Abstract
Estimating the biomass of phytoplankton communities via remote sensing is a key requirement for understanding global ocean ecosystems. Of particular interest is the carbon associated with diatoms given their unequivocal ecological and biogeochemical roles. Satellite‐based algorithms often rely on accessory pigment proxies to define diatom biomass, despite a lack of validation against independent diatom biomass measurements. We used imaging‐in‐flow cytometry to quantify diatom carbon in the western North Atlantic, and compared results to those obtained from accessory pigment‐based approximations. Based on this analysis, we offer a new empirical formula to estimate diatom carbon concentrations from chlorophyll a. Additionally, we developed a neural network model in which we integrated chlorophyll a and environmental information to estimate diatom carbon distributions in the western North Atlantic. The potential for improving satellite‐based diatom carbon estimates by integrating environmental information into a model, compared to models that are based solely on chlorophyll a, is discussed.
Keywords: diatoms, carbon, remote sensing, pigments, cell imagery
Key Points
Field observations show that diatom carbon estimates derived from pigment‐based proxies are higher than those derived from plankton imaging
An updated equation to estimate diatom carbon from in situ or satellite chlorophyll a concentration measurements is provided
Environmental data and plankton imagery are applied in a neural network to map diatom carbon concentrations in the western North Atlantic
1. Introduction
Information on phytoplankton populations at large spatial and temporal scales is needed to assess broad‐scale changes in phytoplankton communities, which are likely to occur in response to changing environmental conditions (Irwin & Oliver, 2009; Rousseaux & Gregg, 2015), and to develop and evaluate regional and global biogeochemical models. Of particular interest are the diatoms, a globally ubiquitous and diverse group of phytoplankton consisting of an estimated 100,000 species (Mann & Vanormelingen, 2013). Some species can form massive blooms and their aggregation and sinking characteristics have been linked to sequestration of carbon to the deep ocean (Honjo & Manganini, 1993; Jin et al., 2006). While diatoms are highly varied in their adaptations to different conditions and their role in the biological carbon pump (Kemp & Villareal, 2018; Tréguer et al., 2018), for the purposes of regional or global assessment they are often considered as one group. Satellite‐based measurements have the potential to provide the large‐scale information on the quantity and distribution of diatoms needed to answer ocean ecosystem and climate research‐related questions.
Previous studies have successfully linked satellite ocean color and reflectance data with phytoplankton groups assessed using flow cytometry (Thyssen et al., 2015; Zubkov and Quartly, 2003). Regarding diatom presence or abundance, information retrieved from remote sensing data include the analysis of multispectral water leaving radiance anomalies (Alvain et al., 2005, 2008; Rêve‐Lamarche et al., 2017), remote sensing reflectance (R rs(λ)) band ratios (Kramer et al., 2018; Sathyendranath et al., 2004), a neural network approach incorporating environmental data (Raitsos et al., 2008), empirical orthogonal functions between R rs(λ) bands and phytoplankton groups (Xi et al., 2020), and a method of differential optical absorption spectroscopy (DOAS) that requires high spectral resolution of water‐leaving radiation measurements in the blue wavelengths (Bracher et al., 2009; Losa et al., 2017; Sadeghi et al., 2012). Phytoplankton pigment concentrations obtained from high performance liquid chromatography (HPLC) measurements are used in the construction and evaluation of the majority of these algorithms. While pigment analysis is invaluable for many applications and has been effective in assessing different phytoplankton groups (e.g., Kramer et al., 2020; Kramer & Siegel, 2019; Swan et al., 2016), it often lacks validation against more direct observations of community composition. For example, fucoxanthin, a photosynthetic carotenoid pigment found in diatoms (Jeffrey & Vesk, 1997), is commonly used as a marker for this group, although it is not unique to it and is present in other common phytoplankton groups, namely prymnesiophytes and silicoflagellates (Jeffrey & Vesk, 1997; Roy et al., 2011). Moreover, pigment ratios within a population can vary in response to the availability of light and nutrients, and as a result of diel changes in pigment synthesis (Becker et al., 2020; Goericke & Montoya, 1998; Organelli et al., 2017).
Historically available data on phytoplankton assemblages typically do not have the spatial and temporal coverage needed for obtaining sufficient matches with remote sensing data, with the exception of continuous plankton recorder (CPR) data. CPR data have been applied to improve and develop algorithms for the detection of diatoms from remote sensing data (Raitsos et al., 2008; Rêve‐Lamarche et al., 2017); however, the data are semi‐quantitative. In addition, the CPR data likely underestimate diatom biomass given the 270 μm mesh size it utilizes (Richardson et al., 2006). Advances in plankton imaging now allow measurements of cell concentrations and cell biovolumes from which estimates of carbon per‐cell can be derived. We used imaging‐in‐flow cytometry and HPLC measurements from the western North Atlantic to compare cell imagery‐based and accessory pigment‐based estimates of diatom carbon. Based on these measurements, we propose a new empirical relationship between diatom carbon and chlorophyll a (Chl a). We also show how satellite‐derived information on Chl a, temperature, and salinity can be integrated to estimate diatom carbon, and compare results to those from accessory‐pigment and Chl a‐based estimates.
2. Data and Methods
2.1. In Situ Temperature, Salinity, and Pigments
The North Atlantic Aerosol and Marine Ecosystems Study (NAAMES, 2015–2018) was conducted in the western North Atlantic onboard the R/V Atlantis, and encompassed different seasons and stages of the phytoplankton annual cycle (Behrenfeld et al., 2019). In‐situ data used in this study were obtained both from instruments deployed for continuous (flowthrough) measurement of surface temperature, salinity, and Chl a concentrations, and from discrete water samples collected for the analysis of HPLC pigment concentrations (Figure 1a). The methods and processing protocols of these data types are provided in Text S1 in Supporting Information S1. Note that throughout the manuscript, Chl a from HPLC refers to as “total Chl” and is defined as the sum of the concentrations of monovinyl Chl a + divinyl Chl a + chlorophyllide a + Chl a allomers and epimers.
Figure 1.
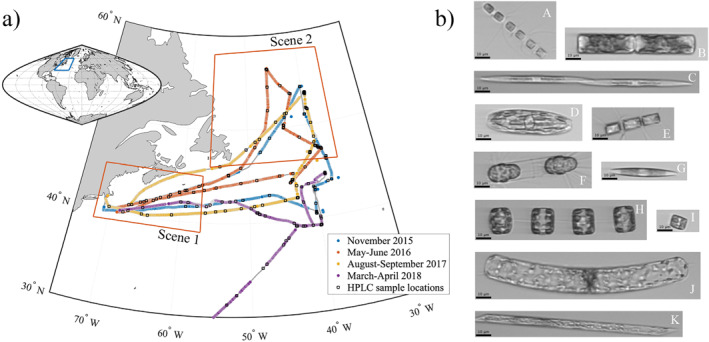
(a) The four NAAMES cruise tracks in the western North Atlantic Ocean. Colored dots show IFCB sample locations; blue = NAAMES01 in November 2015; orange = NAAMES02 in May‐June 2016; yellow = NAAMES03 in August‐September 2017; and purple = NAAMES04 in March‐April 2018 (total n = 4,328). Black squares show locations of water samples taken for HPLC analysis (n = 205). Scenes 1 and 2 from NAAMES02 further analyzed with satellite data are outlined in red boxes. (b) Example diatom images collected underway during NAAMES02. Black 10 μm scale bars on each image are equivalent. Genera/categories of each image: A—Chaetoceros sp., B—likely Guinardia sp., C—Pseudo‐nitzschia sp., D—order Naviculales, E—Chaetoceros sp., F—Corethron sp., G—unidentified pennate, H—Thalassiosira sp., I—unidentified centric, J—Guinardia sp., K—Rhizosolenia sp.
2.2. Diatom Carbon Estimated From Plankton Imagery
Phytoplankton cells were imaged with an Imaging FlowCytobot (IFCB, McLean Research Laboratories, Inc.; Olson & Sosik, 2007) with a 150 μm Nitex mesh attached to the intake (Text S2 in Supporting Information S1). Images were classified into the highest taxonomic category possible based on morphology using the EcoTaxa platform (Picheral et al., 2017; https://ecotaxa.obs-vlfr.fr/), and then grouped more broadly into 18 categories, including a category for all combined diatoms. A deep learning classification network was trained and tested using the classified images, and then applied to all images within the data set (Text S2 in Supporting Information S1). The network identified diatoms with 90% accuracy, and in addition was found to correct some mislabeled classified images due to human error (Text S2 in Supporting Information S1). In total, 336,872 diatom cells or chains were identified from 4,328 IFCB samples across all four NAAMES cruises (Figure 1b; Figure S1 in Supporting Information S1). When cell counts were low, multiple samples were combined to increase sample volume and reduce statistical counting uncertainty, as in Chase et al. (2020); details are provided in Text S2 in Supporting Information S1. Following the procedure to combine samples, the final number of IFCB samples used in the present analysis is 1,449. Biovolume (μm3) was calculated for each diatom cell or chain (Moberg & Sosik, 2012; Sosik & Olson, 2007; https://github.com/hsosik/ifcb-analysis/tree/features_v3), and converted to diatom carbon (Cdiat) following the diatom‐specific formula reported by Menden‐Deuer and Lessard (2000):
| (1) |
On average, diatoms have a lower carbon per volume compared to other phytoplankton (see figure 5 in Menden‐Deuer & Lessard, 2000), which may be the result of large vacuoles often present in diatoms (Strathmann, 1967). We calculated uncertainties in Equation 1 using the confidence intervals for the coefficients reported in table 4 of Menden‐Deuer and Lessard (2000). A recent study by McNair et al. (2021) showed that the conversion from cell volume to carbon is not greatly affected by the method used to estimate cell volume. Total diatom carbon concentrations for a given sample were normalized to the sample water volume, resulting in diatom carbon concentration per sample (Cdiat_IFCB, units of mg m−3).
2.3. Diatom Carbon Estimated From Accessory Pigments
The fraction of diatom contribution to total Chl a was estimated using a method often referred to as the diagnostic pigment analysis (DPA), which assigns accessory pigments to phytoplankton groups. The application of accessory “diagnostic” pigments was originally used to define phytoplankton size classes (Uitz et al., 2006; Vidussi et al., 2001), and has subsequently been used to define phytoplankton taxonomic groups (Hirata et al., 2011; Losa et al., 2017; Soppa et al., 2014). Using a global data set, the fraction of total Chl a attributed to diatoms was defined by Hirata et al. (2011) as:
| (2) |
where the summed weighted diagnostic pigments are defined as = 1.41Fuco + 1.41Peri + 1.27Hexa + 0.35Buta + 0.6Allo + 1.01Chlb + 0.86Zea (Uitz et al., 2006), and where Fucocorr = Fuco – (Fuco/Hexa)baseline * Hexa (Hirata et al., 2011). Pigment abbreviations are as follows: Fuco = fucoxanthin, Peri = peridinin, Hexa = 19′‐hexanoyloxyfucoxanthin, Allo = alloxanthin, Buta = 19′‐butanoyloxyfucoxanthin, Chlb = total chlorophyll b, and Zea = zeaxanthin. The (Fuco/Hexa)baseline is defined as the median of the Fuco/Hexa ratio across all 205 HPLC samples. We also applied the equations in Losa et al. (2017):
| (3a) |
| (3b) |
where the weights for accessory pigments are the values from their global study: = 1.27Fuco + 2.43Peri + 1.07Hexa + 0.0Buta + 2.06Allo + 1.30Chlb + 2.36Zea, and the contribution of fucoxanthin to nanoplankton (Fuconano) in Equation 3b is defined in Losa et al. (2017) as:
| (4) |
Additionally, we tested the application of the CHEMTAX program (Mackey et al., 1996) to estimate the relative contribution of phytoplankton groups to total Chl a (Text S3 and Figures S2–S3 in Supporting Information S1).
To compare cell imagery‐ and pigment‐based estimates of diatom biomass in units of mg C m−3, we calculated Chl a concentrations of diatoms by multiplying fDiat by Chl a, and then assumed a constant cellular carbon‐to‐chlorophyll (C:Chl) ratio (Equation (5a), (5b), (5c)). Given that the C:Chl ratio varies (Behrenfeld et al., 2016; Jackson et al., 2017; Sathyendranath et al., 2009), we used mean, minimum, and maximum values of C:Chl for diatoms to calculate a range of possible values for pigment‐derived diatom carbon (Cdiat_Pigments) as follows:
| (5a) |
| (5b) |
| (5c) |
where Chl a concentrations were obtained from HPLC analysis, and C:Chl values were reported in Sathyendranath et al. (2009). We applied Equation (5a), (5b), (5c) to fDiat defined in both Equations 2 and (3a), (3b).
To increase the number of matches between Cdiat_Pigments and Cdiat_IFCB, we estimated diatom biomass from Chl a measurements using the empirical relationships proposed by Hirata et al. (2011), Losa et al. (2017), and Soppa et al. (2014). All equations are provided in Table S1 in Supporting Information S1. Chl a concentrations were obtained from spectral particulate absorption measurements (Text S1 in Supporting Information S1), and converted results to units of carbon with the same C:Chl constants shown in Equation (5a), (5b), (5c).
2.4. Neural Network‐Based Estimates of Diatom Carbon
Neural networks provide a tool to model complex relationships between a desired target quantity (here, diatom carbon) and multiple input parameters (here, temperature, salinity, and Chl a). We trained a shallow neural network model by using Cdiat_IFCB from the entire NAAMES02 cruise track (631 IFCB samples comprising 209,261 diatom images collected between 11 May and 4 June 2016) as the target parameter (Text S4, Figure S4 in Supporting Information S1). Chl a, surface water temperature and surface salinity were provided as input parameters. These input parameters were chosen as they are significantly correlated with Cdiat_IFCB (Figure S5 in Supporting Information S1; see also Brun et al., 2015), and can be derived from remote sensing data.
To assess distributions of diatom carbon across the study area, two sections of the ship track that had relatively low cloud cover were chosen and denoted Scene 1 (39–45°N and 57–72°W) and Scene 2 (46–58°N and 36–56°W) (Figure 1a). The network model was applied to remote sensing data from the two scenes using satellite‐based products for Chl a, temperature, and salinity (Text S5 in Supporting Information S1).
3. Results and Discussion
3.1. Comparison of Pigment‐ and Cell Imagery‐Derived Cdiat
Across the diverse environmental conditions sampled during the four NAAMES campaigns, values of Cdiat_Pigments were higher than Cdiat_IFCB regardless of the model used. The median bias value was 142%, which amounts to a median difference of 2.4 mg C m−3 for the model of Losa et al. (2017), and 138% and 0.9 mg C m−3 for the model of Hirata et al. (2011) (Figure 2a). This is likely explained in part by the presence of other fucoxanthin‐containing phytoplankton groups including prymnesiophytes, silicoflagellates, and pelagophytes (Jeffrey & Vesk, 1997; Roy et al., 2011), as well as some dinoflagellate types (Yoon et al., 2002). Representatives of these groups were detected in the NAAMES IFCB samples across both the nano‐ and microphytoplankton size classes (Chase et al., 2020). The model of Losa et al. (2017) considers the fucoxanthin found in nanoplankton (Equations (3a), (3b), (4)) and similarly Hirata et al. (2011) apply a method to correct the representation of diatoms using fucoxanthin by removing a portion of it as a function of Hexa. However, the discrepancy between cell imagery‐ and accessory pigment‐based estimates of diatoms remains, and can likely be attributed to the approximations made when defining diatoms using simple accessory pigment ratios. The CHEMTAX model, a more complex application of accessory pigment ratios, also predicts higher diatom carbon concentrations compared to imagery‐based results (Text S3 and Figures S2–S3 in Supporting Information S1). The degree of overestimation varies, depending on the initial pigment ratio and phytoplankton group inputs used in the CHEMTAX model. Initial pigment ratios and phytoplankton groups suggested by van de Poll et al. (2013) show the lowest deviation (Figure S3b in Supporting Information S1). We note that as the choice of phytoplankton groups and initial pigment ratios produces noticeably different CHEMTAX results (Figures S2–S3 in Supporting Information S1), potential use of the CHEMTAX approach is best suited to scenarios where a priori knowledge of the phytoplankton communities exists.
Figure 2.
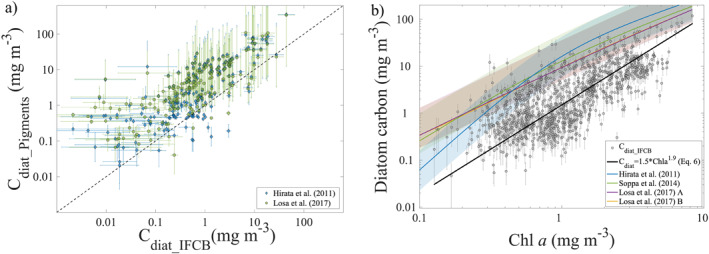
(a) Diatom carbon estimated from IFCB imagery (x‐axis; Section 2.2) and from accessory pigment‐based methods (y‐axis; Equations (2), (3a), (3b), (4), (5a), (5b), (5c)). Y‐axis error bars show the range of possible values when converting from accessory pigment‐based Chl a values to diatom carbon (Equation (5a), (5b), (5c), Section 2.3). (b) Chl a versus diatom carbon estimated from IFCB imagery across all four NAAMES cruises (gray dots, n = 1,449), and using the equations in Table S1 in Supporting Information S1 (colored lines). Shaded areas around the lines represent the range of values in diatom carbon with minimum and maximum C:Chl values applied (Section 2.3). Black line shows the fit described by Equation (5a), (5b), (5c) of this study. Cdiat_IFCB error bars in both (a) and (b) represent the combined uncertainty in particle biovolume estimates, uncertainties in the conversion from cell volume to carbon, and statistical counting errors.
Estimates of Chl a from IFCB imagery, in combination with conventional flow cytometry data, compare well with Chl a from HPLC for the NAAMES study (see figure 3a in Chase et al., 2020), but Cdiat_IFCB, could potentially underestimate diatom carbon due to the 150 μm mesh at the sampling intake that prevents very large cells and chains of cells from entering the flow cell. However, some large chains do enter the instrument as they tend to orient themselves with their major axis parallel to the flow, and in the NAAMES data set, the major axis length of diatoms ranged between approximately 6 and 200 μm (Figure S9 in Supporting Information S1). Specifically, 4.3% of all images classified as diatoms have a major axis length >150 μm. Although small nano‐diatoms are often identifiable as diatoms in IFCB imagery due to their symmetrical shapes and rectangular cross‐section, the IFCB does not comprehensively image particles smaller than approximately 6–9 μm (sensitive to instrument settings), and thus they are excluded from our analysis. In some regions, small nanoplanktonic diatoms can contribute significantly to total particulate organic carbon, as has been shown in the Mediterranean Sea (up to 26%; Leblanc et al., 2018). However, both global ocean and region‐specific databases (e.g., the North Atlantic) suggest that small nano‐diatoms (<5 μm) generally account for a low percentage of total diatom abundance in the open ocean (Leblanc et al., 2012, 2018). The potential underestimation of diatoms due to omission of the largest and smallest cells implies that at times and places when there are significant concentrations of the largest and smallest diatom cells, estimates based on the IFCB imagery may represent a lower bound in diatom carbon concentration.
3.2. A Revised Empirical Model for Cdiat
Estimates of diatom carbon from published Chl a‐based models (Table S1 in Supporting Information S1) were higher than those from IFCB imagery (Figure 2b). This is expected due to approximations made when using accessory pigments as a proxy for diatoms. Based on the NAAMES imagery and Chl a data, we propose a new empirical relationship between diatom carbon () and Chl a (both in units of mg m−3):
| (6) |
where coefficient uncertainty values are shown in parentheses. Uncertainties for the coefficients were calculated using a bootstrapping method, where the 1,449 data points used to derive the fit (Figure 2b) are iteratively subsampled (10,000 iterations; subsampled with replacement). The standard deviations of the results of this bootstrapping, for each of the two coefficients, are then defined as the uncertainty values. Equation (2), (5a), (5b), (5c), (6) provides a relationship between Chl a and diatom carbon that is based on direct measurements of diatom concentrations and therefore may be more suitable for use than previously developed models that are solely based on relationships with accessory pigment. Equation (2), (5a), (5b), (5c), (6), however, does not capture the variability observed in the imagery data as a function of Chl a concentrations (Figure 2b). It also indicates that while Chl a‐based methods can provide a reasonable estimate of diatom carbon on average over an ocean basin, they have high uncertainty when predicting diatom biomass at any given time and location. This should be considered during application of any Chl a‐based equation as a tool to assess diatom distributions.
3.3. Diatom Carbon Models Applied to Remote Sensing Data
We applied three approaches to estimate diatom carbon (Equations (2), (5a), (5b), (5c), (6); and the neural network model (Section 2.4)) from satellite data available for May 2016, and compared the resulting distribution maps. The application of Equations 2 and (5a), (5b), (5c) results in overall higher diatom carbon concentrations compared to the other two approaches (Figures 3a and 4a). Diatom carbon concentrations derived from Equation (5a), (5b), (5c) and the neural network model, both based on the IFCB cell imagery data, show a similar order of magnitude in diatom carbon but differ in their spatial patterns, notably with more (sub)mesoscale features highlighted in the results from the neural network model (Figures 3b and 3c, 4b and 4c). Comparison of in situ measurements to satellite‐modeled diatom carbon in the highly dynamic North Atlantic region is challenging, as a result of changes in physical and biological features on the timescale of hours to days and the need to use composite multi‐day satellite data (Figures S8 and S10 in Supporting Information S1). As with the application of any remote sensing algorithm, the ability of the satellite data to accurately represent in situ conditions is ultimately a limiting factor. More detailed analyses of satellite subpixel variability in regards to phytoplankton community composition are warranted.
Figure 3.
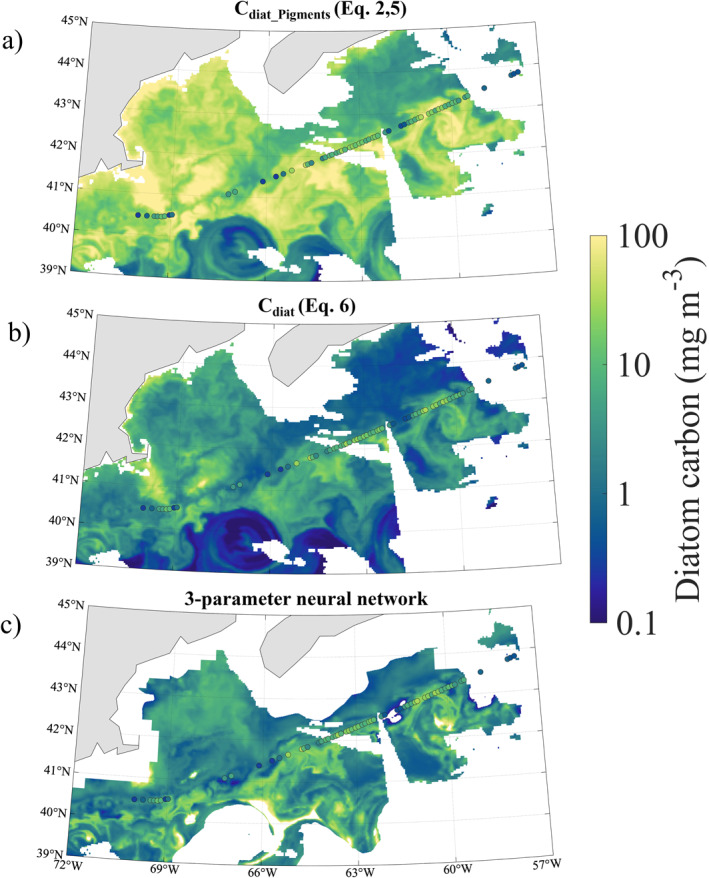
Scene 1 diatom carbon estimated from satellite data, with in situ imagery‐based data points from May 11 to 13, 2016 overlain. (a) Cdiat estimated using fDiatH11 from Hirata et al. (2011) (Equation (5a), (5b), (5c)) converted to units of carbon (mg m−3) (Equation (5a), (5b), (5c)). Cdiat mean, median, and SD are 19.9, 10.2, and 23.1 mg C m−3, respectively. (b) Cdiat estimated using Equation (5a), (5b), (5c) of this study. Cdiat mean, median, and SD = 3.5, 1.5, and 6.7 mg C m−3, respectively. (c) Diatom carbon estimated with the three‐parameter neural network model described in this study (Section 2.4). Cdiat mean, median, and SD = 5.6, 2.3, and 9.7 mg C m−3, respectively. Regions of missing data not seen in the other two panels are the result of neural network inputs from satellite data falling outside the range of in situ data used to train the neural network.
Figure 4.
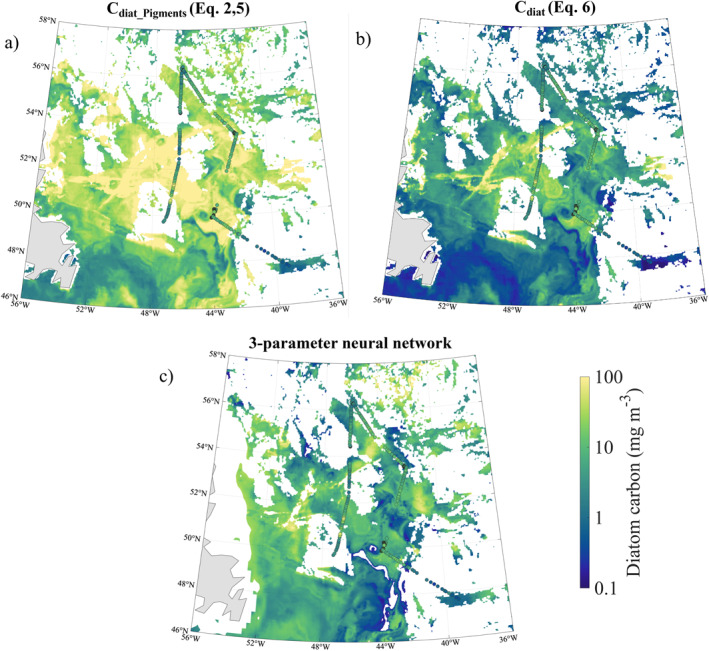
Scene 2 diatom carbon estimated from satellite data. Panels as in Figure 3, but for the Scene 2 region with imagery‐based in situ data from May 16 to 23, 2016. (a) Cdiat mean, median, and SD are 16.6, 7.0, and 22.6 mg C m−3, respectively. (b) Cdiat mean, median, and SD = 4.1, 1.1, and 10.3 mg C m−3, respectively. (c) Cdiat mean, median, and SD = 6.5, 2.7, and 10.8 mg C m−3, respectively.
In the western North Atlantic, relationships between diatom carbon and Chl a, temperature, and salinity are highly variable (Figure S4 in Supporting Information S1), precluding a simple and robust model for predicting diatom carbon at a given time and location within the region using these three inputs. However, our results demonstrate that further work with neural networks, which have previously shown promise (Palacz et al., 2013; Raitsos et al., 2008), could be beneficial to resolve finer spatial scale distributions of phytoplankton communities. In addition to the seasonal variability of the North Atlantic, there is high spatial variability in Chl a concentration, temperature, and salinity (Figures S6, S7 in Supporting Information S1). This underscores the need to better understand conditions and mechanisms determining diatom abundance in the North Atlantic, consistent with recent studies that highlight and work toward explaining the unexpected lack of large bloom‐forming diatoms in the region (Behrenfeld et al., 2021; Bolaños et al., 2020).
Uncertainties must be accounted for, both in the data and in the model approach used. The average uncertainty in Cdiat using the neural network (Section 2.4; Figures 3c and 4c) is 65% (Text S4 in Supporting Information S1). This value takes into account the data used for developing the neural network model, and the accuracy of the model itself (Figure S4 in Supporting Information S1). Calculated uncertainties for the parameters in Equation (5a), (5b), (5c) and a description of their calculation are provided (Section 3.2). Previously published algorithms to estimate diatoms from Chl a report statistics of best fit lines, but not uncertainties based on data used in algorithm development and/or in the uncertainty modeled fit parameters. We emphasize the need for defining and propagating uncertainties when estimating diatom and other phytoplankton group quantities from pigments both in situ and using remote sensing data and interpreting the observed patterns.
4. Conclusions
We provide here an updated model for estimating diatom carbon from Chl a, and highlight the need for independent measurements of diatom biomass, which is critical to quantify and thus remove biases associated with different measurement types. Our results also illustrate the potential for combining satellite data from multiple platforms in a neural network framework. The inclusion of ancillary environmental information such as water temperature and salinity can improve phytoplankton group modeling efforts (Figure S10 and Table S2 in Supporting Information S1; see also Brewin et al., 2019; Xi et al., 2021). Looking forward, the upcoming NASA Plankton, Aerosol, Cloud, and ocean Ecosystem (PACE) mission will include polarimetry and hyperspectral ocean color instruments that can potentially be combined with other remotely sensed or modeled data. Accessory phytoplankton pigments can be estimated from hyperspectral R rs(λ) (Chase et al., 2017; Kramer et al., 2022), and this information could help constrain neural network results, as different major phytoplankton groups contain different accessory pigment assemblages. Estimating diatoms and other phytoplankton groups using remote sensing techniques requires in situ data collected for validation, using a variety of methods, as well as critical attention to uncertainties and potential biases. The study presented here aims to address these needs while working toward the goal of phytoplankton community composition assessment from space.
Supporting information
Supporting Information S1
Acknowledgments
The authors are grateful to Michael Behrenfeld and Chris Hostetler for their leadership of the NAAMES expedition, as well as the crew and scientists who helped with data collection, with special thanks to Sasha Kramer and Nicholas Baetge. Thank you to Adriana Zingone for assistance with diatom taxonomic identification. The authors appreciate the helpful feedback from M. Thyssen and one anonymous reviewer, which helped improve the manuscript. The authors state no financial conflicts of interest. Funding for this work was provided by NASA grants #NNX15AE67G and #80NSSC20M0202. A. Chase is supported by a Washington Research Foundation Postdoctoral Fellowship.
Chase, A. P. , Boss, E. S. , Haëntjens, N. , Culhane, E. , Roesler, C. , & Karp‐Boss, L. (2022). Plankton imagery data inform satellite‐based estimates of diatom carbon. Geophysical Research Letters, 49, e2022GL098076. 10.1029/2022GL098076
Data Availability Statement
Data presented in this paper are available at the NASA SeaBASS repository (https://seabass.gsfc.nasa.gov/naames; DOI: https://doi.org/10.5067/SeaBASS/NAAMES/DATA001). IFCB images are viewable on the EcoTaxa platform (https://ecotaxa.obs-vlfr.fr/) and image feature data following classification with the neural network are available at DOI: https://doi.org/10.5281/zenodo.6595852.
References
References
- Alvain, S. , Moulin, C. , Dandonneau, Y. , & Bréon, F. M. (2005). Remote sensing of phytoplankton groups in case 1 waters from global SeaWiFS imagery. Deep‐Sea Research ‐ Part. I, 52(11), 1989–2004. 10.1016/j.dsr.2005.06.015 [DOI] [Google Scholar]
- Alvain, S. , Moulin, C. , Dandonneau, Y. , & Loisel, H. (2008). Seasonal distribution and succession of dominant phytoplankton groups in the global ocean: A satellite view. Global Biogeochemical Cycles, 22(3). [Google Scholar]
- Becker, K. W. , Harke, M. J. , Mende, D. R. , Muratore, D. , Weitz, J. S. , DeLong, E. F. , et al. (2020). Combined pigment and metatranscriptomic analysis reveals synchronized diel patterns of phenotypic light response across domains in the open ocean. The ISME Journal, 15, 520–533. 10.1038/s41396-020-00793-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrenfeld, M. J. , Halsey, K. H. , Boss, E. , Karp‐Boss, L. , Milligan, A. J. , & Peers, G. (2021). Thoughts on the evolution and ecological niche of diatoms. Ecological Monographs, 91(3), e01457. 10.1002/ecm.1457 [DOI] [Google Scholar]
- Behrenfeld, M. J. , Malley, R. T. O. , Boss, E. S. , Westberry, T. K. , Graff, J. R. , Halsey, K. H. , et al. (2016). Revaluating ocean warming impacts on global phytoplankton. Nature Climate Change, 6, 323–330. 10.1038/nclimate2838 [DOI] [Google Scholar]
- Behrenfeld, M. J. , Moore, R. H. , Hostetler, C. A. , Graff, J. , Gaube, P. , Russell, L. M. , et al. (2019). The North Atlantic Aerosol and Marine Ecosystem Study (NAAMES): Science motive and mission overview. Frontiers in Marine Science, 6, 122. 10.3389/fmars.2019.00122 [DOI] [Google Scholar]
- Bolaños, L. M. , Karp‐Boss, L. , Choi, C. J. , Worden, A. Z. , Graff, J. R. , Haëntjens, N. , et al. (2020). Small phytoplankton dominate Western North Atlantic biomass. The ISME Journal, 14(7), 1663–1674. 10.1038/s41396-020-0636-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher, A. , Vountas, M. , Dinter, T. , Burrows, J. P. , Röttgers, R. , & Peeken, I. (2009). Quantitative observation of cyanobacteria and diatoms from space using PhytoDOAS on SCIAMACHY data. Biogeosciences, 6(5), 751–764. 10.5194/bg-6-751-2009 [DOI] [Google Scholar]
- Brewin, R. J. W. , Ciavatta, S. , Sathyendranath, S. , Skákala, J. , Bruggeman, J. , Ford, D. , & Platt, T. (2019). The influence of temperature and community structure on light absorption by phytoplankton in the North Atlantic. Sensors, 19, 4182. 10.3390/s19194182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, P. , Vogt, M. , Payne, M. R. , Gruber, N. , O’Brien, C. J. , Buitenhuis, E. T. , et al. (2015). Ecological niches of open ocean phytoplankton taxa. Limnology & Oceanography, 60(3), 1020–1038. 10.1002/lno.10074 [DOI] [Google Scholar]
- Chase, A. P. , Boss, E. , Cetinić, I. , & Slade, W. (2017). Estimation of phytoplankton accessory pigments from hyperspectral reflectance spectra: Toward a global algorithm. Journal of Geophysical Research: Oceans, 122(12), 9725–9743. 10.1002/2017JC012859 [DOI] [Google Scholar]
- Chase, A. P. , Kramer, S. J. , Haëntjens, N. , Boss, E. S. , Karp‐Boss, L. , Edmondson, M. , & Graff, J. R. (2020). Evaluation of diagnostic pigments to estimate phytoplankton size classes. Limnology and Oceanography: Methods, 18(10), 570–584. 10.1002/lom3.10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goericke, R. , & Montoya, J. P. (1998). Estimating the contribution of microalgal taxa to chlorophyll a in the field—Variations of pigment ratios under nutrient‐ and light‐limited growth. Marine Ecology Progress Series, 169, 97–112. 10.3354/meps169097 [DOI] [Google Scholar]
- Hirata, T. , Hardman‐Mountford, N. J. , Brewin, R. J. W. , Aiken, J. , Barlow, R. , Suzuki, K. , et al. (2011). Synoptic relationships between surface chlorophyll‐a and diagnostic pigments specific to phytoplankton functional types. Biogeosciences, 8(2), 311–327. 10.5194/bg-8-311-2011 [DOI] [Google Scholar]
- Honjo, S. , & Manganini, S. J. (1993). Annual biogenic particle fluxes to the interior of the North Atlantic Ocean; studied at 34°N 21°W and 48°N 21°W. Deep‐Sea Research Part II, 40(1–2), 587–607. 10.1016/0967-0645(93)90034-K [DOI] [Google Scholar]
- Irwin, A. J. , & Oliver, M. J. (2009). Are ocean deserts getting larger? Geophysical Research Letters, 36(18), L18609. 10.1029/2009GL039883 [DOI] [Google Scholar]
- Jackson, T. , Sathyendranath, S. , & Platt, T. (2017). An exact solution for modeling photoacclimation of the carbon‐to‐chlorophyll ratio in phytoplankton. Frontiers in Marine Science, 4, 283. 10.3389/fmars.2017.00283 [DOI] [Google Scholar]
- Jeffrey, S. W. , & Vesk, M. (1997). Introduction to marine phytoplankton and their pigment signatures. In Jeffrey S. W., Mantoura R. F. C., & Wright S. W. (Eds.), Phytoplankton pigments in oceanography: Guidelines to modern methods (pp. 33–84). UNESCO. [Google Scholar]
- Jin, X. , Gruber, N. , Dune, J. P. , Sarmiento, J. L. , & Armstrong, R. A. (2006). Diagnosing the contributions of phytoplankton functional groups to the production and export of particulate organic carbon, CaCO3, and opal from global nutrient and alkalinity distributions. Global Biogeochemical Cycles, 20(2), 1. 10.1029/2005GB002532 [DOI] [Google Scholar]
- Kemp, A. E. S. , & Villareal, T. A. (2018). The case of the diatoms and the muddled mandalas: Time to recognize diatom adaptations to stratified waters. Progress in Oceanography, 167, 138–149. 10.1016/j.pocean.2018.08.002 [DOI] [Google Scholar]
- Kramer, S. J. , Roesler, C. S. , & Sosik, H. M. (2018). Bio‐optical discrimination of diatoms from other phytoplankton in the surface ocean: Evaluation and refinement of a model for the Northwest Atlantic. Remote Sensing of Environment, 217, 126–143. 10.1016/j.rse.2018.08.010 [DOI] [Google Scholar]
- Kramer, S. J. , & Siegel, D. A. (2019). How can phytoplankton pigments be best used to characterize surface ocean phytoplankton groups for ocean color remote sensing algorithms? Journal of Geophysical Research: Oceans, 124, 7557–7574. 10.1029/2019JC015604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, S. J. , Siegel, D. A. , & Graff, J. R. (2020). Phytoplankton community composition determined from co‐variability among phytoplankton pigments from the NAAMES field campaign. Frontiers in Marine Science, 7, 215. 10.3389/fmars.2020.00215 [DOI] [Google Scholar]
- Kramer, S. J. , Siegel, D. A. , Maritorena, S. , & Catlett, D. (2022). Modeling surface ocean phytoplankton pigments from hyperspectral remote sensing reflectance on global scales. Remote Sensing of Environment, 270, 112879. 10.1016/j.rse.2021.112879 [DOI] [Google Scholar]
- Leblanc, K. , Arístegui, J. , Armand, L. , Assmy, P. , Beker, B. , Bode, A. , et al. (2012). A global diatom database ‐ Abundance, biovolume and biomass in the world ocean. Earth System Science Data, 4(1), 149–165. 10.5194/essd-4-149-2012 [DOI] [Google Scholar]
- Leblanc, K. , Quéguiner, B. , Diaz, F. , Cornet, V. , Michel‐Rodriguez, M. , Durrieu de Madron, X. , et al. (2018). Nanoplanktonic diatoms are globally overlooked but play a role in spring blooms and carbon export. Nature Communications, 9(953), 1–12. 10.1038/s41467-018-03376-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losa, S. N. , Soppa, M. A. , Dinter, T. , Wolanin, A. , Brewin, R. J. W. , Bricaud, A. , et al. (2017). Synergistic exploitation of hyper‐ and multi‐spectral precursor sentinel measurements to determine phytoplankton functional types (SynSenPFT). Frontiers in Marine Science, 4, 203. 10.3389/fmars.2017.00203 [DOI] [Google Scholar]
- Mackey, M. , Mackey, D. , Higgins, H. W. , & Wright, S. W. (1996). CHEMTAX ‐ A program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Marine Ecology Progress Series, 144, 265–283. 10.3354/meps144265 [DOI] [Google Scholar]
- Mann, D. G. , & Vanormelingen, P. (2013). An inordinate fondness? The number, distributions, and origins of diatom species. The Journal of Eukaryotic Microbiology, 60(4), 414–420. 10.1111/jeu.12047 [DOI] [PubMed] [Google Scholar]
- McNair, H. , Hammond, C. N. , & Menden‐Deuer, S. (2021). Phytoplankton carbon and nitrogen biomass estimates are robust to volume measurement method and growth environment. Journal of Plankton Research, 43(2), 103–112. 10.1093/plankt/fbab014 [DOI] [Google Scholar]
- Menden‐Deuer, S. , & Lessard, E. J. (2000). Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnology & Oceanography, 45(3), 569–579. 10.4319/lo.2000.45.3.0569 [DOI] [Google Scholar]
- Moberg, E. A. , & Sosik, H. M. (2012). Distance maps to estimate cell volume from two‐dimensional plankton images. Limnology and Oceanography: Methods, 10, 278–288. 10.4319/lom.2012.10.278 [DOI] [Google Scholar]
- Olson, R. J. , & Sosik, H. M. (2007). A submersible imaging‐in‐flow instrument to analyze nano‐ and microplankton: Imaging FlowCytobot. Limnology and Oceanography: Methods, 5, 195–203. 10.4319/lom.2007.5.195 [DOI] [Google Scholar]
- Organelli, E. , Nuccio, C. , Lazzara, L. , Uitz, J. , Bricaud, A. , & Massi, L. (2017). On the discrimination of multiple phytoplankton groups from light absorption spectra of assemblages with mixed taxonomic composition and variable light conditions. Applied Optics, 56(14), 3952. 10.1364/AO.56.003952 [DOI] [PubMed] [Google Scholar]
- Palacz, A. P. , John, M. A. , Brewin, R. J. W. , Hirata, T. , & Gregg, W. W. (2013). Distribution of phytoplankton functional types in high‐nitrate, low‐chlorophyll waters in a new diagnostic ecological indicator model. Biogeosciences, 10(11), 7553–7574. 10.5194/bg-10-7553-2013 [DOI] [Google Scholar]
- Picheral, M. , Colin, S. , & Irisson, J.‐O. (2017). EcoTaxa, a tool for the taxonomic classification of images. Retrieved from http://ecotaxa.obs-vlfr.fr
- Raitsos, D. E. , Lavender, S. J. , Maravelias, C. D. , Haralabous, J. , Richardson, A. J. , & Reid, P. C. (2008). Identifying four phytoplankton functional types from space: An ecological approach. Limnology & Oceanography, 53(2), 605–613. 10.4319/lo.2008.53.2.0605 [DOI] [Google Scholar]
- Rêve‐Lamarche, A.‐H. , Alvain, S. , Racault, M.‐F. , Dessailly, D. , Guiselin, N. , Jamet, C. , et al. (2017). Estimation of the potential detection of diatom assemblages based on ocean color radiance anomalies in the North Sea. Frontiers in Marine Science, 4, 1–20. 10.3389/fmars.2017.00408 [DOI] [Google Scholar]
- Richardson, A. J. , Walne, A. W. , John, A. W. G. , Jonas, T. D. , Lindley, J. A. , Sims, D. W. , et al. (2006). Using continuous plankton recorder data. Progress in Oceanography, 68(1), 27–74. 10.1016/j.pocean.2005.09.011 [DOI] [Google Scholar]
- Rousseaux, C. S. , & Gregg, W. W. (2015). Recent decadal trends in global phytoplankton composition. Global Biogeochemical Cycles, 29, 1674–1688. 10.1002/2015GB005139 [DOI] [Google Scholar]
- Roy, S. , Llewellyn, C. A. , Egeland, E. S. , & Johnsen, G. (Eds.) (2011)., Phytoplankton pigments: Characterization, chemotaxonomy and applications in oceanography. Cambridge University Press. [Google Scholar]
- Sadeghi, A. , Dinter, T. , Vountas, M. , Taylor, B. , Altenburg‐Soppa, M. , & Bracher, A. (2012). Remote sensing of coccolithophore blooms in selected oceanic regions using the PhytoDOAS method applied to hyper‐spectral satellite data. Biogeosciences, 9(6), 2127–2143. 10.5194/bg-9-2127-2012 [DOI] [Google Scholar]
- Sathyendranath, S. , Stuart, V. , Nair, A. , Oka, K. , Nakane, T. , Bouman, H. , et al. (2009). Carbon‐to‐chlorophyll ratio and growth rate of phytoplankton in the sea. Marine Ecology Progress Series, 383, 73–84. 10.3354/meps07998 [DOI] [Google Scholar]
- Sathyendranath, S. , Watts, L. , Devred, E. , Platt, T. , Caverhill, C. , & Maass, H. (2004). Discrimination of diatoms from other phytoplankton using ocean‐colour data. Marine Ecology Progress Series, 272, 59–68. 10.3354/meps272059 [DOI] [Google Scholar]
- Slade, W. H. , Boss, E. , Dall’olmo, G. , Langner, M. R. , Loftin, J. , Behrenfeld, M. J. , et al. (2010). Underway and moored methods for improving accuracy in measurement of spectral particulate absorption and attenuation. Journal of Atmospheric and Oceanic Technology, 27(10), 1733–1746. 10.1175/2010JTECHO755.1 [DOI] [Google Scholar]
- Soppa, M. A. , Hirata, T. , Silva, B. , Dinter, T. , Peeken, I. , Wiegmann, S. , & Bracher, A. (2014). Global retrieval of diatom abundance based on phytoplankton pigments and satellite data. Remote Sensing, 6(10), 10089–10106. 10.3390/rs61010089 [DOI] [Google Scholar]
- Sosik, H. M. , & Olson, R. J. (2007). Automated taxonomic classification of phytoplankton sampled with imaging in‐flow cytometry. Limnology and Oceanography: Methods, 5, 204–216. 10.4319/lom.2007.5.204 [DOI] [Google Scholar]
- Strathmann, R. R. (1967). Estimating the organic carbon content of phytoplankton from cell volume or plasma volume. Limnology & Oceanography, 12(3), 411–418. 10.4319/lo.1967.12.3.0411 [DOI] [Google Scholar]
- Swan, C. M. , Vogt, M. , Gruber, N. , & Laufkoetter, C. (2016). A global seasonal surface ocean climatology of phytoplankton types based on CHEMTAX analysis of HPLC pigments. Deep‐Sea Research Part I Oceanographic Research Papers, 109, 137–156. 10.1016/j.dsr.2015.12.002 [DOI] [Google Scholar]
- Thyssen, M. , Alvain, S. , Lefèbvre, A. , Dessailly, D. , Rijkeboer, M. , Guiselin, N. , et al. (2015). High‐resolution analysis of a North Sea phytoplankton community structure based on in situ flow cytometry observations and potential implication for remote sensing. Biogeosciences, 12(13), 4051–4066. 10.5194/bg-12-4051-2015 [DOI] [Google Scholar]
- Tréguer, P. , Bowler, C. , Moriceau, B. , Dutkiewicz, S. , Gehlen, M. , Aumont, O. , et al. (2018). Influence of diatom diversity on the ocean biological carbon pump. Nature Geoscience, 11(1), 27–37. 10.1038/s41561-017-0028-x [DOI] [Google Scholar]
- Uitz, J. , Claustre, H. , Morel, A. , & Hooker, S. B. (2006). Vertical distribution of phytoplankton communities in open ocean: An assessment based on surface chlorophyll. Journal of Geophysical Research, 111, C08005. 10.1029/2005JC003207 [DOI] [Google Scholar]
- Van de Poll, W. H. , Kulk, G. , Timmermans, K. R. , Brussaard, C. P. D. , Van Der Woerd, H. J. , Kehoe, M. J. , et al. (2013). Phytoplankton chlorophyll a biomass, composition, and productivity along a temperature and stratification gradient in the Northeast Atlantic Ocean. Biogeosciences, 10(6), 4227–4240. 10.5194/bg-10-4227-2013 [DOI] [Google Scholar]
- Vidussi, F. , Claustre, H. , Manca, B. B. , Luchetta, A. , & Marty, J.‐C. (2001). Phytoplankton pigment distribution in relation to upper thermocline circulation in the eastern Mediterranean Sea during winter. Journal of Geophysical Research, 106(C9), 19939–19956. 10.1029/1999jc000308 [DOI] [Google Scholar]
- Xi, H. , Losa, S. N. , Mangin, A. , Garnesson, P. , Bretagnon, M. , Demaria, J. , et al. (2021). Global chlorophyll a concentrations of phytoplankton functional types with detailed uncertainty assessment using multi‐sensor ocean color and sea surface temperature satellite products. Journal of Geophysical Research: Oceans, 126, e2020JC017127. 10.1029/2020jc017127 [DOI] [Google Scholar]
- Xi, H. , Losa, S. N. , Mangin, A. , Soppa, M. A. , Garnesson, P. , Demaria, J. , et al. (2020). Global retrieval of phytoplankton functional types based on empirical orthogonal functions using CMEMS GlobColour merged products and further extension to OLCI data. Remote Sensing of Environment, 240, 111704. 10.1016/j.rse.2020.111704 [DOI] [Google Scholar]
- Yoon, H. S. , Hackett, J. D. , & Bhattacharya, D. (2002). A single origin of the peridinin‐ and fucoxanthin‐containing plastids in dinoflagellates through tertiary endosymbiosis. Proceedings of the National Academy of Sciences of the United States of America, 99(18), 11724–11729. 10.1073/pnas.172234799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov, M. V. , & Quartly, G. D. (2003). Ultraplankton distribution in surface waters of the Mozambique Channel ‐ Flow cytometry and satellite imagery. Aquatic Microbial Ecology, 33(2), 155–161. 10.3354/ame033155 [DOI] [Google Scholar]
References From the Supporting Information
- Boss, E. , Haëntjens, N. , Ackleson, S. , Balch, B. , Chase, A. , Dall’Olmo, G. , et al. (2019). Inherent optical property measurements and protocols: Best practices for the collection and processing of ship‐based underway flow‐through optical data. In Neeley A. R. & Mannino A. (Eds.), IOCCG protocol Series, IOCCG ocean optics and biogeochemistry protocols for satellite ocean colour sensor validation (Vol 40). IOCCG. 10.25607/OBP-664 [DOI] [Google Scholar]
- Chase, A. , Boss, E. , Zaneveld, R. , Bricaud, A. , Claustre, H. , Ras, J. , et al. (2013). Decomposition of in situ particulate absorption spectra. Methods in Oceanography, 7, 110–124. [Google Scholar]
- Chase, A. P. , Kramer, S. J. , Haëntjens, N. , Boss, E. S. , Karp‐Boss, L. , Edmondson, M. , & Graff, J. R. (2020). Evaluation of diagnostic pigments to estimate phytoplankton size classes. Limnology and Oceanography: Methods, 18(10), 570–584. 10.1002/lom3.10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, T. , Hardman‐Mountford, N. J. , Brewin, R. J. W. , Aiken, J. , Barlow, R. , Suzuki, K. , et al. (2011). Synoptic relationships between surface chlorophyll‐a and diagnostic pigments specific to phytoplankton functional types. Biogeosciences, 8(2), 311–327. 10.5194/bg-8-311-2011 [DOI] [Google Scholar]
- Hooker, S. B. , Van Heukelem, L. , Thomas, C. S. , Claustre, H. , Ras, J. , Schlüter, L. , et al. (2009). The third SeaWiFS HPLC analysis round‐robin experiment SeaHARRE‐3. NASA/TM‐2009‐215849. December, 2009.
- Hu, C. , Lee, Z. , & Franz, B. (2012). Chlorophyll a algorithms for oligotrophic oceans: A novel approach based on three‐band reflectance difference. Journal of Geophysical Research, 117(C1), C01011. 10.1029/2011JC007395 [DOI] [Google Scholar]
- Losa, S. N. , Soppa, M. A. , Dinter, T. , Wolanin, A. , Brewin, R. J. W. , Bricaud, A. , et al. (2017). Synergistic exploitation of hyper‐ and multi‐spectral precursor sentinel measurements to determine phytoplankton functional types (SynSenPFT). Frontiers in Marine Science, 4, 203. 10.3389/fmars.2017.00203 [DOI] [Google Scholar]
- Mackey, M. , Mackey, D. , Higgins, H. W. , & Wright, S. W. (1996). CHEMTAX ‐ A program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Marine Ecology Progress Series, 144, 265–283. 10.3354/meps144265 [DOI] [Google Scholar]
- Picheral, M. , Colin, S. , & Irisson, J.‐O. (2017). EcoTaxa, a tool for the taxonomic classification of images. Retrieved from http://ecotaxa.obs-vlfr.fr
- Slade, W. H. , Boss, E. , Dall’olmo, G. , Langner, M. R. , Loftin, J. , Behrenfeld, M. J. , et al. (2010). Underway and moored methods for improving accuracy in measurement of spectral particulate absorption and attenuation. Journal of Atmospheric and Oceanic Technology, 27(10), 1733–1746. 10.1175/2010JTECHO755.1 [DOI] [Google Scholar]
- Soppa, M. A. , Hirata, T. , Silva, B. , Dinter, T. , Peeken, I. , Wiegmann, S. , & Bracher, A. (2014). Global retrieval of diatom abundance based on phytoplankton pigments and satellite data. Remote Sensing, 6(10), 10089–10106. 10.3390/rs61010089 [DOI] [Google Scholar]
- Swan, C. M. , Vogt, M. , Gruber, N. , & Laufkoetter, C. (2016). A global seasonal surface ocean climatology of phytoplankton types based on CHEMTAX analysis of HPLC pigments. Deep‐Sea Research Part I Oceanographic Research Papers, 109, 137–156. 10.1016/j.dsr.2015.12.002 [DOI] [Google Scholar]
- Van de Poll, W. H. , Kulk, G. , Timmermans, K. R. , Brussaard, C. P. D. , Van Der Woerd, H. J. , Kehoe, M. J. , et al. (2013). Phytoplankton chlorophyll a biomass, composition, and productivity along a temperature and stratification gradient in the Northeast Atlantic Ocean. Biogeosciences, 10(6), 4227–4240. 10.5194/bg-10-4227-2013 [DOI] [Google Scholar]
- Van Heukelem, L. , & Thomas, C. S. (2001). Computer‐assisted high‐performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments. Journal of Chromatography A, 910(1), 31–49. 10.1016/S0378-434700)00603-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information S1
Data Availability Statement
Data presented in this paper are available at the NASA SeaBASS repository (https://seabass.gsfc.nasa.gov/naames; DOI: https://doi.org/10.5067/SeaBASS/NAAMES/DATA001). IFCB images are viewable on the EcoTaxa platform (https://ecotaxa.obs-vlfr.fr/) and image feature data following classification with the neural network are available at DOI: https://doi.org/10.5281/zenodo.6595852.


