CT) was set at 10 times the standard deviation of the mean baseline emission between cycles 3 and 15 of amplification. The quantity of DNA and cDNA in each reaction was determined from the CT value with reference to a standard curve generated by amplification of known amounts of target DNA or RNA (converted to cDNA). As reported previously for the IS6110 DNA TaqMan assay (5), the standard curves for the RNA targets were linear over 5 orders of magnitude, with an intra- or interassay coefficient of variation of less than 10% with standards.
TABLE 1.
Sequences of primers and probes used for quantification of specific DNA and RNA products in bacillary extracts
| Target | PCR primer sequences (5′ to 3′)a | TaqMan probe sequence |
|---|---|---|
| IS6110 DNA | F: GGC TGT GGG TAG CAG ACC | TGT CGA CCT GGG CAG GGT TCG |
| R: CGG GTC CAG ATG GCT TGC | ||
| 16S rRNA | F: GAG TGG CGA AGC GGT GAG TAA CA | TCC ACC ACA AGA CAT GCA TCC CGT G |
| R: CAC CCC ACC AAC AAG CTG ATA GG | ||
| RT primer: CCG CAC GCT CAC AG | ||
| 85B mRNA (alpha antigen) | F: TCA GGG GAT GGG GCC TAG CC | TCG AGT GAC CCG GCA TGG GAG CGT |
| R: GCT TGG GGA TCT GCT GCG TA | ||
| RT primer: GCT TGG GGA TCT GCT GCG TA | ||
| Acr mRNA (alpha-crystallin homologue) | F: ATC CCG GCC CAC CTT CGA CA R: AGC ACC TAC CGG CAG CGA CA | GCA AGT CCT TCT GCT CGG TGC GCT C |
| RT primer: GAA TGC CCT TGT CGT | ||
| AroB mRNA (3-dehydroquinate synthase) | F: AGG TCG CCG TCG TGC ATC AG R: CCG ATG CGG CCC AAC ACC TC | TCG AGA TCC CCG ACG CCG AGG C |
| RT primer: GCC TGG ACC ACT |
F, forward; R, reverse.
(ii) PCR conditions.
PCR conditions were nearly identical for all of the assays. The 50-μl reaction mixture consisted of 1× PCR Buffer II (Perkin-Elmer), 3 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate (dATP, dCTP, and dGTP), 400 μM dUTP, 0.5 U of uracil DNA glycosylase, 1 U of Taq polymerase (ISC Bioexpress, Kaysville, Utah) or AmpliTaq Gold polymerase for 16S rRNA (Perkin-Elmer), 0.2 μM concentrations of each primer, and a 0.1 μM concentration of the TaqMan probe. The PCR profile consisted of 1 cycle at 50°C (for uracil DNA glycosylase decontamination) for 2 min, 1 cycle at 95°C for 5 min, and 45 cycles of 94°C for 30 s and 68°C for 1 min. PCR primers for all of the targets were designed to hybridize only to M. tuberculosis complex strains. These assays were determined to be specific for M. tuberculosis complex organisms by screening with a panel of 42 mycobacterial species and related genera (data not shown).
(iii) IS6110 PCR.
DNA samples were tested in the ABI quantitative IS6110 PCR assay as previously described (5). These values were used to calculate the number of mycobacterial genomes per sample (the number of IS6110 DNA molecules was divided by 16, which is the number of copies of the IS6110 sequence in M. tuberculosis strain H37Rv [2]).
(iv) RT-PCR.
RNA was reverse transcribed using avian myeloblastosis virus reverse transcriptase in a 20-μl reaction volume for all three mRNA targets simultaneously and in a separate reaction mixture for 16S rRNA as described previously (6). After reverse transcription (RT), the amount of cDNA obtained from each RNA species was measured by PCR with TaqMan probes in separate reactions. The amount of contaminating chromosomal DNA was tested in parallel reactions without reverse transcriptase. No contamination was observed in these samples.
The efficiency of RT was determined for each assay by using in vitro transcripts of cloned M. tuberculosis fbpB, aroB, hspX, and 16S rRNA genes. In vitro transcripts were generated from pBlueScript with the T3 RNA polymerase promoter (Stratagene, La Jolla, Calif.) using a MAXIscript T3 Kit (Ambion, Austin, Tex.) according to the manufacturer's instructions. Transcripts were determined to be of the correct size by denaturing gel electrophoresis and were quantified by spectrophotometry. Dilutions of control transcripts ranging from 102 to 106 molecules/μl in 10 ng of yeast carrier RNA (Ambion)/μl were included in each RT-PCR assay. The efficiency of RT was determined by calculating the ratio of the number of observed to the number of expected molecules per reaction. Typical efficiencies were approximately 10% for fbpB and aroB, 3% for hspX, and 3% for 16S rRNA.
The number of molecules of mRNA per sample was calculated by correcting for the efficiency of RT and the dilution factors involved in the reverse transcriptase reaction and PCR.
Protein analysis.
M. tuberculosis cultures were sonicated as previously described (22, 23) and then centrifuged at 10,000 × g for 5 min before filtration through a 0.2-μm filter (Corning, Corning, N.Y.). Protein concentration was determined by the Bradford assay (Bio-Rad, Hercules, Calif.). Twenty micrograms of protein for each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a Tris-tricine 16.5% polyacrylamide gel (Bio-Rad). The gel was stained and the protein was transferred to a nitrocellulose paper (Schleicher & Schuell, Keene, N.H.) for Western blotting. The primary antibody was either CS49 (16), in which case the secondary antibody was goat anti-mouse horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), or the primary antibody was RS88-13078 (23), in which case the secondary antibody was goat anti-rabbit horseradish peroxidase-conjugated antibody (ICN ImmunoBiologicals, Lisie, Ill.).
RESULTS
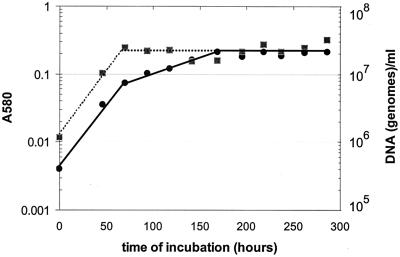
Cultures of M. tuberculosis H37Rv grown under oxygen depletion conditions exhibited the characteristic three-stage optical density pattern (Fig. 1), corresponding to the initial aerobic logarithmic growth phase for about 60 h, followed by a shiftdown to the microaerophilic NRP-1 stage (in which replication stopped but the OD580 continued to rise at a reduced rate) and finally the abrupt deflection at approximately 160 h to the anaerobic plateau of the NRP-2 stage. The increase in genomic DNA content of successive samples (Fig. 1) coincided with the increase in turbidity (OD580) of the cultures up to the time of first deflection into NRP-1; at that point, the DNA content stopped rising, thus confirming termination of replication of bacilli at that deflection. Neither an increase in OD580 nor an increase in DNA content was observed after entry into the anaerobic NRP-2 stage.
FIG. 1.
Optical absorbance (A580) and DNA content (genomes per milliliter) of cultures of M. tuberculosis H37Rv in the slowly stirred 0.5 HSR model. Aerobic growth occurs for approximately 60 h, after which the bacilli shift down to the microaerophilic NRP-1 stage until oxygen is completely depleted, when the culture is about 160 h old and bacilli shift down to the anaerobic NRP-2 stage. DNA synthesis stops abruptly as the bacilli enter NRP-1. This figure represents one of two independent experiments that gave comparable results. Symbols: A580, solid circles; DNA, solid squares.
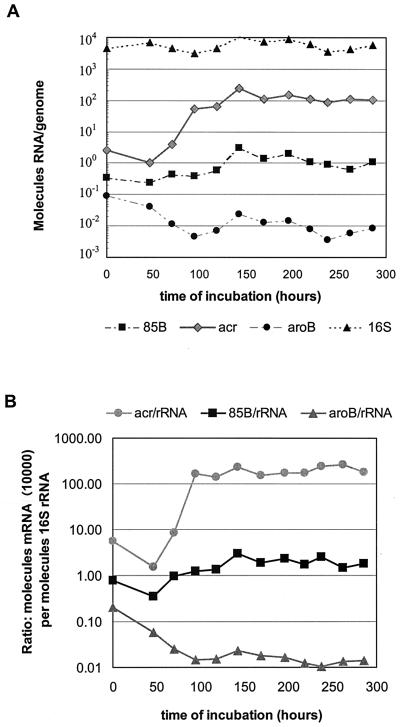
Changes in the amounts of different messages (mRNA) during cultivation, expressed as molecules per genome to compensate for the bacillary replication during the aerobic phase and termination on entry into NRP-1 as well as to correct for any possible losses during centrifugal concentration of the bacterial suspensions, are shown in Fig. 2A. The number of hspX mRNA molecules per genome increased approximately 100-fold upon transition from aerobic log phase growth into the microaerophilic NRP-1 stage and leveled off as the bacilli enter the anaerobic NRP-2 stage. This is in contrast to a stable number (5.5 × 103 ± 2.1 × 103) of 16S rRNA molecules per genome throughout the three growth phases.
FIG. 2.
Amounts of selected RNA species in cultures of M. tuberculosis H37Rv in the slowly stirred 0.5 HSR model. (A) RNA expressed as number of molecules per genome. (B) mRNA expressed as number of molecules (×10,000) per molecule of 16S rRNA (mRNA/rRNA). The ×10,000 factor was used to reflect the approximate numbers of transcripts per cell equivalent.
The amount of message of fbpB, a gene encoding antigen 85B that functions as a mycolyltransferase in cell wall biosynthesis (1), was increased about fivefold in the microaerophilic NRP-1 stage over that seen aerobically (Fig. 2A) and remained at this level in the anaerobic NRP-2 state. The number of molecules of aroB mRNA, which encodes dehydroquinate synthase, decreased an order of magnitude as the bacilli shifted into the microaerophilic NRP-1 stage (Fig. 2A), and this lower amount remained constant in the anaerobic NRP-2. The changes in numbers of molecules per genome of hspX, fbpB, and aroB mRNA during cultivation with limited or no oxygen were supported when expressed as a ratio of mRNA to 16S rRNA content to compensate for the lesser stability of RNA compared to that of DNA and control for the efficiency of RNA isolation (Fig. 2B).
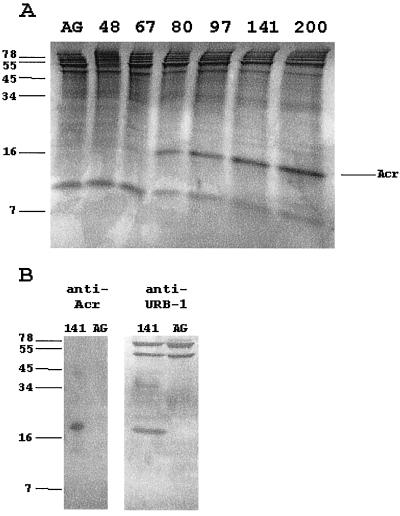
To further investigate the dramatic induction of hspX mRNA at the point of shiftdown into the NRP-1 state, we examined Acr protein expression throughout the time course of the oxygen depletion model. No Acr protein was observed in protein profiles from aerobic cultures or those before microaerophilic shiftdown but was first seen in the 67-h sample at about the time of entry into NRP-1 (Fig. 3A). The amount of Acr protein increased as the culture progressed further through NRP-1 into NRP-2. Acr expression within a few hours after the initiation of the shift of the culture into NRP-1 coincides with the induction of hspX mRNA. A Western blot with anti-Acr antibody confirmed the identity of the Acr band (data not shown).
FIG. 3.
Protein analysis of M. tuberculosis. (A) SDS-PAGE analysis of protein extracts from M. tuberculosis stained with Coomassie blue. Cultures were incubated for the indicated times in slowly stirred sealed tubes. AG indicates shaking aerobic incubation for 80 h. The alpha-crystallin homologue is indicated. (B) Immunoblot analysis of the Acr protein. Proteins from either aerobic (AG) or 141-h-old hypoxic cultures were probed with either monoclonal anti-Acr (CS49) or polyclonal anti-URB-1 (RS88-13078) antibody.
To determine if the Acr protein and the URB-1 protein were identical, Western blot analysis was done with anti-URB-1 polyclonal antibody and the Acr monoclonal antibody. The Acr antibody identified a single protein (Fig. 3B), while the URB-1 antibody reacted with several proteins. URB-1 was originally identified as an antigen absent in aerobically growing cells but present after hypoxic incubation (23). There is a single protein band present in the NRP-1 culture but missing from the aerobic culture that separates with the same approximate molecular weight as the Acr protein, suggesting that URB-1 and Acr are the same protein.
DISCUSSION
The data presented here indicate a very abrupt induction of the hspX mRNA at the point of termination of replication and DNA synthesis of tubercle bacilli that corresponds to their entry into microaerophilic NRP-1; i.e., when dissolved oxygen levels have dropped below 10% air saturation equivalence and rapidly approached 1% of their initial saturation level (20). This coincides with the point of rapid acceleration of expression of the enzyme responsible for reductive amination of glyoxylate (glycine dehydrogenase) (20) and of the respiratory nitrate reductase (21). Furthermore, the ongoing rise in concentration of hspX mRNA appears to terminate just before the culture becomes sufficiently anaerobic to enter NRP-2, which is below 0.06% air saturation equivalence (20). In view of the relative instability of the hspX mRNA, with a half-life of only 2 min (14), the persistent plateau level of this transcript suggests equilibrium between decay and low-level synthesis throughout NRP-1 and well into NRP-2. The Acr protein, on the other hand, is relatively stable (25) and was found to be at high levels into NRP-2. Its accumulation, as seen on SDS-PAGE, mirrors the transcription of hspX mRNA. Further support for our observations was recently reported by Boon et al. (1a), who used the slowly stirred shiftdown model to examine dormancy-induced proteins of Mycobacterium bovis BCG. Levels of Acr protein dramatically increased as the BCG entered the hypoxic state and remained elevated throughout the hypoxic stationary phase. This steady-state level of Acr correlated with an increase in the steady-state level of hspX mRNA, suggesting that Acr is regulated at the transcriptional level.
There is an apparent discrepancy between our observations on the timing of transcription and translation of hspX and those reported by Hu and Coates (14). Using a modification of the unstirred settling hypoxic shiftdown model originally described by Wayne and Lin (17, 22), they concluded that the induction of hspX mRNA started during aerobic logarithmic growth and terminated during the microaerophilic stage. On the other hand, levels of Acr protein were not detected until oxygen was severely depleted (25, 26). Thus, Hu and Coates hypothesized an inverse relationship between transcriptional and posttranscriptional control of Acr protein levels. No growth curve data were presented in their report to establish the physiologic stages of the cultures used for the hspX mRNA assays, but based on data from the two papers cited earlier (17, 22), they concluded that a 4-day-old unstirred culture was still in mid-log growth phase. However, it is not the temporal age of a culture in the hypoxic model that determines the physiologic stage but rather the cell density and associated rate of depletion of dissolved oxygen (20). In the original work with unstirred cultures the medium was inoculated with about 1.2 × 106 CFU/ml (17, 22), whereas Hu and Coates started with unstirred cultures representing a 1:10 dilution of a 10-day-old culture, suggesting that their initial inoculum was at least an order of magnitude greater. In a subsequent paper in which their modified settling model was used again, Hu and colleagues recorded (15) a 200-fold depletion of dissolved oxygen in the bottom of the culture after only 24 h of incubation. Thus, a significant fraction of the population of bacilli at or near the bottom of the container by the fourth day was subjected to severe hypoxic conditions, so the whole unstirred 4-day-old culture comprised a physiologically mixed population in a self-generated oxygen gradient (17). Our data demonstrate that the hypoxic component of the population would have been responsible for the presence of the hspX mRNA at that time in the Hu and Coates model, and from this we may infer that the half-life reported by Hu and Coates (14) does correspond to that of the hypoxically induced transcript.
Although the most dramatic finding in this study is the significant increase in hspX mRNA, there was also a small increase in transcription of the fbpB mRNA. This gene encodes the 85B mycolyl transferase protein that is involved in cell wall biosynthesis (1), prompting speculation that this product may play a role in the cell wall thickening demonstrated by electron microscopy in cells of tubercle bacilli in microaerophilic NRP-1 but not in old nonreplicating cultures that were subjected to continuing aeration (3). It is also of interest that aroB mRNA decreases as the bacilli enter limiting oxygen conditions. With its role as a biosynthetic enzyme in the shikimate pathway (9), one could speculate that as the tubercle bacilli shift down to the nonreplicating phase there is decreasing need for de novo protein synthesis, thus the observed decrease in aroB mRNA.
Acr has been shown, through the use of a reporter gene construct, to be induced upon entrance of M. tuberculosis into the macrophage-like THP-1 cell line, and tubercle bacilli whose hspX genes have been disrupted are attenuated for growth within those cells (26). It has also been reported (25) that tubercle bacilli engineered to overexpress the Acr protein grew much slower than the wild strain in culture. Yuan and associates hypothesized that expression of Acr was responsible for this low growth rate and that down-regulation of this gene was necessary to achieve replication of M. tuberculosis at its normal rate (25). Wayne and Lin (22) noted that resumption of aeration of cultures in the NRP state resulted in dilution of the URB-1 antigen, which corresponds to Acr, with freshly synthesized protein; the loss of tolerance of the bacilli to anaerobic conditions paralleled the reduction in specific activity of Acr in the bacilli during this aerobic shiftup.
At least 85% of a series of tuberculosis patients have been shown to have antibody to the Acr protein of M. tuberculosis, demonstrating that it is ubiquitous in infected individuals (16). Since inflammatory, granulomatous, and necrotic lesions have hypoxic regions (22a), the hypoxic shiftdown to NRP seen with this organism in vitro suggests that this probably occurs in the human host as well. Within this context, monitoring the ratio of concentration of the Acr mRNA to that of the 85B mRNA in tissues of experimentally infected animals or sputum specimens of tuberculosis patients may be a useful means for assessing the temporal relationship between the emerging immunologic response of the host and changes in the physiologic state of the tubercle bacilli. It should be noted that Garbe and colleagues (8) have reported that precursors to nitric oxide, which is produced by macrophages, can also induce production of Acr by tubercle bacilli; details of the physiologic state of the bacilli used to induce that effect were not presented. Nevertheless, there may indeed be alternative mechanisms by which the protective Acr is induced.
Our results are consistent with previous studies showing that Acr protein is not detected until aerobic growth has terminated (1a, 25, 26). The induction of Acr during hypoxic shiftdown, its function as a chaperone (26), its location in the cell wall (3), the negative effect of its overexpression on growth rate (26), and the correlation of its cellular concentration with tolerance of tubercle bacilli to abrupt anaerobic shock (22) all suggest a critical role for this product in the ability of M. tuberculosis to persist without replicating in hostile regions of the host's tissues. An understanding of the mechanisms of induction of factors associated with the hypoxic shiftdown tubercle bacilli should contribute to the development of strategies for preventing that persistence.
ACKNOWLEDGMENTS
The study reported here was supported by contract NO-AI-45244 from the National Institutes of Health, Tuberculosis Research Unit, and by the Medical Research Service of the Department of Veterans Affairs.
We are grateful to Chris Hemphill, Robert Pruss, Sandra Sudberg, and Maria Winters for expert technical assistance. We thank John Spencer and John Belisle for the anti-Acr antibody and technical advice.
REFERENCES
- 1.Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 1a.Boon C, Li R, Qi R, Dick T. Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J Bacteriol. 2001;183:2672–2676. doi: 10.1128/JB.183.8.2672-2676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham O, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor T, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desjardin, L. E. Isolation of Mycobacterium tuberculosis RNA from sputum. In S. H. Gillespie (ed.), Antibiotic resistance methods and protocols. Humana Press, Totowa, N.J., in press.
- 5.Desjardin L E, Chen Y, Perkins M D, Teixeira L, Cave M D, Eisenach K D. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 in sputum during treatment for tuberculosis. J Clin Microbiol. 1998;36:1964–1968. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desjardin L E, Perkins M D, Teixeira L, Cave M D, Eisenach K D. Alkaline decontamination of sputum specimens adversely affects stability of mycobacterial mRNA. J Clin Microbiol. 1996;34:2435–2439. doi: 10.1128/jcm.34.10.2435-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desjardin L E, Perkins M D, Wolski K, Haun S, Teixeira L, Chen Y, Johnson J L, Ellner J J, Dietze R, Bates J, Cave M D, Eisenach K D. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Respir Crit Care Med. 1999;160:203–210. doi: 10.1164/ajrccm.160.1.9811006. [DOI] [PubMed] [Google Scholar]
- 8.Garbe T R, Hibler N S, Deretic V. Response to reactive nitrogen intermediates in Mycobacterium tuberculosis: induction of the 16-kilodalton α-crystallin homolog by exposure to nitric oxide donors. Infect Immun. 1999;67:460–465. doi: 10.1128/iai.67.1.460-465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garbe T R, Servos S, Hawkins A, Demetriadis G, Young D, Dougan G, Charles I. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Mol Gen Genet. 1991;228:382–392. doi: 10.1007/BF00260631. [DOI] [PubMed] [Google Scholar]
- 10.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 11.Harth G, Lee B Y, Horwitz M A. High-level heterologous expression and secretion in rapidly growing nonpathogenic mycobacteria of four major Mycobacterium tuberculosis extracellular proteins considered to be leading vaccine candidates and drug targets. Infect Immun. 1998;65:2321–2328. doi: 10.1128/iai.65.6.2321-2328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 13.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′*3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y M, Coates A R M. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J Bacteriol. 1999;181:1380–1387. doi: 10.1128/jb.181.5.1380-1387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y M, Butcher P D, Mangan J A, Rajandream M-A, Coates A R M. Regulation of hmp gene transcription in Mycobacterium tuberculosis: effects of oxygen limitation and nitrosative and oxidative stress. J Bacteriol. 1999;181:3486–3493. doi: 10.1128/jb.181.11.3486-3493.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B-Y, Hefta S A, Brennan P J. Characterization of the major membrane protein of virulent Mycobacterium tuberculosis. Infect Immun. 1992;60:2066–2074. doi: 10.1128/iai.60.5.2066-2074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne L G. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis. 1976;114:807–811. doi: 10.1164/arrd.1976.114.4.807. [DOI] [PubMed] [Google Scholar]
- 18.Wayne L G. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 19.Wayne L G, Diaz G A. Autolysis and secondary growth of Mycobacterium tuberculosis in submerged culture. J Bacteriol. 1967;93:1374–1381. doi: 10.1128/jb.93.4.1374-1381.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wayne L G, Hayes L G. An in vitro model for the sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wayne L G, Hayes L G. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber Lung Dis. 1998;79:127–132. doi: 10.1054/tuld.1998.0015. [DOI] [PubMed] [Google Scholar]
- 22.Wayne L G, Lin K-Y. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Wayne, L. G., and C. D. Sohaskey. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol., in press. [DOI] [PubMed]
- 23.Wayne L G, Sramek H A. Antigenic differences between extracts of actively replicating and synchronized resting cells of Mycobacterium tuberculosis. Infect Immun. 1979;24:363–370. doi: 10.1128/iai.24.2.363-370.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkinson R J, Wilkinson K A, De Smet K A L, Haslov K, Pasvol G, Singh M, Svarcova I, Ivanyi J. Human T- and B-cell reactivity to the 16 kDa α-crystallin protein of Mycobacterium tuberculosis. Scand J Immunol. 1998;48:403–409. doi: 10.1046/j.1365-3083.1998.00420.x. [DOI] [PubMed] [Google Scholar]
- 25.Yuan Y, Crane D D, Barry C E., III Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Y, Crane D D, Simpson R M, Zhu Y-Q, Hickey M J, Sherman D R, Barry C E., III The 16-kDa α-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc Natl Acad Sci USA. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]